Abstract
Saccharomyces cerevisiae is by far the most widely used yeast in oenology. However, during the last decade, several other yeasts species has been purposed for winemaking as they could positively impact wine quality. Some of these non-conventional yeasts (Torulaspora delbrueckii, Metschnikowia pulcherrima, Pichia kluyveri, Lachancea thermotolerans, etc.) are now proposed as starters culture for winemakers in mixed fermentation with S. cerevisiae, and several others are the subject of various studies (Hanseniaspora uvarum, Starmerella bacillaris, etc.). Along with their biotechnological use, the knowledge of these non-conventional yeasts greatly increased these last 10 years. The aim of this review is to describe the last updates and the current state-of-art of the genetics of non-conventional yeasts (including S. uvarum, T. delbrueckii, S. bacillaris, etc.). We describe how genomics and genetics tools provide new data into the population structure and biodiversity of non-conventional yeasts in winemaking environments. Future challenges will lie on the development of selection programs and/or genetic improvement of these non-conventional species. We discuss how genetics, genomics and the advances in next-generation sequencing will help the wine industry to develop the biotechnological use of non-conventional yeasts to improve the quality and differentiation of wines.
Keywords: non-conventional yeast, non-Saccharomyces, wine, enology, oenology, microsatellite
Introduction
In oenology, alcoholic fermentation is generally performed by Saccharomyces cerevisiae yeast, the “conventional” wine yeast. Currently, the winemakers have the choice between hundreds of S. cerevisiae starters that have been selected for various characteristics including their ability to complete alcoholic fermentation in oenological conditions, their low release of off-flavor compounds, their positive impact on wine aromas, etc., (Pretorius, 2000; Marullo and Dubourdieu, 2010). The growing demand for more diversified wines or for specific characteristics (low ethanol content, etc.) has led to the exploration of new species for winemaking. These non-conventional yeasts may contribute to the wine's flavor and taste by producing a broad range of secondary metabolites and extracellular enzymes (Hong and Park, 2013; Ciani et al., 2014; Wang et al., 2015). Some species could be interesting for alcohol level reduction in wine (Masneuf-Pomarede et al., 2010; Bely et al., 2013) or for greater fermentative ability in harsh conditions due to enhanced fructophily (Sutterlin, 2010; Magyar and Tóth, 2011). It has to be noted that, as only some Saccharomyces species (i.e., S. cerevisiae, S. uvarum, and some interspecific hybrids) are able to consume all the sugar contained in grape must, non-Saccharomyces yeasts must be used in co- or sequential-fermentation with a Saccharomyces spp. able to secure AF completion (Jolly et al., 2006; Bely et al., 2013).
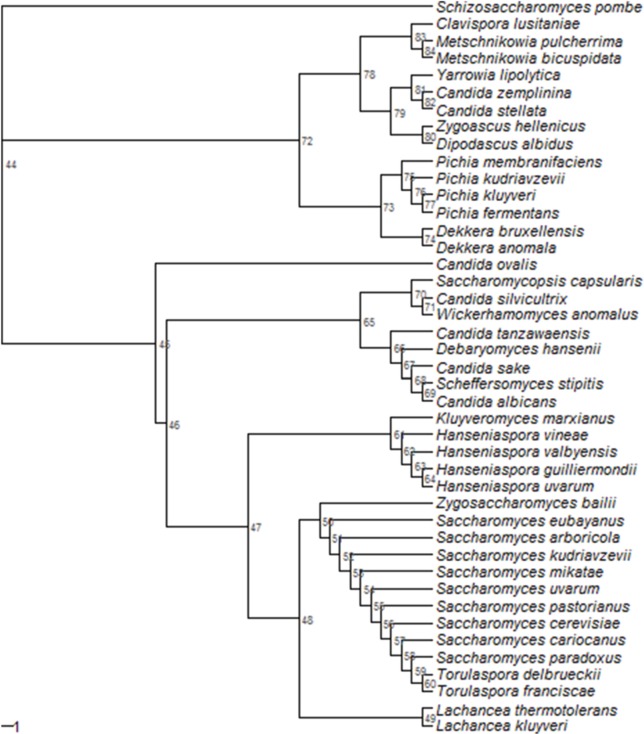
The wine industry currently proposes starters of a few non-conventional yeasts (Torulaspora delbrueckii, Metschnikowia pulcherrima, Pichia kluyveri, Lachancea thermotolerans, etc.), while several other species (Hanseniaspora uvarum, Starmerella bacillaris, etc.) are the subject of various studies to assess both positive contribution (Table 1) and negative impact (if any) on wine quality (Bely et al., 2013; Maturano et al., 2015). These non-conventional yeasts are widely distributed amongst the Saccharomycetales (Figure 1). In order to evaluate the oenological potential of a given species, several strains are usually compared for phenotypes of interest like fermentation ability (Renault et al., 2009) or glycerol production (Magyar and Tóth, 2011). However, in most cases, neither the relationships between the tested strains are described, nor the genetic structuration of the species is known. This lack of genetic knowledge is clearly detrimental, since we are not able to determine whether the phenotypic diversity described is representative of the species or not. The recent advances in next-generation sequencing (NGS) have triggered the development of genomic and genetic tools for some of these non-conventional yeasts, but the field is still in its infancy. The objective of this paper is thus to review the current state-of-art of the genetics of non-conventional wine yeasts and to discuss the future prospects and challenges from an oenological viewpoint.
Table 1.
Comparison of wine yeast species.
| Species/synonym (anamorph) | Features of interest in winemaking | Genome size | Full nuclear genome sequence | Basic ploidy level | Sporulation/zygote formation | Heterozygositya | Ecological niches | Genetic subgroups | Genetic diversity from winemaking environmentsb |
|---|---|---|---|---|---|---|---|---|---|
| Saccharomyces cerevisiae | AF completion | Nucleus: 12.0 Mb, 16 chromosomes (Goffeau et al., 1996). Mitochondrion: 85 Kb (Foury et al., 1998). | Several hundred sequences: lab strain S288c (Goffeau et al., 1996), wine strains EC1118 (Novo et al., 2009) and AWRI1631 (Borneman et al., 2008), the 100-genomes strains (Strope et al., 2015), etc. | Diploid, occasional tetraploid associated with specific environments (Albertin et al., 2009; Al Safadi et al., 2010) | 4 spores per ascus. Zygotes readily observed. (Kurtzman et al., 2011) |
75.1–81.9% (308/410 clones, 136/166 clones) (Legras et al., 2007; Muller and McCusker, 2009) | Wild environments: fruit, plant, insect, soil. Anthropic environments: wine, other distilled and traditional fermented beverages, food fermentation, dairy product, bioethanol. Lab environments. Clinical environments. (Fay and Benavides, 2005; Legras et al., 2007; Kvitek et al., 2008; Diezmann and Dietrich, 2009; Schacherer et al., 2009; Wang et al., 2012) |
Wild and domestic populations associated with wine, beer, bread, etc. (Fay and Benavides, 2005; Legras et al., 2007; Almeida et al., 2015), multiple domestication events (Schacherer et al., 2009). | 0.39–0.65 (Albertin et al., 2014b); 0.00–1.00 (Schuller et al., 2012); 0.27–0.35 (Hall et al., 2011) |
| Saccharomyces uvarum | AF completion (Masneuf-Pomarede et al., 2010); reduced ethanol production (Bely et al., 2013); psychrophilism (Masneuf-Pomarede et al., 2010); Acetate ester production (Masneuf-Pomarede et al., 2010) | Nucleus: 11.5 Mb, 16 chromosomes (Almeida et al., 2014). | More than 50 genomes of which CBS7001T (Cliften et al., 2003; Almeida et al., 2014) | Diploid | 4 spores per ascus. Zygotes readily observed. (Kurtzman et al., 2011) |
0% (0/40 strains) (Masneuf-Pomarede et al., 2007) | Wild environments: plant. Anthropic environments: wine, cider. (Almeida et al., 2014) |
Wild and domestic populations associated with wine and cider (Almeida et al., 2014) | 0.00–0.62 (Masneuf-Pomarede et al., 2007) |
| Torulaspora delbrueckii (Candida colliculosa) | Volatile acidity reduction (Bely et al., 2008); Aroma and complexity (Ciani and Maccarelli, 1998; Renault et al., 2009; Azzolini et al., 2012) | Nucleus: 9.2–11.5 Mb, 8 chromosomes (Gordon et al., 2011; Gomez-Angulo et al., 2015). Mitochondrion: 28–45 Kb (Wu et al., 2015). | 2 genomes: CBS 1146T and NRRL Y-50541 (Gordon et al., 2011; Gomez-Angulo et al., 2015) | Unclear, could be diploid (Albertin et al., 2014a) | One spore per ascus, occasional 2–3 spores/ascus (Kurtzman et al., 2011; Albertin et al., 2014a). | 26.4% (29/110 strains) (Albertin et al., 2014a) | Wild environments: fruit, plant, insect, soil. Anthropic environments: wine, other distilled and traditional fermented beverages, food fermentations, dairy products. (Albertin et al., 2014a) |
Wild and domestic populations associated with wine and other bioprocesses, geographical clustering for wild populations (Albertin et al., 2014a). | 0.35–1.00 (Albertin et al., 2015) |
| Hanseniaspora uvarum (Kloeckera apiculate) | Aroma (Rojas et al., 2001) | Nucleus: 8.08–9.08 Mb, 8 to 9 chromosomes (Esteve-Zarzoso et al., 2001). Mitochondrion: 11Kb (Pramateftaki et al., 2006). | 2 genomes: DSM 2768 and 34–9 (NCBI1) | Unclear, could be diploid (Albertin et al., 2016) | One, seldom two spores per ascus (Kreger-van Rij, 1977). Zygotes described3. | 82.6% (95/115 strains) (Albertin et al., 2016) | Wild environments: fruit, plant, insect, bird, mollusc, shrimp, soil. Anthropic environments: wine, other distilled and traditional fermented beverages. (Grangeteau et al., 2015; Albertin et al., 2016) |
Geographical and temporal clustering (Albertin et al., 2016). | 1.00 (but low number of strains per sample) (Albertin et al., 2016). |
