Abstract
In metazoans, multiple RNA-binding proteins, including the shuttling serine/arginine-rich (SR)-splicing factors, function as adapters for mRNA nuclear export by interacting with the export receptor TAP/nuclear export factor 1 (NXF1). Yet, it is unclear how interactions between adapters and TAP are regulated. Here, we demonstrate that the SR proteins 9G8 and ASF/SF2 exhibit higher affinity for TAP/NXF1 when hypophosphorylated. 9G8 is recruited to the pre-mRNA in a hyperphosphorylated form but becomes hypophosphorylated during splicing both in vivo and in vitro. TAP preferentially binds spliced mRNA–protein complexes compared with pre-mRNA–protein complexes. Thus, the phosphorylation state of the SR protein adapters may underlie the selectivity of TAP-mediated export of spliced mRNA.
In eukaryotic cells, pre-mRNA and mRNA molecules exist complexed with proteins to form ribonucleoprotein (RNP) particles. RNPs are subjected to substantial remodeling as pre-mRNAs undergo splicing and polyadenylation before mRNA export through the nuclear pore to the cytoplasm (1). Export-competent mRNPs contain at least two critical components: adapter proteins that bind directly to the mRNA cargo and the export receptor TAP (Mex67p in yeast) (2, 3). Consistent with its central role in mRNA export, TAP, in concert with its cofactor p15 (Mtr-2 in yeast) (4), interacts with both adapter proteins and components of the nuclear pore complex to effect transport. RNA export factor 1 (REF1)/Aly (Yra1p in yeast) is an adapter that is recruited to mRNA by specific transcription elongation complexes that include the DEAD (D-E-A-D in the single-letter code for Asp-Glu-Ala-Asp)-box RNA helicase UAP56 (Sub2p in yeast) (5, 6). In yeast, Mex67p, Yra1p, and Sub2p are all required for mRNA export (2, 3). However, in Drosophila cells and Caenorhabditis elegans, only TAP and UAP56 are essential whereas REF1 is not (7–9). The lack of a requirement for REF1 in metazoans was rationalized by the existence of another class of adapters for mRNA export, serine/arginine-rich (SR) proteins, that shuttle between the nucleus and the cytoplasm, such as 9G8, ASF/SF2, and SRp20 (10). 9G8 and SRp20 can be UV-crosslinked to both nuclear and cytoplasmic polyadenylated RNAs in mammalian cells (11), and 9G8 was found associated with microinjected adenovirus mRNAs in both the nucleus and cytoplasm of Xenopus oocytes (12). In yeast, the RNA-binding protein Npl3p is required for mRNA export and shuttles between the nucleus and the cytoplasm in association with polyadenylated mRNA (13). The shuttling behavior of Npl3p depends on its phosphorylation state (14).
In metazoans, SR proteins comprise a family of essential splicing factors that are often functionally interchangeable (15). They bind multiple sites, including exonic enhancers, in pre-mRNA, and some remain bound to spliced mRNA (12, 15). SR proteins possess a C-terminal region rich in serine/arginine dipeptides that is the target of extensive phosphorylation and has been linked to splicing activity (15). Specifically, in vitro, active splicing complexes fail to form in the presence of protein phosphatases (16), and splicing is arrested before catalysis when thio-phosphorylated (phosphatase resistant) ASF/SF2 (17) or phosphatase inhibitors (16) are present. These results, together with the observation that spliced mRNAs are preferentially exported, suggested that SR protein dephosphorylation occurring during splicing may trigger the recruitment of TAP.
Here, we have tested this hypothesis by examining the role of phosphorylation of two shuttling SR proteins, 9G8 and ASF/SF2, in their interaction with the export receptor TAP. We find that dephosphorylation indeed enhances the binding of these SR proteins to TAP. Moreover, the act of splicing both in vivo and in vitro is accompanied by dephosphorylation of SR proteins associated with mRNA.
