Abstract
Prion diseases are infectious neurodegenerative disorders linked to the accumulation in the central nervous system of the abnormally folded prion protein (PrP) scrapie (PrPsc), which is thought to be the infectious agent. Once present, PrPsc catalyzes the conversion of naturally occurring cellular PrP (PrPc) to PrPsc. Prion infection is usually initiated in peripheral organs, but the mechanisms involved in infectious spread to the brain are unclear. We found that both PrPc and PrPsc were actively released into the extracellular environment by PrP-expressing cells before and after infection with sheep prions, respectively. Based on Western blot with specific markers, MS, and morphological analysis, our data revealed that PrPc and PrPsc in the medium are associated with exosomes, membranous vesicles that are secreted upon fusion of multivesicular endosomes with the plasma membrane. Furthermore, we found that exosomes bearing PrPsc are infectious. Our data suggest that exosomes may contribute to intercellular membrane exchange and the spread of prions throughout the organism.
Infectious prion diseases include Kuru and variant Creutzfeldt–Jakob disease in humans, scrapie in sheep, and bovine spongiform encephalopathy in cattle (1, 2). In these diseases, infectious prions enter the host through the gastrointestinal tract and migrate to the spleen, after which they cause pathology in the central nervous system (3). Different cell types, including immune cells, contribute to the replication and transfer of infectious prions from peripheral sites of replication to the brain (4). The mechanisms underlying this intercellular transfer are not elucidated (2), but close cell contact may be involved (5). Nevertheless, cell-free conversion data (6) indicate that additional pathways involving non-cell-associated forms of infectious agent may participate in the propagation of prions. Consistent with this notion, the culture medium of scrapie-infected GT1 cells was infectious (7), suggesting that PrPsc may be released from cells and induce transconformation of PrPc in neighboring cells. Noninfected PrP-expressing cells may also have the ability to release PrPc, given that PrPc has been shown to be transferred between cells (8). Thus, release of PrPc and PrPsc by PrP-expressing cells may provide for a potential cellular mechanism underlying propagation and replication of prions. In this study, we further explored the possibility that PrPsc and PrPc may occur in a non-cell-associated form and analyzed their nature in the culture medium of infected and noninfected cell cultures. Our studies indicate that PrPsc and PrPc are associated with exosomes, secreted intralumenal contents of multivesicular bodies (MVB). These findings open the possibility that exosomes may provide for intercellular carriers of both PrPc and PrPsc.
Materials and Methods
Cells, Reagents, and Antibodies. Rov cells are derived from the RK13 cell line and express the ovine VRQ allele of PrP in a doxycycline-dependent manner (9). Mov cells are immortalized neuroglial cells isolated from mice expressing ovine PrP (10). Doxycycline (Sigma) was used at 1 μg/ml. Ex vivo infection of Rov and Mov cells was done as described in ref. 9. The antibodies used were transferrin receptor (Zymed), Tsg101 (M-19, Santa Cruz Biotechnology), Hsc70, grp94, and calnexin (SPA-815, SPA-850, and SPA-865, Stressgen Biotechnologies, Victoria, Canada), flotillin 1 (BD Biosciences), rabbit anti-FITC (Molecular Probes), and PrP [3B5, 8G8, and SAF 84; SPI/CEA (SPI-BIO/Commissariat a L'Energie Atomique), Saclay, France]. FITC-coupled cholera toxin B subunit was from Sigma. Protein A coupled to gold (PAG) and BSA conjugated to 5-nm gold particles (BSAG) were from the Department of Cell Biology, Utrecht University (Utrecht, The Netherlands).
Differential Ultracentrifugation and Exosome Isolation. Cell culture media were centrifuged twice for 5 min at 3,000 × g and 4,500 × g, respectively and ultracentrifuged at 10,000 × g for 30 min and at 100,000 × g for 1 h. For centrifugation onto 2.3 M sucrose cushions, samples were pelleted by ultracentrifugation (1 h, 100,000 × g) on a sucrose cushion; the interface was collected and recovered by a final ultracentrifugation after dilution with PBS. Continuous sucrose gradients were performed as reported in ref. 11.
Western Blotting. Cell culture supernatants and cell lysates were digested with proteinase K (PK) for 2 h at 37°C (2 μg of PK for 500 μg of protein). Pefabloc (4 mM) was added, and aggregated PK-resistant PrP was collected by centrifugation at 14,000 × g for 20 min. Pellets were resuspended in sample buffer, subjected to 10% SDS/PAGE electrophoresis, and transferred to nitrocellulose membranes (9). Western blots were revealed by enhanced chemiluminescence (Amersham Pharmacia).
Dot Blot Analysis. Density gradient fractions dot-blotted onto nitrocellulose (Schleicher and Schuell) were incubated with horse-radish peroxidase-coupled cholera toxin B subunit to detect GM1 (12). Horseradish peroxidase was revealed by enhanced chemiluminescence.
Electron Microscopy. Isolated membranes. Membranes were pelleted by ultracentrifugation on a sucrose cushion as described above. Samples were deposited on Formvar-carbon-coated electron microscopy grids, fixed with 2% paraformaldehyde or a mixture of 2% paraformaldehyde and 0.125% glutaraldehyde, and single or double ImmunoGold-labeled with antibodies, followed by the addition of PAG. Labeling with anti-Tsg101 antibody was done after and during permeabilization with 0.1% saponin for 30 min.
