Abstract
Transfer of organelle DNA into the nuclear genome has been significant in eukaryotic evolution, because it appears to be the origin of many nuclear genes. Most studies on organelle DNA transfer have been restricted to evolutionary events but experimental systems recently became available to monitor the process in real time. We designed an experimental screen to detect plastid DNA (ptDNA) transfers to the nucleus in whole plants grown under natural conditions. The resultant genotypes facilitated investigation of the evolutionary mechanisms underlying ptDNA transfer and nuclear integration. Here we report the characterization of nuclear loci formed by integration of newly transferred ptDNA. Large, often multiple, fragments of ptDNA between 6.0 and 22.3 kb in size are incorporated into chromosomes at single Mendelian loci. The lack of chloroplast transcripts of comparable size to the ptDNA integrants suggests that DNA molecules are directly involved in the transfer process. Microhomology (2–5 bp) and rearrangements of ptDNA and nuclear DNA were frequently found near integration sites, suggesting that nonhomologous recombination plays a major role in integration. The mechanisms of ptDNA integration appear similar to those of biolistic transformation of plant cells, but no sequence preference was identified near junctions. This article provides substantial molecular analysis of real-time ptDNA transfer and integration that has resulted from natural processes with no involvement of cell injury, infection, and tissue culture. We highlight the impact of cytoplasmic organellar genome mobility on nuclear genome evolution.
Intracellular transfer of ancestral organelle DNA into the nuclear genome has been a driving force in eukaryote genome evolution. The transfer of DNA fragments from captured ancestral prokaryotes to the eukaryotic host genome has led to the acquisition of thousands of erstwhile prokaryote genes by the nucleus and the diminution of organelle genomes to small remnants (1). Nonfunctional organelle DNA fragments are present in nearly all eukaryote nuclear genomes with some of the largest integrants representing entire chloroplast or mitochondrial genomes (2–7). Sequence comparisons of nuclear and organelle genomes suggest that the transfer of organelle DNA is an ongoing process (1–3).
Organelle DNA transfer to the nuclear genome may use either RNA-mediated or direct DNA transfer processes. RNA-mediated transfer of mitochondrial sequences to the nucleus has been inferred in higher plants because some organelle-derived nuclear genes more closely resemble edited mitochondrial mRNAs than mtDNA (8–10). An RNA-mediated process presumably requires reverse transcription to precede DNA integration into the nuclear genome. However, many nuclear mitochondrial DNA sequences (numts) and the analogous nuclear integrants of plastid DNA (nupts) are longer than any known organellar transcripts, arguing that DNA is directly involved in the relocation events (11–15).
Once organelle DNA or cDNA is transferred to the nucleus, it may be integrated into nuclear chromosomes by mechanisms yet to be defined. Nonhomologous recombination is the dominant mechanism for the integration of heterologous DNA into nuclear DNA of higher eukaryotes by biolistics and via Agrobacterium transformation (16–18). The involvement of this mechanism also has been inferred in the nuclear integration of organelle DNA sequences (14), although the interpretation of such analyses is complicated by the presence of many preexisting organelle sequences in the nucleus and potential postintegrative rearrangement and sequence decay. With the advent of experimental systems to detect de novo plastid DNA (ptDNA) transfer (19, 20), direct studies of the nuclear integration mechanisms are possible for the first time.
We investigated plastid-to-nucleus DNA transfer in the progeny of a higher plant, by inserting into the tobacco plastome two marker genes: aminoglycoside 3′-adenyltransferase (aadA) and a neomycin phosphotransferase gene containing a nuclear intron from the potato STLS gene (neoSTLS2). Seventeen independent kanamycin-resistant (kr) plants were obtained, and each of these plants possessed the native tobacco plastome and de novo nupt(s) (19). Here we report the characterization of some of the de novo nupts, providing insight into mechanisms that promote ptDNA integration into nuclear DNA.
Materials and Methods
Plant Material. Two transplastomic lines, tp7 and tp17, and 17 kr lines containing de novo nupts (19) were used.
Nucleic Acid Hybridization. DNA and RNA blot analyses were performed as described in refs. 12 and 21.
PCR Amplification of Tobacco Genomic DNA. Inverse PCR (iPCR) (19) and long-range PCR were used to amplify genomic DNA sequences flanking de novo nupts. An Extend Long Template PCR kit (Boehringer Mannheim) was used in long-range PCR according to the manufacturer's recommendations. Primer locations in the transplastome are shown in Fig. 1A, and primer sequences are listed in Table 1. Primer combinations and their respective targets are as follows: neoF1 and neoR2 for the amplification of junction sequence (J1b) located on a 2.1-kb EcoRI fragment of kr1, PtF6 and 35SR for the nupt sequence adjacent to the XbaI site in the 10.9-kb neo-hybridizing XbaI fragment of kr1, PtF4 and 35SR for amplification of both junctions J5-2a and J5-2b from a 5.4-kb neo-hybridizing XbaI fragment (kr5-2) of kr5 and J7a from a 5.0-kb neo-hybridizing XbaI fragment of kr7, neoF2 and 35SR for J10a from a 2.8-kb neo-hybridizing SacI fragment of kr10, and aadAF and PtR1 for J14a from a 5.7-kb aadA-hybridizing XbaI fragment of kr14. Primer pairs P1 and P2 were used for amplification of the kr5-1 fragment of kr5, and P1 and P3 were used for amplification of the kr10 nupt fragment.
