Abstract
Aggregation of misfolded proteins is a characteristic of several neurodegenerative diseases. The huntingtin amino-terminal fragment with extended polyglutamine repeat forms aggregates closely associated with chaperones both in the cytoplasm and the nucleus. Because each cellular compartment contains distinct chaperones and because the molecular mechanisms controlling polyglutamine aggregation are largely unknown, we decided to investigate the influence of different cellular environments on the aggregation of this pathological protein. Here, we show that aggregation of a protein containing a polyglutamine stretch of pathological length is abolished when its expression is targeted to the endoplasmic reticulum. Once retrogradely transported outside the endoplasmic reticulum, the aggregation-prone polyglutamine-containing protein recovers its ability to aggregate. When expressed in the mitochondria, a protein containing 73 glutamines is entirely soluble, whereas the nucleocytosolic equivalent has an extremely high tendency to aggregate. Our data imply that polyglutamine aggregation is a property restricted to the nucleocytosolic compartment and suggest the existence of compartment-specific cofactors promoting or preventing aggregation of pathological proteins.
Several neurodegenerative diseases including Alzheimer's disease, Parkinson's disease, Huntington's diseases (and related polyglutamine disorders), amyotrophic lateral sclerosis, and prion disorders share striking common features despite diverse clinical symptoms and transmission mode (1, 2). The hallmark of these disorders is the progressive neuronal loss associated with deposition of aggregated protein of aberrant conformation (1–6). Despite considerable differences in primary sequence, the pathological conformers of the disease-specific proteins share similar important structural features (5, 7). Numerous genetic, cell biology, and biochemical data support the model whereby neurodegeneration is caused by a toxic gain of function of the mutated protein. The molecular mechanisms controlling the initiation of the conformational transition from a soluble, functional conformation to an aggregated pathological one are so far unknown. Quality control pathways have evolved to protect cells from the deleterious effect of misfolded proteins. Molecular chaperones refold abnormally folded polypeptides or direct them to degradation machinery (8). Failure to refold or to degrade aberrant proteins leads to their aggregation. Neurones, as postmitotic cells, are particularly sensitive to misfolding injury. In addition, the accumulation of aberrant proteins increases with age, when quality control might become less efficient, as illustrated by a decreased activity of the proteasome degradation machinery (9). Together, these observations may provide a rationale for the late onset of neurodegenerative diseases.
One of the paradigmatic misfolding diseases is Huntington's disease, an autosomal dominant disease caused by a CAG repeat expansion in the first exon of the huntingtin gene, which translates into polyglutamine and causes aggregation of the mutated protein in patient brains (10), transgenic mice (11), and cellular models of Huntington's disease (12–14). Polyglutamine repeat (polyQ)-containing polypeptides of a pathological length were proposed to adopt an aberrant structure causing their aggregation and subsequent cellular dysfunction (15). This model received support from numerous investigators and is reinforced with the conditional expression of the amino-terminal region of huntingtin in transgenic mice showing that aggregate formation and appearance of pathological symptoms are closely linked (16). More recently, polyQ aggregation was shown to directly contribute to cellular dysfunction (17, 18). Pathological polyQ-containing proteins form aggregates closely associated with chaperones both in the cytosol and in the nucleus. Because each cellular compartment contains distinct chaperones and because the mechanism controlling polyQ aggregation is unknown, we addressed the expression of huntingtin amino-terminal fragment to the endoplasmic reticulum (ER) and mitochondria and found that polyQ aggregation is dramatically influenced by the subcellular environment. Our results suggest the existence of cellular factors dramatically modulating pathological protein aggregation.
Materials and Methods
Expression Plasmids, Cell Culture, and Transfections. Htt17 and Htt73 are, respectively, 171-17 and 171-73 from ref. 19. Htt17 encodes the first 163 aa of wild-type huntingtin followed by a hemagglutinin epitope in pcDNA1/Amp vector (Invitrogen), and Htt73 is the mutant version with 73 CAG. The first 19 amino acids of hBIP (signal peptide, SP) were fused to Htt17 or Htt73 to generate SP-Htt17 and SP-Htt73 (20). A KDEL sequence was added carboxyl-terminally to generate SP-HttKDEL. 293T cells were maintained in DMEM supplemented with 10% FBS and transfected in six-well plates with 1 μg of the indicated expression plasmids by using the calcium phosphate method, leading routinely to >70% transfection efficiency. NG108-15 cells were cultured as described in ref. 13 and transfected with FuGENE (Roche Molecular Biochemicals). Protein extracts were prepared 48 h later by boiling in 180 μl of Laemmli buffer, 20 μl of which was analyzed by huntingtin immunoblot as described below; 5 μl of such extracts was analyzed on GRP94 immunoblot.
