Abstract
Type 1 diabetes (T1D) is an autoimmune disease characterized by T cell-mediated destruction of the pancreatic islet beta cells. Multiple genetic loci contribute to disease susceptibility in humans, with the most responsible locus being the major histocompatibility complex (MHC). Certain MHC alleles are predisposing, including the common HLA-A∗02:01. After the MHC, the locus conferring the strongest susceptibility to T1D is the regulatory region of the insulin gene, and alleles associated with reduced thymic insulin expression are predisposing. Mice express two insulin genes, Ins1 and Ins2. While both are expressed in beta cells, only Ins2 is expressed in the thymus. We have developed an HLA-A∗02:01-transgenic NOD-based T1D model that is heterozygous for a functional Ins2 gene. These mice exhibit reduced thymic insulin expression and accelerated disease in both genders. Immune cell populations are not grossly altered, and the mice exhibit typical signs of islet autoimmunity, including CD8 T cell responses to beta cell peptides also targeted in HLA-A∗02:01-positive type 1 diabetes patients. This model should find utility as a tool to uncover the mechanisms underlying the association between reduced thymic insulin expression and T1D in humans and aid in preclinical studies to evaluate insulin-targeted immunotherapies for the disease.
1. Introduction
Type 1 diabetes (T1D) is an autoimmune disease characterized by T cell-mediated destruction of the pancreatic islet beta cells. Multiple genetic loci contribute to T1D susceptibility in humans, with the most responsible locus being the major histocompatibility complex (MHC) [1]. The ability of certain class II MHC genes to influence disease risk has long been appreciated [2, 3]. Multiple studies have also revealed an association with certain class I MHC alleles, including the common HLA-A∗02:01 [4–13]. These findings are not surprising, given that CD4 and CD8 T cell responses to a variety of beta cell antigens, including insulin, are observed in T1D patients [14].
After the MHC, the locus that confers the strongest susceptibility to T1D in humans is the variable number of tandem repeats (VNTR) regulatory region of the insulin gene [1, 15]. VNTR alleles with the smallest number of repeats, designated as class I, predispose to T1D [16, 17], while the longer class III alleles have a dominant protective effect [15, 18]. Class III VNTR alleles are associated with thymic insulin RNA levels that are increased two- to threefold compared to class I alleles [19], leading to the hypothesis that impaired negative selection of insulin-specific T cells in individuals with class I VNTR alleles explains their predisposition to T1D [19, 20]. While findings from a single human study are consistent with this idea [21], the development of a mouse model for T1D that incorporates the reduced, but not abolished, thymic insulin expression observed in patients would allow for more rigorous future testing of this hypothesis.
The NOD mouse is the primary rodent model used for studying T1D [22]. Unlike humans, mice express two insulin genes, Ins1 and Ins2. While both genes are expressed in beta cells [23], Ins2 expression predominates in the thymus [24–27], with little [24] to no [25–27] detectable thymic Ins1 expression. Ins2-deficient (Ins2KO) NOD mice develop diabetes at an accelerated rate [28–30], as do HLA-A∗02:01-transgenic Ins2KO NOD mice [28], and both Ins2-deficient strains have increased insulin-specific islet-infiltrating CD8 T cells compared to their wild-type (WT) counterparts [28]. While these Ins2KO mouse strains highlight the importance of thymic insulin expression, they do not accurately represent a human patient, where thymic insulin expression is diminished but still present [19, 20]. Here we have developed an HLA-A∗02:01-transgenic NOD-based T1D model that is heterozygous (het) for the Ins2KO allele, resulting in thymic insulin expression that is decreased but not eliminated. The mice develop accelerated disease compared to Ins2WT mice, and this is true regardless of gender. Immune cell populations are not grossly altered, and the mice exhibit typical signs of islet autoimmunity, including CD8 T cell responses to beta cell peptides also targeted in HLA-A∗02:01-positive T1D patients. This model should find utility as a tool to uncover the mechanisms underlying the association between class I VNTR alleles and T1D in humans. It should also aid in preclinical studies to evaluate insulin-targeted immunotherapies for the disease.