Methods
Antibodies and Plasmids. Antibodies specific for 9G8, ASF/SF2, and SC35 (12) and the anti-hnRNP A1 (18) were gifts from J. Stévenin (Institut de Génétique et de Biologie Moleculaire et Cellulaire, Illkirch, France) and G. Dreyfuss (University of Pennsylvania School of Medicine, Philadelphia), respectively. Mouse preimmune IgG (Upstate Biotechnology, Lake Placid, NY) and the monoclonal anti-Flag affinity agarose beads (Sigma) were purchased. Plasmids AdML (19), pcDNA-Flag, pcDNA-Flag-TAP (12), and pGEX-4T1-TAP 1-231 for generating GST-TAP231 (12) have been described. Plasmid pXJ42-9G8 for synthesizing 9G8 in vitro was a gift from J. Stévenin.
Phosphatase Treatment and Binding Assays. For phosphatase treatment, 5 μl of HeLa nuclear extracts were mixed with 2 μl of alkaline phosphatase (AP, 1 unit/μl, Boehringer), 1 μl of 10× AP buffer, and 2 μl of buffer D (12) in a final volume of 10 μl, followed by incubation at 30°C for 10 min. To inhibit dephosphorylation, phosphatase inhibitors β-glycerophosphate and potassium fluoride were also included in the reaction at a final concentration of 20 mM each. For binding assays, GST or GST-TAP231 (5 μg each) prebound on 5 μl of glutathione beads (Amersham Pharmacia) were incubated with HeLa nuclear extract (20 μl) in 300 μl of binding buffer (10 mM Tris·HCl, pH 8.0/150 mM NaCl/10% glycerol/0.2% Triton X-100/1 mM DTT/0.25 mM PMSF/1× protease inhibitor mixture (Calbiochem)/20 mM β-glycerophosphate/20 mM potassium fluoride/20 μg/ml RNase A) at 4°C for 3 h. Beads were washed six times with binding buffer, and bound proteins were eluted with SDS/PAGE sample buffer, followed by 10% SDS/PAGE and Western blotting analysis (12).
Anti-Flag immunoprecipitations followed by Western blotting analysis were performed as described (12).
HeLa Cell Fractionation and RNP Purification. HeLa cell fractionation and RNP purification were performed essentially as described by Mili et al. (20). For RNA analysis, immunopurified complexes were treated with 0.2 mg/ml proteinase K (Amresco, Euclid, OH) in 0.5% SDS, 50 mM NaCl, 50 mM Tris·Cl (pH 7.5), and 5 mM EDTA (pH 8.0), followed by phenol extraction, ethanol precipitation, and RQ1 DNase (Promega) treatment (20). Reverse transcription was performed by using SuperScript II RNase H- Reverse Transcriptase (Invitrogen) with oligo(dT)20 primers (Invitrogen) according to the manufacturer's directions. PCR used the same sets of primers specific for the human β-actin RNA (20) and 2.5 units of Taq polymerase (Roche), for 30 cycles of 1 min at 94°C, 30 s at 55°C, and 1 min at 72°C. PCR products were resolved on 1% agarose gels.
For immunopurification of Flag-TAP-associated RNPs, 5 × 106 HeLa cells transfected with pcDNA-Flag or pcDNA-Flag-TAP plasmid were rinsed with cold PBS, resuspended in 1 ml of sonication buffer consisting of 10 mM Tris·HCl (pH 7.4), 100 mM NaCl, 2.5 mM MgCl2, 0.5% (vol/vol) Triton X-100, 1 mM DTT, and 1× protease inhibitor mixture, then sonicated on ice for 5 s by using a microtip sonicator (Branson Sonifier 250) set at output 20. The sonicated material was loaded on top of a sucrose cushion (30% sucrose/10 mM Tris·HCl, pH 7.4/100 mM NaCl/2.5 mM MgCl2) and centrifuged at 4,000 × g for 15 min (20). Thirty microliters of the anti-Flag affinity agarose beads were added to the supernatant, and binding was carried out for 1 h at 4°C. RNAs extracted from bound RNPs, as well as from 2% of the binding supernatants, were analyzed as above.
In Vitro Splicing and Binding Assays. 9G8 was synthesized by using the TnT T7-coupled in vitro transcription and translation reticulocyte system (Promega) in the presence of 35S-l-methione (for 35S-labeled 9G8) or unlabeled methionine (for unlabeled 9G8) according to the manufacturer's protocol. A control reaction in the absence of the 9G8-encoding plasmid pXJ42-9G8 was carried out in parallel. After the reaction, the mixtures were passed through MicroSpin G-25 columns (Amersham Pharmacia) to remove unincorporated methionine and directly added to in vitro splicing reactions.