Antibodies, Filter Retardation Assay, and Immunoblotting. Monoclonal 2B4 is described in ref. 21. Calnexin polyclonal serum and GRP94 antibody were purchased from Stressgen, and ubiquitin rabbit antiserum was from Sigma. For filter retardation assay performed as described in ref. 22, 5–20 and 30–50 μl of protein extracts were loaded on cellulose acetate for 2B4 and ubiquitin immunoblots, respectively. Duplicates loaded on nitrocellulose membrane of 0.2 μM pore, probed with huntingtin antibody, constitute an accurate loading control only when comparing Htt derivatives of similar size. Immunoblotting procedure was as described in ref. 21. For SDS/PAGE followed by immunoblot, equal loading of whole cell extracts was verified by Ponceau red staining of the membrane.
Immunocytochemistry. Immunostaining of Htt derivatives was performed as described in ref. 21. Where indicated, cells were labeled with 25 nM MitoTracker (Molecular Probes) 20 min before fixation.
Proteasome Inhibition and Cycloheximide Treatment. Sixteen hours posttransfection, culture media were changed, and MG-132 or lactacystin (Calbiochem) was added at the indicated doses for 16 h. Proteasome inhibitors were solubilized in DMSO. Final DMSO concentration on cells is <0.05% vol/vol, a dose where no effect on huntingtin aggregation was observed (data not shown). To estimate the half-life of the various Htt derivatives, the amount of plasmids was adjusted to allow comparable expression levels of the different proteins. Cells were treated with 50 μg/ml cycloheximide 36 h posttransfection for the indicated time (Figs. 1d and 2d).
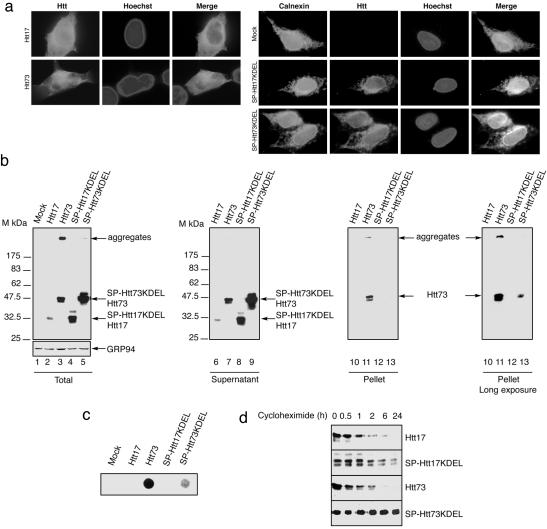
Fig. 1.
Targeting Htt73 expression to the ER markedly reduces its ability to aggregate. (a) Immunostaining of Htt derivatives in 293T transfected cells. Cells expressing Htt17 or Htt73 constructs were stained for huntingtin. SP-Htt17KDEL, SP-Htt73KDEL, or mock-transfected cells were costained for huntingtin and calnexin, an ER marker, 16 h posttransfection. Nuclei were revealed with Hoechst. Note that Htt73 forms multiple foci that precede the formation of large aggregates. (b) Immunoblots of 293T cell extracts 48 h posttransfection of the indicated constructs revealed with huntingtin and GRP94 antibodies. 293T cells lysates shown in lanes 2–5 were centrifuged to separate soluble (lanes 6–9) from insoluble material and revealed by immunoblot. SDS-insoluble pellets were solubilized by using formic acid before immunoblot (lanes 10–13). (c) Filter retardation assay with similar lysates as shown in b, lanes 2–5, revealed by immunoblot with huntingtin antibody. (d) Immunoblot of Htt17-, Htt73-, SP-Htt17KDEL-, and SP-Htt73KDEL-transfected cell extracts untreated or treated with 50 μg/ml cycloheximide for the indicated time.
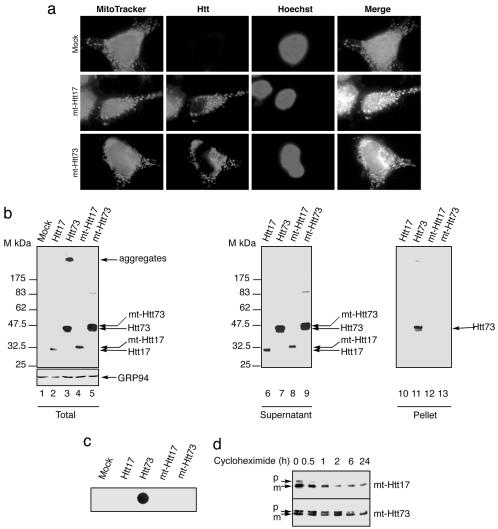
Fig. 2.