| Hanseniaspora guillermondii (Kloeckera apis) | Acetate ester production (Rojas et al., 2001; Moreira et al., 2008; Viana et al., 2008) | Nucleus: 8 to 9 chromosomes (Esteve-Zarzoso et al., 2001). | – | – | Four spores per ascus (Barnett et al., 2000). Zygotes described3. | – | Wild environments: fruit, soil. Anthropic environments: wine. |
– | – |
| Hanseniaspora vinae (Kloeckera africana) | Acetate ester production (Viana et al., 2011) | Nucleus: 11.4 Mb, 5 chromosomes (Esteve-Zarzoso et al., 2001; Giorello et al., 2014). | 1 genome: T02/19AF (Giorello et al., 2014) | – | One, seldom two spores per ascus (Kreger-van Rij, 1977). | – | Anthropic environments: wine. | – | – |
| Starmerella bacillaris (Candida zemplinina) | Fructophily (Magyar and Tóth, 2011; Tofalo et al., 2012; Englezos et al., 2015); reduced ethanol production (Di Maio et al., 2012; Bely et al., 2013; Giaramida et al., 2013); glycerol production (Di Maio et al., 2012; Giaramida et al., 2013; Zara et al., 2014); Aroma release (Andorrà et al., 2012); other characteristics (Mangani et al., 2011; Sadoudi et al., 2012; Tofalo et al., 2012; Domizio et al., 2014; Magyar et al., 2014) | Nucleus: 3 chromosomes (Sipiczki, 2004). Mitochondrion: 23 Kb (Pramateftaki et al., 2008). | – | Unclear, could be haploid (Masneuf-Pomarede et al., 2015) | No evidence of sporulation ability (Masneuf-Pomarede et al., 2015) | 0.01% (1/163) (Masneuf-Pomarede et al., 2015) | Rare in wild environments. Anthropic environments: grape and wine. (Masneuf-Pomarede et al., 2015) | No evidence of domestication event, geographical clustering. (Masneuf-Pomarede et al., 2015) | 0.90–0.97 (Masneuf-Pomarede et al., 2015) |
| Candida stellata/Torulopsis stellata | Glycerol production (Ciani and Maccarelli, 1998); Fructophily (Magyar and Tóth, 2011) | Nucleus: 3 chromosomes (Sipiczki, 2004) | – | – | No evidence of sporulation ability | – | Anthropic environments:wine (Csoma and Sipiczki, 2008) | – | – |
| Lachancea thermotolerans /Kluyveromyces thermotolerans | Glycerol overproduction (Comitini et al., 2011); Acetate ester production (Comitini et al., 2011); reduction of volatile acidity (Comitini et al., 2011) | Nucleus: 10.4 Mb, 8 chromosomes (Malpertuy et al., 2000). Mitochondrion: 21.9–25.1 Kb (Talla et al., 2005; Freel et al., 2014). | 1 genome: CBS 6340T (Malpertuy et al., 2000) | Controversial: haploid (Freel et al., 2014) or diploid (Souciet et al., 2009) | One to four spores per ascus (Barnett et al., 2000). Zygotes described3. | – | Wild environments: fruit, plant. Anthropic environments:wine and agave fermentations (Freel et al., 2014) | Geographical clustering (Freel et al., 2014) | – |
| Lachancea kluyveri | NA | Nucleus: 11.3 Mb, 8 chromosomes (Souciet et al., 2009). Mitochondrion: 49–53.7 Kb piskur 1998; 51.5 (Jung et al., 2012) | 1 genome: NCYC 543T (Souciet et al., 2009) | Diploid, occasional triploid (Freel et al., 2014) | – | – | Wild environments: soil, insect, plant (Jung et al., 2012). | Geographical clustering (Jung et al., 2012) | – |
| Debaryomyces hansenii/Pichia hansenii (Candida famata) | Enzymatic activities (Yanai and Sato, 1999) | Nucleus: 11–46-12.18 Mb, 7 chromosomes (Dujon et al., 2004) Mitochondrion: 29.5 Kb (Dujon et al., 2004) | 2 genomes: CBS 767 and MTCC 234 (Dujon et al., 2004; Kumar et al., 2012) | Haploid (Breuer and Harms, 2006) | One (occasionally two) spores per ascus (Barnett et al., 2000). Zygotes described (Breuer and Harms, 2006) | – | Wild environments:ocean. Anthropic environments: cheese, grape. | – | – |
| Pichia kluyveri/Hanseluna kluyveri | Aromas (Anfang et al., 2009) | Mitochondrion: 43.1 Kb (CBS 7907)1. | – | Diploid (Starmer et al., 1992) | Four spores per ascus (Barnett et al., 2000). Zygotes described (Starmer et al., 1992) | – | Wild environments: fruit, insect. Anthropic environments: wine. (Starmer et al., 1992) | – | – |
| Pichia kudriavzevii/Issatchenkia orientalis (Candida krusei) | Under assessment (Clemente-Jimenez et al., 2004; Wang and Liu, 2013; Steensels and Verstrepen, 2014) | Nucleus: 10.18–12.94 Mb (Chan et al., 2012). | 3 genomes:SD108, M12, NBRC 1279 (Chan et al., 2012) | Diploid | One or two spores per ascus (Barnett et al., 2000). Zygotes described3. | – | Wild environments: plant. Anthropic environments: wine, other traditional fermented beverages, food fermentation, dairy product. (Chan et al., 2012) | – | – |
| Pichia membranifaciens (Candida valida) | Esters production (Viana et al., 2008) | Nucleus: 11.58 Mb2, between 2 and 8 chromosomes (Naumov and Naumova, 2009) | 1 genome2 | – | One to four spores per ascus (Barnett et al., 2000). | – | Wild environments: plant. Anthropic environments: AF and food spoilage yeast. | – | – |
| Pichia fermentans (Candida lambica) | Aromas (Clemente-Jimenez et al., 2005) | Maybe 2 chromosomes (Miller et al., 1989). | – | – | Two to four spores per ascus (Barnett et al., 2000). Zygotes described3. | – | Wild environments: plant, water, soil. Anthropic environments:wine, brewery. Clinical environments. | – | – |
| Pichia anomala/Hanseluna anomala (Candida pelliculosa) | Aromas (Rojas et al., 2001; Domizio et al., 2011a,b); killer against Dekkera/Brettanomyces (Comitini et al., 2004) | Nucleus: 26.55 Mb, 6 chromosomes (Friel et al., 2005). | 1 genome: NRRL Y-3661 | Diploid | One to four spores per ascus (Barnett et al., 2000). Zygotes described3. | – | Wild environments:soil, water, plant, animal. Anthropic environments:wine, fermentation contaminant, ensilage (Kurtzman et al., 2011) | – | – |
| Metschnikowia pulcherrima/Torulopsis pulcherrima (Candida pulcherrima) | Aromas and esters production (Clemente-Jimenez et al., 2004; Parapouli et al., 2010; Zott et al., 2011; Sadoudi et al., 2012) | – | – | Diploid | One to two spores (Barnett et al., 2000). | – | Wild environments: plant. Anthropic environments: wine | – | – |
| Zygosaccharomyces bailii | Fructophily (Sutterlin, 2010) | Nucleus: 10.27–21.14 Mb, 5 to 13 chromosomes (Mira et al., 2014) | 2 genomes: CLIB 213T and ISA 1307 (NCBI1) | Haploid and diploid strains (Rodrigues et al., 2003) | One to four spores per ascus (Barnett et al., 2000). | – | Wild environments: fruit, tree. Anthropic environment: food spoilage | – | – |
Proportion of strains with heterozygous microsatellite loci
Genetic diversity (0 means fully clonal population and 1 means fully diversified population)
Web sites: NCBI1, http://www.ncbi.nlm.nih.gov/genome/; JGI2, http://genome.jgi.doe.gov/; UCDAVIS3, http://wineserver.ucdavis.edu/industry/enology/winemicro/wineyeast/diversity.html.
Figure 1.
Phylogeny of 41 species of Saccharomycetales on the basis of 18S ribosomal DNA sequence. Multiple sequence alignment (1951 bases) was performed by Clustal Omega (EMBL-EBI website). Genetic distance was computed using the K80 Kimura model (Kimura, 1980), phylogenetic tree was built using Neighbor joining clustering method and bootstrapping (1000 replicates) was used to assess the robustness of the nodes by means of R package ape (Paradis et al., 2004). Schizosaccharomyces pombe was used as outgroup species. The following sequences and strains (mostly type strains) were used: AB000642.1|Dipodascus albidus IFO 1984; AB013504.1|C. tanzawaensis JCM 1648; AB018175.1|C. stellata JCM 9476; AB023473.1|M. pulcherrima IFO 1678; AB040997.1|S. kudriavzevii IFO 1802; AB040998.1|S. mikatae IFO 1815; AB054561.1|C. silvicultrix JCM 9831; AB013529.1|C. sake JCM 2951; AF548094.1|S. cerevisiae CBS 1171; AJ271813.1|S. cariocanus UFRJ 50816; AY046254.1|H. valbyensis NRRL Y-1626; AY046256.1|H. guilliermondii NRRL Y-1625; AY046257.1|H. uvarum NRRL Y-1614; AY046258.1|H. vineae NRRL Y-17529; S. bacillaris CBS 9494; EF550365.1|P. membranifaciens NRRL Y-2026; EF550372.1|P. fermentans Y-1619; EF550389.1|P. kluyveri NRRL Y-11519; EF550396.1|D. anomala NRRL Y-17522; EF550479.1|Wickerhamomyces anomalus NRRL Y-366; EU011714.1|C. ovalis NRRL Y-17662; EU011734.1|D. bruxellensis NRRL Y-12961; EU348783.1|C. albicans NRRL Y-12983; FJ153136.1|L. thermotolerans NRRL Y-8284; FJ153143.1|T. franciscae NRRL Y-6686; GU266277.1|S. arboricola AS 2.3317; GU597328.1|Zygoascus hellenicus CBS 5839; HQ651939.1|Scheffersomyces stipitis ATCC 58376; JQ698884.1|Saccharomycopsis capsularis NRRL Y-17639; JQ698900.1|Clavispora lusitaniae NRRL Y-11827; JQ698910.1|Debaryomyces hansenii NRRL Y-7426; JQ698926.1|Yarrowia lipolytica NRRL YB-423; JQ698936.1|Schizosaccharomyces pombe NRRL Y-12796; M55528.1|P. kudriavzevii MUCL 29849; S. eubayanus FM1318; S. uvarum CBS7001; X69846.1|M. bicuspidata MUCL 31145; X89523.1|L. marxianus CBS 712; X91083.1|Zygosaccharomyces bailii NCYC 1416; X97805.1|S. pastorianus NCYC 392; X97806.1|S. paradoxus CBS 432; X98120.1|T. delbrueckii CBS 1146; Z75580.1|L. kluyveri NCYC 543.