Samples were contrasted and embedded in a mixture of methylcellulose and uranyl acetate and viewed under a CM120 electron microscope (Philips, Eindhoven, The Netherlands).
ImmunoGold labeling on ultrathin cryosections. Cells were fixed with a mixture of 2% paraformaldehyde and 0.125% glutaraldehyde in 0.2 M phosphate buffer, pH 7.4, for 2 h at room temperature. Cells were processed for ultrathin cryosectioning, ImmunoGold-labeled, and contrasted as described in ref. 13. PrP was detected with PrP antibodies (8G8) and PAG coupled to 10- or 15-nm gold particles (PAG10 or PAG15), as indicated in the figures. In one set of experiments, before fixation cells were pulsed for 10 min at 37°C with BSAG, washed at 4°C, and chased for 30 min at 37°C. GM1 was visualized on sections with FITC-coupled cholera toxin B subunit. FITC was detected with a rabbit anti-FITC antibody.
MS Analysis. SDS/PAGE separation and protein digestion. Mov cell lysates and exosomal membranes isolated after floatation on sucrose gradients were loaded onto 12% SDS/polyacrylamide gels. After staining with Coomassie blue (R250, Bio-Rad), the gel was regularly cut into 45 slices of ≈1 mm. Gel slices were reduced and alkylated by using DTT and iodoacetamide, respectively, and subjected to digestion with trypsin (Sigma) by following the protocol published in ref. 14 modified by an overnight digestion at 30°C. Extracted peptides were dried and resolubilized in buffer [95/5 (vol/vol) water/acetonitrile]. Typically one-third of the digestion product of a gel slice was used per liquid chromatography-tandem MS (MS/MS) analysis.
Liquid chromatography-MS/MS analysis. Protein digests were concentrated and separated on an LC Packings system (Dionex) coupled to the nanoelectrospray II ionization interface of a QSTAR/Pulsar i (Applied Biosystems). The MS/MS data from the different experiments was searched twice by using mascot software (Matrix Science, London) on an internal server, first without taxonomic restriction to reveal the presence of proteins of interest and mammalian contaminants, then against the National Center for Biotechnology Information nr Mus database from (National Library of Medicine, Bethesda).
Mouse Bioassay. Culture media of infected Rov cells were harvested 5 days after culture, centrifuged for 5 min at 4,500 × g, and then ultracentrifuged for 1 h at 100,000 × g. Pellets were either resuspended directly in 150 μl of 10% brain homogenate from mice, nullizygous for the mouse Prnp gene (Prnp0/0), or first floated onto a 2.3 M sucrose cushion, washed with PBS by ultracentrifugation, and then resuspended in diluted brain homogenate. The infectivity assays were performed on transgenic mice overexpressing the ovine Prnp gene (VRQ allele) and nullizygous for the mouse Prnp gene (Prnp0/0) (15). Animals were infected intracerebrally with 20 μlof inoculum (127 strain). Inoculated mice were examined for neurological dysfunction every 2 days until clinical signs of scrapie were detected. The brains of all diseased animals were examined for the presence of PK-resistant PrP by immunoblotting and by histological examination so as to confirm the diagnosis of scrapie (data not shown).
Results
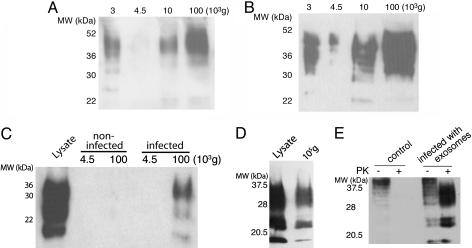
Infected Cells Release PrPsc. We made use of two cell systems that actively replicate sheep prions after contact with scrapie brain extracts (9, 10). Rov cells are derived from rabbit epithelial cells engineered to express the ovine PrP (9). Mov cells are neuroglial cells derived from transgenic mice expressing ovine PrP (10). To investigate whether infected cells release PrP, the culture medium of infected Rov (Fig. 1A) and Mov cells (Fig. 1B) was submitted to sequential centrifugation steps with increasing centrifugal forces (see Fig. 1 legend). Some PrP was present in the 10,000 × g pellet, but the maximal amounts were collected at 100,000 × g, indicating the release of PrP in part of a relatively large complex. To evaluate the presence of PrPsc, pellets obtained after ultracentrifugation at 4,500 × g and 100,000 × g were submitted to proteolysis by PK before Western blot analysis. PK treatment distinguishes between PrPc and abnormally folded PrPsc because the latter has an unusual resistance to proteases (1). The pelleted material from the culture medium of infected cells contained PK-resistant PrP (Fig. 1 C and D), indicating the presence of PrPsc. Quantitation of the band intensities in the lysate and supernatants indicated that ≈1% of the cell-associated PrPsc was released. PrPsc was not detected in the pellets from culture medium of noninfected Mov (Fig. 1C) or Rov cells (data not shown), confirming PrPsc release only from infected cells.
Fig. 1.