Splicing reactions were carried out in a volume of 40 μl (for splicing plus binding) or 20 μl (for splicing only) including 50% nuclear extract, 25 mM creatine phosphate, 0.5 mM ATP, 2.4 mM MgCl2, 20% reticulocyte lysate and 5 × 104 cpm (or an equal volume of unlabeled) splicing substrate, with or without 15 μM of the indicated 2′-O-methylated RNA oligonucleotides (21). Splicing reactions were routinely incubated at 30°C for 90 min. RNAs were then extracted, resolved on 8% denaturing polyacrylamide gels, and detected by autoradiography. For splicing plus binding assays, GST or GST-TAP231 (5 μg each) prebound on 5-μl glutathione beads were added after splicing in 300 μl of binding buffer (10 mM Tris·HCl, pH 8.0/150 mM NaCl/10% glycerol/0.2% Triton X-100/1 mM DTT/0.25 mM PMSF/1× protease inhibitor mixture/20 mM β-glycerophosphate/20 mM potassium fluoride/5 units of RNase inhibitor) and incubated at 4°C for 1 h. Beads were washed six times with binding buffer. RNAs were extracted from bound RNPs and from the supernatants and analyzed as above. Alternatively, after 90 min of splicing, reactions were treated with proteinase K, followed by phenol extraction and ethanol precipitation of RNAs. RNA pellets were then dissolved in 40 μl of water and incubated with beads under the same conditions as above.
For splicing inhibition experiments, oligonucleotides were added to the reaction and preincubated at 30°C for 20 min, followed by addition of the splicing substrate and incubation at 30°C for 70 min. SDS sample buffer was then added, products were resolved by 10% SDS/PAGE, and 35S-labeled 9G8 was visualized by autoradiography.
Results
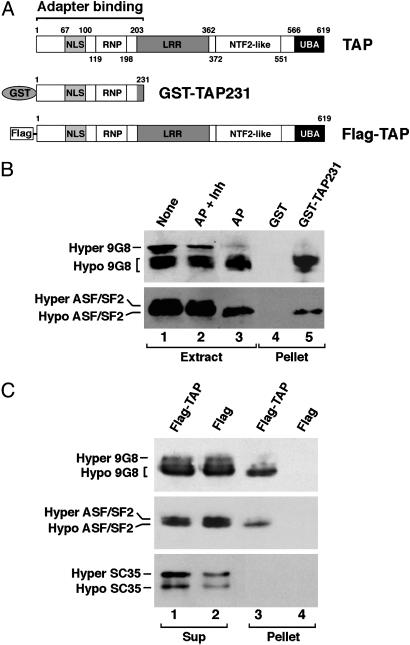
TAP Preferentially Binds Hypophosphorylated 9G8 and ASF/SF2 Both in Vitro and in Vivo. To ask whether the phosphorylation state of SR proteins modulates interaction with TAP, HeLa cell nuclear extracts were incubated in the presence of RNase with GST-TAP231 or GST (Fig. 1A) prebound to glutathione beads. GST-TAP231 (containing the N-terminal region of TAP) has been shown to be sufficient for interaction with adapter proteins (12). SDS/PAGE revealed two major phosphorylated forms of 9G8 and ASF/SF2 that we arbitrarily designate as “Hyper” and “Hypo” in HeLa nuclear extracts (Fig. 1B, lane 1). The intensity of the Hyper forms significantly decreased when the extract was treated with AP (lane 3) but remained when exposed to both phosphatase and inhibitors (lane 2). The separation between the hyper- and hypophosphorylated forms of ASF/SF2 seen is not as great as that reported by Misteli (22), most likely because of cell type differences; the phosphorylation profile of 9G8 from HeLa cells differs from that of HEK293 cells (compare Fig. 1 B to C). Importantly, GST-TAP231 preferentially selected the Hypo forms from the extract (compare lane 5 with lane 1, Fig. 1B), indicating that hypophosphorylated 9G8 and ASF/SF2 display higher affinity for TAP than their hyperphosphorylated counterparts.
Fig. 1.