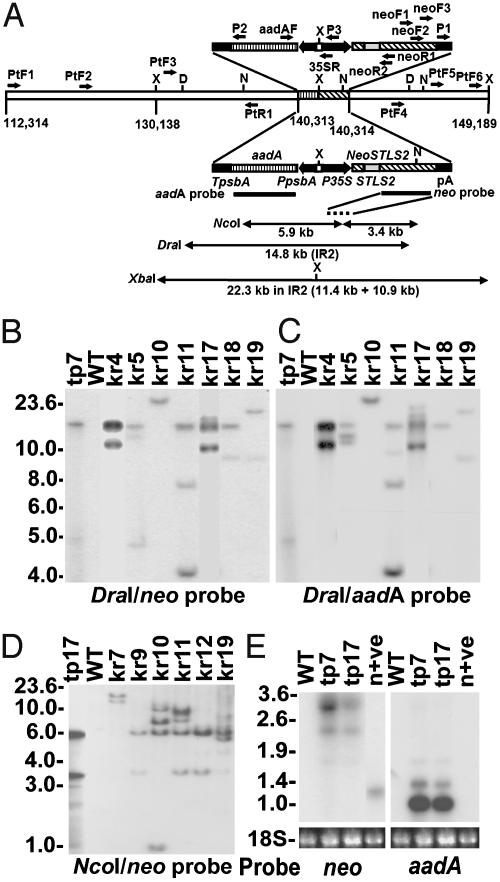
Fig. 1.
Molecular analysis of new ptDNA integrants. (A) Description of the second inverted repeat region (IR2) of transplastome, primer locations (single-headed arrows), and diagrammatic features of aadA and neoSTLS2 in the transplastome of tp7 and tp17. Two identical transgenic regions in IR2 shown above and below the modified wild-type (WT) plastome line are used to depict primer locations (above) and features of two marker genes (below). Promoters (filled arrow boxes, pointing to the direction of transcription), terminators (black boxes), neoSTLS2 coding regions (boxes with diagonal lines), STLS2 intron (gray box), aadA coding regions (box with vertical lines), hybridization probes (thick black lines), and transplastomic DraI (D) NcoI (N) and XbaI (X) fragments (double-headed arrows). Nucleotide positions are numbered according to the native plastome sequence (GenBank accession no. Z00044). (B and C) Southern analysis of DraI-digested DNA from seven kr plants, WT, and a transplastomic control (tp7) hybridized with the neo (B) or aadA (C) probe. (D) Southern analysis of NcoI-digested DNA from six kr plants, WT, and a transplastomic control (tp17) with the neo probe. Total cellular DNA (5 μg) was used in each lane except for tp7 and tp17 (0.005 μg). (E) RNA gel blot analysis of two independent but identical transplastomic lines, tp7 and tp17, WT, and a nuclear neoSTLS2 control (n+ve). Total leaf RNA (3 μg) was hybridized with either neo or aadA probes. 18S RNA stained with ethidium bromide shows RNA loadings. Molecular sizes (kb) are indicated to the left of B, D, and E.
Table 1. Primer sequences used for PCR amplification.
| Primer | Primer location* | Sequence (5′ → 3′) | GenBank accession no. |
|---|---|---|---|
| neoF1 | 1-20 | GGACCGCTATCAGGACATAG | AJ616346 |
| neoF2 | 1-20 | GCTATCAGGACATAGCGTTG | AJ616350 |
| neoF3 | 120-139 | CCTTCTATCGCCTTCTTGAC | AJ616346 |
| neoR1 | 3,311-3,292 | CAGATCATCCTGATCGACAA | AF485783 |
| neoR2 | 1,413-1,431 | GGGCTGCACATCAACCAAA | AJ616346 |
| 35SR | 1,739-1,720 | CAATAGCCCTCTGGTCTTCT | AJ616348 |
| aadAF | 1-20 | GAAGTTTCCAAAAGGTCGTT | AJ616351 |
| PtF1 | 112,314-112,342 | GCACGATTATATGACCAATCA | Z00044 |
| PtF2 | 125,525-126,545 | TGGATTTATCAGTCGAAGCAG | Z00044 |
| PtF3 | 130,457-130,475 | GCTGTTGAAACCAGAAGAG | Z00044 |
| PtF4 | 142,329-142,348 | CCAAGGACTCAATCGTATGG | Z00044 |
| PtF5 | 145,408-145,427 | GACTGGACGAAACCAAGAAA | Z00044 |
| PtF6 | 148,503-148,552 | CCAGATCATGAATGTTTGGA | Z00044 |
| PtR1 | 136,886-136,906 | CGAACAGCTTGATGCACTATC | Z00044 |
| P1 | 149-177 | GGGACCCGCAAAAATCACCAGTCTCTCTC | AJ616350 |
| P2 | 2,072-2,044 | TCGCGCGCAGATCAGTTGGAAGAATTTGT | U12812 |
| P3 | 1,510-1,539 | CGAGAGTGTCGTGCTCCACCATGTTGACCT | AJ616350 |
| kr14F | 662-679 | GGCTTGGACGTCACTTCA | AJ616351 |
Nucleotide location is specific for each quoted accession number. For clarity, where primers are located in the inverted repeats, numbers refer only to the second inverted repeat of the tobacco plastome
Confirmation of the Origin of iPCR and Long-Range PCR Products. Based on the sequences obtained from iPCR and long-range PCR products, new primers were designed to specifically amplify each junction sequence from total DNA of the respective kr plant and its progeny. Kanamycin-resistance status was assayed as described in ref. 19. The primers used for junction confirmation are listed in Table 1, and their locations in the transplastome are shown in Fig. 1 A except for the nuclear sequence Kr14F. The targeted junction sequences of these primers are J5-1a and J5-1b for PtF5 and PtF1, J5-2a and J5-2b for neoF3 and PtF3, J7a for PtF4 and PtF2, J10a for neoF2 and PtR1, and J14a for Kr14F and PtR1.