The cell culture medium of infected cells contains infectious PrP. (A) The culture medium from 5 × 106 infected Rov cells was submitted to differential centrifugation (11). The medium was submitted to a first centrifugation for 5 min at 4,500 × g to remove cells in suspension. The supernatant was ultracentrifuged at 10,000 × g for 30 min to remove cell debris, and, finally, the last supernatant was ultracentrifuged at 100,000 × g for 1 h. The resulting pellets of each centrifugation step were analyzed by Western blot for PrP. (B) The culture medium of 5 × 106 infected Mov cells was analyzed for PrP as in A. (C) After 5 days of culture, the media of 2 × 107 control Mov cells or scrapie-infected Mov cells were harvested and centrifuged for 5 min at 4,500 × g to remove potentially dissociated cells; the supernatant was then recentrifuged at 100,000 × g for 1 h. The pellets were digested with PK and analyzed for the presence of PK-resistant PrPsc by Western blotting with SAF 84. The lysate comprised Mov cells. (D) The medium of 3 × 107 infected Rov cells was centrifuged and analyzed as in C. The lysate comprised Rov cells. (E) Cell culture medium from 3 × 107 infected Rov cells was ultracentrifuged as above. The pellet was resuspended and incubated for 7 days with uninfected Rov cells. Cultures were grown for several weeks and monitored for the accumulation of cell-associated PK-resistant PrP. Total PrP was obtained by methanol precipitation of undigested cell lysates (lanes 1 and 3, 25 μg of proteins). PrP was isolated from PK-digested cell lysates (lanes 2 and 4, 250 μg of proteins). MW indicates molecular mass in all figures.
We next asked whether the PrPsc contained in the cell medium is infectious. Noninfected Rov cells were incubated in the presence of resuspended 100,000 × g pellets from cell culture media of infected Rov cells. The cell cultures were then grown and analyzed periodically for the presence of PrPsc (see Materials and Methods). PrPsc was not detected after only a few passages (data not shown), ruling out the possibility that the PrPsc signal originated from the input material. By contrast, a clear PrPsc signal was observed thereafter (Fig. 1E). The presence of newly formed PrPsc in late passage-infected Rov cultures indicated that pelletable PrPsc released by the donor cells elicited conversion of endogenous PrP to PrPsc in the recipient cells. To further confirm the presence of prion infectivity in the cell medium, the pelletable fraction from infected Rov cells was inoculated in transgenic mice that express ovine PrP and are highly susceptible to sheep prions (15). In an additional set of experiments, mice were inoculated with a PrP-containing “floating” fraction from a discontinuous sucrose gradient on which the 100,000 × g pellets were fractionated by centrifugation and floatation (see below). All inoculated mice died as a consequence of acute, typical neurological disorders (Table 1). The amount of infectivity released in the conditioned medium corresponds to ≈1% of the cell-associated infectivity, consistent with the amount of biochemically detectable PrPsc in the conditioned medium (Table 1 compared with cell lysate infectivity) (data not shown). These findings demonstrate that the pelletable, floating fraction of PrPsc released by infected Rov cells is infectious.
Table 1. Bioassay of medium conditioned by infected Rov cultures.
| Material | Incubation period,* days | No. affected/no. inoculated |
|---|---|---|
| Control (sample diluent) | >200 | 0/5 |
| 100,000 × g | 76.8 ± 2.6 | 5/5 |
| 100,000 × g pellet floated on 2.3 M sucrose | 78 ± 1.2 | 5/5 |
Mean ± SEM after intracerebral injection of tg338 mice (15)
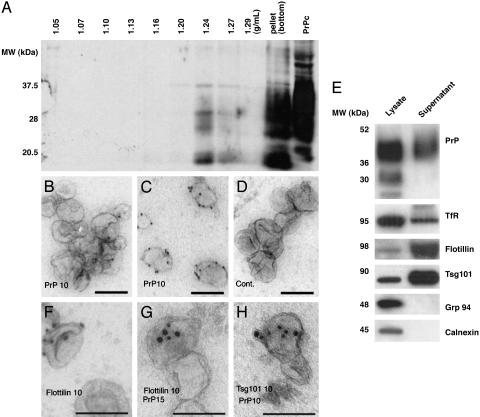
PrPsc Is Released from Cells in Association with Exosome-Like Vesicles. To further characterize the nature of the infectious fraction released from cells, the resuspended 100,000 × g pellets obtained from supernatants of infected Rov cells were analyzed by floatation in a continuous sucrose density gradient. After centrifugation, the density gradient fractions were submitted to PK treatment and analyzed by Western blot for PrPsc. Equivalent experiments were also done with Mov cell supernatants (data not shown). Interestingly, although most of PK-resistant PrP remained in the bottom of the gradient, a fraction of PK-resistant PrP floated to an equilibrium density of ≈1.24 g/ml (Fig. 2A), indicating that secreted PrPsc may be associated with membranes. Immunoelectron microscopy (IEM) analysis revealed the presence of PrP on membrane vesicles, often aggregated with a mean size of 50–90 nm (Fig. 2B). Because the antibodies used do not distinguish between PrPsc and PrPc, labeling was also performed after denaturation by guanidium, a treatment used to specifically detect PrPsc (16). Labeling for PrP increased at least 2-fold, in agreement with the presence of PrPsc (Fig. 2C). Moreover, IEM analysis of the PrPsc-containing pellet that sedimented in the bottom of the sucrose gradient showed the presence of aggregates of amyloid-like fibers often trapping small membrane vesicles (data not shown). These data suggest that at least a fraction of released PrPsc is present on membrane vesicles.
Fig. 2.