Basic genetic knowledge of wine yeasts
As a model organism, the genomic outline of S. cerevisiae is well-known: its genome size is around 12 Mb organized in 16 chromosomes, with a mitochondrial genome of 85 Kb (Table 1). The genome sequences of several hundreds of strains of various origins are available, and much more sequences are produced easily using NGS technology and subsequently assembled even by lab with moderate bioinformatics skills. The population genomics of S. uvarum has been improved recently with the sequencing of more than 50 strains of various origins (Almeida et al., 2014). The type strain CBS7001T has a genome size of 11.5 Mb and 16 chromosomes (Cliften et al., 2003). By contrast, such basic knowledge (genome size, chromosome number, etc.) is available only for a small number of non-conventional wine species: T. delbrueckii has a genome of 9–11 Mb distributed on eight chromosomes; L. thermotolerans has a 10.4 Mb genome with eight chromosomes. Other wine yeast species usually have genome size ranging from 8 to 12 Mb, with chromosomes number unknown yet (P. kluyveri, M. pulcherrima, etc.). Moreover, there is still a lack of reference genome sequence for several non-conventional wine yeasts of interest like S. bacillaris, P. fermentans, etc., (Table 1). Disparities exist also for the mitochondrial genome, with full sequences available for some species like L. thermotolerans or H. uvarum, and partial sequences for other species (C. stellata, P. membranifaciens, etc.). Thus, although the genomic data of non-conventional wine yeast greatly increased this last decade, there is still a lot of work to achieve in this field.
The life-cycle of wine yeasts
The life cycle of Saccharomyces wine species is well-known: both S. cerevisiae and S. uvarum are diploid species that divide asexually by mitosis. They are able to enter meiosis and form asci containing generally four haploid spores (tetrads). While haploid cells can undergo mitosis, the haploid level is generally transient and crosses between haploid spores of opposite mating types are readily observed, leading to diploid zygote formation. Moreover, haploid cells are usually able to switch mating type at mitosis (homothallism). The physical proximity between mother and daughter haploid cells of opposite mating type usually results in high level of inbreeding (Ruderfer et al., 2006; Cubillos et al., 2009; Warringer et al., 2011). Variations in this breeding system were described for S. cerevisiae like near-dioecy or higher level of outcrossing, but seemed quite rare and associated with environmental specificities (Knop, 2006; Al Safadi et al., 2010; Murphy and Zeyl, 2010).
By comparison, the precise life-cycle of most non-Saccharomyces yeasts is unknown yet. Sporulation was observed for most non-conventional yeast, albeit forming non-tetrad asci in many cases (T. delbrueckii, D. hansenii, H. vinae, etc., Table 1). No evidence of sporulation ability was recorded to date for Starmerella/Candida species. Data regarding the occurrence of sexual reproduction is usually scarce for most non-Saccharomyces yeasts, so classical genetic manipulations are impossible to date. To circumvent this limitation, both intra and inter specific hybridizations by protoplast fusion can be achieved as demonstrated in the past (Ball, 1984; Pina et al., 1986).
The basic ploidy level is also usually unresolved (Table 1): T. delbrueckii has been considered as a haploid species for a long time, but the detection of several strains harboring several loci with two alleles (26.4% of strains showing heterozygosity), its ability to sporulate and the presence of mating type genes is more congruent with a diploid status (Albertin et al., 2014a). Conversely, for S. bacillaris, the proportion of heterozygous strains was almost null (0.01%). This, combined with its inability to sporulate, is more consistent with an hypothesis of an haploid status (Masneuf-Pomarede et al., 2015) but has still to be formally demonstrated. Finally, despite its fully sequenced genome, the ploidy status of L. thermotolerans is controversial: haploid or diploid depending on the authors (Souciet et al., 2009; Freel et al., 2014). In conclusion, the biological life-cycle of many non-Saccharomyces yeasts remains to be elucidated.
Ecology of wine yeast
Most wine yeasts can colonize several ecological niches, including wine-related environments like grape, must, winery equipment and premise (Table 1). Moreover, many of them can be isolated from other human-associated processes (brewery, bakery, dairy, bioethanol, distillery, etc.) and also from wild substrates (soil, insect, plant, etc.). Isolation from clinical specimens is rarely described yet possible (yeasts being opportunistic microorganisms), and most wine yeasts are Generally Recognized As Safe (GRAS). Dissemination and transfer between the different ecological reservoirs could be performed through insects (Parle and Di Menna, 1966; Stefanini et al., 2012; Palanca et al., 2013), but also through human activities like material exchanges, etc., (Goddard et al., 2010). Indeed, although most wine yeasts are described as ubiquitous from an ecological viewpoint, some species have a restricted substrate range. This is the case of H. guillermondii and Starmerella species for example, which are very rarely isolated from non-wine-related substrates (Masneuf-Pomarede et al., 2015). Thus, the study of most wine yeast should consider not only wine strains but also isolates from other technological processes and substrates in order to assess their biodiversity.
Adaptation to winemaking environments and evolutionary mechanisms
Wine environments are particularly harsh and inconstant: winemaking is a seasonal practice, so that yeasts present at the surface of grape berries at harvest suddenly have to survive in grape must containing high sugar concentrations, usually with sulfur dioxide content. Moreover, from an ecological viewpoint, the ensuing alcoholic fermentation is a rapidly fluctuating ecosystem: within a few days, grape must is depleted of nitrogen nutrients, while ethanol concentration and temperature increase steadily thanks to Saccharomyces spp. metabolism, thus conferring a fitness advantage for Saccharomyces spp. over the other wine yeasts (Goddard, 2008; Salvadó et al., 2011). In addition, the range of temperature can be quite high, with either short-term variations (daily variations) or long-term evolution (seasonal variations). As a result, within wine yeast species, some strains show specific wine-adaptation (Steensels and Verstrepen, 2014) like sulphite resistance (Divol et al., 2012), ethanol tolerance (García-Ríos et al., 2014), low pH adaptation (Pretorius, 2000), temperature adaptation (Naumov et al., 2000), etc. The underlying adaptive mechanisms vary greatly from one species to another: in S. cerevisiae, molecular approaches identified allelic variations as molecular causes of adaptation to the winemaking process (Aa et al., 2006; Marullo et al., 2007; Ambroset et al., 2011; Salinas et al., 2012; Jara et al., 2014). At the chromosome level, translocations were shown to be responsible for adaptation to sulfite (Zimmer et al., 2014). Polyploidy and hybridization are also major evolutionary processes that probably triggered adaptation to wine environments (Borneman et al., 2012; Erny et al., 2012) and are currently explored for biotechnological application (Timberlake et al., 2011; Plech et al., 2014; Blein-Nicolas et al., 2015; da Silva et al., 2015). Large genomic introgressions were evidenced in S. uvarum strains associated with human-driven fermentations, suggesting a link between introgressions and domestication (Almeida et al., 2014). Various horizontal gene transfers were also evidenced for wine S. cerevisiae strains (Novo et al., 2009), and were shown to favor adaptation to the nitrogen-limited wine fermentation environment (Marsit et al., 2015). Other evolutionary mechanisms were described (Dujon et al., 2004; Barrio et al., 2006; Scannell et al., 2007), and it is highly probable that further investigations will allow the identification of additional adaptation processes in wine yeasts. In particular, it could be interesting to focus on transposon families and their possible implication in environmental adaptation (Zeyl, 2004; Liti et al., 2005; Sarilar et al., 2015), to explore the impact of mitochondrial genome variation regarding adaptation to wine environments and practices (Picazo et al., 2015; Wu et al., 2015) or to describe the landscape of gene duplication and prion involvement in fitness issues (Landry et al., 2006; Jarosz et al., 2014). However, to date, most of these data were obtained from Saccharomyces species and could now be obtained from non-Saccharomyces of interest.
Population genetics of yeast species associated with winemaking
Within a given species, the colonization of different ecosystems can led to the evolutionary differentiation of the subpopulations, in relationship with their adaptation to environmental specificities. This is the case of S. cerevisiae species that shows genetic subgroups of wild and domestic strains associated with human activities like wine, bread, beer, sake, etc., (Fay and Benavides, 2005; Liti et al., 2009; Sicard and Legras, 2011; Almeida et al., 2015), that probably originated through multiple domestication events (Schacherer et al., 2009). In a recent study, Almeida et al. (2014) showed that S. uvarum was also divided in genetic subgroups, one of domestic strains used in both winemaking and cidermaking and associated with the northern hemisphere, while others subgroups were composed of wild isolates from South America and Australasia. The current hypothesis is that a Patagonian “wild” sub-population gave rise to the domestic subpopulation through a recent bottleneck (Almeida et al., 2014). Another wine species was recently described as domesticated: T. delbrueckii is also divided in genetic subgroups of wild and domestic strains (Albertin et al., 2014a). Moreover, the wine/grape-related group showed an increase ability to ferment sugar in oenological condition, confirming the occurrence of phenotypic domestication (Albertin et al., 2015). By contrast, no hint of domestication was recorded to date for S. bacillaris and H. uvarum whose genetic diversity is shaped by geographical localization and/or time variation (Masneuf-Pomarede et al., 2015; Albertin et al., 2016).