TAP preferentially binds hypophosphorylated 9G8 and ASF/SF2. (A) Schematic of the domain structures of TAP (2, 29) and the recombinant proteins used in the binding assays. (B) TAP binding to 9G8 and ASF/SF2 in vitro. Bacterially expressed and affinity-purified GST-TAP231 (lane 5) or GST alone (lane 4) prebound on glutathione beads was incubated with HeLa cell nuclear extract. Bound (Pellet) fractions were resolved by 10% SDS/PAGE, followed by Western blot analysis by using anti-9G8 (Upper) or anti-ASF/SF2 (Lower) antibodies. Parallel extract samples not treated (None, lane 1), treated with alkaline phosphatase plus phosphatase inhibitors (AP + Inh, lane 2), or treated with AP (AP, lane 3) indicate the positions of differentially phosphorylated forms of 9G8 (Upper) and ASF/SF2 (Lower). (C) TAP binding to 9G8 and ASF/SF2 in vivo. HEK293 cells were transiently transfected with plasmids pcDNA-Flag-TAP expressing a Flag-tagged full-length TAP or pcDNA-Flag alone. The Flag-TAP or Flag was immunoprecipitated from the transfected cell extracts with anti-Flag antibodies. Five percent of the supernatants (Sup, lanes 1 and 2) and the precipitated proteins (Pellet, lanes 3 and 4) were analyzed by Western blotting by using anti-9G8 (Top), anti-ASF/SF2 (Middle), or anti-SC35 (Bottom) antibodies.
We next tested whether selective interactions of TAP with hypophosphorylated SR proteins also occur in vivo. A Flag-tagged full-length TAP protein or Flag alone (Fig. 1 A) was expressed in HEK293 cells by transient transfection. Associated proteins purified from transfected cell extracts with anti-Flag antibodies were analyzed by Western blotting (Fig. 1C). The tagged TAP did not bind the nonshuttling SR-splicing factor SC35 (10), consistent with previous results (12). However, as in the in vitro experiments (Fig. 1B), full-length Flag-TAP, but not Flag alone, preferentially selected hypophosphorylated forms of 9G8 and ASF/SF2 from the extract (Fig. 1C Top and Middle).
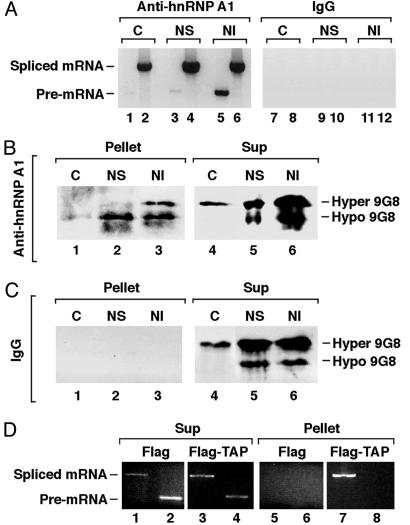
9G8 Associated with Spliced mRNP Is Predominantly Hypophosphorylated in Vivo. The phosphorylation state of proteins associated with mRNA in different stages of maturation in vivo can be investigated employing a powerful method pioneered by Piñol-Roma and colleagues (20). It uses antibodies to hnRNP A1, an abundant mRNA-binding protein that associates with pre-mRNA, nuclear-spliced mRNA, and cytoplasmic mRNA (20, 23) to isolate RNPs. Before immunoprecipitation, HeLa cells are permeabilized and fractionated into cytoplasmic (C) and two nuclear fractions: nuclear soluble (NS), containing free mRNP, and nuclear insoluble (NI), containing mainly chromatin-bound pre-mRNP. To confirm the selectivity of the fractionation method, RNAs extracted from the three RNP fractions were reverse transcribed and PCR amplified by using primers specific for β-actin RNA (20). In agreement with previously published results (20), RNPs from the cytoplasmic and the NS fractions contained primarily spliced mRNA (Fig. 2A, lanes 2 and 4) whereas those from the NI (chromatin) fraction contained both spliced and pre-mRNA (lanes 5 and 6). The control IgG precipitates did not contain detectable levels of β-actin RNA (lanes 7–12).
Fig. 2.