PrPsc released by infected cells is associated with exosome-like vesicles. (A)A continuous 0.25–2.5 M sucrose gradient was loaded on top of the 100,000 × g pellet isolated from Rov-infected cell culture medium (3 × 107 cells) and ultracentrifuged to equilibrium. Fractions were PK-digested and analyzed by Western blotting for PrPsc. In the right lane, a lysate (non-PK-digested) from noninfected cells was probed for PrPc. (B–D and F–H) IEM analysis of membranes that were collected from the culture medium by centrifugation on a sucrose cushion. (B and C) ImmunoGold labeling for PrP before (B) and after (C) guanidium (3 M, 5 min) treatment. (D) Control (Cont.) with irrelevant antibody. (F and G) Single and double IEM for flotillin or flotillin and PrP, respectively. (H) Double IEM for Tsg101 and PrP on permeabilized exosomes. (E) Cell lysates and exosomes collected from the culture media of infected cells by centrifugation on a sucrose cushion were analyzed by Western blotting. IEM and Western blotting for PrP was performed with 8G8. TfR, transferrin receptor. (Scale bar, 100 nm.)
The morphology and size of the membrane vesicles bearing PrPsc are reminiscent of exosomes, 50- to 90-nm vesicles of endosomal origin that are released by many cell types into the extracellular environment upon fusion of MVB with the cell surface (17, 18). To explore whether membrane vesicles secreted by infected cells had characteristics of exosomes, cell lysates and PrP-containing membranes isolated from the cell culture medium of infected Rov cells after floatation in sucrose gradients were probed for the presence of proteins known to be enriched in exosomes (Fig. 2E) (17, 18). To exclude the possibility of organelle release as a consequence of cell lysis, the samples were tested for the presence of calnexin and Grp94, an integral membrane and luminal marker, respectively, of the endoplasmic reticulum. These markers were detected in the cell lysate but not in the PrP-containing membrane fraction. As compared with cell lysates, the PrP-containing fraction was enriched in Tsg101, a protein involved in the biogenesis of MVBs and present in exosomes from dendritic cells (19). The transferrin receptor was detected in the PrP-enriched membranes to some extent, consistent with its presence in exosomes from certain cell types, such as differentiating reticulocytes (20, 21), but was not as enriched relative to cell lysates as Tsg101. Flotillin, present in reticulocyte exosomes (22), was also found to be enriched in the PrP-rich membranes. The presence of PrP, Tsg101, and flotillin in membrane vesicles that sedimented from the culture media was confirmed by IEM (Fig. 2 B, E, F, G, and H, respectively). Further supporting the release of specific membrane structures by infected cells rather than general membrane structures resulting from cell lysis is the finding that the Coomassie blue-stained pattern of content proteins fractionated by SDS/PAGE differed between total cell lysates and supernatants (see Fig. 4B). Taken together, these observations indicate that steady state sheep prion-infected cells release infectious PrPsc by means of membrane vesicles with features of exosomes.
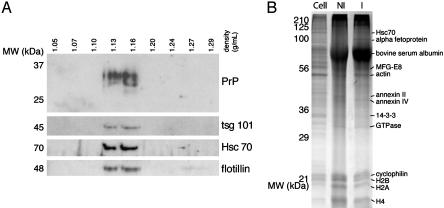
Fig. 4.
PrPc is associated with exosomes. (A) A continuous 0.25–2.5 M sucrose gradient was loaded on top of the 100,000 × g pellet obtained from Mov cell culture medium (4 × 106 cells) and ultracentrifuged to equilibrium. Fractions were analyzed by Western blotting for PrP, Tsg101, Hsc70, and flotillin. (B) Equivalent protein loads of cell lysates (Cell). The 100,000 × g pellet of noninfected (NI) or infected (I) cell culture was fractionated by SDS/PAGE (12%) and stained with Coomassie blue. Some major proteins that were clearly stained and identified by MS (Table 2) are indicated.
PrPc Is Associated with Exosomal Membranes. The above results strongly indicate that a fraction of PrPsc released by infected cells is associated with exosome-like vesicles. These results could potentially reflect PrPsc association with exosomes and release as a consequence of the infectious status of the cell. We therefore tested whether PrPc is also associated with and secreted by means of exosomes.
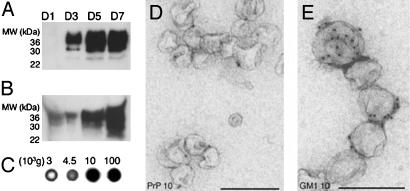
To investigate the presence of PrPc in the cell culture supernatant of Rov and Mov cells, the cell culture medium was submitted to sequential centrifugation steps with increasing centrifugal forces as described above (11). The amount of PrPc pelletable at 100,000 × g increased with time, indicative of a constitutive release (Fig. 3A). As with PrPsc from infected cells, the bulk of PrPc was retrieved from the conditioned media of uninfected cells with maximal efficiency at 100,000 × g (Fig. 3B), although a significant amount of PrPc could also be recovered at 10,000 × g. Consistent with an association of PrPc with membranes, the glycolipid GM1 (12), a typical marker of membrane microdomains, was detected in similar relative proportions in the pellets by dot blot analysis with horse-radish peroxidase-conjugated cholera toxin (Fig. 3C). Analysis of the 100,000 × g pellets by electron microscopy indicated that they consist of small membrane vesicles with a mean diameter of 50–90 nm that can be ImmunoGold-labeled for PrP and GM1 (Fig. 3 D and E, respectively). Analysis of the 10,000 × g pellet revealed aggregated vesicles labeling for both PrP and GM1 (data not shown), explaining why some PrP sedimented at this relatively low centrifugal force. The presence of PrPc in GM1-positive membrane vesicles in the cell culture supernatants indicated that native PrPc, like infectious PrPsc, is constitutively released from cells associated with vesicles.