Targeting Htt73 to mitochondria abolishes its aggregation. (a) Photomicrographs of mock-, mt-Htt17-, or mt-Htt73-transfected 293T cells labeled with MitoTracker and stained for Htt. Hoechst stains all nuclei. (b) Immunoblots of 293T cell extracts 48 h posttransfection of the indicated constructs revealed with huntingtin and GRP94 antibodies. 293T cells lysates shown in lanes 2–5 were centrifuged to separate soluble (lanes 6–9) from insoluble material and revealed by immunoblot. SDS-insoluble pellets were solubilized by using formic acid before immunoblot (lanes 10–13). (c) Filter retardation assay of similar lysates revealed by immunoblot with huntingtin antibody. (d) Huntingtin immunoblots of cells extracts from 293T cells transfected with mt-Htt17 and mt-Htt73 either left untreated or treated with 50 μg/ml cycloheximide for the indicated time. p and m indicate the precursor and the mature form of the mt-Htt proteins.
Results
Htt73 Expressed in the ER Is Soluble. In the present study, we used, as a well characterized model, the amino-terminal region of wild-type huntingtin with 17Q and the corresponding region of mutant huntingtin containing a repeat of 73Q, Htt17, and Htt73, respectively. The extent of aggregation of the mutant protein was analyzed in different subcellular compartments. Htt proteins were addressed to the ER by fusing the SP sequence of BIP, an ER chaperone (20) to Htt17 and Htt73, generating SP-Htt17 and SP-Htt73, respectively. A sequence coding for the ER retention sequence KDEL (23) was added to the chimeras SP-Htt to generate SP-Htt17KDEL and SP-Htt73KDEL. Immunocytochemical studies performed on 293T cells 16 h posttransfection reveal that Htt17 and Htt73 proteins exhibit a diffuse nucleocytoplasmic location with small inclusions in Htt73-transfected cells, whereas SP-HttKDEL derivatives colocalize with the ER marker calnexin (Fig. 1a). Htt73 aggregates are readily observable by immunostaining 48 h posttransfection, as reported by others (12), whereas both SP-Htt17KDEL and SP-Htt73KDEL proteins are predominantly located in vesicular structures emanating from the ER (data not shown). On immunoblots of total cellular lysates of 293T cells obtained 48 h posttransfection, Htt17 derivatives are detected at a position corresponding to ≈32 kDa, whereas 73Q-containing proteins migrate at ≈47 kDa (Fig. 1b). In addition, in cell extracts derived from Htt73-transfected cells, a band is detected at a position corresponding to the top of the gel, a feature of SDS-resistant aggregates. Strikingly, aggregation of SP-Htt73KDEL proteins is almost abolished as compared with Htt73. Because SP-Htt17KDEL and SP-Htt73KDEL expression levels exceed by ≈7-fold their Htt counterparts, it seems unlikely that the marked reduction of SP-Htt73KDEL aggregation results from insufficient concentration. Total cellular extracts were subsequently fractionated by microcentrifugation as described in ref. 21. The supernatant of this fractionation contains soluble proteins (Fig. 1b, lanes 6–9). The pellets, containing insoluble material mostly composed of Htt aggregates, were dissolved by formic acid (24) and subjected to immunoblotting (Fig. 1b, lanes 10–13). The formic acid-treated Htt73 proteins are readily observable, whereas the signal corresponding to the SP-Htt73KDEL solubilized material is detected only after very long exposure of the immunoblot (Fig. 1b, rightmost blot) confirming our former observation. To get insight on the ratio of soluble versus aggregated Htt73 derivatives, we used a filter retardation assay (22). In this quantitative assay, the signal corresponding to Htt73 is >10-fold higher than the signal corresponding to SP-Htt73KDEL (Fig. 1c). This suggests that addressing Htt73 to the ER markedly reduces aggregation. The ER folding capacity is tightly controlled and can be massively augmented in response to accumulation of misfolded proteins (25). SP-HttKDEL overexpression does not induce an ER stress response monitored here by a GRP94 immunoblot (Fig. 1b), suggesting that in the ER a protein containing expanded polyQ is not recognized as misfolded. We next examined whether aggregation of SP-Htt73KDEL could be hindered by means of an increased turnover. 293T cells were transfected with the indicated constructs and collected at various times after blockade of new protein synthesis by using cycloheximide. Immunoblots were performed on total cellular extracts (Fig. 1d). The measured half-life of both Htt17 and Htt73 is ≈45 min (Fig. 1d). Because SP-HttKDEL proteins are noticeably more stable than the Htt proteins, it seems unlikely that SP-Htt73KDEL aggregation is reduced by means of an increased turnover (Fig. 1d). Because the ER environment is different from the cytosol regarding redox conditions and ions, SP-HttKDEL expressing cells were challenged with Thapsigargin treatment, provoking a calcium release from this reservoir and thereby a dramatic change in the ER milieu (26). This treatment did not alter SP-Htt73KDEL solubility (data not shown).
