Abstract
This data article presents an analysis of structural water molecules in the high affinity interaction between a potent tumor growth inhibiting antibody (fragment), J22.9-xi, and the tumor marker antigen CD269 (B cell maturation antigen, BCMA). The 1.89 Å X-ray crystal structure shows exquisite details of the binding interface between the two molecules, which comprises relatively few, mostly hydrophobic, direct contacts but many indirect interactions over solvent waters. These are partly or wholly buried in, and therefore part of, the interface. A partial description of the structure is included in an article on the tumor inhibiting effects of the antibody: “Potent anti-tumor response by targeting B cell maturation antigen (BCMA) in a mouse model of multiple myeloma”, Mol. Oncol. 9 (7) (2015) pp. 1348–58.
Keywords: Crystal structure, Fab fragment, BCMA, Binding interface, High affinity, Water molecules
Specifications Table
| Subject area | Chemistry, Biology, Cancer immunology |
| More specific subject area | Structural biology |
| Type of data | Parameter table for structurally observed water molecules; figures depicting positions of these waters in a binding interface |
| How data was acquired | Analysis of the refined X-ray crystallographic structure. |
| Data format | Table, figures. |
| Experimental factors | Fab fragments from anti-CD269 (BCMA) IgGs co-crystallized with antigen and the structure solved to high (1.89 Å) resolution |
| Experimental features | Interface interactions in the refined structure were assessed and the contribution of water to the binding evaluated. |
| Data source location | Protein Data Bank |
| Data accessibility | 〈http://www.rcsb.org/pdb/home/home.do〉 (PDB code: 4ZFO) |
Value of the data
-
•
The antibody targets the very restrictively expressed tumor marker BCMA for Multiple Myeloma and potently inhibits tumor growth in mice
-
•
The antibody has an exceptionally high affinity to BCMA (picomolar)
-
•
The high resolution crystal structure of the antibody-BCMA complex reveals a network of water molecules in the binding interface
-
•
The structure shows how very high affinity can be achieved with a minimal number of direct protein-protein contacts
1. Data, experimental design, materials and methods
Multiple Myeloma (MM) is an incurable malignancy of antibody-secreting B cells (plasma cells) with a mean life expectancy of 5 years from diagnosis [1]. We generated a chimeric mouse/human antibody (J22.9-xi) against CD269 (BCMA), a plasma membrane receptor expressed nearly exclusively in plasma cells; the antibody shows potent cell killing activity on MM cells in vitro and anti-tumor activity in vivo. Surface Plasmon Resonance Spectrometry (SPR) measurements gave an exceptionally high affinity of J22.9-xi for BCMA of 50 pM [2]. Although higher affinity antibody:antigen interactions have been achieved by protein engineering/selection techniques [3], the J22.9-xi interaction exceeds the low nanomolar affinity range typical of therapeutic antibodies [4], [5] (see also, for example, Herceptin at 5 nM [6] and Rituximab at 8 nM [7]) by approximately 100-fold.
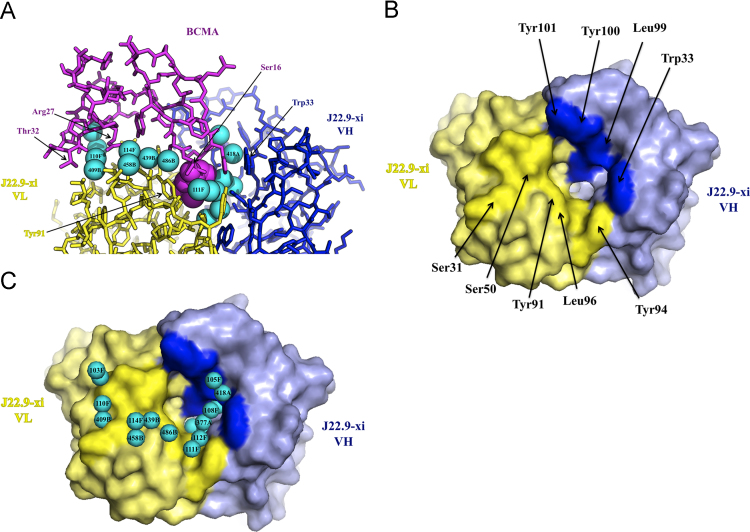
A protruding loop in the antigen BCMA (the DxL loop) with Leu17 at its apex is critical for binding to the native BCMA ligands, B cell Activating Factor of the TNF Family (BAFF) and A PRoliferation Inducing Ligand (APRIL) [8], both of which have a hydrophobic cavity that packs tightly around Leu17. Our 1.89 Å X-ray structure of a J22.9-xi Fab fragment in complex with BCMA shows that J22.9-xi also targets the DxL loop (Fig. 1A). However, the Leu17 binding cavity in J22.9-xi is substantially larger than the Leu17 side chain and provides only limited hydrophobic interactions between Leu17 and the VL of the antibody, primarily with Tyr91; the remainder of the binding cavity is filled with 6 water molecules that pack against the side of Leu17 opposite the Tyr91 interaction (Fig. 1A, B and C). This high affinity interaction comprises few minor direct contacts between the molecules (4 direct hydrogen bonds and 21 mostly single atom van der Waals contacts) but at least 50 hydrogen bonds involving waters “bridging” interactions between main chain and side chain atoms; 17 of these water molecules are directly in the binding interface (Table 1). Thus, the structure shows how high affinity binding can be achieved with only a small number of direct interactions between the binding partners.