Fig. 3.
Noninfected cells release PrPc in association with membrane vesicles. (A) Kinetics of PrP release by Mov cells. Culture media were collected after 1, 3, 5, or 7 days and centrifuged at 4,500 × g for 5 min and subsequently for 60 min at 100,000 × g. Pellets were analyzed by Western blotting for PrP. (B) The PrP content of uninfected Mov cell culture medium was analyzed by Western blot on pellets obtained after differential centrifugation. (C) GM1 content of the same fractions shown in B was analyzed by dot blot with cholera toxin. (D and E) IEM analysis of the 100,000 × g pellet for PrP and GM1. IEM and Western blotting for PrP was done with 8G8. (Scale bar, 200 nm.)
Released Membrane Vesicles Have the Hallmarks of Exosomes. The observations that noninfected cells release PrPc by means of membrane vesicles enabled us to further characterize biochemically the membrane vesicles to which PrPc associates. First, 100,000 × g pellets from Mov cell supernatants were analyzed by floatation on a continuous sucrose density gradient as described above for PrPsc. Supporting the observation that released PrPc is associated with membrane vesicles, the total amount of PrP floated at an equilibrium density of ≈1.14 g/ml (Fig. 4A). This floating density is reminiscent of that found for exosomes secreted by B cells (1.13 g/ml), dendritic cells (1.14 g/ml), and intestinal epithelial cells (1.19 g/ml) (11, 23, 24). Unlike PrPsc recovered from the supernatants of infected cells, no detectable PrPc was found at the bottom of the gradient, and the floatation density of the membrane vesicles was slightly lower than that observed for PrPsc (see Fig. 2). The higher floating density found for PrPsc could be because of protein aggregation and the presence of amyloid trapping exosomes (see above). The PrPc-containing fractions were then tested for the presence of components present in exosomes (17, 18). As shown in Fig. 4A, the exosome constituents Hsc70, Tsg101, and flotillin float at the same densities as PrPc. To finally demonstrate the exosomal nature of the PrP-enriched vesicles and to rule out the possibility that the vesicles with which PrPc associates contain cellular contaminants, their protein composition was evaluated by MS (see Materials and Methods, Table 2, and Table 3, which is published as supporting information on the PNAS web site). As analyzed by SDS/PAGE and subsequent Coomassie blue staining, the protein profiles of the 100,000 × g pellets from both noninfected and infected Mov cell supernatants are consistently different from those of the cell lysates and, importantly, are similar to each other (Fig. 4B). Interestingly, the large majority of the proteins sequenced have been shown to be present in exosomes secreted by other cells (19, 24, 25). These common proteins are likely to be involved in cell adhesion (e.g., MFGE8 and integrins), membrane fusion (e.g., annexins and rab proteins), and exosome biogenesis (e.g., annexin II and Hsc70) (17, 18). Thus the PrPc-enriched vesicles are, in all likelihood, bona fide endosome-derived exosomes.
Table 2. MS analysis.
| Classifications | Proteins |
|---|---|
| Targeting/adhesion | Integrins β1, α3, α7, and αV |
| MFG-E8, lactadherin | |
| Chaperones | |
| hsc70 and hsp84 | |
| Membrane fusion | Annexins A1, A2, A3, A4, A5, A6, and A11 |
| Arf3, Arf6, and Arf5 | |
| Rab5c, Rab7, and Rab10 | |
| RabGDI | |
| Rap1A and Rap2B | |
| Cytoskeleton | Actin |
| Cofillin 1 | |
| Moesin | |
| Tubulins α1, α2, β5, β3, and α6 | |
| Signal transduction | 14-3-3 ξ, γ, ε |
| Gβ1 and Gi2α | |
| Enzymes | Enolase 1 |
| GAPDH | |
| Pyruvate kinase | |
| Histones | H2B, H2A, and H4 |
| Others | Translation elongation factor 1α |
| lamp2 | |
| C3 |
Proteins found from MS/MS analysis that are common to exosomes from different cell types. Classification was made based on the putative role of proteins according to Thery et al. (18). The other proteins that were sequenced and accession numbers for the National Center for Biotechnology Information nr database are available in Table 3.
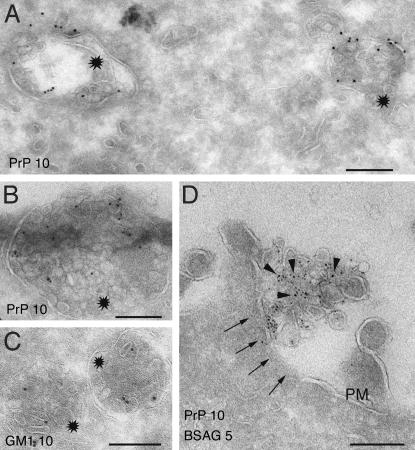
Exosomes correspond to the intraluminal vesicles of MVBs that are released into the extracellular environment upon fusion of these organelles with the cell surface (20). To ascertain the intracellular origin of the PrP-positive exosomes we performed IEM on ultrathin cryosections of Rov (Fig. 5A) and Mov (Fig. 5B) cells by using anti-PrP antibodies. These analyses revealed the presence of PrPc in MVBs. The glycolipid GM1 probed with cholera toxin was also present in the internal vesicles of MVBs (Fig. 5C). We also obtained evidence for exocytic fusion of these endosomes with the cell surface (Fig. 5D), reminiscent to that described in B lymphocytes and dendritic cells (11, 26). The presence of GM1 and PrP in the membrane vesicles recovered from the cell culture supernatants (see Fig. 3), together with their subcellular localization in the intralumenal endosomal vesicles with the same morphology and size, corroborate the exosomal raft-like nature of the PrP-rich membrane vesicles.