9G8 is hypophosphorylated in spliced mRNPs in vivo.(A) Agarose gels showing the RT-PCR products of endogenous β-actin RNA (20) in RNPs immunoprecipitated by anti-hnRNP A1 antibodies (lanes 1–6) or by a control mouse IgG (lanes 7–12) from the cytoplasmic (C), NS, and NI fractions. In the odd lanes, a set of primers corresponding to the second intron and the third exon of the human β-actin gene, respectively, was used for the pre-mRNA. In the even lanes, a set of primers corresponding to the second and the fifth exons of the gene, respectively, was used for the spliced mRNA. (B and C) Results of representative Western blot analyses of 9G8 in RNPs immunoprecipitated by anti-hnRNP A1 antibody (B, lanes 1–3) or control mouse IgG (C, lanes 1–3) from the indicated fractions. One percent of the corresponding supernatants were analyzed on the right. The efficiency of hnRNP A1 precipitation was 10% (data not shown). (D) TAP preferentially associates with spliced mRNP in vivo. HeLa cells were transiently transfected with plasmids pcDNA-Flag-TAP expressing Flag-TAP or pcDNA-Flag alone. RNPs from the transfected cell lysates were isolated by using anti-Flag antibody. RT-PCR products of RNAs extracted from the purified RNPs (Pellet, lanes 5–8) and from 2% of the supernatants (Sup, lanes 1–4) were resolved on 1% agarose gels. Primers for PCR were as in A.
The phosphorylation profile of the SR protein 9G8 associated with the three RNP fractions, as well as the corresponding supernatants, was then analyzed by Western blotting. 9G8 and hnRNP A1 have been shown not to interact with each other directly. Both hyper- and hypophosphorylated 9G8 was detected in the RNPs from the NI (chromatin) fraction (Fig. 2B, lane 3). In striking contrast, 9G8 in the cytoplasmic and the NS mRNPs was largely hypophosphorylated (Fig. 2B, lanes 1 and 2), even though hyperphosphorylated 9G8 predominated in these supernatants (lanes 4 and 5). No 9G8 was detected in the control IgG precipitates (Fig. 2C, lanes 1–3), nor in the anti-hnRNP A1 precipitates treated with RNase A (data not shown). The presence of hyperphosphorylated 9G8 in the insoluble nuclear fraction (Fig. 2B, lane 3) is most likely due to its association with pre-mRNA because phosphorylation of SR proteins is necessary for the early steps of spliceosome assembly (22, 24). Together, these results suggest that 9G8 undergoes partial dephosphorylation during or after splicing in vivo.
TAP Preferentially Binds Spliced mRNP Both in Vivo and in Vitro. To ask whether 9G8 dephosphorylation correlates with enhanced association of TAP with spliced mRNP in vivo, we examined HeLa cells transiently expressing a Flag-tagged TAP protein or Flag alone. Although both unspliced and spliced RNA were present in the transfected cell lysates (Fig. 2D, lanes 1–4), after selection with anti-Flag antibody, analysis of bound RNAs by RT-PCR revealed that Flag-TAP selected only spliced β-actin RNA (compare lane 7 with lane 8).
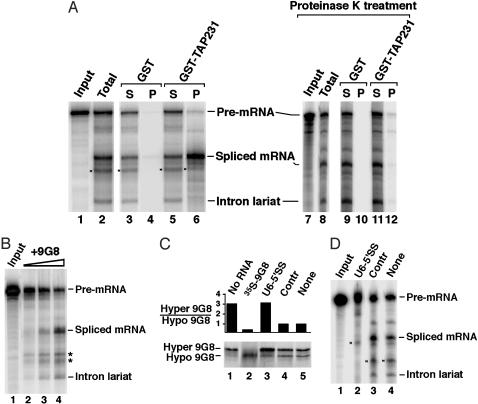
To confirm further the preferential association of TAP with spliced mRNP, we carried out in vitro splicing followed by binding assays. 32P-labeled adenovirus pre-mRNA substrate was incubated with HeLa nuclear extract under splicing conditions. Reaction products were exposed to glutathione beads coated with GST-TAP231 or GST, and RNAs extracted from both the supernatants and the pellets were resolved by denaturing gel electrophoresis. As shown in Fig. 3A, the spliced mRNP more efficiently bound to GST-TAP231 than the pre-mRNP or complexes containing the intron lariat (compare lane 6 with lane 5). When reactions were treated with proteinase K before the binding assay, the association was abolished (compare lane 12 with lane 6), demonstrating that TAP binding to spliced mRNP is mediated by protein–protein interactions rather than direct RNA binding.