DNA Sequence Analysis. Products of PCR and iPCR were separated by gel electrophoresis, purified by using a QIAEX I Gel Extraction kit (Qiagen, Valencia, CA), and sequenced by using Applied Biosystems sequencing technology.
Results
Single and Multiple ptDNA Integrants at New Nuclear Loci. Southern analysis of DNA from 16 kr plants with neo and aadA probes revealed simple and more complex hybridization patterns after XbaI digestion (19). Nine kr plants appear to contain single-copy inserts, and seven kr plants appear to include multiple copy inserts. Total cellular DNA of the latter (kr4, kr5, kr10, kr11, kr17, kr18, and kr19) was digested with DraI to determine the number of ptDNA integrants because there are no DraI sites within the aadA or neoSTLS2 genes (Fig. 1 A). Fig. 1 B and C shows that two or three DraI fragments hybridize both neo and aadA probes in five kr plants (kr4, kr5, kr11, kr17, and kr19), indicating the presence of multiple nuclear integrants of newly transposed ptDNA. One plant (kr18) contained a neo-specific DraI fragment in addition to a second fragment, which hybridized both neo and aadA probes (Fig. 1 B and C), and another plant (kr10) contained a unique DraI fragment that hybridized both aadA and neo probes. The DraI fragment of kr10 was larger in size than that of the transplastome (Fig. 1 A–C), suggesting either a rearrangement of the transposed ptDNA or that the size of the ptDNA integrant is smaller than the DraI fragment of the transplastome. Subsequent analysis revealed the presence of multiple neo and aadA sequences in kr10 (see Figs. 4 and 5, which are published as supporting information on the PNAS web site).
Four DraI fragments that hybridized only to either the neo or aadA probe were observed in three kr plants (kr5, kr11, and kr18) with multiple integrants (Fig. 1 B and C). In kr5, a 4.8-kb DraI fragment hybridized only to the neo probe, and an 11-kb DraI fragment hybridized only to the aadA probe. In kr11, a 9.7-kb DraI fragment hybridized weakly to the aadA probe only, and a 9.0-kb DraI fragment hybridized only to the neo probe in kr18. These results suggest that some separation of aadA and neoSTLS2 sequences occurred during the integration process.
To determine whether the multiple integrants behave as single genetic loci, DNA samples from 12 progeny of each kr plant were analyzed by Southern analysis (Table 2 and Fig. 4). These data demonstrate that different nupts may be highly variable in sequence organization and consist of complex, rearranged, multiple copies of ptDNA at single loci. However, evidence for meiotic recombination may be forthcoming after analyses of larger progenies.
Table 2. Molecular analysis of ptDNA integrants in the nuclear genome of tobacco.
| Corresponding transplastome (5′-ptDNA - aadA - neoSTLS2 - ptDNA-3′)
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Plant | No. of integrants | Minimum size 5′ ptDNA | 5′ ptDNA termination | aadA | neoSTLS2 | 3′ ptDNA termination | Minimum size 3′ ptDNA | Special feature | Size of integrant, kb | Locus no. |
| kr1 | 1 | 132,624 (AJ495859) | + | + | 140,369 (AJ616346) | Two ptDNA fragments (no filler) | >21.8 | 1 | ||
| - | - | 328 bp (AJ616346) | + | >149,189 | ||||||
| kr3 | 1 | >130,138 | + | + | >149,189 | >22.3 | 1 | |||
| kr4 | 2 | >130,138 | + | + | >149,189 | >22.3 | 1 | |||
| >136,831 | + | + | >143,256 | >9.3 | ||||||
| kr5 | 4 | >130,515 | + | + | >142,414 | >14.8 | 1 | |||
| ND | +/- | +/- | ND | ND | ||||||
| - | - | - | +/- | ND | ND | |||||
| ND | +/- | - | - | - | ND | |||||
| kr6 | 1 | >138,541 | + | + | 141,675 | - | > 6.0 | 1 | ||
| kr7 | 1 | >138,541 | + | + | 143,030 (AJ616349) | - | Two ptDNA fragments with no filler | > 7.5 | 1 | |
| - | - | - | - | - | 126,736 to 126,183 (AJ616349) | |||||
| kr8 | 1 | >136,831 | + | + | 143,256 | - | > 9.3 | 1 | ||
| kr9 | 1 | >130,138 | + | + | 149,189 | - | >22.3 | 1 | ||
| kr10 | 1 | ND | ND | + | 140,892 (AJ616350) | - | Two ptDNA fragments with no filler | > 7.2 | 1 | |
| - | 136,727 (AJ616350) | + | +/- | ND | ||||||
| kr11 | 4 | >130,515 | + | + | >142,414 | >14.8 | ND | |||
| ND | +/- | +/- | ND | ND | ||||||
| ND | +/- | +/- | ND | ND | ||||||
| ND | +/- | - | - | - | ND | |||||
| kr12 | 1 | >130,138 | + | + | >149,189 | >22.3 | 1 | |||