Fig. 5.
PrPc localizes to MVBs. (A and B). Ultrathin cryosections of Rov (A) and Mov (B) cells were ImmunoGold-labeled for PrP (PAG10). PrP localizes to both the limiting membrane and the internal vesicles of MVBs (stars). (C) Ultrathin cryosections of Mov cells were labeled with cholera toxin. Note the presence of GM1 in MVBs (stars), similar to PrP. (D) Before fixation, cells were allowed to internalize BSAG (see Materials and Methods). Exocytic fusion (arrows) is defined by the presence of externalized BSAG associated with the extracellular vesicles labeled for PrP (arrowheads). PM, plasma membrane. (Scale bar, 200 nm.)
Discussion
Through the analysis of two distinct cellular models expressing ovine PrP, we obtained evidence that PrPc and PrPsc are released by cells in association with membrane vesicles. The protein composition, biochemical properties, morphology, and size of the membrane vesicles bearing PrP is similar to exosomes, 50- to 90-nm vesicles of endosomal origin that are released into the extracellular environment upon fusion of MVBs with the cell surface (17, 18). Exosomes are secreted by many cell types, including dendritic cells, B cells, mast cells, platelets, reticulocytes, melanoma, and intestinal epithelial cells (11, 20, 23, 24, 27–30). The protein and lipid composition of exosomes, as determined by IEM, Western blot analysis, and MS/MS, is distinct from that of plasma membrane and reflects their endosomal origin. Exosomes are enriched in cell type-specific proteins, including MHC class I and II in dendritic cells and B cells, and in ubiquitous proteins, including the chaperones Hsc70, Hsc90, subunits of trimeric G proteins, Tsg101, cytoskeletal proteins, and tetraspanins (17–19, 25). These latter components are thought to play a role in the biogenesis of exosomes in endosomes or exosome adhesion to target cells (17, 18). The presence of raft components, such as GM1, is consistent with the lipid composition of exosomes, with a particular enrichment in cholesterol and sphingomyelin (25, 31), two lipids also present in plasma membrane rafts (32). In addition, emphasizing the raft-like nature of the membrane environment of the non-cell-associated PrP, flotillin is also detected in PrP-rich exosomes. Flotillin was first identified as a resident membrane protein of caveolae (33) and was recently shown to be present in lipid rafts in late endosomes (34) and in exosomes from reticulocytes (22). PrPc and PrPsc are glycosylphosphatidylinositol-anchored proteins known to partition into lipid rafts (2, 35). Other glycosylphosphatidylinositol-anchored proteins have already been shown to be efficiently incorporated into exosomes (22, 36), which is also consistent with the association of prions with exosomes.
In some cell types, PrPc travel through caveolae and caveolin-containing electron-lucent compartments named caveosomes in route to MVBs and lysosomes (37). The localization of PrP to multivesicular endosomes and the late endosomal derivation of exosomes is consistent with this recent study and other studies showing that PrPc (38) and PrPsc localize to late endosomes and lysosomes (16). It is not clear, however, how the caveolar pathway mix with late endocytic organelles to transfer PrP and other raft-associated proteins, such as flotillin, to intraluminal vesicles of MVBs (37).
We now show that one fate of these endo/lysosomal compartments is exocytic fusion, having as a consequence the release into the extracellular environment of exosomes bearing PrPc and PrPsc. Accumulating data of the past recent years lead to the idea that exosomes provide for a new mode of intercellular communication (17, 18). In addition to their ability to stimulate T cell proliferation and antitumor immune responses (11, 26), exosomes have been shown to participate in the transfer of MHC class II peptide complexes among dendritic cells (39) and may be responsible for the acquisition of MHC class II molecules by follicular dendritic cells (40). Thus, exosomes may mediate exchange of membranes between cells and as such could be involved in the recently observed intercellular transfer of PrPc (8). The molecular machinery implicated in MVB fusion with the cell surface and the subcellular mechanisms involved in exosome transmission between cells is far from being unraveled and needs to be carefully addressed in different cellular models of relevance to PrPsc transmission.
Our study indicates that exosome-associated PrPsc is capable of transferring infectivity in vitro and in vivo but does not allow a determination of whether exosomal membranes participate in the conversion of PrPc into PrPsc. However, recent studies indicate that the transconformation of PrPc to PrPsc requires membrane exchange between cells by an unknown mechanism (6). Hypothetically, PrP conversion may be initiated as a consequence of the binding of PrPsc bearing exosomes to acceptor cells. In this context, it is noteworthy that the topology of exosomal membranes is identical to that of the plasma membrane (17). Alternatively, but not mutually exclusive, exosomes captured by target cells could fuse with the cell surface or be internalized by an unclear mode of entry to induce conversion of PrPc at the cell surface and/or in endocytic compartments, respectively. In addition, it is tempting to hypothesize that the raft-like nature of exosomal membranes (25, 31) may be a favorable environment for transconformation and amyloid fiber formation (41).