Biodiversity in winemaking conditions
Several molecular methods were developed in order to perform intra-specific discrimination, like pulsed field electrophoresis, RAPD-PCR fingerprinting, tandem repeat-tRNA, Fourier transform infrared spectroscopy, RFLP, etc., (Barquet et al., 2012; Tofalo et al., 2013, 2014; Pfliegler et al., 2014; Grangeteau et al., 2015). However, these approaches do not allow the establishment of the genetic relationships within a given species and subsequent population genetics studies. An alternative is the use of microsatellite genotyping. It has been successfully applied to S. cerevisiae (Legras et al., 2005; Richards et al., 2009), S. uvarum (Masneuf-Pomarede et al., 2009), T. delbrueckii (Albertin et al., 2014a), S. bacillaris (Masneuf-Pomarede et al., 2015), H. uvarum (Albertin et al., 2016) as well as to the spoilage wine yeast Brettanomyces bruxellensis (Albertin et al., 2014c), and is currently developed for additional wine species like Meyerozyma guilliermondii (Wrent et al., 2015). In addition to population genetic clustering, microsatellites allow measuring the genetic diversity of a given species in specific conditions. In S. cerevisiae, the genetic diversity varied greatly, from 0 (fully clonal populations) to 1 (fully diversified population, Table 1). The precise impact of S. cerevisiae diversity (or absence of diversity) on wine quality is still debated/studied (Egli et al., 1998; Howell et al., 2006; King et al., 2008) and the direct link between microbial diversity and wine complexity should be considered with caution. S. uvarum and T. delbrueckii showed also a large range of diversity (0.35–1 and 0–0.62). By contrast, other species show systematic high diversity (>0.9 for H. uvarum or S. bacillaris), suggesting that they are not under selective pressure in winemaking environments (Masneuf-Pomarede et al., 2015; Albertin et al., 2016).
Future challenges
Definite progresses in the genetics of non-conventional yeasts were made in the last decade. However, there is still a great lack of data compared to the conventional wine yeast S. cerevisiae. Such knowledge is nowadays within reach thanks to the NGS revolution (Solieri et al., 2013). NGS allows the development of genome-assisted approaches like whole genome sequencing and resequencing, transcriptome profiling, ChIP-sequencing to identify DNA-structure, etc., (Solieri et al., 2013). De novo sequencing is greatly needed as some wine species still lack of nuclear and mitochondrial reference genomes (S. bacillaris, P. fermentans, M. pulcherrima, etc.). However, de novo assembly is sometimes difficult to conduct due to high heterozygosity level or sequence repeat, and led to draft genome with high number of contigs or scaffolds. For example, H. uvarum DSM 2768 genome displays 335 contigs, P. kudriavzevii M12 has 621 scaffolds, and P. anomala NRRL Y-366 shows 1932 scaffolds. Thus, the first aim of non-conventional wine yeast studies should be the completion of robust genomic sequences. Then, additional genome sequencing could be performed: genome re-sequencing using NGS captures individual genotypes and allows population genetics and ecologic studies within species. Such comparative genomics approaches were successfully applied to S. cerevisiae (Liti et al., 2009) and S. uvarum (Almeida et al., 2014), and could now address non-Saccharomyces yeasts of technological interest. In addition to intraspecific genomics, comparative genomics between yeast species is particularly useful to understand genome evolution (Liti and Louis, 2005). The identification of specific metabolic pathways, gene duplications or functions between species may increase our appreciation of adaptation's mechanisms and their biotechnological interest (Blein-Nicolas et al., 2015). It has to be noted that several species genetically close to wine yeasts show no peculiar affinity with winemaking environment (Figure 1). This is the case of S. paradoxus: despite being the most closely related species to S. cerevisiae, S. paradoxus is essentially associated with wild environments and particularly trees (Sniegowski et al., 2002; Johnson et al., 2004). Comparative genomics of wine vs. non-wine yeast species could thus increase our knowledge of the common genomic requirement for grape/wine colonization, if any. Finally, NGS technologies have greatly improved genome-assisted approaches aiming at detecting genetic variants associated with phenotypes in S. cerevisiae (Ehrenreich et al., 2010). In particular, QTL-seq or genome-wide association studies (GWAS) could now be applied to non-conventional yeasts depending on whether classical breeding is possible (QTL-seq) or not (GWAS). These fields are blank pages waiting to be filled in the next future of oenology microbial research.
The use of mixed-cultures, combining both non-conventional yeasts and one Saccharomyces species able to complete AF, is increasing in winemaking. Thus, another challenge lies in understanding yeast-yeast interactions and their underlying mechanisms (Ciani et al., 2010; Ciani and Comitini, 2015). Indeed, several types of yeast-yeast interactions have been described in enological conditions: competition for nutriments, release of toxic compounds (Fleet, 2003), and even “quorum-sensing” like mechanisms (Nissen and Arneborg, 2003; Nissen et al., 2003; Renault et al., 2013). Understanding these complex interactions is of first importance as the combination of some yeast strains seems condemned to failure: for example, cell-cell contact was recently shown to be involved in the death of strains of T. delbrueckii and L. thermotolerans during mixed-culture alcoholic fermentation with S. cerevisiae (Renault et al., 2013; Kemsawasd et al., 2015). In some cases, yeast death was associated with the release of metabolites or killer toxin (Pérez-Nevado et al., 2006; Albergaria et al., 2010; Branco et al., 2015; Ramírez et al., 2015). The precise impact of such interactions regarding wine quality and aromas is still unclear (Ciani et al., 2006), but will have to be considered to control and optimize complex mixed oenological fermentation.
Finally, in addition to NGS-assisted approaches and interactions studies, another prospect in the field of non-conventional wine yeast lies in classical genetic approaches: indeed, one of the limits of the previously detailed approaches is their low ability in elucidating the basic life-cycle of wine yeasts, particularly regarding the occurrence and control of sexual reproduction. Still, classical breeding is one of the key issues for genetic improvement of industrial strains of S. cerevisiae (Pretorius, 2000; Giudici et al., 2005; Marullo et al., 2006; Steensels et al., 2014) and represents a technological barrier that must be overcome for actual improvement of non-Saccharomyces wine yeasts. There is an important need for traditional sporulation assays, spore microdissection attempts, subsequent segregant analyses, breeding assays, etc. In addition, genetic transformation of non-conventional wine yeasts would be a welcomed tool for subsequent functional studies (Pacheco et al., 2009; Roberts and Oliver, 2011). These classical approaches are time-consuming and necessitate traditional yeast-manipulation know-how, sometimes viewed as old-fashioned and therefore neglected. However, these old approaches are essential for our future understanding of the genetics of non-conventional wine yeast, and are complementary to the more en vogue NGS-assisted approaches.
Funding
This work was funded by the European Union's Seventh Framework Programme (FP7/2007–2013) under grant agreement n° 315065 WILDWINE project: “Multi-strain indigenous Yeast and Bacterial starters for “Wild-ferment” wine production.”
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aa E., Townsend J. P., Adams R. I., Nielsen K. M., Taylor J. W. (2006). Population structure and gene evolution in Saccharomyces cerevisiae. FEMS Yeast Res. 6, 702–715. 10.1111/j.1567-1364.2006.00059.x [DOI] [PubMed] [Google Scholar]
- Albergaria H., Francisco D., Gori K., Arneborg N., Gírio F. (2010). Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. 10.1007/s00253-009-2409-6 [DOI] [PubMed] [Google Scholar]
- Albertin W., Chasseriaud L., Comte G., Panfili A., Delcamp A., Salin F., et al. (2014a). Winemaking and bioprocesses strongly shaped the genetic diversity of the ubiquitous yeast Torulaspora delbrueckii. PLoS ONE 9:e94246. 10.1371/journal.pone.0094246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Marullo P., Aigle M., Bourgais A., Bely M., Dillmann C., et al. (2009). Evidence for autotetraploidy associated with reproductive isolation in Saccharomyces cerevisiae: towards a new domesticated species. J. Evol. Biol. 22, 2157–2170. 10.1111/j.1420-9101.2009.01828.x [DOI] [PubMed] [Google Scholar]
- Albertin W., Marullo P., Peltier E., Windholtz S., Bely M., Masneuf-Pomarede I. (2015). Biodiversity of Wine Yeasts: New Insights from Population Genetics. Oeno2015. [Google Scholar]
- Albertin W., Miot-Sertier C., Bely M., Marullo P., Coulon J., Moine V., et al. (2014b). Oenological prefermentation practices strongly impact yeast population dynamics and alcoholic fermentation kinetics in Chardonnay grape must. Int. J. Food Microbiol. 178, 87–97. 10.1016/j.ijfoodmicro.2014.03.009 [DOI] [PubMed] [Google Scholar]
- Albertin W., Miot-Sertier C., Bely M., Mostert T. T., Colonna-Ceccaldi B., Coulon J., et al. (2016). Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering. Front. Microbiol. 6:1569 10.3389/fmicb.2015.01569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertin W., Panfili A., Miot-Sertier C., Goulielmakis A., Delcamp A., Salin F., et al. (2014c). Development of microsatellite markers for the rapid and reliable genotyping of Brettanomyces bruxellensis at strain level. Food Microbiol. 42, 188–195. 10.1016/j.fm.2014.03.012 [DOI] [PubMed] [Google Scholar]