| kr13 | 1 | >130,138 | + | + | >149,189 | >22.3 | 1 | |||
| kr14 | 1 | - | 136,498 (AJ616351) | + | + | >140,951 | > 7.3 | 1 | ||
| kr16 | 1 | >136,831 | + | + | >143,256 | > 9.3 | 1 | |||
| kr17 | 3 | >130,515 | + | + | >142,414 | >14.8 | 1 | |||
| ND | +/- | +/- | ND | ND | ||||||
| ND | +/- | +/- | ND | ND | ||||||
| kr18 | 2 | >130,515 | + | + | >142,414 | >14.8 | 1 | |||
| - | - | - | + (AJ517486) | 141,486 (AJ517486) | - | 2.9 | ||||
| kr19 | 2 | >136,831 | + | + | >143,256 | > 9.3 | 1 | |||
| ND | +/- | +/- | ND | ND | ||||||
The number of integrants was determined by DraI-digested total cellular DNA hybridized separately to neo and aadA probes as shown in Fig. 1 B and C. Minimum size estimates of integrants were determined by presence or absence of aadA- or neo-hybridizing restriction fragments with the same size as the parental transplastome. DNAs were restricted with the following enzymes and the size of contiguous transplastomic restriction fragments indicated in parentheses by nucleotide locations in the second inverted repeat of the tobacco plastome sequence (Genbank accession no. Z00044); DraI (130,515-142,414), EcoRI (138,541 or 141,675), EcoRV (140,951), NcoI (136,831 or 143,256), or XbaI (130,138 or 149,189). The minimum sizes for the 5′ and 3′ ends of each integrant, where known, are shown in columns labeled as minimum size 5′ and 3′ ptDNA. For integrants in which the end of the transplastomic fragment has been precisely determined by cloning and sequencing, the terminal 5′ and 3′ nucleotide positions are provided with sequence accession numbers (columns labeled 5′ and 3′ ptDNA termination). Presence of complete aadA (1206 bp) and neoSTLS2 (1696 bp) sequences are indicated by “+”, partial sequences of these genes are indicated by “+/-,” and a complete absence is indicated by “-.” ND, not determined. The number of loci was determined by segregation analysis of 12 progeny.
Further molecular analyses were undertaken on nine kr plants anticipated to contain simple, single integrants. To determine whether the XbaI fragments that hybridize both neo and aadA probes in nuclear DNA from these plants are part of single continuous nupts, NcoI digestion and Southern analysis were used. The neo probe hybridized to two predicted NcoI fragments (5.9 and 3.4 kb) in the transplastome (Fig. 1 A and D, lane tp17). The identification of equivalent fragments in the nuclear genomes of kr plants suggests that the aadA and neoSTLS2 were transferred together on a continuous stretch of ptDNA. Seven kr plants (kr9 and kr12 shown in Fig. 1D together with kr3, kr8, kr13, kr14, and kr16 in ref. 19), with single neo- and aadA-hybridizing XbaI fragments, possessed both 5.9- and 3.4-kb neo-hybridizing NcoI fragments, except for kr14, which contained the 5.9-kb fragment and a fragment larger than 3.4 kb (19). The 5.9-kb NcoI fragment in kr14 suggests a single ptDNA integrant containing the complete aadA and a large portion of neoSTLS2 (Fig. 2). The remaining two plants, kr7 (Fig. 1D) and kr6 (19), contained two neo-hybridizing NcoI fragments differing in size from those of the transplastome. PCR analyses with primers located within aadA (aadAF) and neoSTLS2 (neoR1) amplified the predicted products from DNA of these two plants (Table 2 and data not shown), indicating that each harbors a nupt containing both aadA and neoSTLS2.
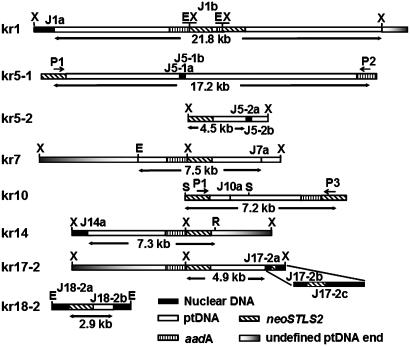
Fig. 2.
Sequence structures of eight ptDNA integrants. Restriction sites are E(EcoRI), R (EcoRV), S (SacI), and X (XbaI). To specify different nupt fragments in the same kr plant, a hyphenated number was used for kr5-1, kr5-2, kr17-2, and kr18-2. Individual junctions were specified by the plant or fragment number with an additional letter (a, b, and c) if more than one junction was present. For clarity, direct linkage of two disparate chloroplast sequences in kr7 and kr10 is highlighted with a vertical black line. Primer sites (single-headed arrows) for long-range PCR are indicated. Different filled boxes represent the origin of the sequences.
Taken together, these results demonstrate that both single and multiple ptDNA segments insert into the tobacco nuclear genome at similar frequencies (Table 2) and that complex rear-rangements of integrant sequences occur in some cases.