2. Materials and Methods
2.1. Mice
NOD.β 2mKO.HHD mice [31] transgenically express a single-chain chimeric HLA-A∗02:01 molecule in which human β2-microglobulin (β 2m) is covalently linked to the α1 and α2 domains of HLA-A∗02:01. The α3, transmembrane, and cytoplasmic portions of the molecule are derived from H-2Db. Mouse class I MHC molecules are not expressed in these mice due to the murine β 2m deficiency. NOD.Ins2KO mice have been described [29]. The two strains were intercrossed to transfer the Ins2KO allele to the NOD.β 2mKO.HHD strain. The resulting progeny were bred as appropriate to obtain Ins2WT and Ins2het NOD.β 2mKO.HHD mice for our studies. Female β 2mKO mice breed poorly in our hands and so were rarely used for this purpose. Similarly, NOD and NOD.Ins2KO mice were intercrossed and the resulting progeny bred as appropriate to obtain Ins2WT and Ins2het NOD mice. The HHD transgene and the WT and KO β 2m and Ins2 alleles were identified by PCR using the following primer pairs: HHD, 5′-CTTCATCGCAGTGGGCTAC-3′ and 5′-CGGTGAGTCTGTGAGTGGG-3′; β 2MWT, 5′-GAAACCCCTCAAATTCAAGTATACTCA-3′ and 5′-GACGGTCTTGGGCTCGGCCATACT-3′; β 2mKO, 5′-GAAACCCCTCAAATTCAAGTATACTCA-3′ and 5′-TCGAATTCGCCAATGACAAGACGCT-3′; Ins2WT, 5′-GGCAGAGAGGAGGTGCTTTG-3′ and 5′-AGAAAACCACCAGGGTAGTTAGC-3′; Ins2KO, 5′-GGCAGAGAGGAGGTGCTTTG-3′ and 5′-ATTGACCGTAATGGGATAGG-3′. All animal experiments were approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
2.2. Measurement of Thymic Ins2 RNA
Female Ins2WT and Ins2het NOD.β2mKO.HHD mice (four each) were sacrificed and thymus was harvested. Total thymic RNA was isolated using the RNeasy Midi Kit (Qiagen, Valencia, CA) and treated with DNase I (Qiagen) to eliminate DNA contamination. 1.5–2.3 μg of RNA was reverse-transcribed to cDNA using random hexadeoxynucleotides and oligo dT primers (Invitrogen). Equal amounts of cDNA were mixed with SYBR Green PCR Master Mix (Qiagen) and each Ins2 primer (5′-CTTCTTCTACACACCCATGTCC-3′ and 5′-TCTACAATGCCACGCTTCTG-3′) or primers for the U6 normalization control (5′-CTCGCTTCGGCAGCACATATACTA-3′ and 5′-ACGAATTTGCGTGTCATCCTTGCG-3′) and brought to a final volume of 25 μL. Real-time quantitative RT-PCR was performed in triplicate using an iQ5 Optical System (Bio-Rad, Hercules, CA). Amplification was carried out as follows: a single denaturing step at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec, 59°C for 30 sec, and 72°C for 30 sec, followed by a final extension step of 72°C for 3 min. Results were analyzed using the Relative Expression Software Tool (REST) [32, 33].
2.3. Type 1 Diabetes Assessment
Glucosuria was monitored weekly using Diastix reagent strips (Bayer, Elkhart, IN). Mice were considered diabetic after two consecutive positive tests, and the date of the first positive test was recorded as the time of onset of disease.
2.4. Flow Cytometry
Splenocytes from NOD.β 2mKO.HHD and NOD.β 2mKO.HHD.Ins2het mice were analyzed by flow cytometry. Cells were stained with anti-B220, anti-CD11c, anti-CD4, and anti-CD8 (all from BD Biosciences, San Jose, CA). In some samples, cells were stained with anti-CD25 (BD Biosciences), fixed and permeabilized with fixation/permeabilization buffer (eBioscience, San Diego, CA), and stained with anti-Foxp3 (eBioscience).
2.5. Pancreas Histology
To assess insulitis in female NOD.β 2mKO.HHD and NOD.β 2mKO.HHD.Ins2het mice at 4 and 8 weeks of age, pancreata were fixed in Bouin's solution, embedded in paraffin, and sectioned at nonoverlapping levels. Sections were stained with aldehyde fuchsine to readily visualize granulated beta cells and counterstained with hematoxylin and eosin for detection of leukocytes. Islets were scored as previously described [34]: 0, no insulitis; 1, local insulitis without infiltration of islet itself; 2, less than 25% infiltration; 3, 25–75% infiltration; or 4, greater than 75% infiltration. An insulitis index was calculated by adding the scores of all islets and dividing by four times the number of islets scored. A minimum of 20 islets per mouse were evaluated. Diabetic mice were assigned an insulitis index of 1.
2.6. Islet Isolation and Culture of Islet-Infiltrating T Cells
Islets were isolated from female NOD.β 2mKO.HHD.Ins2het mice at 8 weeks of age by collagenase P perfusion of the common bile duct as previously described [35]. Islets were handpicked using a micromanipulator and a dissecting microscope and up to 50 islets were transferred per well to 24-well plates in 500 μL R-10 medium (RPMI 1640 (Invitrogen, Carlsbad, CA) containing 10% FBS, 1 mM sodium pyruvate, 28 μM β-mercaptoethanol, 1x nonessential amino acids (Invitrogen)) with 50 U/mL recombinant human IL-2 (PeproTech, Rocky Hill, NJ). Cells were cultured for 7 days at 37°C in 5% CO2, at which point the majority of the cells are expected to be CD8 T cells [35].
2.7. IFN-γ ELISPOT Assay
Human HLA-A∗02:01-positive T2 cells [36], deficient for the transporter associated with antigen processing, were cultured at 26°C overnight prior to use. ELISPOT plates (Millipore MAHA S4510, Billerica, MA) were coated with anti-mouse IFNγ antibody (BD Biosciences) and blocked with 1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO). T2 cells were plated at 2 × 104 cells/well and pulsed with 10 μM of the indicated peptides for 1 hour at 26°C. Cultured islet-infiltrating T cells from NOD.β 2mKO.HHD.Ins2het mice were added at 2 × 104 cells/well in 50 μL R-10. Cells were incubated for 40 hours at 37°C. Wells were then washed with 0.05% Tween 20/PBS and biotinylated anti-mouse IFNγ detection antibody (BD Biosciences) was added for 2 hours at 37°C. After washing, streptavidin-alkaline phosphatase (Zymed Laboratories, Carlsbad, CA) was added and incubated for 1 hour at 37°C. Wells were washed and spots were developed using 5-bromo-4-chloro-3-indolyl-phosphate/nitro-blue tetrazolium substrate (Sigma-Aldrich). Spots were counted using an automated ELISPOT reader system (Autoimmun Diagnostika, Strassberg, Germany). Responses are reported as a stimulation index, which is defined as spot number in response to the test peptide divided by spot number in response to an irrelevant HIV-derived HLA-A∗02:01-binding peptide (SLYNTVATL) [37]. The cutoff for positivity is a stimulation index greater than 2 and a test peptide spot number greater than 5 per 1 × 105 T cells [38].