Htt73 Expressed in the Mitochondria Is Entirely Soluble. Because the ER is a peculiar environment, we next examined Htt73 aggregation properties when targeted to another cellular compartment, the mitochondria. mt-Htt17 and mt-Htt73 chimeras were generated by fusing the first 35 aa of human aconitase 2 to Htt17 or Htt73. Aconitase 2 mitochondrial targeting sequence was chosen because aconitase 2 mRNA has been found in the vicinity of mitochondria, a property that could favor the rapidity of protein import in this organelle (27). This caveat was important because we aimed to import polyQ-containing proteins of pathological length in the mitochondria before their aggregation in the cytosol. Subcellular location of the mt-Htt constructs was analyzed by immunocytochemistry performed on 293T transfected cells (Fig. 2a). In mt-Htt17- or mt-Htt73-transfected cells, the huntingtin antibody labels a compartment that is colabeled with the mitochondrion-selective probe MitoTracker. Immunoblot analysis of total cellular extracts reveals that in contrast to Htt73, which gives rise to two bands corresponding to the soluble protein migrating at an estimated molecular mass of 47 kDa, with the aggregated one incapable of penetrating the gel, Htt17, mt-Htt17, and mt-Htt73 are exclusively soluble and migrate, respectively, at ≈32 and 47 kDa (Fig. 2b). In mitochondria, a protein containing a 73 polyQ is entirely SDS-soluble in contrast to Htt73. This observation is confirmed by sedimentation experiments. Whereas Htt73 is present both in the SDS-soluble fraction (Fig. 2b, lane 7) and in the formic acid-solubilized pellet (Fig. 2b, lane 11), Htt17, mt-Htt17, and mt-Htt73 are exclusively detected in the soluble fractions (Fig. 2b, lanes 6 and 10, 8 and 12, and 9 and 13, respectively). Htt73 aggregates are retained by filtration on cellulose acetate membrane, as revealed by the immunoblot of the membrane (Fig. 2c), whereas Htt17, mt-Htt17, and mt-Htt73 flow through, confirming their solubility. The stability of mt-Htt derivatives was evaluated by inhibiting new protein synthesis with cycloheximide. Similarly to the SP-HttKDEL derivatives, the half-life of mt-Htt proteins is extended compared to their Htt counterparts (Figs. 1d and 2d). Note that in untreated cells mt-Htt17 and mt-Htt73 are both revealed as two bands corresponding most likely to the precursor protein (p) containing the uncleaved mitochondrial targeting sequence and the mature protein (m) of faster mobility. In the course of protein synthesis inhibition, we observed disappearance of the precursor (Fig. 2d), further confirming that mt-Htt proteins are addressed to the mitochondria. Because mt-Htt proteins are expressed at a higher level than Htt proteins and because the cytosol is a compartment of a greater volume than the mitochondrial matrix, the concentration of mt-Htt73 seems, therefore, higher in the mitochondria than the concentration of Htt73 in the nucleocytoplasmic compartment. Targeting Htt73 to mitochondria hinders its aggregation.