- Almeida P., Barbosa R., Zalar P., Imanishi Y., Shimizu K., Turchetti B., et al. (2015). A population genomics insight into the mediterranean origins of wine yeast domestication. Mol. Ecol. 24, 5412–5427. 10.1111/mec.13341 [DOI] [PubMed] [Google Scholar]
- Almeida P., Gonçalves C., Teixeira S., Libkind D., Bontrager M., Masneuf-Pomarède I., et al. (2014). A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 5:4044. 10.1038/ncomms5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Safadi R., Weiss-Gayet M., Briolay J., Aigle M. (2010). A polyploid population of Saccharomyces cerevisiae with separate sexes (dioecy). FEMS Yeast Res. 10, 757–768. 10.1111/j.1567-1364.2010.00660.x [DOI] [PubMed] [Google Scholar]
- Ambroset C., Petit M., Brion C., Sanchez I., Delobel P., Guérin C., et al. (2011). Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3 1, 263–281. 10.1534/g3.111.000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andorrà I., Berradre M., Mas A., Esteve-Zarzoso B., Guillamón J. M. (2012). Effect of mixed culture fermentations on yeast populations and aroma profile. LWT Food Sci. Technol. 49, 8–13. 10.1016/j.lwt.2012.04.008 [DOI] [Google Scholar]
- Anfang N., Brajkovich M., Goddard M. R. (2009). Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon Blanc. Aust. J. Grape Wine Res. 15, 1–8. 10.1111/j.1755-0238.2008.00031.x [DOI] [Google Scholar]
- Azzolini M., Fedrizzi B., Tosi E., Finato F., Vagnoli P., Scrinzi C., et al. (2012). Effects of Torulaspora delbrueckii and Saccharomyces cerevisiae mixed cultures on fermentation and aroma of Amarone wine. Eur. Food Res. Technol. 235, 303–313. 10.1007/s00217-012-1762-3 [DOI] [Google Scholar]
- Ball C. (1984). Genetics and Breeding of Industrial Microorganisms. Seattle, WA: CRC Press. [Google Scholar]
- Barnett J. A., Payne R. W., Yarrow D. (2000). Yeasts: Characteritics and Identification. Cambridge, UK: Cambridge University Press. [Google Scholar]
- Barquet M., Martín V., Medina K., Pérez G., Carrau F., Gaggero C. (2012). Tandem repeat-tRNA (TRtRNA) PCR method for the molecular typing of non-Saccharomyces subspecies. Appl. Microbiol. Biotechnol. 93, 807–814. 10.1007/s00253-011-3714-4 [DOI] [PubMed] [Google Scholar]
- Barrio E., Gonzalez S., Arias A., Belloch C., Querol A. (2006). Molecular mechanisms involved in the adaptive evolution of industrial yeasts, in Yeasts in Food and Beverages, eds Querol A., Fleet G. (Berlin; Heildeberg: Springer-Verlag; ), 153–174. [Google Scholar]
- Bely M., Renault P., da Silva T., Masneuf-Pomarede I., Albertin W., Moine V., et al. (2013). Non-conventional yeasts and alcohol level reduction, in Alcohol Level Reduction in Wine (Vigne et Vin Publications Internationales; ), 33–37. [Google Scholar]
- Bely M., Stoeckle P., Masneuf-Pomarède I., Dubourdieu D. (2008). Impact of mixed Torulaspora delbrueckii-Saccharomyces cerevisiae culture on high-sugar fermentation. Int. J. Food Microbiol. 122, 312–320. 10.1016/j.ijfoodmicro.2007.12.023 [DOI] [PubMed] [Google Scholar]
- Blein-Nicolas M., Albertin W., da Silva T., Valot B., Balliau T., Masneuf-Pomarede I., et al. (2015). A systems approach to elucidate heterosis of protein abundances in yeast. Mol. Cell. Proteomics 14, 2056–2071. 10.1074/mcp.M115.048058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Desany B. A., Riches D., Affourtit J. P., Forgan A. H., Pretorius I. S., et al. (2012). The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 12, 88–96. 10.1111/j.1567-1364.2011.00773.x [DOI] [PubMed] [Google Scholar]
- Borneman A. R., Forgan A. H., Pretorius I. S., Chambers P. J. (2008). Comparative genome analysis of a Saccharomyces cerevisiae wine strain. FEMS Yeast Res. 8, 1185–1195. 10.1111/j.1567-1364.2008.00434.x [DOI] [PubMed] [Google Scholar]
- Branco P., Viana T., Albergaria H., Arneborg N. (2015). Antimicrobial peptides (AMPs) produced by Saccharomyces cerevisiae induce alterations in the intracellular pH, membrane permeability and culturability of Hanseniaspora guilliermondii cells. Int. J. Food Microbiol. 205, 112–118. 10.1016/j.ijfoodmicro.2015.04.015 [DOI] [PubMed] [Google Scholar]
- Breuer U., Harms H. (2006). Debaryomyces hansenii–an extremophilic yeast with biotechnological potential. Yeast 23, 415–437. 10.1002/yea.1374 [DOI] [PubMed] [Google Scholar]
- Chan G. F., Gan H. M., Ling H. L., Rashid N. A. (2012). Genome sequence of Pichia kudriavzevii M12, a potential producer of bioethanol and phytase. Eukaryot. Cell 11, 1300–1301. 10.1128/EC.00229-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M., Beco L., Comitini F. (2006). Fermentation behaviour and metabolic interactions of multistarter wine yeast fermentations. Int. J. Food Microbiol. 108, 239–245. 10.1016/j.ijfoodmicro.2005.11.012 [DOI] [PubMed] [Google Scholar]
- Ciani M., Canonico L., Oro L., Comitini F. (2014). Sequential fermentation using non-Saccharomyces yeasts for the reduction of alcohol content in wine. BIO Web Conf. 3:02015 10.1051/bioconf/20140302015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani M., Comitini F. (2015). Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 1, 1–6. 10.1016/j.cofs.2014.07.001 [DOI] [Google Scholar]
- Ciani M., Comitini F., Mannazzu I., Domizio P. (2010). Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 10, 123–133. 10.1111/j.1567-1364.2009.00579.x [DOI] [PubMed] [Google Scholar]
- Ciani M., Maccarelli F. (1998). Oenological properties of non-Saccharomyces yeasts associated with wine-making. World J. Microbiol. Biotechnol. 14, 199–203. 10.1023/A:1008825928354 [DOI] [Google Scholar]
- Clemente-Jimenez J. M., Mingorance-Cazorla L., Martıìnez-Rodrıìguez S., Heras-Vázquez F. J. L., Rodrıìguez-Vico F. (2004). Molecular characterization and oenological properties of wine yeasts isolated during spontaneous fermentation of six varieties of grape must. Food Microbiol. 21, 149–155. 10.1016/S0740-0020(03)00063-7 [DOI] [Google Scholar]
- Clemente-Jimenez J. M., Mingorance-Cazorla L., Martínez-Rodríguez S., Las Heras-Vázquez F. J., Rodríguez-Vico F. (2005). Influence of sequential yeast mixtures on wine fermentation. Int. J. Food Microbiol. 98, 301–308. 10.1016/j.ijfoodmicro.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., et al. (2003). Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 301, 71–76. 10.1126/science.1084337 [DOI] [PubMed] [Google Scholar]
- Comitini F., Gobbi M., Domizio P., Romani C., Lencioni L., Mannazzu I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multistarter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28, 873–882. 10.1016/j.fm.2010.12.001 [DOI] [PubMed] [Google Scholar]
- Comitini F., De Ingeniis J., Pepe L., Mannazzu I., Ciani M. (2004). Pichia anomala and Kluyveromyces wickerhamii killer toxins as new tools against Dekkera/Brettanomyces spoilage yeasts. FEMS Microbiol. Lett. 238, 235–240. 10.1111/j.1574-6968.2004.tb09761.x [DOI] [PubMed] [Google Scholar]
- Csoma H., Sipiczki M. (2008). Taxonomic reclassification of Candida stellata strains reveals frequent occurrence of Candida zemplinina in wine fermentation. FEMS Yeast Res. 8, 328–336. 10.1111/j.1567-1364.2007.00339.x [DOI] [PubMed] [Google Scholar]
- Cubillos F. A., Vásquez C., Faugeron S., Ganga A., Martínez C. (2009). Self-fertilization is the main sexual reproduction mechanism in native wine yeast populations. FEMS Microbiol. Ecol. 67, 162–170. 10.1111/j.1574-6941.2008.00600.x [DOI] [PubMed] [Google Scholar]
- da Silva T., Albertin W., Dillmann C., Bely M., la Guerche S., Giraud C., et al. (2015). Hybridization within Saccharomyces Genus Results in Homoeostasis and Phenotypic Novelty in Winemaking Conditions. PLoS ONE 10:e0123834. 10.1371/journal.pone.0123834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezmann S., Dietrich F. S. (2009). Saccharomyces cerevisiae: population Divergence and Resistance to Oxidative Stress in Clinical, Domesticated and Wild Isolates. PLoS ONE 4:e5317. 10.1371/journal.pone.0005317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Maio S., Genna G., Gandolfo V., Amore G., Ciaccio M., Oliva D. (2012). Presence of Candida zemplinina in sicilian musts and selection of a strain for wine mixed fermentations. S. Afr. J. Enol. Viticult. 33, 80–87. [Google Scholar]
- Divol B., du Toit M., Duckitt E. (2012). Surviving in the presence of sulphur dioxide: strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 95, 601–613. 10.1007/s00253-012-4186-x [DOI] [PubMed] [Google Scholar]
- Domizio P., Liu Y., Bisson L. F., Barile D. (2014). Use of non-Saccharomyces wine yeasts as novel sources of mannoproteins in wine. Food Microbiol. 43, 5–15. 10.1016/j.fm.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Domizio P., Romani C., Comitini F., Gobbi M., Lencioni L., Mannazzu I., et al. (2011a). Potential spoilage non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Ann. Microbiol. 61, 137–144. 10.1007/s13213-010-0125-1 [DOI] [Google Scholar]
- Domizio P., Romani C., Lencioni L., Comitini F., Gobbi M., Mannazzu I., et al. (2011b). Outlining a future for non-Saccharomyces yeasts: Selection of putative spoilage wine strains to be used in association with Saccharomyces cerevisiae for grape juice fermentation. Int. J. Food Microbiol. 147, 170–180. 10.1016/j.ijfoodmicro.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Dujon B., Sherman D., Fischer G., Durrens P., Casaregola S., Lafontaine I., et al. (2004). Genome evolution in yeasts. Nature 430, 35–44. 10.1038/nature02579 [DOI] [PubMed] [Google Scholar]