3. Results
3.1. Accelerated Diabetes Development in NOD.β 2mKO.HHD.Ins2het Mice
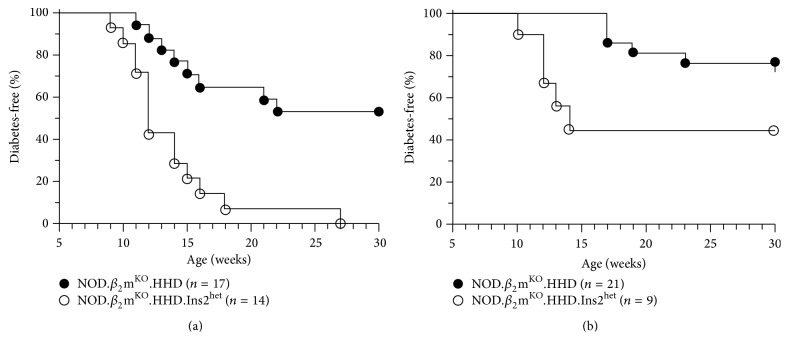
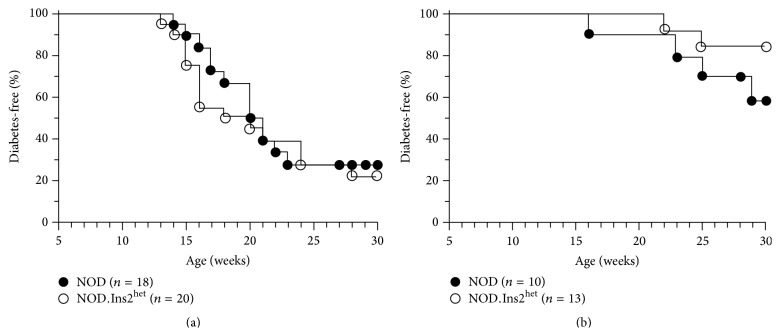
A previous study had demonstrated that Ins2het mice of mixed, but primarily C57BL/6, background experience a reduction in thymic insulin expression of approximately 40% [24]. To develop a mouse model of T1D having reduced thymic insulin quantity, and also expressing the human class I MHC molecule HLA-A∗02:01, we generated NOD.β 2mKO.HHD.Ins2het mice. Using quantitative RT-PCR, we similarly found a reduction in thymic insulin expression of 35% in female Ins2het compared to Ins2WT mice (n = 4 mice of each genotype). Diabetes development in NOD.β 2mKO.HHD.Ins2het and NOD.β 2mKO.HHD mice of both genders was then compared. Both female (Figure 1(a)) and male NOD.β 2mKO.HHD.Ins2het mice (Figure 1(b)) demonstrated accelerated diabetes development compared to their Ins2WT counterparts. Female NOD.β 2mKO.HHD.Ins2het mice developed diabetes as early as 9 weeks of age and all were diabetic by 27 weeks (Figure 1(a)). The first onset of diabetes in Ins2WT female mice was at 11 weeks, and only 47% developed diabetes by 30 weeks. As also seen in standard NOD males [39, 40], diabetes development was slowed and overall incidence was reduced in NOD.β 2mKO.HHD males (Figure 1(b)) compared to females. However, Ins2het males exhibited an earlier onset of disease compared to Ins2WT males (10 weeks versus 17 weeks), and a larger percentage (56% versus 24%) had developed diabetes by 30 weeks of age (Figure 1(b)). Thus, both genders of NOD.β 2mKO.HHD.Ins2het mice faithfully model the circumstance in humans where reduced thymic insulin expression is predisposing to T1D [16, 17]. Note that this is not what we observed in the case of NOD.Ins2het mice, where both female (Figure 2(a)) and male Ins2het mice (Figure 2(b)) exhibit a diabetes profile that is statistically indistinguishable from that of NOD mice.
Figure 1.
Diabetes development in NOD.β 2mKO.HHD and NOD.β 2mKO.HHD.Ins2het mice. (a) Female and (b) male NOD.β 2mKO.HHD (filled circles) and NOD.β 2mKO.HHD.Ins2het mice (open circles) were followed weekly for diabetes development. (a) p = 0.0002, Mantel-Cox. (b) p = 0.04, Mantel-Cox.
Figure 2.
Diabetes development in NOD and NOD.Ins2het mice. (a) Female and (b) male NOD (filled circles) and NOD.Ins2het mice (open circles) were followed weekly for diabetes development. (a) p = 0.63 (not significant), Mantel-Cox. (b) p = 0.19 (not significant), Mantel-Cox.