Fig. 3.
TAP-mRNP association and splicing-dependent 9G8 hypophosphorylation in vitro. (A) TAP binds spliced mRNP after splicing in vitro. 32P-labeled adenovirus splicing substrate (AdML) (19) (Input) was incubated with HeLa nuclear extract under splicing conditions. Glutathione beads precoated with GST-TAP231 or GST alone were added to the total reaction (lanes 3–6) or to a proteinase K-treated reaction (lanes 9–12). Radioactive RNAs selected by the bead-bound proteins (P, lanes 4, 6, 10, and 12) or from 5% (S, lanes 3 and 5) or 20% of the corresponding supernatants (lanes 9 and 11) were resolved on 8% denaturing polyacrylamide gels and visualized by PhosphorImager. Total, 5% of the splicing reaction before the binding assays. The bands marked * in this panel and in the following panels were nonspecific degradation products. (B) Adenovirus splicing substrate (lane 1) was incubated under splicing conditions in HeLa nuclear extract deficient in splicing activity (lane 2). Splicing was stimulated in a dose-dependent manner by addition of 9G8 made by in vitro translation (lanes 3 and 4). (C) In vitro splicing was performed with in vitro translated 35S-labeled 9G8 and unlabeled splicing substrate. Quantitations (Upper) were the average of two independent experiments, where the observed ratios differed by no more than 15%. Splicing reactions were in the absence (lane 1) or presence (lanes 3–5) of the splicing substrate and with the addition of the U6-5′SS oligonucleotide (CUCUGUAUCGUUCCAAUUUU, lane 3), a control (Contr) oligonucleotide (UUUCCAGUAGCUGAA, lane 4), or no oligonucleotide (lane 5). Three microliters of rabbit reticulocyte lysate containing the 35S-labeled 9G8 used were loaded in lane 2. (D) A splicing reaction comparable to C contained an equivalent amount of in vitro translated but unlabeled 9G8 whereas the splicing substrate was 32P-labeled. The identities of the spliced products are marked on the right in A, B, and D.
Dephosphorylation of 9G8 Occurs Before the First Catalytic Step of Splicing in Vitro. To investigate when during splicing 9G8 dephosphorylation occurs, we performed in vitro splicing in the presence or absence of blocking oligonucleotides (22). Splicing involves the sequential assembly of complexes E, A, B, and C, with the catalytic reactions taking place in the C complex (15). We compared the 2′-O-methylated RNA oligonucleotide U6-5′SS, which is known to block splicing at the A to B transition to a control oligonucleotide (Contr) (21). We used HeLa cell nuclear extracts whose splicing activity depends on the addition of 35S-labeled 9G8 made by in vitro translation (Fig. 3B). When added to the HeLa nuclear extract, the labeled 9G8 was mainly hypophosphorylated (Fig. 3C, lane 2) but became largely hyperphosphorylated upon incubation under splicing conditions even in the absence of substrate (lane 1). We confirmed that, under the same conditions (data not shown), endogenous SR proteins in the extract were likewise converted to hyperphosphorylated forms. When splicing was blocked by the 2′-O-methyl oligonucleotide U6-5′SS (Fig. 3D, lane 2), the same ratio (≈3:1) of hyper- to hypophosphorylated 9G8 was observed (Fig. 3C, compare lane 3 with lane 1). In striking contrast, the ratio decreased to one (Fig. 3C, lanes 4 and 5) when splicing proceeded (Fig. 3D, lanes 3 and 4). These results argue that dephosphorylation of 9G8 depends on active splicing and occurs before the first catalytic step, consistent with published reports (16, 17).