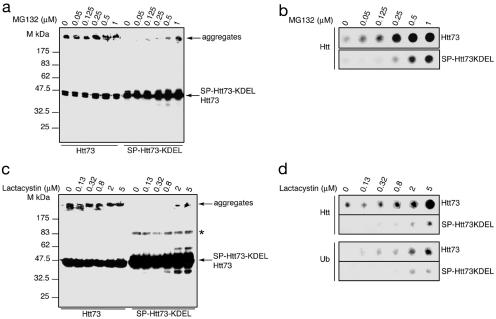
Proteasome Inhibition Provokes Aggregation of SP-Htt73KDEL. In light of these findings, we reexamined the meaning of the trace of aggregation observed by overexpressing the SP-Htt73KDEL chimera (Fig. 1 b and c). When ER proteins have to be degraded, they are subjected to retrograde transport into the cytosol, where they are ubiquitinated and subsequently degraded by the proteasome, a process referred to ER-associated degradation, or ERAD (28, 29). We speculated that SP-Htt73KDEL aggregates might arise not in the ER, where the protein would be soluble, but in the cytosol, if and when the protein would be retrogradely transported out of the ER. Despite the obvious stability of the SP-HttKDEL derivatives (Fig. 1d), we hypothesized that some of these derivatives might return in the cytosol and be targeted to proteasomal degradation. Compromising proteasome activity would then increase the likelihood of the occurrence of SP-Htt73KDEL aggregates. 293T cells expressing either Htt73 or SP-Htt73KDEL were treated with MG-132 or lactacystin. In agreement with previous reports, proteasome inhibition increases the amount of Htt73 aggregation (30, 31) (Fig. 3). This effect is more obvious in the quantitative filter retardation assay (Fig. 3 b and d). Both MG-132 and lactacystin induce SP-Htt73KDEL aggregation in a concentration-dependent manner (Fig. 3). The trace amount of SP-Htt73KDEL aggregation in untreated cells (Fig. 1 b and c) is thereby likely due to the retrograde transport of the protein in the cytosol. Immunoblotting a cellulose acetate membrane on which Htt73 and SP-Htt73KDEL aggregates were retained with an antibody reacting with ubiquitin reveals that they are ubiquitinated (Fig. 3d). This result suggests that both Htt73 and SP-Htt73KDEL aggregates form in a cellular compartment where the ubiquitin-proteasome system is found, namely in the cytosol or in the nucleus (32). Abrogation of SP-Htt73KDEL aggregation in the ER is reversible, indicating that the protein has only transiently lost its aggregation propensity because of its mislocation.
Fig. 3.
Proteasome inhibition induces SP-Htt73KDEL aggregation. Increasing doses of MG-132 (a) or lactacystin (c) augment Htt73 aggregation and induce SP-Htt73KDEL aggregation. Immunoblots with the 2B4 antibody of lysates from 293T cells transfected with the indicated expression vectors and either left untreated or treated with 0.05, 0.125, 0.25, 0.5, and 1 μM MG-132 or 0.13, 0.32, 0.8, 2, and 5 μM lactacystin 16 h posttransfection and for 16 h. A band of ≈90 kDa is revealed on the Htt73 and SP-Htt73KDEL immunoblots either after long exposure (data not shown) or with high Htt proteins amount (*). (b and d) Cell extracts presented in a and c were analyzed by filter retardation assay and revealed by immunoblot with huntingtin or ubiquitin antibody.
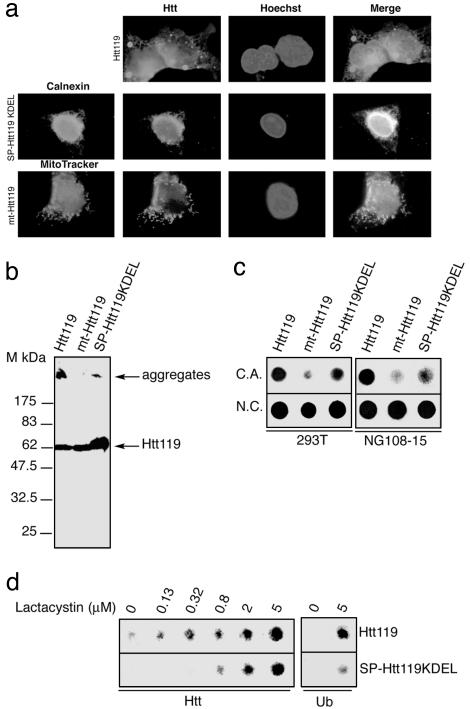
Because huntingtin aggregation increases with the length of the polyQ, we tested the aggregation extent of Htt derivatives containing 119Q in different compartments. SP-Htt119KDEL and mt-Htt119 chimeras were constructed. Immunocytochemical analysis reveals that Htt119 aggregates are observable as early as 16 h posttransfection. SP-Htt119KDEL and mt-Htt119 locate in the expected compartments, the ER and the mitochondria, respectively (Fig. 4a). Htt119 aggregates are retained at the top of the gel, whereas the soluble Htt119 protein migrates similarly to a 62-kDa protein (Fig. 4b). SP-Htt119KDEL is highly expressed but has a reduced propensity to aggregate when compared with Htt119. mt-Htt119 seems completely soluble. Filter retardation assay analysis confirms that aggregation of SP-Htt119KDEL is markedly reduced compared with Htt119, and mt-Htt119 is mostly soluble (Fig. 4c). Aggregation of SP-Htt119KDEL is augmented by treatment with the proteasome inhibitor lactacystin (Fig. 4d), and both aggregated Htt119 and SP-Htt119KDEL proteins are ubiquitinated (Fig. 4d). The trace of mt-Htt119 aggregate might arise in the cytosol prior to the import of the protein. We next analyzed the aggregation of Htt119, mt-Htt119, and SP-Htt119KDEL in a well characterized cellular model of Huntington's disease, the neuronal-like NG108-15 cells (13). The observations made by using 293T cells are fully recapitulated in NG108-15 cells (Fig. 4c) (data not shown). The results shown here indicate that polyQs of pathological length lose their aggregation ability when targeted to the ER or the mitochondria.