3.2. Immune Cell Populations Are Not Grossly Altered in NOD.β 2mKO.HHD.Ins2het Mice
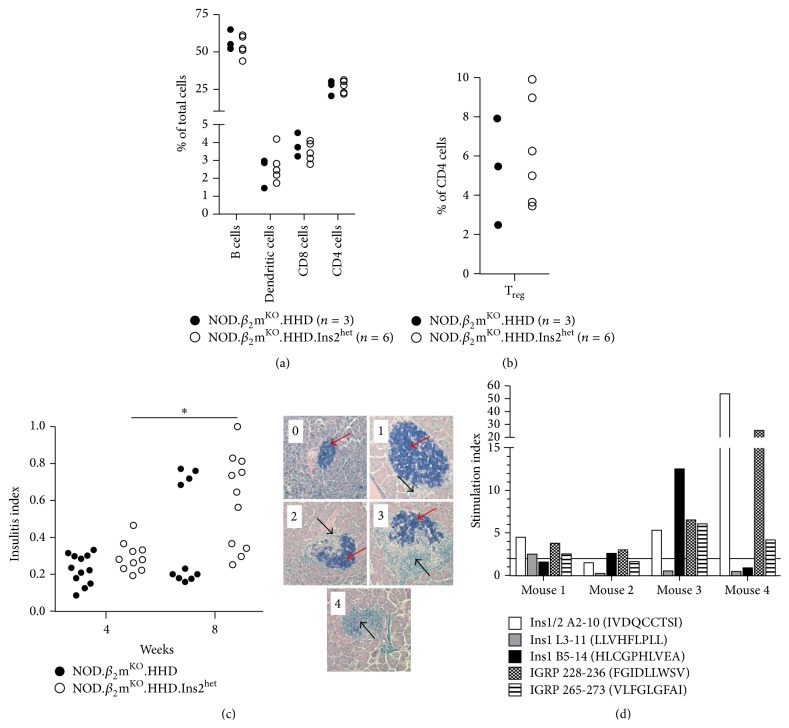
To verify that the accelerated diabetes development observed in NOD.β 2mKO.HHD.Ins2het mice could not be attributed to a gross alteration in immune cell populations, we examined the splenocyte composition of 8-week-old female nondiabetic NOD.β 2mKO.HHD and NOD.β 2mKO.HHD.Ins2het mice (Figure 3(a)). It was previously shown that NOD.β 2mKO.HHD mice have a reduced CD8 T cell population and elevated B and CD4 T cells compared to standard NOD mice [31]. This was also true for NOD.β 2mKO.HHD.Ins2het mice, and no differences were observed in any of the cell types analyzed as a percentage of total cells. To investigate whether a reduction in regulatory T cells (Treg) might contribute to disease pathogenesis in the Ins2het mice, NOD.β 2mKO.HHD and NOD.β 2mKO.HHD.Ins2het splenocytes were monitored for expression of the characteristic Treg cell phenotype, CD4+CD25+Foxp3+. No difference was observed in Treg cells as a percentage of CD4 T cells (Figure 3(b)). These results indicate that the accelerated diabetes development seen in NOD.β 2mKO.HHD.Ins2het mice is the result of neither an altered immune cell composition nor reduced Treg cells, at least at the level investigated here, that is, without regard to antigenic specificity.
Figure 3.
Splenocyte composition, insulitis, and autoreactive CD8 T cell specificities in NOD.β 2mKO.HHD.Ins2het mice. (a) and (b) Splenocytes from 8-week-old female NOD.β 2mKO.HHD (filled circles) and NOD.β 2mKO.HHD.Ins2het mice (open circles) were analyzed by flow cytometry. Each symbol represents an individual mouse. (c) Female NOD.β 2mKO.HHD (filled circles) and NOD.β 2mKO.HHD.Ins2het mice (open circles) were sacrificed at 4 and 8 weeks of age and insulitis indices were determined as described in Materials and Methods and plotted. Each symbol represents an individual mouse. ∗ p = 0.0037 (Mann-Whitney U). Representative islets from a single NOD.β 2mKO.HHD.Ins2het mouse are also shown. In these images, beta cells appear dark purple and are denoted by red arrows, while the more lightly stained infiltrating immune cells are marked by black arrows. The number on each image indicates the insulitis score of the islet shown. (d) Islet-infiltrating cells from 8-week-old female NOD.β 2mKO.HHD.Ins2het mice were tested for reactivity to the indicated HLA-A∗02:01-restricted insulin and IGRP epitopes by IFN-γ ELISPOT. Stimulation index was calculated by dividing the number of spots detected for a given peptide by the number of spots detected with an irrelevant HIV-derived HLA-A∗02:01-binding peptide. A stimulation index greater than 2 was considered a positive response.
3.3. NOD.β 2mKO.HHD.Ins2het Mice Exhibit Typical Signs of Islet Autoimmunity
In mixed background mice carrying zero, one, or two copies of the Ins2 gene, pancreatic insulin content is indistinguishable [24]. Furthermore, Ins2KO mice perform identically to their Ins2WT counterparts in intraperitoneal glucose tolerance tests [41]. Thus, we hypothesized that the diabetes observed in NOD.β 2mKO.HHD.Ins2het mice was of an autoimmune nature, as is the case for the NOD.β 2mKO.HHD parent strain [31], and not a deficiency in pancreatic insulin production due to the presence of only one functional copy of the Ins2 gene. To verify this, histological sections of pancreata from female mice at 4 and 8 weeks of age were examined. All mice studied exhibited some degree of insulitis, which progressed significantly with age (Figure 3(c)), and islets showing a wide range of immune cell infiltration and beta cell destruction were observed (Figure 3(c)).