- Egli C. M., Edinger W. D., Mitrakul C. M., Henick-Kling T. (1998). Dynamics of indigenous and inoculated yeast populations and their effect on the sensory character of Riesling and Chardonnay wines. J. Appl. Microbiol. 85, 779–789. 10.1046/j.1365-2672.1998.00521.x [DOI] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., Shapiro J. A., et al. (2010). Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464, 1039–1042. 10.1038/nature08923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englezos V., Rantsiou K., Torchio F., Rolle L., Gerbi V., Cocolin L. (2015). Exploitation of the non-Saccharomyces yeast Starmerella bacillaris (synonym Candida zemplinina) in wine fermentation: physiological and molecular characterizations. Int. J. Food Microbiol. 199, 33–40. 10.1016/j.ijfoodmicro.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Erny C., Raoult P., Alais A., Butterlin G., Delobel P., Matei-Radoi F., et al. (2012). Ecological success of a group of Saccharomyces cerevisiae/Saccharomyces kudriavzevii hybrids in the Northern European wine making environment. Appl. Environ. Microbiol. 8, 3256–3265. 10.1128/AEM.06752-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Zarzoso B., Peris-Torán M., Ramón D., Querol A. (2001). Molecular characterisation of Hanseniaspora species. Antonie van Leeuwenhoek 80, 85–92. 10.1023/A:1012268931569 [DOI] [PubMed] [Google Scholar]
- Fay J. C., Benavides J. A. (2005). Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 1, 66–71. 10.1371/journal.pgen.0010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet G. H. (2003). Yeast interactions and wine flavour. Int. J. Food Microbiol. 86, 11–22. 10.1016/S0168-1605(03)00245-9 [DOI] [PubMed] [Google Scholar]
- Foury F., Roganti T., Lecrenier N., Purnelle B. (1998). The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett. 440, 325–331. 10.1016/S0014-5793(98)01467-7 [DOI] [PubMed] [Google Scholar]
- Freel K. C., Friedrich A., Hou J., Schacherer J. (2014). Population genomic analysis reveals highly conserved mitochondrial genomes in the yeast species Lachancea thermotolerans. Genome Biol. Evol. 6, 2586–2594. 10.1093/gbe/evu203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friel D., Vandenbol M., Jijakli M. H. (2005). Genetic characterization of the yeast Pichia anomala (strain K), an antagonist of postharvest diseases of apple. J. Appl. Microbiol. 98, 783–788. 10.1111/j.1365-2672.2004.02520.x [DOI] [PubMed] [Google Scholar]
- García-Ríos E., Gutiérrez A., Salvadó Z., Arroyo-López F. N., Guillamon J. M. (2014). The fitness advantage of commercial wine yeasts in relation to the nitrogen concentration, temperature, and ethanol content under microvinification conditions. Appl. Environ. Microbiol. 80, 704–713. 10.1128/AEM.03405-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaramida P., Ponticello G., Di Maio S., Squadrito M., Genna G., Barone E., et al. (2013). Candida zemplinina for production of wines with less alcohol and more glycerol. S. Afr. J. Enol. Viticult. 34, 204–211. 10.1128/AEM.06768-11 [DOI] [Google Scholar]
- Giorello F. M., Berna L., Greif G., Camesasca L., Salzman V., Medina K., et al. (2014). Genome sequence of the native apiculate wine yeast Hanseniaspora vineae T02/19AF. Genome Announc 2, e00530–e00614. 10.1128/genomeA.00530-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudici P., Solieri L., Pulvirenti A. M., Cassanelli S. (2005). Strategies and perspectives for genetic improvement of wine yeasts. Appl. Microbiol. Biotechnol. 66, 622–628. 10.1007/s00253-004-1784-2 [DOI] [PubMed] [Google Scholar]
- Goddard M. R. (2008). Quantifying the complexities of Saccharomyces cerevisiae's ecosystem engineering via fermentation. Ecology 89, 2077–2082. 10.1890/07-2060.1 [DOI] [PubMed] [Google Scholar]
- Goddard M. R., Anfang N., Tang R., Gardner R. C., Jun C. (2010). A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ. Microbiol. 12, 63–73. 10.1111/j.1462-2920.2009.02035.x [DOI] [PubMed] [Google Scholar]
- Goffeau A., Barrell B. G., Bussey H., Davis R. W., Dujon B., Feldmann H., et al. (1996). Life with 6000 genes. Science 274, 546; 563–547. 10.1126/science.274.5287.546 [DOI] [PubMed] [Google Scholar]
- Gomez-Angulo J., Vega-Alvarado L., Escalante-García Z., Grande R., Gschaedler-Mathis A., Amaya-Delgado L., et al. (2015). Genome sequence of Torulaspora delbrueckii NRRL Y-50541, isolated from mezcal fermentation. Genome Announc. 3, e00438–e00515. 10.1128/genomeA.00438-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon J. L., Armisén D., Proux-Wéra E., ÓhÉigeartaigh S. S., Byrne K. P., Wolfe K. H. (2011). Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. U.S.A. 108, 20024–20029. 10.1073/pnas.1112808108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grangeteau C., Gerhards D., Rousseaux S., von Wallbrunn C., Alexandre H., Guilloux-Benatier M. (2015). Diversity of yeast strains of the genus Hanseniaspora in the winery environment: what is their involvement in grape must fermentation? Food Microbiol. 50, 70–77. 10.1016/j.fm.2015.03.009 [DOI] [PubMed] [Google Scholar]
- Hall B., Durall D. M., Stanley G. (2011). Population dynamics of Saccharomyces cerevisiae during spontaneous fermentation at a British Columbia Winery. Am. J. Enol. Vitic. 62, 66–72. 10.5344/ajev.2010.10054 [DOI] [Google Scholar]
- Hong Y.-A., Park H.-D. (2013). Role of non-Saccharomyces yeasts in Korean wines produced from campbell early grapes: potential use of Hanseniaspora uvarum as a starter culture. Food Microbiol. 34, 207–214. 10.1016/j.fm.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Howell K. S., Cozzolino D., Bartowsky E. J., Fleet G. H., Henschke P. A. (2006). Metabolic profiling as a tool for revealing Saccharomyces interactions during wine fermentation. FEMS Yeast Res. 6, 91–101. 10.1111/j.1567-1364.2005.00010.x [DOI] [PubMed] [Google Scholar]
- Jara M., Cubillos F. A., García V., Salinas F., Aguilera O., Liti G., et al. (2014). Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS ONE 9:e86533. 10.1371/journal.pone.0086533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarosz D. F., Brown J. C., Walker G. A., Datta M. S., Ung W. L., Lancaster A. K., et al. (2014). Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158, 1083–1093. 10.1016/j.cell.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L. J., Koufopanou V., Goddard M. R., Hetherington R., Schäfer S. M., Burt A. (2004). Population genetics of the wild yeast Saccharomyces paradoxus. Genetics 166, 43–52. 10.1534/genetics.166.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly N. P., Augustyn O. P. H., Pretorius I. S. (2006). The role and use of non-Saccharomyces yeasts in wine production. S. Afr. J. Enol. Viticult. 27, 15–39. [Google Scholar]
- Jung P. P., Friedrich A., Reisser C., Hou J., Schacherer J. (2012). Mitochondrial genome evolution in a single protoploid yeast species. G3 2, 1103–1111. 10.1534/g3.112.003152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemsawasd V., Branco P., Almeida M. G., Caldeira J., Albergaria H., Arneborg N. (2015). Cell-to-cell contact and antimicrobial peptides play a combined role in the death of Lachanchea thermotolerans during mixed-culture alcoholic fermentation with Saccharomyces cerevisiae. FEMS Microbiol. Lett. 362:fnv103. 10.1093/femsle/fnv103 [DOI] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- King E. S., Swiegers J. H., Travis B., Francis I. L., Bastian S. E. P., Pretorius I. S. (2008). Coinoculated fermentations using Saccharomyces yeasts affect the volatile composition and sensory properties of Vitis vinifera L. cv. Sauvignon Blanc Wines. J. Agric. Food Chem. 56, 10829–10837. 10.1021/jf801695h [DOI] [PubMed] [Google Scholar]
- Knop M. (2006). Evolution of the hemiascomycete yeasts: on life styles and the importance of inbreeding. Bioessays 28, 696–708. 10.1002/bies.20435 [DOI] [PubMed] [Google Scholar]
- Kreger-van Rij N. J. W. (1977). Ultrastructure of Hanseniaspora ascospores. Antonie Van Leeuwenhoek 43, 225–232. 10.1007/BF00395677 [DOI] [PubMed] [Google Scholar]
- Kumar S., Randhawa A., Ganesan K., Raghava G. P. S., Mondal A. K. (2012). Draft genome sequence of salt-tolerant yeast Debaryomyces hansenii var. hansenii MTCC 234. Eukaryotic Cell 11, 961–962. 10.1128/EC.00137-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman C. P., Fell J. W., Boekhout T. (2011). The Yeasts: A Taxonomic Study. Amsterdam: Elsevier. [Google Scholar]
- Kvitek D. J., Will J. L., Gasch A. P. (2008). Variations in Stress Sensitivity and Genomic Expression in Diverse, S. cerevisiae Isolates. PLoS Genet. 4:e1000223. 10.1371/journal.pgen.1000223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry C. R., Oh J., Hartl D. L., Cavalieri D. (2006). Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 366, 343–351. 10.1016/j.gene.2005.10.042 [DOI] [PubMed] [Google Scholar]
- Legras J. L., Merdinoglu D., Cornuet J. M., Karst F. (2007). Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 16, 2091–2102. 10.1111/j.1365-294X.2007.03266.x [DOI] [PubMed] [Google Scholar]
- Legras J. L., Ruh O., Merdinoglu D., Karst F. (2005). Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 102, 73–83. 10.1016/j.ijfoodmicro.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., James S. A., et al. (2009). Population genomics of domestic and wild yeasts. Nature 458, 337–341. 10.1038/nature07743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Louis E. J. (2005). Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59, 135–153. 10.1146/annurev.micro.59.030804.121400 [DOI] [PubMed] [Google Scholar]