Fig. 4.
Targeting of Htt119 to the ER or mitochondria markedly reduces its ability to aggregate. (a) Immunocytochemical detection of Htt derivatives in 293T cells transfected with the indicated constructs, 16 h posttransfection. Cells expressing SP-Htt73KDEL were costained with an anti-calnexin antibody. Cells transfected with mt-Htt119 construct were labeled with MitoTracker. Nuclei are stained with Hoechst. (b) Immunoblots with the 2B4 antibody of lysates from 293T cells 48 h posttransfection with the indicated constructs. (c) Filter retardation assay on cellulose acetate membrane (C.A.) of lysates from 293T or neuronal-like NG108-15 cells transfected with the indicated constructs. (Lower) Samples were spotted in duplicate on a nitrocellulose (N.C.) membrane, 0.2-μm pore size, as loading control. (d) Filter retardation assay with cells extracts of 293T cells transfected with Htt119 and SP-Htt119KDEL either left untreated or treated with or 0.13, 0.32, 0.8, 2, and 5 μM lactacystin 16 h posttransfection for 16 h and revealed by immunoblot with huntingtin or ubiquitin antibody.
Discussion
Targeting mutant huntingtin polypeptide to the ER and mitochondria led us to discover that aggregation of polyQ-containing proteins of pathological length depends on cellular environment. The cellular system used in our study composed of the amino-terminal polyQ-containing region of huntingtin expressed in 293T cells is a very robust one because the expression levels of the various Htt derivatives is extremely high. Preventing aggregation of a polyQ-containing protein of pathological length while achieving high expression levels is thus a remarkable finding. In addition, the compartmentalized polyglutamine propensity to aggregate is not restricted to 293T cells, because our findings are fully reproduced in neuronal-like NG108-15 cells. Because both SP and mitochondrial targeting sequence presequences are cleaved upon import in the ER and mitochondria, respectively (20, 33) (Fig. 2d), it is unlikely that their addition is responsible for the solubility of the polyQ-containing proteins of pathological length in the ER and mitochondria. More importantly, the ER-targeted SP-Htt73KDEL and SP-Htt119KDEL-derived proteins aggregate when they are retrotranslocated to the cytosol, indicating that these artificially created proteins are fully aggregation-competent.
Our findings imply the existence of cellular factors dramatically modulating polyQ aggregation. Antiaggregation factors might prevent polyQ aggregation in the ER and the mitochondria. Such factors would be present under basal conditions because expression of SP-HttKDEL proteins does not lead to a detectable ER stress induction (Fig. 1b). Alternatively, cytosolic cofactors might be required to assist or even catalyze expanded polyQ aggregation. Identification of factors modulating polyQ aggregation becomes of first importance in the comprehension of the heretofore mysterious origin of this process and because such factors will serve as therapeutic targets. Whether protein sequestered within polyQ aggregates, such as chaperones or transcription factors (4), participates in aggregate formation is one path to explore. Remarkably, in yeast, deletion of the chaperone-encoding gene Hsp104 completely prevents aggregation of a protein containing as many as 103 glutamines (34). In addition, deletion of HSP104 irreversibly cures all three yeast prions (35). Alteration of other chaperone expressions modulates polyQ aggregation and prion conversion and propagation to a more subtle extent (35). Whereas no HSP104 orthologue (36) has been identified so far in mammals, our results prompt us to suggest the existence of a functional analogue.