We previously identified several HLA-A∗02:01-restricted beta cell epitopes, derived from the autoantigens insulin and islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) that are recognized by islet-infiltrating T cells from NOD.β 2mKO.HHD mice [31, 42]. To further confirm the autoimmune nature of the diabetes observed in NOD.β 2mKO.HHD.Ins2het mice, islets from 8-week-old females were cultured for 7 days and T cell reactivity to the previously identified beta cell epitopes was monitored by IFNγ ELISPOT. All mice harbored autoreactive T cells specific for at least two epitopes (Figure 3(d)), further confirming the autoimmune nature of their disease. A subset of these epitopes (Ins B5–14, IGRP 228–236, and IGRP 265–273) have previously been shown to be recognized by CD8 T cells in HLA-A∗02:01-positive T1D patients [43–46], supporting the clinical relevance of the model.
4. Discussion
Insulin is an important autoantigen recognized by T cells in both human T1D and the NOD mouse model of the disease [47]. Reduced thymic insulin expression is associated with susceptibility to T1D in patients [16, 17, 19, 20], suggesting that impaired negative selection of T cells specific for insulin is responsible for this predisposition. Here we have developed and characterized NOD.β 2mKO.HHD.Ins2het mice as a model of T1D that incorporates reduced thymic insulin. We find that, as in patients, disease is accelerated (Figure 1), and we suggest these mice as a new diabetes model that can be used to better understand this phenomenon. The NOD.β 2mKO.HHD.Ins2het mice present advantages over other disease models that have been described for this purpose. For example, thymic insulin expression is abolished in NOD.Ins2KO and NOD.β 2mKO.HHD.Ins2KO mice, and both exhibit accelerated T1D [28–30] and increased insulin-specific islet-infiltrating CD8 T cells [28] when compared to their Ins2WT counterparts. While these findings suggest the importance of thymic insulin expression, Ins2KO models do not accurately represent patients, where thymic insulin expression is reduced, but not eliminated [19, 20]. As for NOD.Ins2het mice, in our hands neither females nor males show accelerated disease (Figure 2). Two earlier studies of NOD.Ins2het mice also showed no effect on disease in males [29, 30], and only one of the two showed acceleration in females [29]. In contrast, both male and female NOD.β 2mKO.HHD.Ins2het mice show enhanced disease (Figure 1). Indeed, the female and male incidence curves are nearly overlapping until 15 weeks of age (cf. Figures 1(a) and 1(b)). Thus, future mechanistic studies could realistically be performed using both genders. These studies should include the quantification of insulin-specific effector T cells and Treg and analysis of their phenotype and function. The recently described ability to isolate insulin-specific CD4 T cells from NOD mouse strains using enrichment with peptide/MHC tetramer reagents will facilitate this work [48].
In NOD mice, establishment of immunological tolerance to insulin can lead to prevention of T1D [49–51] and remission of established disease [52]. Because of these findings, there is great interest in immunological interventions for human T1D that seek to manipulate the T cell response to insulin [53]. The NOD.β 2mKO.HHD.Ins2het mouse strain should be considered as an additional preclinical model to be used to evaluate such therapies, as it incorporates aspects of the human disease that are not represented in standard NOD mice, including reduced thymic insulin expression. In humans, VNTR alleles associated with diminished thymic insulin have been shown to alter the frequency and avidity of insulin-specific T cells [21], both of which could reasonably influence the outcome of therapies designed to manipulate the immune response to insulin. Given that human insulin-specific CD8 T cells have been shown to have cytotoxic activity against islets [54], an additional advantage of the NOD.β 2mKO.HHD.Ins2het mouse model is the expression of the T1D-predisposing human class I MHC allele HLA-A∗02:01 [4, 6, 8, 12], which we have shown as supporting the development of T cells specific for HLA-A∗02:01-restricted insulin epitopes in these mice (Figure 3(d)). In terms of insulin-specific CD4 T cells, the class II MHC allele expressed in the NOD.β 2mKO.HHD.Ins2het mice is I-Ag7, which is structurally similar to the human T1D-predisposing HLA-DQ8 [55, 56]. Indeed, I-Ag7 and HLA-DQ8 are capable of presenting similar peptides [57–59]. The NOD.β 2mKO.HHD.Ins2het mouse therefore has a variety of potential uses as a humanized model of T1D, including CD8 and CD4 T cell epitope identification, analysis of the relationship between thymic insulin expression and tolerance, and the evaluation of antigen-specific immunotherapies, particularly those targeting the immune response to insulin.
5. Conclusions
NOD.β 2mKO.HHD.Ins2het mice represent a model for T1D that incorporates the reduced, but not abolished, thymic insulin expression observed in patients. This model should find utility in investigations to probe the mechanisms underlying the association between reduced thymic insulin expression and T1D in humans. It will also be an important tool for T cell epitope discovery and for the preclinical evaluation of insulin-targeted immunotherapies for the disease.