New Nupts Are Generally Larger than the Transcripts of Selectable Markers Present in the Chloroplast. Most newly transposed nupts analyzed were derived from the inverted repeat regions of the plastome linked to aadA and/or neoSTLS2. This was expected because plants containing new nupts were identified by their resistance to kanamycin. To determine the length of the de novo nupts, the sizes of neo- and/or aadA-hybridizing fragments were measured after digestion of total cellular DNA with DraI (Fig. 1 B and C) and NcoI (Fig. 1D) (19) and compared with corresponding fragments of the transplastome. Four kr plants (kr5, kr11, kr17, and kr18) contained at least one continuous integrant of plastid origin that was identical in size to the DraI fragment present in the transplastome of the parent line tp7 (Fig. 1 B and C), indicating that each contains at least one nupt larger than 14.8 kb (Table 2). Likewise, three kr plants (kr8, kr16, and kr19) contained at least one continuous integrant of plastid origin larger than 9.3 kb, consisting of two neo-hybridizing NcoI fragments (5.9 + 3.4 kb), and five other plants (kr3, kr4, kr9, kr12, and kr13) contained at least one continuous integrant of plastid origin larger than 22.3 kb, encompassing the aadA-hybridizing XbaI fragment (11.4 kb) and the neo-hybridizing XbaI fragment (10.9 kb) of the transplastome (Fig. 1 A and Table 2). Further digestions with EcoRI, EcoRV, and SacI in combination with PCR were used to determine the minimum sizes of de novo nupts in kr6, kr7, kr10, kr14, kr18, and kr1 because the sizes of some nupts in these kr plants were different from the transplastomic neo-hybridizing NcoI fragments (Fig. 1D) (19). The results of all these analyses indicated that each of the 17 kr plants contained at least one integrant larger than 6.0 kb with the longest integrant exceeding 22.3 kb in size (Table 2).
Northern analysis of total cellular RNA from transplastomic lines tp7 and tp17 by using a double-stranded DNA neo probe revealed 3.3- and 2.2-kb transcripts that are larger than the correctly processed 1.2-kb transcripts present in RNA of a nuclear neoSTLS2 control line (Fig. 1E). These plastid-associated neoSTLS2 transcripts are presumably the products of transcriptional read-through from the 35S promoter or the promoter(s) of adjacent plastomic gene(s). Two transcripts (1.0 and 1.4 kb) were detected in RNA of both transplastomic lines, using a double-stranded DNA probe for aadA (Fig. 1E). No transcripts hybridized to both aadA and neoSTLS2 probes, indicating that the transplastomic plants do not produce RNA molecules that encompass both genes. Therefore, we conclude that these large, de novo nupts were unlikely to have originated via an RNA intermediate. However, it is possible that smaller, RNA-mediated transfer events may also have occurred, but such events are less likely to include an entire neoSTLS2 gene and, hence, may not be detected in the screen (19).
ptDNA Integration Involves Nonhomologous Recombination and Sequence Rearrangement. To gain further insight into the nature of the integration events, a total of 14 nuclear genomic DNA junctions were obtained by iPCR or long-range PCR from seven kr plants. The authenticity of the iPCR and long-range PCR products, which contained seven new junction (J) sequences (J5-1a, J5-1b, J5-2a, J5-2b, J7a, J10a, and J14a) from four kr plants was established by monitoring cosegregation of the junctions with the kanamycin-resistance phenotype in the appropriate family (see Figs. 2 and 5). The confirmation of the origin of the iPCR products containing four additional junction sequences (J1b, J17-2b, J17-2c, and J18-2a) in three kr plants (kr1, kr17, and kr18) are described in ref. 19.
Eleven of the 14 junctions identified (Fig. 2) were transplastomic/nuclear junctions (J1a, J5-1a, J5-1b, J5-2a, J5-2b, J14a, J17-2a, J17-2b, J17-2c, J18-2a, and J18-2b) and two (J1b and J10a) were transplastomic/transplastomic junctions. The components of J7a remain uncertain because one of two ptDNA fragments involved had no direct physical linkage to either aadA or neoSTLS2 sequences. Therefore, although the two disjunct ptDNA fragments could have inserted at the same time, it is possible that the insertion was into a preexisting nupt.
A number of junction sites arose from nonhomologous recombination and/or by rearrangements of the transplastomic fragments in the nucleus as illustrated in Fig. 2. Two pieces of ptDNA from disparate regions of the plastome are directly joined together in kr7 and kr10, and one aadA- and neoSTLS2-containing ptDNA fragment is joined to a partial duplication of the same fragment in kr1. The two nupt fragments (kr5-1 and kr5-2) in kr5 consist of two distinct pieces of ptDNA joined together by short (55 and 25 bp, respectively) sequences of unknown origin. A 2.9-kb nupt (kr18-2) was found to contain only the neoSTLS2 gene. These molecular data confirm Southern and genetic analysis and establish the existence of both simple integrants and complex rearrangements of one or more ptDNA fragments at single loci within the kr plant set.