The behavior of the SP-HttKDEL derivatives strikingly resembles the one of the mammalian prion protein PrP, which conversion into an aberrant conformation occurs in the cytosol (37–41). PrP is a protein that naturally traffics through the secretory route and that appears in the cytosol by retrograde translocation out of the ER (38, 39). The striking common properties of the extended polyQ-containing SP-HttKDEL proteins and PrP protein further underscore common mechanistic features in aggregate formation of pathological polyQ-containing proteins and PrP conversion into pathogenic conformation. A protein X, yet to be identified, was proposed to assist PrP conversion to the scrapie conformation (42). The results presented here might, therefore, apply to other misfolding diseases such as prion disorders and Alzheimer's and Parkinson's diseases, and an aggregation cofactor might be common to the aforementioned misfolding diseases. As recently suggested, an RNA might also participate in the aggregation of proteins involved in neurodegenerative diseases (43). This work provides a robust and simple experimental system for identification of pathological protein aggregation modulators.
Acknowledgments
We thank Bruno Goud and Franck Perez (Institut Curie, Paris) for insightful discussions and reagents, Frédéric Saudou (Institut Curie, Orsay, France) for 171-17 and 171-73 constructs, Didier Devys (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for pTL-Flag125 construct, Liliana Paslaru for help with antibodies, Elisabeth Scheer for advice with the transfections, Anne Levesqueau for excellent technical assistance, and Xavier Jacq for valuable comments on the manuscript. This work was supported by Groupement d'Intérêt Scientifique Infections à Prions, and B.D. is a recipient of an MRT fellowship. L.B.-H. and Y.T. are supported by the Hereditary Disease Foundation.
Abbreviations: SP, signal peptide; ER, endoplasmic reticulum; polyQ, polyglutamine repeat.
References
- 1.Taylor, J. P., Hardy, J. & Fischbeck, K. H. (2002) Science 296, 1991-1995. [DOI] [PubMed] [Google Scholar]
- 2.Sherman, M. Y. & Goldberg, A. L. (2001) Neuron 29, 15-32. [DOI] [PubMed] [Google Scholar]
- 3.Orr, H. T. (2001) Genes Dev. 15, 925-932. [DOI] [PubMed] [Google Scholar]
- 4.Sakahira, H., Breuer, P., Hayer-Hartl, M. K. & Hartl, F. U. (2002) Proc. Natl. Acad. Sci. USA 99, Suppl. 4, 16412-16418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soto, C. (2003) Nat. Rev. Neurosci. 4, 49-60. [DOI] [PubMed] [Google Scholar]
- 6.Selkoe, D. J. (2003) Nature 426, 900-904. [DOI] [PubMed] [Google Scholar]
- 7.Temussi, P. A., Masino, L. & Pastore, A. (2003) EMBO J. 22, 355-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wickner, S., Maurizi, M. R. & Gottesman, S. (1999) Science 286, 1888-1893. [DOI] [PubMed] [Google Scholar]
- 9.Carrard, G., Bulteau, A. L., Petropoulos, I. & Friguet, B. (2002) Int. J. Biochem. Cell Biol. 34, 1461-1474. [DOI] [PubMed] [Google Scholar]
- 10.DiFiglia, M., Sapp, E., Chase, K. O., Davies, S. W., Bates, G. P., Vonsattel, J. P. & Aronin, N. (1997) Science 277, 1990-1993. [DOI] [PubMed] [Google Scholar]
- 11.Davies, S. W., Turmaine, M., Cozens, B. A., DiFiglia, M., Sharp, A. H., Ross, C. A., Scherzinger, E., Wanker, E. E., Mangiarini, L. & Bates, G. P. (1997) Cell 90, 537-548. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, J. K., Schilling, G., Peters, M. F., Herring, W. J., Sharp, A. H., Kaminsky, Z., Masone, J., Khan, F. A., Delanoy, M., Borchelt, D. R., et al. (1998) Hum. Mol. Genet. 7, 783-790. [DOI] [PubMed] [Google Scholar]
- 13.Lunkes, A. & Mandel, J. L. (1998) Hum. Mol. Genet. 7, 1355-1361. [DOI] [PubMed] [Google Scholar]