Acknowledgments
This work was supported by the National Institutes of Health (R01 DK094327, R01 DK064315, and R03 AI119225 to Teresa P. DiLorenzo; T32 GM007288 and F30 DK103368, which supported Jennifer Schloss; P60 DK020541, which supports the Diabetes Research Center of the Albert Einstein College of Medicine; and P30 CA013330, which supports the flow cytometry facility). Teresa P. DiLorenzo is the Diane Belfer, Cypres and Endelson Families Faculty Scholar in Diabetes Research.
Ethical Approval
All animal experiments were approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Pociot F., Akolkar B., Concannon P., et al. Genetics of type 1 diabetes: what's next? Diabetes. 2010;59(7):1561–1571. doi: 10.2337/db10-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cucca F., Lampis R., Congia M., et al. A correlation between the relative predisposition of MHC class II alleles to type 1 diabetes and the structure of their proteins. Human Molecular Genetics. 2001;10(19):2025–2037. doi: 10.1093/hmg/10.19.2025. [DOI] [PubMed] [Google Scholar]
- 3.Todd J. A., Bell J. I., McDevitt H. O. HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature. 1987;329(6140):599–604. doi: 10.1038/329599a0. [DOI] [PubMed] [Google Scholar]
- 4.Fennessy M., Metcalfe K., Hitman G. A., et al. A gene in the HLA class I region contributes to susceptibility to IDDM in the finnish population. Diabetologia. 1994;37(9):937–944. doi: 10.1007/bf00400951. [DOI] [PubMed] [Google Scholar]
- 5.Honeyman M. C., Harrison L. C., Drummond B., Colman P. G., Tait B. D. Analysis of families at risk for insulin-dependent diabetes mellitus reveals that HLA antigens influence progression to clinical disease. Molecular Medicine. 1995;1(5):576–582. [PMC free article] [PubMed] [Google Scholar]
- 6.Howson J. M. M., Walker N. M., Clayton D., Todd J. A. Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes, Obesity and Metabolism. 2009;11(supplement 1):31–45. doi: 10.1111/j.1463-1326.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakanishi K., Kobayashi T., Murase T., Naruse T., Nose Y., Inoko H. Human leukocyte antigen-A24 and -DQA1∗0301 in Japanese insulin-dependent diabetes mellitus: independent contributions to susceptibility to the disease and additive contributions to acceleration of β-cell destruction. Journal of Clinical Endocrinology and Metabolism. 1999;84(10):3721–3725. doi: 10.1210/jcem.84.10.6045. [DOI] [PubMed] [Google Scholar]
- 8.Nejentsev S., Howson J. M., Walker N. M., et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450(7171):887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nejentsev S., Reijonen H., Adojaan B., et al. The effect of HLA-B allele on the IDDM risk defined by DRB1∗04 subtypes and DQB1∗0302. Diabetes. 1997;46(11):1888–1892. doi: 10.2337/diab.46.11.1888. [DOI] [PubMed] [Google Scholar]
- 10.Noble J. A., Valdes A. M., Bugawan T. L., Apple R. J., Thomson G., Erlich H. A. The HLA class I A locus affects susceptibility to type 1 diabetes. Human Immunology. 2002;63(8):657–664. doi: 10.1016/s0198-8859(02)00421-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noble J. A., Valdes A. M., Varney M. D., et al. HLA class I and genetic susceptibility to type 1 diabetes: results from the type 1 diabetes genetics consortium. Diabetes. 2010;59(11):2972–2979. doi: 10.2337/db10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robles D. T., Eisenbarth G. S., Wang T., et al. Identification of children with early onset and high incidence of anti-islet autoantibodies. Clinical Immunology. 2002;102(3):217–224. doi: 10.1006/clim.2001.5171. [DOI] [PubMed] [Google Scholar]
- 13.Tait B. D., Colman P. G., Morahan G., et al. HLA genes associated with autoimmunity and progression to disease in type 1 diabetes. Tissue Antigens. 2003;61(2):146–153. doi: 10.1034/j.1399-0039.2003.00013.x. [DOI] [PubMed] [Google Scholar]
- 14.Di Lorenzo T. P., Peakman M., Roep B. O. Translational mini-review series on type 1 diabetes: systematic analysis of T cell epitopes in autoimmune diabetes. Clinical & Experimental Immunology. 2007;148(1):1–16. doi: 10.1111/j.1365-2249.2006.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett S. T., Lucassen A. M., Gough S. C. L., et al. Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nature Genetics. 1995;9(3):284–292. doi: 10.1038/ng0395-284. [DOI] [PubMed] [Google Scholar]
- 16.Bell G. I., Horita S., Karam J. H. A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes. 1984;33(2):176–183. doi: 10.2337/diab.33.2.176. [DOI] [PubMed] [Google Scholar]
- 17.Hitman G. A., Tarn A. C., Winter R. M., et al. Type 1 (insulin-dependent) diabetes and a highly variable locus close to the insulin gene on chromosome 11. Diabetologia. 1985;28(4):218–222. doi: 10.1007/BF00282236. [DOI] [PubMed] [Google Scholar]
- 18.Bennett S. T., Wilson A. J., Cucca F., et al. IDDM2-VNTR-encoded susceptibility to type 1 diabetes: dominant protection and parental transmission of alleles of the insulin gene-linked minisatellite locus. Journal of Autoimmunity. 1996;9(3):415–421. doi: 10.1006/jaut.1996.0057. [DOI] [PubMed] [Google Scholar]