- Liti G., Peruffo A., James S. A., Roberts I. N., Louis E. J. (2005). Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22, 177–192. 10.1002/yea.1200 [DOI] [PubMed] [Google Scholar]
- Magyar I., Nyitrai-Sárdy D., Leskó A., Pomázi A., Kállay M. (2014). Anaerobic organic acid metabolism of Candida zemplinina in comparison with Saccharomyces wine yeasts. Int. J. Food Microbiol. 178, 1–6. 10.1016/j.ijfoodmicro.2014.03.002 [DOI] [PubMed] [Google Scholar]
- Magyar I., Tóth T. (2011). Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 28, 94–100. 10.1016/j.fm.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Malpertuy A., Llorente B., Blandin G., Artiguenave F., Wincker P., Dujon B. (2000). Genomic exploration of the hemiascomycetous yeasts: 10. Kluyveromyces thermotolerans. FEBS Lett. 487, 61–65. 10.1016/S0014-5793(00)02281-X [DOI] [PubMed] [Google Scholar]
- Mangani S., Buscioni G., Collina L., Bocci E., Vincenzini M. (2011). Effects of microbial populations on anthocyanin profile of sangiovese wines produced in Tuscany, Italy. Am. J. Enol. Vitic. 62, 487–494. 10.5344/ajev.2011.11047 [DOI] [Google Scholar]
- Marsit S., Mena A., Bigey F., Sauvage F. X., Couloux A., Guy J., et al. (2015). Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol. Biol. Evol. 32, 1695–1707. 10.1093/molbev/msv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P., Aigle M., Bely M., Masneuf-Pomarède I., Durrens P., Dubourdieu D., et al. (2007). Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 7, 941–952. 10.1111/j.1567-1364.2007.00252.x [DOI] [PubMed] [Google Scholar]
- Marullo P., Bely M., Masneuf-Pomarède I., Pons M., Aigle M., Dubourdieu D. (2006). Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 6, 268–279. 10.1111/j.1567-1364.2006.00034.x [DOI] [PubMed] [Google Scholar]
- Marullo P., Dubourdieu D. (2010). Yeast selection for wine flavour modulation, in Managing Wine Quality, Vol. 2, edReynolds A. G. (Cambridge: Woodhead Publishing; ), 293–345. [Google Scholar]
- Masneuf-Pomarede I., Bely M., Marullo P., Lonvaud-Funel A., Dubourdieu D. (2010). Reassessment of phenotypic traits for Saccharomyces bayanus var. uvarum wine yeast strains. Int. J. Food Microbiol. 139, 79–86. 10.1016/j.ijfoodmicro.2010.01.038 [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I., Juquin E., Miot-Sertier C., Renault P., Laizet Y. H., Salin F., et al. (2015). The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. 15:fov045. FEMS Yeast Res. 10.1093/femsyr/fov045 [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I., Le Jeune C., Durrens P., Lollier M., Aigle M., Dubourdieu D. (2007). Molecular typing of wine yeast strains Saccharomyces bayanus var. uvarum using microsatellite markers. Syst. Appl. Microbiol. 30, 75–82. 10.1016/j.syapm.2006.02.006 [DOI] [PubMed] [Google Scholar]
- Masneuf-Pomarede I., Salin F., Coton E., Coton M., Le Jeune C., Lonvaud-Funel A., et al. (2009). First insight into the genetic structure of S. bayanus var. uvarum revealed by microsatellite markers analysis, in 27th ISSY (Paris: ). [Google Scholar]
- Maturano Y. P., Mestre M. V., Esteve-Zarzoso B., Nally M. C., Lerena M. C., Toro M. E., et al. (2015). Yeast population dynamics during prefermentative cold soak of Cabernet Sauvignon and Malbec wines. Int. J. Food Microbiol. 199, 23–32. 10.1016/j.ijfoodmicro.2015.01.005 [DOI] [PubMed] [Google Scholar]
- Miller M., Kock J. L. F., Pretorius G. H. J., Coetzee D. J. (1989). The value of orthogonal-field-alternation gel electrophoresis and other criteria in the taxonomy of the Genus Pichia Hansen emend. Kurtzman. Syst. Appl. Microbiol. 12, 191–202. 10.1016/S0723-2020(89)80014-1 [DOI] [Google Scholar]
- Mira N. P., Münsterkötter M., Dias-Valada F., Santos J., Palma M., Roque F. C., et al. (2014). The genome sequence of the highly acetic acid-tolerant Zygosaccharomyces bailii-derived interspecies hybrid strain ISA1307, isolated from a sparkling wine plant. DNA Res. 21, 299–313. 10.1093/dnares/dst058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira N., Mendes F., Guedes de Pinho P., Hogg T., Vasconcelos I. (2008). Heavy sulphur compounds, higher alcohols and esters production profile of Hanseniaspora uvarum and Hanseniaspora guilliermondii grown as pure and mixed cultures in grape must. Int. J. Food Microbiol. 124, 231–238. 10.1016/j.ijfoodmicro.2008.03.025 [DOI] [PubMed] [Google Scholar]
- Muller L. A. H., McCusker J. H. (2009). Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 18, 2779–2786. 10.1111/j.1365-294X.2009.04234.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy H. A., Zeyl C. W. (2010). Yeast sex: surprisingly high rates of outcrossing between asci. PLoS ONE 5:e10461. 10.1371/journal.pone.0010461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov G. I., Masneuf I., Naumova E. S., Aigle M., Dubourdieu D. (2000). Association of Saccharomyces bayanus var. uvarum with some French wines: genetic analysis of yeast populations. Res. Microbiol. 151, 683–691. 10.1016/S0923-2508(00)90131-1 [DOI] [PubMed] [Google Scholar]
- Naumov G. I., Naumova E. S. (2009). Chromosomal differentiation of the sibling species Pichia membranifaciens and Pichia manshurica. Microbiology 78, 214–217. 10.1134/S002626170902012X [DOI] [PubMed] [Google Scholar]
- Nissen P., Arneborg N. (2003). Characterization of early deaths of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch. Microbiol. 180, 257–263. 10.1007/s00203-003-0585-9 [DOI] [PubMed] [Google Scholar]
- Nissen P., Nielsen D., Arneborg N. (2003). Viable Saccharomyces cerevisiae cells at high concentrations cause early growth arrest of non-Saccharomyces yeasts in mixed cultures by a cell–cell contact-mediated mechanism. Yeast 20, 331–341. 10.1002/yea.965 [DOI] [PubMed] [Google Scholar]
- Novo M., Bigey F., Beyne E., Galeote V., Gavory F., Mallet S., et al. (2009). Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. U.S.A. 106, 16333–16338. 10.1073/pnas.0904673106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacheco A., Almeida M. J., Sousa M. J. (2009). Improved gene disruption method for Torulaspora delbrueckii. FEMS Yeast Res. 9, 158–160. 10.1111/j.1567-1364.2008.00452.x [DOI] [PubMed] [Google Scholar]
- Palanca L., Gaskett A. C., Günther C. S., Newcomb R. D., Goddard M. R. (2013). Quantifying variation in the ability of yeasts to attract Drosophila melanogaster. PLoS ONE 8:e75332. 10.1371/journal.pone.0075332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E., Claude J., Strimmer K. (2004). APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. 10.1093/bioinformatics/btg412 [DOI] [PubMed] [Google Scholar]
- Parapouli M., Hatziloukas E., Drainas C., Perisynakis A. (2010). The effect of Debina grapevine indigenous yeast strains of Metschnikowia and Saccharomyces on wine flavour. J. Indust. Microbiol. Biotechnol. 37, 85–93. 10.1007/s10295-009-0651-7 [DOI] [PubMed] [Google Scholar]
- Parle J. N., Di Menna M. E. (1966). The source of yeasts in New Zealand wines. N.Z. J. Agric. Res. 9, 98–107. 10.1080/00288233.1966.10418122 [DOI] [Google Scholar]
- Pérez-Nevado F., Albergaria H., Hogg T., Girio F. (2006). Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 108, 336–345. 10.1016/j.ijfoodmicro.2005.12.012 [DOI] [PubMed] [Google Scholar]
- Pfliegler W. P., Horváth E., Kallai Z., Sipiczki M. (2014). Diversity of Candida zemplinina isolates inferred from RAPD, micro/minisatellite and physiological analysis. Microbiol. Res. 169, 402–410. 10.1016/j.micres.2013.09.006 [DOI] [PubMed] [Google Scholar]
- Picazo C., Gamero-Sandemetrio E., Orozco H., Albertin W., Marullo P., Matallana E., et al. (2015). Mitochondria inheritance is a key factor for tolerance to dehydration in wine yeast production. Lett. Appl. Microbiol. 60, 217–222. 10.1111/lam.12369 [DOI] [PubMed] [Google Scholar]
- Pina A., Calderón I. L., Benítez T. (1986). Intergeneric hybrids of Saccharomyces cerevisiae and Zygosaccharomyces fermentati obtained by protoplast fusion. Appl. Environ. Microbiol. 51, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plech M., de Visser J. A., Korona R. (2014). Heterosis is prevalent among domesticated but not wild strains of Saccharomyces cerevisiae. G3 4, 315–323. 10.1534/g3.113.009381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramateftaki P. V., Kouvelis V. N., Lanaridis P., Typas M. A. (2006). The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 6, 77–90. 10.1111/j.1567-1364.2005.00018.x [DOI] [PubMed] [Google Scholar]
- Pramateftaki P. V., Kouvelis V. N., Lanaridis P., Typas M. A. (2008). Complete mitochondrial genome sequence of the wine yeast Candida zemplinina: intraspecies distribution of a novel group-IIB1 intron with eubacterial affiliations. FEMS Yeast Res. 8, 311–327. 10.1111/j.1567-1364.2007.00332.x [DOI] [PubMed] [Google Scholar]
- Pretorius I. S. (2000). Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast 16, 675–729. [DOI] [PubMed] [Google Scholar]
- Ramírez M., Velázquez R., Maqueda M., López-Piñeiro A., Ribas J. C. (2015). A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 6:983. 10.3389/fmicb.2015.00983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault P. E., Albertin W., Bely M. (2013). An innovative tool reveals interaction mechanisms among yeast populations under oenological conditions. Appl. Microbiol. Biotechnol. 97, 4105–4119. 10.1007/s00253-012-4660-5 [DOI] [PubMed] [Google Scholar]
- Renault P., Miot-Sertier C., Marullo P., Hernández-Orte P., Lagarrigue L., Lonvaud-Funel A., et al. (2009). Genetic characterization and phenotypic variability in Torulaspora delbrueckii species: potential applications in the wine industry. Int. J. Food Microbiol. 134, 201–210. 10.1016/j.ijfoodmicro.2009.06.008 [DOI] [PubMed] [Google Scholar]