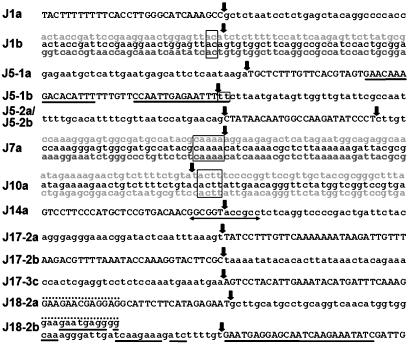
A range of sequence features were observed near the junction sites. Sequence alignments of three transplastomic/transplastomic junctions (from J1b, J7a, and J10a) revealed 2–5 bp of homology near the junctions (Fig. 3). A 2-bp homology was also found at the J5-1b transplastomic/nuclear junction (Fig. 3). Such microhomology is a feature often associated with nonhomologous integration in experimental transformation of plant nuclear DNA (17, 22). A 12-bp palindromic sequence is present at the transplastomic/nuclear junction of J14a (Fig. 3), which may have promoted ptDNA insertion.
Fig. 3.
Sequence features adjacent to 14 junctions in de novo nupts. Nucleotide sequences containing a junction(s) in 5′ to 3′ orientation are labeled on the left, and junctions are indicated by vertical arrows. Sequences of nuclear origin are represented by uppercase letters, and sequences of transplastome origin are represented by lowercase letters. Homologous sequences from different regions of the transplastome (top and bottom lines in light gray) were aligned with de novo nupt sequences (middle line) in J1b, J7a, and J10a to identify nucleotide homology (boxed). A double-headed arrow marks the palindrome sequence in J14a. Physically linked sequences of J5-1a and J5-1b are separated into two lines because of space limitations. The filler DNA of 55 nucleotides (uppercase letters) between J5-1a and J5-1b contains two sequences of 16 nucleotides (underlined) identical to ptDNA. Short filler DNA of 25 nucleotides (uppercase letter) also was found between J5-2a and J5-2b. The contiguous sequence containing J18-2b is shown as two separated lines to highlight its similarity to the 13 nucleotides near J18-2a (dotted lines above letters). The extreme 5′ sequence of 13 nucleotides is shown in the upper line with a dotted line above letters that joins the 5′ end of the lower line as one contiguous string.
All transplastomic DNA sequences adjacent to junction sites terminated at different, apparently random, positions of the transplastome with no insertional preference for AT- or T-rich sequences as previously observed in biolistic- and/or Agrobacterium-mediated DNA integration (22–26).
More complex sequence features were observed at J18-2a and J18-2b junctions. The first 25-bp of nuclear DNA downstream of J18-2b (the uppercase letters underlined in Fig. 3) were similar to the adjacent 3′ ptDNA sequence of the nupt (the lowercase letters underlined in Fig. 3). The ptDNA sequence at this junction differed from the nuclear counterpart by a 9-bp insertion/deletion and two nucleotide substitutions (the residues not underlined in Fig. 3). A further 13-bp nuclear sequence upstream (dotted line above the residues in Fig. 3) also was similar to the 3′ terminus of the ptDNA sequence of kr18-2 except for two nucleotide substitutions, suggesting that the synthesis of short segments from neighboring ptDNA or nuclear DNA may have been involved in the integration process (27, 28). In addition, an inverted 188-bp sequence derived from neoSTLS2 with some rearrangements (a 6-bp deletion of the neoSTLS2 sequence and a 10-bp insertion of an unknown sequence) was located 463 bp downstream of J17-2a (Fig. 2) (EMBL accession no. AJ517467). The 5′ end of this sequence started at the neoSTLS2 intron (J17-2b in Fig. 3), and its 3′ end (J17-2c) terminated in the 19-bp imperfect palindrome of the CaMV 35S promoter at the exact nucleotide previously identified as a hot spot for recombination in rice transformation experiments (17).
Discussion
DNA Molecules Are Involved in ptDNA Transposition. Sequence analysis of ptDNA integrants in the tobacco nuclear genome resulting from 17 independent transfer events suggests that, for the most part, the process involves DNA molecules directly. Several lines of evidence support this conclusion, including the relatively large size of the majority of the integrants (>6.0 to >22.3 kb), which contain both aadA and neoSTLS2 genes that are frequently flanked by extensive ptDNA sequences (Table 2). In addition, there are no detectable transcripts containing linked aadA and neoSTLS2 sequences (Fig. 1E). These results make RNA an unlikely intermediate molecule in the transposition process. Although RNA-mediated transfer has been inferred in some functional gene transfers from mitochondria in higher plants (8–10), there is little evidence of RNA-mediated ptDNA transfer to the nucleus or mitochondrion.
Mechanisms of ptDNA Integration into the Nuclear Genome. The variety of organization of de novo nupts (Table 2 and Figs. 2 and 3) reveals the diversity of events that must have occurred before or during nuclear integration of ptDNA. These include nonhomologous recombination between transferred ptDNA fragments (kr1, kr5, and kr10); scrambling of transplastomic DNA sequences in the nuclear genomic DNA (kr17-2); incorporation of filler DNA between disjunct regions of ptDNA (kr5-1), from neighboring ptDNA (kr18-2), or from an unknown source (kr5-2); formation of complex nuclear loci with multiple ptDNA integrants (Table 2); and the presence of microhomology (2–5 bp) near the transplastomic junctions (J1b, J5-1b, J7a, and J10a). The junction sequences display many of the hallmarks of nonhomologous recombination, and their organization is reminiscent of nuclear integrants after biolistic and T-DNA delivery (27, 29–32), although they show no sequence preference for the AT- and T-rich sequences observed in both biolistic and T-DNA transformation (22–26). We have not been able to obtain a complete picture of any native integration sites before nupt insertion, although several nuclear junctions were obtained. The acquisition of the preinsertion sites is severely hampered by sequence complexity at the target site (e.g., repeat sequences flanking the new nupt fragments), the preexistence of abundant nupts in tobacco (12), sequence deletions/insertions at the preinsertion sites (16, 33), and multiple new ptDNA fragments involved in the same integration event.