- 19.Vafiadis P., Bennett S. T., Todd J. A., et al. Insulin expression in human thymus is modulated by INS VNTR alleles at the IDDM2 locus. Nature Genetics. 1997;15(3):289–292. doi: 10.1038/ng0397-289. [DOI] [PubMed] [Google Scholar]
- 20.Pugliese A., Zeller M., Fernandez A., Jr., et al. The insulin gene is transcribed in the human thymus and transcription levels correlate with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nature Genetics. 1997;15(3):293–297. doi: 10.1038/ng0397-293. [DOI] [PubMed] [Google Scholar]
- 21.Durinovic-Belló I., Wu R. P., Gersuk V. H., Sanda S., Shilling H. G., Nepom G. T. Insulin gene VNTR genotype associates with frequency and phenotype of the autoimmune response to proinsulin. Genes and Immunity. 2010;11(2):188–193. doi: 10.1038/gene.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driver J. P., Serreze D. V., Chen Y.-G. Mouse models for the study of autoimmune type 1 diabetes: a NOD to similarities and differences to human disease. Seminars in Immunopathology. 2011;33(1):67–87. doi: 10.1007/s00281-010-0204-1. [DOI] [PubMed] [Google Scholar]
- 23.Wentworth B. M., Schaefer I. M., Villa-Komaroff L., Chirgwin J. M. Characterization of the two nonallelic genes encoding mouse preproinsulin. Journal of Molecular Evolution. 1986;23(4):305–312. doi: 10.1007/bf02100639. [DOI] [PubMed] [Google Scholar]
- 24.Chentoufi A. A., Polychronakos C. Insulin expression levels in the thymus modulate insulin-specific autoreactive T-cell tolerance: the mechanism by which the IDDM2 locus may predispose to diabetes. Diabetes. 2002;51(5):1383–1390. doi: 10.2337/diabetes.51.5.1383. [DOI] [PubMed] [Google Scholar]
- 25.Heath V. L., Moore N. C., Parnell S. M., Mason D. W. Intrathymic expression of genes involved in organ specific autoimmune disease. Journal of Autoimmunity. 1998;11(4):309–318. doi: 10.1006/jaut.1998.0210. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama M., Babaya N., Miao D., Sikora K., Elliott J. F., Eisenbarth G. S. Thymic expression of mutated B16:a preproinsulin messenger RNA does not reverse acceleration of NOD diabetes associated with insulin 2 (thymic expressed insulin) knockout. Journal of Autoimmunity. 2005;25(3):193–198. doi: 10.1016/j.jaut.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 27.Pléau J.-M., Esling A., Geutkens S., Dardenne M., Homo-Delarche F. Pancreatic hormone and glutamic acid decarboxylase expression in the mouse thymus: a real-time PCR study. Biochemical and Biophysical Research Communications. 2001;283(4):843–848. doi: 10.1006/bbrc.2001.4884. [DOI] [PubMed] [Google Scholar]
- 28.Jarchum I., DiLorenzo T. P. Ins2 deficiency augments spontaneous HLA-A∗0201-restricted T cell responses to insulin. Journal of Immunology. 2010;185(2):658–665. doi: 10.4049/jimmunol.0903414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriyama H., Abiru N., Paronen J., et al. Evidence for a primary islet autoantigen (preproinsulin 1) for insulitis and diabetes in the nonobese diabetic mouse. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(18):10376–10381. doi: 10.1073/pnas.1834450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thébault-Baumont K., Dubois-Laforgue D., Krief P., et al. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. The Journal of Clinical Investigation. 2003;111(6):851–857. doi: 10.1172/jci200316584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takaki T., Marron M. P., Mathews C. E., et al. HLA-A∗0201-restricted T cells from humanized NOD mice recognize autoantigens of potential clinical relevance to type 1 diabetes. The Journal of Immunology. 2006;176(5):3257–3265. doi: 10.4049/jimmunol.176.5.3257. [DOI] [PubMed] [Google Scholar]
- 32.Pfaffl M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9, article e45) doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfaffl M. W., Horgan G. W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Research. 2002;30(9, article e36) doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Serreze D. V., Chapman H. D., Varnum D. S., et al. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new ‘speed congenic’ stock of NOD.Igμ(null) mice. Journal of Experimental Medicine. 1996;184(5):2049–2053. doi: 10.1084/jem.184.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarchum I., Takaki T., DiLorenzo T. P. Efficient culture of CD8+ T cells from the islets of NOD mice and their use for the study of autoreactive specificities. Journal of Immunological Methods. 2008;339(1):66–73. doi: 10.1016/j.jim.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salter R. D., Howell D. N., Cresswell P. Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics. 1985;21(3):235–246. doi: 10.1007/bf00375376. [DOI] [PubMed] [Google Scholar]