Discussion
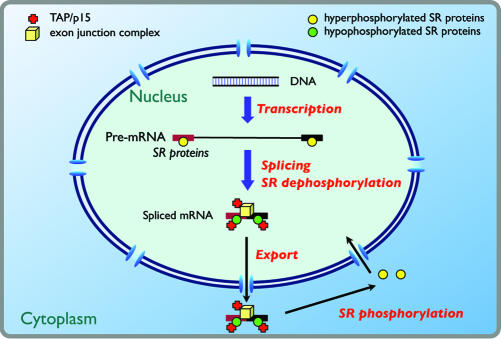
Our findings suggest that dephosphorylation of SR protein adapters are critical for determining the selectivity and efficiency of TAP-dependent mRNA export (Fig. 4). The effects could be both direct and indirect. First, dephosphorylation directly increases the affinity of 9G8, ASF/SF2, and presumably other shuttling SR proteins for TAP (Fig. 1 B and C). Second, dephosphorylation of bound SR proteins during splicing could alter protein–protein interactions, leading to conformational changes of the mRNP such that it becomes competent for TAP binding (Figs. 2D and 3A). Previously, in vitro assays have shown that phosphorylation of ASF/SF2 enhances its binding to U1-70K, a U1 snRNP-specific protein essential for the initiation of splicing, but decreases its association with itself and another SR protein (24). The splicing-dependent dephosphorylation of SR protein adapters may explain why shuttling SR proteins directly tethered to a reporter pre-mRNA fail to stimulate export (data not shown).
Fig. 4.
A model for the role of SR protein dephosphorylation in mRNA export. Hyperphosphorylated SR proteins are recruited to pre-mRNA molecules at exonic enhancers (15). During splicing, SR proteins are hypophosphorylated but remain associated with the spliced mRNP. Together with the exon junction complex (EJC), which contains REF1 (ref. 30 and references therein), they recruit multiple copies of TAP, thereby increasing the efficiency of export of spliced versus unspliced mRNP. In the cytoplasm, rephosphorylation of the SR protein adapters results in their dissociation from mRNP complexes and in their recycling to the nucleus.
We observed that the minor fraction of cytoplasmic 9G8 that is RNA-associated is hypophosphorylated whereas overall cytoplasmic 9G8 is hyperphosphorylated (Fig. 2B). Thus, the cytoplasmic rephosphorylation of shuttling SR protein adapters may trigger their disassembly from TAP and the exported mRNA cargo and contribute to their reimport into the nucleus. Indeed, phosphorylation stimulates the nuclear import of mammalian SR proteins (25), as well as the dissociation of Npl3p from mRNA in the cytoplasm of yeast (14). It was previously proposed that catalytic removal of mRNP proteins by the Dbp5 RNA helicase and their rebinding to import receptors at the cytoplasmic face of the nuclear pore promote the disassembly and remodeling of exported mRNPs in the cytoplasm (2, 3, 26). Our findings add another layer of regulation (SR protein rephosphorylation) to the directionality of the export process and the cytoplasmic remodeling of mRNPs.
The compartmentalized phosphorylation of export factors is conserved among different export pathways and organisms. For instance, it has been elegantly demonstrated that both phosphorylation and dephosphorylation of the export mediator phosphorylated adaptor for RNA export (PHAX) are essential for the chromosome region maintenance 1 (CRM1)-dependent nuclear export of U1 small nuclear RNA in the Xenopus oocyte (27). Similarly, cytoplasmic phosphorylation and subsequent nuclear dephosphorylation of Npl3p at least partially by means of the Glc7p phosphatase have been recently found to be required for mRNA export in yeast (28). More detailed definition of the splicing step during which dephosphorylation of SR proteins occurs, as well as the identification of specific phosphatases involved in the regulation of TAP/SR protein interactions, will further enhance our understanding of the molecular mechanism of mRNA export.
Acknowledgments
We thank J. Stévenin and G. Dreyfuss for antibodies and plasmids; N. Conrad, T. Hirose, A. Rebane, and K. Tycowski for critical reading of the manuscript; and W.-Y. Tarn for sharing unpublished results. This work was supported by a National Institutes of Health grant (to J.A.S.) and funds from the Department of Obstetrics, Gynecology, and Reproductive Sciences of the Yale School of Medicine (Y.H.). J.A.S. is an investigator of the Howard Hughes Medical Institute.
Abbreviations: RNP, ribonucleoprotein; SR, serine/arginine-rich; AP, alkaline phosphatase; NI, nuclear insoluble; REF1, RNA export factor 1.