There is increasing evidence that monocytes and bone marrow-derived and follicular dendritic cells accumulate infectious prions and play a key role in the onset of disease (4). Most of these cells actively secrete exosomes (17, 18), and PrPc is present in exosomes secreted by bone marrow-derived dendritic cells (our unpublished observations). Future studies are needed to determine whether exosomes secreted by these cells participate in the propagation of this infectious agent in vivo.
MVBs are well known to be critical intermediates in the endocytic pathway (42). This and numerous other recent studies indicate that cells may exploit the nature of endosome-derived exosomes to communicate with each other in normal and pathological situations, providing for a novel route of cell-to-cell communication and therefore of pathogen transmission. Unraveling the cellular mechanisms and the molecular components involved in exocytic fusion of PrP-containing MVBs with the cell surface and transfer of exosomal membranes between cells will certainly shed light on the possible contribution of exosomes in prion propagation.
Supplementary Material
Acknowledgments
We thank SPI/CEA, M. Moudjou (Institut National de la Recherche Agronomique) for providing antibodies, P. Debey for support in the course of this work, W. Stoorvogel and M. Sachse for helpful discussions, and M. S. Marks for critical reading of the manuscript. This work was supported by Institut Curie (G.R., D.L., and W.F.), Centre National de la Recherche Scientifique (G.R. and M.V.), Institut National de la Recherche Agronomique (D.V. and H.L.), and a Ministère de la Recherche GIS Prion Grant. B.F. was supported by the Ministère National de la Recherche et de la Technologie and a fellowship from the Carrefour International Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BSAG, BSA conjugated to 5-nm gold particles; IEM, immunoelectron microscopy; MS/MS, tandem MS; MVB, multivesicular bodies; PAG, protein A coupled to gold; PK, proteinase K; PrP, prion protein; PrPc, cellular PrP; PrPsc, scrapie PrP.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363-13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris, D. A. (1999) Clin. Microbiol. Rev. 12, 429-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissmann, C., Enari, M., Klöhn, P. C., Rossi, D. & Flechsig, E. (2002) Proc. Natl. Acad. Sci. USA 99, 16378-16383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glatzel, M. & Aguzzi, A. (2000) Microbes Infect. 2, 613-619. [DOI] [PubMed] [Google Scholar]
- 5.Kanu, N., Imokawa, Y., Drechsel, D. N., Williamson, R. A., Birkett, C. R., Bostock, C. J. & Brockes, J. P. (2002) Curr. Biol. 12, 523-530. [DOI] [PubMed] [Google Scholar]
- 6.Baron, G. S., Wehrly, K., Dorward, D. W., Chesebro, B. & Caughey, B. (2002) EMBO J. 21, 1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schatzl, H. M., Laszlo, L., Holtzman, D. M., Tatzelt, J., DeArmond, S. J., Weiner, R. I., Mobley, W. C. & Prusiner, S. B. (1997) J. Virol. 71, 8821-8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu, T., Li, R., Pan, T., Liu, D., Petersen, R. B., Wong, B. S., Gambetti, P. & Sy, M. S. (2002) J. Biol. Chem. 277, 47671-47678. [DOI] [PubMed] [Google Scholar]
- 9.Vilette, D., Andreoletti, O., Archer, F., Madelaine, M. F., Vilotte, J. L., Lehmann, S. & Laude, H. (2001) Proc. Natl. Acad. Sci. USA 98, 4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Archer, F., Bachelin, C., Andreoletti, O., Besnard, N., Perrot, G., Langevin, C., Le Dur, A., Vilette, D., Baron-Van Evercooren, A., Vilotte, J. L. & Laude, H. (2004) J. Virol. 78, 482-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo, G., Nijman, H. W., Stoorvogel, W., Liejendekker, R., Harding, C. V., Melief, C. J. & Geuze, H. J. (1996) J. Exp. Med. 183, 1161-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyningen, S. V. (1974) Science 183, 656-657.4810267 [Google Scholar]
- 13.Raposo, G., Kleijmeer, M. J., Posthuma, G., Slot, J. W. & Geuze, H. J. (1997) in Weirs Handbook of Experimental Immunology, eds. Herzenberg, L. A., Weir, D., Herzenberg, L. A. & Blackwell, C. (Blackwell, Cambridge, MA), 5th Ed., Vol. 4, pp. 1-11. [Google Scholar]
- 14.Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68, 850-858. [DOI] [PubMed] [Google Scholar]