- 37.Tsomides T. J., Aldovini A., Johnson R. P., Walker B. D., Young R. A., Eisen H. N. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. Journal of Experimental Medicine. 1994;180(4):1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moodie Z., Price L., Gouttefangeas C., et al. Response definition criteria for ELISPOT assays revisited. Cancer Immunology, Immunotherapy. 2010;59(10):1489–1501. doi: 10.1007/s00262-010-0875-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markle J. G. M., Frank D. N., Mortin-Toth S., et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 40.Pozzilli P., Signore A., Williams A. J. K., Beales P. E. NOD mouse colonies around the world—recent facts and figures. Immunology Today. 1993;14(5):193–196. doi: 10.1016/0167-5699(93)90160-m. [DOI] [PubMed] [Google Scholar]
- 41.Leroux L., Desbois P., Lamotte L., et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2 . Diabetes. 2001;50(supplement 1):S150–S153. doi: 10.2337/diabetes.50.2007.s150. [DOI] [PubMed] [Google Scholar]
- 42.Jarchum I., Baker J. C., Yamada T., et al. In vivo cytotoxicity of insulin-specific CD8+ T-cells in HLA-A∗0201 transgenic NOD mice. Diabetes. 2007;56(10):2551–2560. doi: 10.2337/db07-0332. [DOI] [PubMed] [Google Scholar]
- 43.Énée E., Martinuzzi E., Blancou P., Bach J.-M., Mallone R., Endert P. V. Equivalent specificity of peripheral blood and islet-infiltrating CD8+ T lymphocytes in spontaneously diabetic HLA-A2 transgenic NOD mice. Journal of Immunology. 2008;180(8):5430–5438. doi: 10.4049/jimmunol.180.8.5430. [DOI] [PubMed] [Google Scholar]
- 44.Jarchum I., Nichol L., Trucco M., Santamaria P., DiLorenzo T. P. Identification of novel IGRP epitopes targeted in type 1 diabetes patients. Clinical Immunology. 2008;127(3):359–365. doi: 10.1016/j.clim.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallone R., Martinuzzi E., Blancou P., et al. CD8+ T-cell responses identify β-cell autoimmunity in human type 1 diabetes. Diabetes. 2007;56(3):613–621. doi: 10.2337/db06-1419. [DOI] [PubMed] [Google Scholar]
- 46.Unger W. W. J., Pearson T., Abreu J. R. F., et al. Islet-specific CTL cloned from a type 1 diabetes patient cause beta-cell destruction after engraftment into HLA-A2 transgenic NOD/scid/IL2RG null mice. PLoS ONE. 2012;7(11) doi: 10.1371/journal.pone.0049213.e49213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakayama M. Insulin as a key autoantigen in the development of type 1 diabetes. Diabetes/Metabolism Research and Reviews. 2011;27(8):773–777. doi: 10.1002/dmrr.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pauken K. E., Linehan J. L., Spanier J. A., et al. Cutting edge: type 1 diabetes occurs despite robust anergy among endogenous insulin-specific CD4 T cells in NOD mice. Journal of Immunology. 2013;191(10):4913–4917. doi: 10.4049/jimmunol.1301927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.French M. B., Allison J., Cram D. S., et al. Transgenic expression of mouse proinsulin II prevents diabetes in nonobese diabetic mice. Diabetes. 1997;46(1):34–39. doi: 10.2337/diab.46.1.34. [DOI] [PubMed] [Google Scholar]
- 50.Jaeckel E., Lipes M. A., von Boehmer H. Recessive tolerance to preproinsulin 2 reduces but does not abolish type 1 diabetes. Nature Immunology. 2004;5(10):1028–1035. doi: 10.1038/ni1120. [DOI] [PubMed] [Google Scholar]
- 51.Krishnamurthy B., Dudek N. L., McKenzie M. D., et al. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. Journal of Clinical Investigation. 2006;116(12):3258–3265. doi: 10.1172/jci29602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fife B. T., Guleria I., Gubbels Bupp M., et al. Insulin-induced remission in new-onset NOD mice is maintained by the PD-1-PD-L1 pathway. Journal of Experimental Medicine. 2006;203(12):2737–2747. doi: 10.1084/jem.20061577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrison L. C., Wentworth J. M., Zhang Y., et al. Antigen-based vaccination and prevention of type 1 diabetes. Current Diabetes Reports. 2013;13(5):616–623. doi: 10.1007/s11892-013-0415-7. [DOI] [PubMed] [Google Scholar]
- 54.Kronenberg D., Knight R. R., Estorninho M., et al. Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells. Diabetes. 2012;61(7):1752–1759. doi: 10.2337/db11-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corper A. L., Stratmann T., Apostolopoulos V., et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 2000;288(5465):505–511. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- 56.Lee K. H., Wucherpfennig K. W., Wiley D. C. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nature Immunology. 2001;2(6):501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 57.Nakayama M., McDaniel K., Fitzgerald-Miller L., et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(14):4429–4434. doi: 10.1073/pnas.1502967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suri A., Walters J. J., Gross M. L., Unanue E. R. Natural peptides selected by diabetogenic DQ8 and murine I-Ag7 molecules show common sequence specificity. Journal of Clinical Investigation. 2005;115(8):2268–2276. doi: 10.1172/jci25350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wen L., Wong F. S., Sherwin R., Mora C. Human DQ8 can substitute for murine I-Ag7 in the selection of diabetogenic T cells restricted to I-Ag7 . The Journal of Immunology. 2002;168(7):3635–3640. doi: 10.4049/jimmunol.168.7.3635. [DOI] [PubMed] [Google Scholar]