Nonhomologous recombination and random integration of organelle DNA into the nuclear genome have been proposed to account for the presence of microhomology (2–9 bp) in the junctions between more anciently transferred ptDNA (11) and mtDNA (13, 14, 34). Similar microhomology (2–4 bp) was identified at mtDNA/nuclear and mtDNA/mtDNA junctions in yeast chromosomal DNA when double-strand breaks were introduced (2). The essentially random distribution of historically transferred mtDNA in the yeast genome (2) and mtDNA and ptDNA in rice chromosome 10 (6) argues against site-selective integration of organelle DNA.
Insertion of Single Versus Multiple ptDNA Fragments at Single Nuclear Loci. Eleven of 17 kr plants in this study contain a single ptDNA integrant in their nuclear genomes. However, 2 of those 11 plants (kr1 and kr10) contain two separate transplastomic fragments, and another (kr7) may contain a second transplastomic fragment (Table 2 and Fig. 2). Therefore, approximately half of the de novo nupts (8 among 17 events) result from a single ptDNA fragment insertion. The ratio of single to multiple fragments is close to that observed when foreign DNA inserts after biolistics without selection (35) but generally lower than that observed with antibiotic selection (36, 37). It is notable that nearly half of the nupts possess multiple inserts at single nuclear loci as is common in biolistic and T-DNA transformation (31, 38, 39). This finding indicates that the insertion of multiple incoming DNA fragments adjacent to each other at a single site is an inherent cellular process in higher plants and not a consequence of experimental manipulation. The integration process clearly does not discriminate between intracellular organelle DNA and foreign DNA. Consequently, modern experimental gene manipulation utilizes cellular mechanisms that have been operating in endosymbiotic evolution for up to two billion years (1). Further understanding of the mechanisms and pathways involved in DNA integration is necessary to control the complexity of transgene loci.
Significance of ptDNA Transfer and Integration. The ptDNA transfer and integration events investigated in this study took place under natural growth conditions. There was no involvement of agrobacterial infection, biolistic injury to cells, or tissue culture, all of which could perturb the process (31, 40, 41). Therefore, the nupts analyzed here truly reflect natural ptDNA-to-nucleus transfer and provide unique insights into the normal cellular processes responsible for the incorporation of DNA into the nuclear genome (1). The transfer of plastid sequences to the nucleus generates genetic diversity amongst progeny and possibly makes every plant unique when it occurs in somatic cells (20). Although most nupts are nonfunctional initially and gradually decay, a small proportion of nupts is known to evolve into novel functional nuclear genes that account for thousands of those resident in the nucleus (1).
Supplementary Material
Acknowledgments
We thank Paul Whitfeld for discussion during the preparation of the manuscript and J. Roberts for technical assistance in the analysis of J5-1a/J5-1b. This work was supported by the Australian Research Council.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: iPCR, inverse PCR; kr, kanamycin-resistant; mtDNA, mitochondrial DNA; nupt, nuclear integrant of plastid DNA; ptDNA, plastid DNA.
Data deposition: The nucleotide sequences reported in this paper have been deposited in the EMBL database [accession nos. AJ616346 (J1b), AJ616347 (J5-1a and J5-1b), AJ616348 (J5-2a and J5-2b), AJ616349 (J7a), AJ616350 (J10a), and AJ616351 (J14a)].
References
- 1.Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. (2004) Nat. Rev. Genet. 5, 123-135. [DOI] [PubMed] [Google Scholar]
- 2.Ricchetti, M., Fairhead, C. & Dujon, B. (1999) Nature 402, 96-100. [DOI] [PubMed] [Google Scholar]
- 3.Mourier, T., Hansen, A. J., Willerslev, E. & Arctander, P. (2001) Mol. Biol. Evol. 18, 1833-1837. [DOI] [PubMed] [Google Scholar]
- 4.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 5.Yuan, Q., Hill, J., Hsiao, J., Moffat, K., Ouyang, S., Cheng, Z., Jiang, J. & Buell, C. R. (2002) Mol. Genet. Genomics 267, 713-720. [DOI] [PubMed] [Google Scholar]
- 6.The Rice Chromosome 10 Sequencing Consortium (2003) Science 300, 1566-1569. [DOI] [PubMed] [Google Scholar]
- 7.Shahmuradov, I. A., Akbarova, Y. Y., Solovyev, V. V. & Aliyev, J. A. (2003) Plant Mol. Biol. 52, 923-934. [DOI] [PubMed] [Google Scholar]