- 15.Vilotte, J. L., Soulier, S., Essalmani, R., Stinnakre, M. G., Vaiman, D., Lepourry, L., Da Silva, J. C., Besnard, N., Dawson, M., Buschmann, A., et al. (2001) J. Virol. 75, 5977-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinley, M. P., Taraboulos, A., Kenaga, L., Serban, D., Stieber, A., DeArmond, S. J., Prusiner, S. B. & Gonatas, N. (1991) Lab. Invest. 65, 622-630. [PubMed] [Google Scholar]
- 17.Stoorvogel, W., Kleijmeer, M. J., Geuze, H. J. & Raposo, G. (2002) Traffic 3, 321-330. [DOI] [PubMed] [Google Scholar]
- 18.Thery, C., Zitvogel, L. & Amigorena, S. (2002) Nat. Rev. Immunol. 2, 569-579. [DOI] [PubMed] [Google Scholar]
- 19.Thery, C., Boussac, M., Veron, P., Ricciardi-Castagnoli, P., Raposo, G., Garin, J. & Amigorena, S. (2001) J. Immunol. 166, 7309-7318. [DOI] [PubMed] [Google Scholar]
- 20.Pan, B. T., Teng, K., Wu, C., Adam, M. & Johnstone, R. M. (1985) J. Cell Biol. 101, 942-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geminard, C., Nault, F., Johnstone, R. M. & Vidal, M. (2001) J. Biol. Chem. 276, 9910-9916. [DOI] [PubMed] [Google Scholar]
- 22.de Gassart, A., Geminard, C., Fevrier, B., Raposo, G. & Vidal, M. (2003) Blood 102, 4336-4344. [DOI] [PubMed] [Google Scholar]
- 23.Thery, C., Regnault, A., Garin, J., Wolfers, J., Zitvogel, L., Ricciardi-Castagnoli, P., Raposo, G. & Amigorena, S. (1999) J. Cell Biol. 147, 599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Niel, G., Raposo, G., Candalh, C., Boussac, M., Hershberg, R., Cerf-Bensussan, N. & Heyman, M. (2001) Gastroenterology 121, 337-349. [DOI] [PubMed] [Google Scholar]
- 25.Wubbolts, R., Leckie, R. S., Veenhuizen, P. T., Schwarzmann, G., Mobius, W., Hoernschemeyer, J., Slot, J. W., Geuze, H. J. & Stoorvogel, W. (2003) J. Biol. Chem. 278, 10963-10972. [DOI] [PubMed] [Google Scholar]
- 26.Zitvogel, L., Regnault, A., Lozier, A., Wolfers, J., Flament, C., Tenza, D., Ricciardi-Castagnoli, P., Raposo, G. & Amigorena, S. (1998) Nat. Med. 4, 594-600. [DOI] [PubMed] [Google Scholar]
- 27.Wolfers, J., Lozier, A., Raposo, G., Regnault, A., Thery, C., Masurier, C., Flament, C., Pouzieux, S., Faure, F., Tursz, T., Angevin, E., et al. (2001) Nat. Med. 7, 297-303. [DOI] [PubMed] [Google Scholar]
- 28.Skokos, D., Le Panse, S., Villa, I., Rousselle, J. C., Peronet, R., David, B., Namane, A. & Mecheri, S. (2001) J. Immunol. 166, 868-876. [DOI] [PubMed] [Google Scholar]
- 29.Heijnen, H. F., Schiel, A. E., Fijnheer, R., Geuze, H. J. & Sixma, J. J. (1999) Blood 94, 3791-3799. [PubMed] [Google Scholar]
- 30.Andre, F., Schartz, N. E., Movassagh, M., Flament, C., Pautier, P., Morice, P., Pomel, C., Lhomme, C., Escudier, B., Le Chevalier, T., et al. (2002) Lancet 360, 295-305. [DOI] [PubMed] [Google Scholar]
- 31.Mobius, W., van Donselaar, E., Ohno-Iwashita, Y., Shimada, Y., Heijnen, H. F., Slot, J. W. & Geuze, H. J. (2003) Traffic 4, 222-231. [DOI] [PubMed] [Google Scholar]
- 32.Simons, K. & Ikonen, E. (1997) Nature 387, 569-572. [DOI] [PubMed] [Google Scholar]
- 33.Bickel, P. E., Scherer, P. E., Schnitzer, J. E., Oh, P., Lisanti, M. P. & Lodish, H. F. (1997) J. Biol. Chem. 272, 13793-13802. [DOI] [PubMed] [Google Scholar]
- 34.Fivaz, M., Vilbois, F., Thurnheer, S., Pasquali, C., Abrami, L., Bickel, P. E., Parton, R. G. & van der Goot, F. G. (2002) EMBO J. 21, 3989-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naslavsky, N., Stein, R., Yanai, A., Friedlander, G. & Taraboulos, A. (1997) J. Biol. Chem. 272, 6324-6331. [DOI] [PubMed] [Google Scholar]
- 36.Rabesandratana, H., Toutant, J. P., Reggio, H. & Vidal, M. (1998) Blood 91, 2573-2580. [PubMed] [Google Scholar]
- 37.Peters, P. J., Mironov, A., Jr., Peretz, D., van Donselaar, E., Leclerc, E., Erpel, S., DeArmond, S. J., Burton, D. R., Williamson, R. A., Vey, M. & Prusiner, S. B. (2003) J. Cell Biol. 162, 703-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laine, J., Marc, M. E., Sy, M. S. & Axelrad, H. (2001) Eur. J. Neurosci. 14, 47-56. [DOI] [PubMed] [Google Scholar]
- 39.Thery, C., Duban, L., Segura, E., Veron, P., Lantz, O. & Amigorena, S. (2002) Nat. Immunol. 3, 1156-1162. [DOI] [PubMed] [Google Scholar]
- 40.Denzer, K., van Eijk, M., Kleijmeer, M. J., Jakobson, E., de Groot, C. & Geuze, H. J. (2000) J. Immunol. 165, 1259-1265. [DOI] [PubMed] [Google Scholar]
- 41.Ehehalt, R., Keller, P., Haass, C., Thiele, C. & Simons, K. (2003) J. Cell Biol. 160, 113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raposo, G., Fevrier, B., Stoorvogel, W. & Marks, M. S. (2002) Cell Struct. Funct. 27, 443-456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.