Fig. 1.
Water molecules in the J22.9-xi binding site. A. A view of the binding interaction between BCMA (magenta) and the variable domains of the heavy chain (VH, blue) and light chain (VL, yellow). Interface water molecules participating in bridging hydrogen bonding interactions between BCMA and J22.9-xi are depicted as cyan spheres with a radius of 1.4 Å. For clarity only some interface residues (with the corresponding chain color) and some interface waters (with chain numbers directly on the spheres) are labeled. Leu17 in the BCMA DxL loop is shown in space-filling representation. B. A view looking down on the binding cavity in J22.9-xi with BCMA removed. The Fab fragment is shown in surface representation with the heavy chain colored blue and the light chain colored yellow. Some of the residues making direct contacts to BCMA are indicated with arrows and darker coloring. C. View as in (B) with interface waters depicted as cyan spheres and labeled as in (A).
Table 1.
Water interactions in the J22.9-xi:CD269 complex. The interaction partners are read from left to right across the table, with the corresponding H-bond distances and thermal displacement (B) factors of the indicated water molecules listed (J22.9-xi:H2O:CD269). For example, Ser31 from the J22.9-xi light chain participates in a hydrogen bonding interaction with water 409B at a distance of 2.50 Å and water 409B in turn interacts with Thr32 of CD269 at a distance of 2.65 Å. The data in Table 1 were generated using the software LigPlot [8] and COOT [11].
| Light Chain (B) | H2O# | CD269 (F) | Distance (Å) | B (Å2) |
|---|---|---|---|---|
| Ser31 (sc) | 409B | Thr32 (sc, mc) | 2.50, 2.65 (sc) | 24.33 |
| 2.50, 3.15 (mc) | ||||
| 409B, 110F | Arg27 (sc) | 2.65, 2.60, 2.98 | 24.33, 23.47 | |
| 409B, 110F | Ser30 (sc) | 2.50, 2.65, 2.69 | 24.33, 23.47 | |
| Ser31 (mc) | 458B, 114F | Arg27 (sc) | 2.71, 2.75, 2.78 | 26.48, 28.32 |
| Asn32 (sc) | 439B | Asp15 (sc) | 2.67, 2.88 | 19.24 |
| 439B, 114F | Arg27 (sc) | 2.67, 2.94, 2.78 | 19.24, 28.32 | |
| 486B | Ser16 (sc) | 2.80, 3.18 | 15.45 | |
| Tyr36 (sc) | 522B, 336A, 377A | Leu17 (mc) | 3.19, 2.90, 2.77, 2.88 | 14.87, 18.33, 35.33 |
| Ser50 (sc) | 439B | Asp15 (sc) | 3.11, 2.88 | 19.24 |
| Ser52 (sc) | 110F | Ser30 (sc) | 2.73, 2.69 | 23.47 |
| 110F | Arg27 (sc) | 2.73, 2.98 | 23.47 | |
| Ser52 (sc) | 479B, 103F | Ser30 (mc) | 2.78, 2.65, 2.94 | 25.34, 31.49 |
| Ser29 (sc) | 2.78, 2.65, 2.54 | |||
| 110F, 409B | Thr32 (sc, mc) | 2.73, 2.60, 2.65 | 23.47, 24.33 | |
| Gly66 (mc) | 409B | Thr32 (sc) | 2.86, 2.65 | 24.33 |
| 409B, 110F | Arg27 (sc) | 2.86, 2.60, 2.98 | 24.33, 23.47 | |
| 409B, 110F | Ser30 (sc) | 2.86, 2.60, 2.69 | 24.33, 23.47 | |
| Gln89 (sc) | 522B, 336A, 377A | Leu17 (mc) | 2.92, 2.90, 2.77, 2.88 | 18.47, 18.33, 35.33 |
| Tyr91 (mc) | 111F | Ser16 (sc) | 2.89, 2.71 | 32.32 |
| 111F, 112F | Ser16 (mc) | 2.89, 2.79, 2.76 | 32.32, 23.28 | |
| Tyr94 (sc) | 112F | Ser16 (mc) | 3.12, 2.76 | 23.28 |
| 112F, 111F | Ser16 (sc) | 3.12, 2.79, 2.71 | 23.28, 32.32 | |
| Heavy Chain (A) | H2O# | CD269 (F) | Distance (Å) | B (Å2) |
| Trp33 (mc) | 421A, 108F | Leu17 (mc) | 2.98, 2.76, 2.62 | 16.25, 31.34 |
| 418A | Leu18 (mc) | 2.90, 3.45 | 19.66 | |