- 14.Martindale, D., Hackam, A., Wieczorek, A., Ellerby, L., Wellington, C., McCutcheon, K., Singaraja, R., Kazemi-Esfarjani, P., Devon, R., Kim, S. U., et al. (1998) Nat. Genet. 18, 150-154. [DOI] [PubMed] [Google Scholar]
- 15.Perutz, M. F., Johnson, T., Suzuki, M. & Finch, J. T. (1994) Proc. Natl. Acad. Sci. USA 91, 5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto, A., Lucas, J. J. & Hen, R. (2000) Cell 101, 57-66. [DOI] [PubMed] [Google Scholar]
- 17.Yang, W., Dunlap, J. R., Andrews, R. B. & Wetzel, R. (2002) Hum. Mol. Genet. 11, 2905-2917. [DOI] [PubMed] [Google Scholar]
- 18.Sanchez, I., Mahlke, C. & Yuan, J. (2003) Nature 421, 373-379. [DOI] [PubMed] [Google Scholar]
- 19.Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. (1998) Cell 95, 55-66. [DOI] [PubMed] [Google Scholar]
- 20.Munro, S. & Pelham, H. R. (1986) Cell 46, 291-300. [DOI] [PubMed] [Google Scholar]
- 21.Lunkes, A., Lindenberg, K. S., Ben-Haiem, L., Weber, C., Devys, D., Landwehrmeyer, G. B., Mandel, J. L. & Trottier, Y. (2002) Mol. Cell 10, 259-269. [DOI] [PubMed] [Google Scholar]
- 22.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R., Bates, G. P., Davies, S. W., Lehrach, H. & Wanker, E. E. (1997) Cell 90, 549-558. [DOI] [PubMed] [Google Scholar]
- 23.Munro, S. & Pelham, H. R. (1987) Cell 48, 899-907. [DOI] [PubMed] [Google Scholar]
- 24.Hazeki, N., Tukamoto, T., Goto, J. & Kanazawa, I. (2000) Biochem. Biophys. Res. Commun. 277, 386-393. [DOI] [PubMed] [Google Scholar]
- 25.Mori, K. (2000) Cell 101, 451-454. [DOI] [PubMed] [Google Scholar]
- 26.Thastrup, O., Cullen, P. J., Drobak, B. K., Hanley, M. R. & Dawson, A. P. (1990) Proc. Natl. Acad. Sci. USA 87, 2466-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sylvestre, J., Margeot, A., Jacq, C., Dujardin, G. & Corral-Debrinski, M. (2003) Mol. Biol. Cell 14, 3848-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopito, R. R. (1997) Cell 88, 427-430. [DOI] [PubMed] [Google Scholar]
- 29.Tsai, B., Ye, Y. & Rapoport, T. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 246-255. [DOI] [PubMed] [Google Scholar]
- 30.Waelter, S., Boeddrich, A., Lurz, R., Scherzinger, E., Lueder, G., Lehrach, H. & Wanker, E. E. (2001) Mol. Biol. Cell 12, 1393-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jana, N. R., Zemskov, E. A., Wang, G. & Nukina, N. (2001) Hum. Mol. Genet. 10, 1049-1059. [DOI] [PubMed] [Google Scholar]
- 32.Ciechanover, A. & Brundin, P. (2003) Neuron 40, 427-446. [DOI] [PubMed] [Google Scholar]
- 33.Pfanner, N. & Geissler, A. (2001) Nat. Rev. Mol. Cell Biol. 2, 339-349. [DOI] [PubMed] [Google Scholar]
- 34.Krobitsch, S. & Lindquist, S. (2000) Proc. Natl. Acad. Sci. USA 97, 1589-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uptain, S. M. & Lindquist, S. (2002) Annu. Rev. Microbiol. 56, 703-741. [DOI] [PubMed] [Google Scholar]
- 36.Parsell, D. A., Sanchez, Y., Stitzel, J. D. & Lindquist, S. (1991) Nature 353, 270-273. [DOI] [PubMed] [Google Scholar]
- 37.Ma, J. & Lindquist, S. (1999) Nat. Cell Biol. 1, 358-361. [DOI] [PubMed] [Google Scholar]
- 38.Yedidia, Y., Horonchik, L., Tzaban, S., Yanai, A. & Taraboulos, A. (2001) EMBO J. 20, 5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, J. & Lindquist, S. (2001) Proc. Natl. Acad. Sci. USA 98, 14955-14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, J., Wollmann, R. & Lindquist, S. (2002) Science 298, 1781-1785. [DOI] [PubMed] [Google Scholar]
- 41.Ma, J. & Lindquist, S. (2002) Science 298, 1785-1788. [DOI] [PubMed] [Google Scholar]
- 42.Kaneko, K., Zulianello, L., Scott, M., Cooper, C. M., Wallace, A. C., James, T. L., Cohen, F. E. & Prusiner, S. B. (1997) Proc. Natl. Acad. Sci. USA 94, 10069-10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deleault, N. R., Lucassen, R. W. & Supattapone, S. (2003) Nature 425, 717-720. [DOI] [PubMed] [Google Scholar]