- 8.Nugent, J. M. & Palmer, J. D. (1991) Cell 66, 473-481. [DOI] [PubMed] [Google Scholar]
- 9.Wischmann, C. & Schuster, W. (1995) FEBS Lett. 374, 152-156. [DOI] [PubMed] [Google Scholar]
- 10.Adams, K. L., Daley, D. O., Qiu, Y. L., Whelan, J. & Palmer, J. D. (2000) Nature 408, 354-357. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, W. Y. & Scott, N. S. (1989) Theor. Appl. Genet. 77, 625-633. [DOI] [PubMed] [Google Scholar]
- 12.Ayliffe, M. A. & Timmis, J. N. (1992) Theor. Appl. Genet. 85, 229-238. [DOI] [PubMed] [Google Scholar]
- 13.Sun, C. W. & Callis, J. (1993) Plant Cell 5, 97-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanchard, J. L. & Schmidt, G. W. (1995) J. Mol. Evol. 41, 397-406. [DOI] [PubMed] [Google Scholar]
- 15.Lin, X., Kaul, S., Rounsley, S., Shea, T. P., Benito, M. I., Town, C. D., Fujii C. Y., Mason, T., Bowman, C. L., Barnstead, M., et al. (1999) Nature 402, 761-768. [DOI] [PubMed] [Google Scholar]
- 16.Salomon, S. & Puchta, H. (1998) EMBO J. 17, 6086-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohli, A., Griffiths, S., Palacios, N., Twyman, R. M., Vain, P., Laurie, D. A. & Christou, P. (1999) Plant J. 17, 591-601. [DOI] [PubMed] [Google Scholar]
- 18.Gorbunova, V. & Levy, A. A. (1999) Trends Plant Sci. 4, 263-269. [DOI] [PubMed] [Google Scholar]
- 19.Huang, C. Y., Ayliffe, M. A. & Timmis, J. N. (2003) Nature 422, 72-76. [DOI] [PubMed] [Google Scholar]
- 20.Stegemann, S., Hartmann, S., Ruf, S. & Bock, R. (2003) Proc. Natl. Acad. Sci. USA 100, 8828-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, C. Y., Barker, S. J., Langridge, P., Smith, F. W. & Graham, R. D. (2000) Plant Physiol. 124, 415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunaud, V., Balzergue, S., Dubreucq, B., Aubourg, S., Samson, F., Chauvin, S., Bechtold, N., Cruaud, C., DeRose, R., Pelletier, G., et al. (2002) EMBO Rep. 3, 1152-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takano, M., Egawa, H., Ikeda, J.-E. & Wakasa, K. (1997) Plant J. 11, 353-361. [DOI] [PubMed] [Google Scholar]
- 24.Sawasaki, T., Takahashi, M., Goshima, N. & Morikawa, H. (1998) Gene 218, 27-35. [DOI] [PubMed] [Google Scholar]
- 25.Muller, A. E., Kamisugi, Y., Gruneberg, R., Niedenhof, I., Horold, R. J. & Meyer, P. (1999) J. Mol. Biol. 291, 29-46. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu, K., Takahashi, M., Goshima, N., Kawakami, S., Irifune, K. & Morikawa, H. (2001) Plant J. 26, 375-384. [DOI] [PubMed] [Google Scholar]
- 27.Gorbunova, V. & Levy, A. A. (1997) Nucleic Acids Res. 25, 4650-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Windels, P., De Buck, S., Van Bockstaele, E., De Loose, M. & Depicker, A. (2003) Plant Physiol. 133, 2061-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Svitashev, S. K., Pawlowski, W. P., Makarevitch, I., Plank, D. W. & Somers, D. A. (2002) Plant J. 32, 433-445. [DOI] [PubMed] [Google Scholar]
- 30.Makarevitch, I., Svitashev, S. K. & Somers, D. A. (2003) Plant Mol. Biol. 52, 421-432. [DOI] [PubMed] [Google Scholar]
- 31.Kohli, A., Leech, M., Vain, P., Laurie, D. A. & Christou, P. (1998) Proc. Natl. Acad. Sci. USA 95, 7203-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chilton, M. M. & Que, Q. (2003) Plant Physiol. 133, 956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirik, A., Salomon, S. & Puchta, H. (2000) EMBO J. 19, 5562-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner, C., Killoran, C., Thomas, N. S., Rosenberg, M., Chuzhanova, N. A., Johnston, J., Kemel, Y., Cooper, D. N. & Biesecker, L. G. (2003) Hum. Genet. 112, 303-309. [DOI] [PubMed] [Google Scholar]
- 35.Popelka, J. C., Xu, J. & Altpeter, F. (2003) Transgenic Res. 12, 587-596. [DOI] [PubMed] [Google Scholar]
- 36.Cheng, M., Fry, J. E., Pang, S., Zhou, H., Hironaka, C. M., Duncan, D. R., Conner, T. W. & Wan, Y. (1997) Plant Physiol. 115, 971-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dai, S. H., Zheng, P., Marmey, P., Zhang, S. P., Tian, W. Z., Chen, S. Y., Beachy, R. N. & Fauquet, C. (2001) Mol. Breed. 7, 25-33. [Google Scholar]
- 38.Pawlowski, W. P. & Somers, D. A. (1996) Mol. Biotechnol. 6, 17-30. [DOI] [PubMed] [Google Scholar]
- 39.De Neve, M., De Buck, S., Jacobs, A., Van Montagu, M. & Depicker, A. (1997) Plant J. 11, 15-29. [DOI] [PubMed] [Google Scholar]
- 40.Svitashev, S. K. & Somers, D. A. (2001) Genome 44, 691-697. [PubMed] [Google Scholar]
- 41.Hunold, R., Bronner, R. & Hahne, G. (1994) Plant J. 5, 593-604. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.