- Richards K. D., Goddard M. R., Gardner R. C. (2009). A database of microsatellite genotypes for Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 96, 355–359. 10.1007/s10482-009-9346-3 [DOI] [PubMed] [Google Scholar]
- Roberts I. N., Oliver S. G. (2011). The yin and yang of yeast: biodiversity research and systems biology as complementary forces driving innovation in biotechnology. Biotechnol. Lett. 33, 477–487. 10.1007/s10529-010-0482-7 [DOI] [PubMed] [Google Scholar]
- Rodrigues F., Ludovico P., Sousa M. J., Steensma H. Y., Côrte-Real M., Leão C. (2003). The spoilage yeast Zygosaccharomyces bailii forms mitotic spores: a screening method for haploidization. Appl. Environ. Microbiol. 69, 649–653. 10.1128/AEM.69.1.649-653.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas V., Gil J. V., Piñaga F., Manzanares P. (2001). Studies on acetate ester production by non-Saccharomyces wine yeasts. Int. J. Food Microbiol. 70, 283–289. 10.1016/S0168-1605(01)00552-9 [DOI] [PubMed] [Google Scholar]
- Ruderfer D. M., Pratt S. C., Seidel H. S., Kruglyak L. (2006). Population genomic analysis of outcrossing and recombination in yeast. Nat. Genet. 38, 1077–1081. 10.1038/ng1859 [DOI] [PubMed] [Google Scholar]
- Sadoudi M., Tourdot-Maréchal R., Rousseaux S., Steyer D., Gallardo-Chacón J. J., Ballester J., et al. (2012). Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non-Saccharomyces and Saccharomyces yeasts. Food Microbiol. 32, 243–253. 10.1016/j.fm.2012.06.006 [DOI] [PubMed] [Google Scholar]
- Salinas F., Cubillos F. A., Soto D., Garcia V., Bergström A., Warringer J., et al. (2012). The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS ONE 7:e49640. 10.1371/journal.pone.0049640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadó Z., Arroyo-López F. N., Barrio E., Querol A., Guillamón J. M. (2011). Quantifying the individual effects of ethanol and temperature on the fitness advantage of Saccharomyces cerevisiae. Food Microbiol. 28, 1155–1161. 10.1016/j.fm.2011.03.008 [DOI] [PubMed] [Google Scholar]
- Sarilar V., Bleykasten-Grosshans C., Neuvéglise C. (2015). Evolutionary dynamics of hAT DNA transposon families in Saccharomycetaceae. Genome Biol. Evol. 7, 172–190. 10.1093/gbe/evu273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D. R., Butler G., Wolfe K. H. (2007). Yeast genome evolution–the origin of the species. Yeast 24, 929–942. 10.1002/yea.1515 [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L. (2009). Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458, 342–345. 10.1038/nature07670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller D., Cardoso F., Sousa S., Gomes P., Gomes A. C., Santos M. A., et al. (2012). Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS ONE 7:e32507. 10.1371/journal.pone.0032507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard D., Legras J. L. (2011). Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C. R. Biol. 334, 229–236. 10.1016/j.crvi.2010.12.016 [DOI] [PubMed] [Google Scholar]
- Sipiczki M. (2004). Species identification and comparative molecular and physiological analysis of Candida zemplinina and Candida stellata. J. Basic Microbiol. 44, 471–479. 10.1002/jobm.200410449 [DOI] [PubMed] [Google Scholar]
- Sniegowski P. D., Dombrowski P. G., Fingerman E. (2002). Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 1, 299–306. 10.1111/j.1567-1364.2002.tb00048.x [DOI] [PubMed] [Google Scholar]
- Solieri L., Dakal T., Giudici P. (2013). Next-generation sequencing and its potential impact on food microbial genomics. Ann. Microbiol. 63, 21–37. 10.1007/s13213-012-0478-8 [DOI] [Google Scholar]
- Souciet J. L., Dujon B., Gaillardin C., Johnston M., Baret P. V., Cliften P., et al. (2009). Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 19, 1696–1709. 10.1101/gr.091546.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starmer W. T., Ganter P. F., Aberdeen V. (1992). Geographic distribution and genetics of killer phenotypes for the yeast Pichia kluyveri across the United States. Appl. Environ. Microbiol. 58, 990–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J., Snoek T., Meersman E., Picca Nicolino M., Voordeckers K., Verstrepen K. J. (2014). Improving industrial yeast strains: exploiting natural and artificial diversity. FEMS Microbiol. Rev. 38, 947–995. 10.1111/1574-6976.12073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensels J., Verstrepen K. J. (2014). Taming wild yeast: potential of conventional and nonconventional yeasts in industrial fermentations. Annu. Rev. Microbiol. 68, 61–80. 10.1146/annurev-micro-091213-113025 [DOI] [PubMed] [Google Scholar]
- Stefanini I., Dapporto L., Legras J. L., Calabretta A., Di Paola M., De Filippo C., et al. (2012). Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc. Natl. Acad. Sci. U.S.A. 109, 13398–13403. 10.1073/pnas.1208362109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strope P. K., Skelly D. A., Kozmin S. G., Mahadevan G., Stone E. A., Magwene P. M., et al. (2015). The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 25, 762–774. 10.1101/gr.185538.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin K. A. (2010). Fructophilic Yeasts to Cure Stuck Fermentations in Alcoholic Beverages. Ph.D. thesis, University of Stellenbosch. [Google Scholar]
- Talla E., Anthouard V., Bouchier C., Frangeul L., Dujon B. (2005). The complete mitochondrial genome of the yeast Kluyveromyces thermotolerans. FEBS Lett. 579, 30–40. 10.1016/j.febslet.2004.10.106 [DOI] [PubMed] [Google Scholar]
- Timberlake W. E., Frizzell M. A., Richards K. D., Gardner R. C. (2011). A new yeast genetic resource for analysis and breeding. Yeast 28, 63–80. 10.1002/yea.1821 [DOI] [PubMed] [Google Scholar]
- Tofalo R., Perpetuini G., Fasoli G., Schirone M., Corsetti A., Suzzi G. (2014). Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d'Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8S-ITS restriction patterns. Food Microbiol. 39, 7–12. 10.1016/j.fm.2013.10.001 [DOI] [PubMed] [Google Scholar]
- Tofalo R., Perpetuini G., Schirone M., Fasoli G., Aguzzi I., Corsetti A., et al. (2013). Biogeographical characterization of Saccharomyces cerevisiae wine yeast by molecular methods. Front. Microbiol. 4:166. 10.3389/fmicb.2013.00166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tofalo R., Schirone M., Torriani S., Rantsiou K., Cocolin L., Perpetuini G., et al. (2012). Diversity of Candida zemplinina strains from grapes and Italian wines. Food Microbiol. 29, 18–26. 10.1016/j.fm.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Viana F., Belloch C., Vallés S., Manzanares P. (2011). Monitoring a mixed starter of Hanseniaspora vineae–Saccharomyces cerevisiae in natural must: impact on 2-phenylethyl acetate production. Int. J. Food Microbiol. 151, 235–240. 10.1016/j.ijfoodmicro.2011.09.005 [DOI] [PubMed] [Google Scholar]
- Viana F., Gil J. V., Genovés S., Vallés S., Manzanares P. (2008). Rational selection of non-Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits. Food Microbiol. 25, 778–785. 10.1016/j.fm.2008.04.015 [DOI] [PubMed] [Google Scholar]
- Wang C., Liu Y. (2013). Dynamic study of yeast species and Saccharomyces cerevisiae strains during the spontaneous fermentations of Muscat blanc in Jingyang, China. Food Microbiol. 33, 172–177. 10.1016/j.fm.2012.09.014 [DOI] [PubMed] [Google Scholar]
- Wang C., Mas A., Esteve-Zarzoso B. (2015). Interaction between Hanseniaspora uvarum and Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 206, 67–74. 10.1016/j.ijfoodmicro.2015.04.022 [DOI] [PubMed] [Google Scholar]
- Wang Q.-M., Liu W.-Q., Liti G., Wang S.-A., Bai F.-Y. (2012). Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol. Ecol. 21, 5404–5417. 10.1111/j.1365-294X.2012.05732.x [DOI] [PubMed] [Google Scholar]
- Warringer J., Zörgö E., Cubillos F. A., Zia A., Gjuvsland A., Simpson J. T., et al. (2011). Trait variation in yeast is defined by population history. PLoS Genet. 7:e1002111. 10.1371/journal.pgen.1002111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrent P., Rivas E. M., Peinado J. M., de Silóniz M. I. (2015). Development of an affordable typing method for Meyerozyma guilliermondii using microsatellite markers. Int. J. Food Microbiol. 217, 1–6. 10.1016/j.ijfoodmicro.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Wu B., Buljic A., Hao W. (2015). Extensive horizontal transfer and homologous recombination generate highly chimeric mitochondrial genomes in yeast. Mol. Biol. Evol. 32, 2559–2570. 10.1093/molbev/msv127 [DOI] [PubMed] [Google Scholar]
- Yanai T., Sato M. (1999). Isolation and properties of β-glucosidase produced by Debaryomyces hansenii and its application in winemaking. Am. J. Enol. Vitic. 50, 231–235. [Google Scholar]
- Zara G., Mannazzu I., Del Caro A., Budroni M., Pinna M. B., Murru M., et al. (2014). Wine quality improvement through the combined utilisation of yeast hulls and Candida zemplinina/Saccharomyces cerevisiae mixed starter cultures. Aust. J. Grape Wine Res. 20, 199–207. 10.1111/ajgw.12078 [DOI] [Google Scholar]
- Zeyl C. (2004). Capturing the adaptive mutation in yeast. Res. Microbiol. 155, 217–223. 10.1016/j.resmic.2003.12.006 [DOI] [PubMed] [Google Scholar]
- Zimmer A., Durand C., Loira N., Durrens P., Sherman D. J., Marullo P. (2014). QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS ONE 9:e86298. 10.1371/journal.pone.0086298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zott K., Thibon C., Bely M., Lonvaud-Funel A., Dubourdieu D., Masneuf-Pomarede I. (2011). The grape must non-Saccharomyces microbial community: impact on volatile thiol release. Int. J. Food Microbiol. 151, 210–215. 10.1016/j.ijfoodmicro.2011.08.026 [DOI] [PubMed] [Google Scholar]