References
- 1.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195-205. [DOI] [PubMed] [Google Scholar]
- 2.Stutz, F. & Izaurralde, E. (2003) Trends Cell Biol. 13, 319-327. [DOI] [PubMed] [Google Scholar]
- 3.Reed, R. & Hurt, E. (2002) Cell 108, 523-531. [DOI] [PubMed] [Google Scholar]
- 4.Katahira, J., Strasser, K., Podtelejnikov, A., Mann, M., Jung, J. U. & Hurt, E. (1999) EMBO J. 18, 2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straber, K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A. G., Aguilera, A., Struhl, K., Reed, R. & Hurt, E. (2002) Nature 28, 1-4. [DOI] [PubMed] [Google Scholar]
- 6.Luo, M. L., Zhou, Z., Magni, K., Christoforides, C., Rappsilber, J., Mann, M. & Reed, R. (2001) Nature 413, 644-647. [DOI] [PubMed] [Google Scholar]
- 7.Gatfield, D. & Izaurralde, E. (2002) J. Cell Biol. 159, 579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Longman, D., Johnstone, I. L. & Cáceres, J. F. (2003) RNA 9, 881-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.MacMorris, M., Brocker, C. & Blumenthal, T. (2003) RNA 9, 847-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cáceres, J. F., Screaton, G. R. & Krainer, A. R. (1998) Genes Dev. 12, 55-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Y. & Steitz, J. A. (2001) Mol. Cell 7, 899-905. [DOI] [PubMed] [Google Scholar]
- 12.Huang, Y., Gattoni, R., Stévenin, J. & Steitz, J. A. (2003) Mol. Cell 11, 837-843. [DOI] [PubMed] [Google Scholar]
- 13.Lee, M. S., Henry, M. & Silver, P. A. (1996) Genes Dev. 10, 1233-1246. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, W., Siebel, C. W. & Guthrie, C. (2001) RNA 7, 302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graveley, B. (2000) RNA 6, 1197-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mermoud, J. E., Cohen, P. T. W. & Lamond, A. I. (1994) EMBO J. 13, 5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao, W., Jamison, S. F. & Garcia-Blanco, M. A. (1997) RNA 3, 1456-1467. [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett, M., Piñol-Roma, S., Staknis, D., Dreyfuss, G. & Reed, R. (1992) Mol. Cell. Biol. 12, 3165-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou, Z., Luo, M., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature 407, 401-405. [DOI] [PubMed] [Google Scholar]
- 20.Mili, S., Shu, H. J., Zhao, Y. & Piñol-Roma, S. (2001) Mol. Cell. Biol. 21, 7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose, T., Shu, M.-D. & Steitz, J. A. (2003) Mol. Cell 12, 113-123. [DOI] [PubMed] [Google Scholar]
- 22.Misteli, T., Caceres, J. F., Clement, J. Q., Krainer, A. R., Wilkinson, M. F. & Spector, D. L. (1998) J. Cell Biol. 143, 297-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piñol-Roma, S. & Dreyfuss, G. (1992) Nature 355, 730-732. [DOI] [PubMed] [Google Scholar]
- 24.Xiao, S. H. & Manley, J. L. (1997) Genes Dev. 11, 334-344. [DOI] [PubMed] [Google Scholar]
- 25.Lai, M.-C., Lin, R.-I., Huang, S.-Y., Tsai, C.-W. & Tarn, W.-Y. (2000) J. Biol. Chem. 275, 7950-7957. [DOI] [PubMed] [Google Scholar]
- 26.Lei, E. P. & Silver, P. A. (2002) Dev. Cell 2, 261-272. [DOI] [PubMed] [Google Scholar]
- 27.Ohno, M., Segref, A., Bachi, A., Wilm, M. & Mattaj, I. W. (2000) Cell 101, 187-198. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert, W. & Guthrie, C. (2004) Mol. Cell 13, 201-212. [DOI] [PubMed] [Google Scholar]
- 29.Izaurralde, E. (2002) Results Probl. Cell Differ. 35, 133-150. [DOI] [PubMed] [Google Scholar]
- 30.Reichert, V. L., Le Hir, H., Jurica, M. S. & Moore, M. J. (2002) Genes Dev. 16, 2778-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]