| Ser35 (sc) | 421A, 108F | Leu17 (mc) | 2.91, 2.76, 2.62 | 16.25, 31.34 |
| 336A, 377A | ||||
| 2.64, 2.77, 2.88 | 18.33, 35.33 | |||
| Trp47 (sc) | 336A, 377A | Leu17 (mc) | 2.96, 2.77, 2.88 | 18.33, 35.33 |
| Glu50 (sc) | 112F | Ser16 (mc) | 3.21, 2.76 | 23.28 |
| 112F, 111F | Ser16 (sc) | 3.21, 2.79, 2.71 | 23.28, 32.32 | |
| 377A | Leu17 (mc) | 2.75, 2.88 | 35.33 | |
| 377A, 108F | Leu17 (mc) | 2.75, 2.48, 2.62 | 35.33, 31.34 | |
| Leu99 (mc) | 108F | Leu17 (mc) | 3.26, 2.62 | 31.34 |
| Tyr101 (mc) | 105F | Leu18 (mc) | 3.50, 2.57 | 25.20 |
(sc=side chain H-bond; mc=main chain H-bond)
2. Analysis of binding interface
Direct atom to atom contacts between BCMA and the VH and VL of J22.9-xi were identified using the PISA interaction server [9]. Direct and indirect hydrogen bonding interactions involving water were identified and validated (distance and geometry) from the structure using LigPlot [10] and COOT [11].
Acknowledgments
The authors wish to thank the Helmholtz Gemeinschaft for financial support.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2015.12.023.
Appendix A. Supplementary material
Supplementary material
References
- 1.Palumbo A., Anderson K. Multiple myeloma. N. Engl. J. Med. 2011;364:1046–1060. doi: 10.1056/NEJMra1011442. [DOI] [PubMed] [Google Scholar]
- 2.Oden F., Marino S.F., Brand J., Scheu S., Kriegel C., Olal D., Takvorian A., Westermann J., Yilmaz B., Hinz M., Daumke O., Höpken U., Müller G., Lipp M. Potent anti-tumor response by targeting B cell maturation antigen (BCMA) in a mouse model of multiple myeloma. Mol. Oncol. 2015;9(7):1348–1358. doi: 10.1016/j.molonc.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boder E.T., Midelfort K.S., Wittrup K.D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. PNAS. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foote J., Eisen H.N. Kinetic and affinity limits on antibodies produced during immune responses. PNAS. 1995;92:1254–1256. doi: 10.1073/pnas.92.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batista F.D., Neuberger M.S. Affinity Dependence of the b cell response to antigen: a threshhold, a ceiling, and the importance of off-rate. Immunity. 1998;8:751–759. doi: 10.1016/s1074-7613(00)80580-4. [DOI] [PubMed] [Google Scholar]
- 6.〈http://www.fda.gov/downloads/Drugs/DevelopmentApprovalProcess/HowDrugsareDevelopedandApproved/ApprovalApplications/TherapeuticBiologicApplications/ucm092760.pdf〉
- 7.〈http://www.globalrph.com/rituximab.htm〉
- 8.Gordon N.C., Pan B., Hymowitz S.G., Yin J., Kelley R.F., Cochran A.G., Yan M., Dixit V.M., Fairbrother W.J., Starovasnik M.A. BAFF/BLyS receptor 3 comprises a minimal TNF receptor-like module that encodes a highly focused ligand-binding site. Biochemistry. 2003;42:5977–5983. doi: 10.1021/bi034017g. [DOI] [PubMed] [Google Scholar]
- 9.Protein interfaces, surfaces and assemblies׳ service PISA at the European Bioinformatics Institute. 〈http://www.ebi.ac.uk/pdbe/prot_int/pistart.html〉; Krissinel, E., Henrick, K., 2007. ׳Inference of macromolecular assemblies from crystalline state.׳ J. Mol. Biol. 372, pp. 774–97. [DOI] [PubMed]
- 10.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1996;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 11.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material