Phosphorylation of NPR by the protein kinase SnRK2.8 allows nuclear import of NPR1, an essential step in systemic acquired resistance.
Abstract
In plants, necrotic lesions occur at the site of pathogen infection through the hypersensitive response, which is followed by induction of systemic acquired resistance (SAR) in distal tissues. Salicylic acid (SA) induces SAR by activating NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1) through an oligomer-to-monomer reaction. However, SA biosynthesis is elevated only slightly in distal tissues during SAR, implying that SA-mediated induction of SAR requires additional factors. Here, we demonstrated that SA-independent systemic signals induce a gene encoding SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8), which phosphorylates NPR1 during SAR. The SnRK2.8-mediated phosphorylation of NPR1 is necessary for its nuclear import. Notably, although SnRK2.8 transcription and SnRK2.8 activation are independent of SA signaling, the SnRK2.8-mediated induction of SAR requires SA. Together with the SA-mediated monomerization of NPR1, these observations indicate that SA signals and SnRK2.8-mediated phosphorylation coordinately function to activate NPR1 via a dual-step process in developing systemic immunity in Arabidopsis thaliana.
INTRODUCTION
Plants are constantly exposed to a broad spectrum of pathogens in nature. To cope with pathogen attack, plants have developed versatile protection systems. Upon pathogen infection, plants sense conserved pathogen-associated molecular patterns (PAMPs) on the surface of invading pathogens through pattern recognition receptors in plant cells (Fu and Dong, 2013). The pattern recognition receptor subsequently induces PAMP-triggered immunity (PTI) as an active defense system (Zipfel, 2009).
Meanwhile, pathogens produce inhibitors of PTI, which are termed effectors (Fu and Dong, 2013), to suppress PTI for successful infection (Block and Alfano, 2011). When effectors are injected into plant cells, effector-triggered immunity is activated by the recognition of effectors (Nomura et al., 2011), which induces programmed cell death through the hypersensitive response around the infection site to block the spreading of pathogen invasion (Zipfel, 2009; Nomura et al., 2011; Fu and Dong, 2013).
By the recognition of pathogen effectors and PAMP, plants trigger another wave of immune response in distal tissues; this is often called systemic acquired resistance (SAR) (Fu and Dong, 2013). SAR does not trigger cell death but rather activates salicylic acid (SA) biosynthesis in both infected (local) and distal (systemic) tissues (Nandi et al., 2004; Chanda et al., 2011; Kachroo and Robin, 2013; Gao et al., 2014). SA accumulation leads to the induction of genes encoding pathogenesis-related (PR) proteins that mediate a variety of antimicrobial functions (Cameron et al., 1999).
The master regulator of SA-triggered SAR is NONEXPRESSER OF PATHOGENESIS-RELATED GENES1 (NPR1). SA regulates the protein stability of the NPR1 transcriptional cofactor (Spoel et al., 2009). It also mediates the dynamic oligomer-to-monomer reaction of NPR1, which is a prerequisite for its nuclear import (Mou et al., 2003). NPR1 interacts with TGA transcription factors to enhance their DNA binding and transcriptional regulatory activity in inducing PR genes (Després et al., 2000, 2003; Fan and Dong, 2002; Boyle et al., 2009). SA rapidly accumulates to a high level in infected tissues (Nandi et al., 2004; Chanda et al., 2011; Kachroo and Robin, 2013; Gao et al., 2014). However, its biosynthesis is induced only slightly in distal tissues during SAR (Nandi et al., 2004; Chanda et al., 2011; Kachroo and Robin, 2013; Gao et al., 2014), obscuring the necessity for SA in driving the nuclear localization of NPR1. In addition, NPR1 has a molecular mass of 66 kD, which is larger than the average size (42 kD) of nucleo-cytoplasmic proteins (Li et al., 2006). It is therefore anticipated that additional signals would be required for the active nuclear import of NPR1 in distal tissues.
In this work, we demonstrated that SA-independent systemic signals induce a gene encoding a serine/threonine (S/T) kinase SNF1-RELATED PROTEIN KINASE 2.8 (SnRK2.8), which phosphorylates NPR1 in distal leaves. SnRK2.8-mediated phosphorylation is necessary for the nuclear import of NPR1, while SA signals induce the monomerization reaction of NPR1 (Mou et al., 2003; Tada et al., 2008). These observations indicate that the coordinated action of SA signaling and SnRK2.8-mediated phosphorylation underlies a two-step activation scheme of NPR1 in inducing SAR. We propose that plants have acquired the SnRK2.8-mediated activation system to ensure SAR to occur in distal tissues upon pathogen infection.
RESULTS
SnRK2.8 Is Involved in Plant Immune Responses
We recently reported that SnRK2.8 phosphorylates the membrane-associated NAM, ATAF1/2, and CUC2 (NAC) transcription factor NTL6 to trigger its nuclear import (Kim et al., 2012). NTL6 plays a role in cold-induced disease resistance in Arabidopsis thaliana (Seo et al., 2010). We therefore hypothesized that SnRK2.8, and perhaps other SnRK2 members as well, would be associated with immune responses.
To examine the linkage between SnRK2.8 and immune response, Col-0 plants were infected with the avirulent Pseudomonas syringae pv tomato DC3000/avrRpt2 (Pst DC3000/avrRpt2) cells (Whalen et al., 1991; Mudgett and Staskawicz, 1999), and SnRK2 transcription was examined in the infected plants. Whereas SnRK2.8 transcription was marginally elevated in local leaves, it was induced by ∼5-fold in distal leaves (Figure 1A; Supplemental Figure 1A), supporting the notion that SnRK2.8 is involved in the plant systemic immune response. Other SnRK2 genes, such as SnRK2.1, 2.2, 2.3, and 2.9, were suppressed after pathogen infection in distal leaves, suggesting that SnRK2.8 is functionally distinct from other SnRK2 members. SnRK2.8 induction was also observed after infection with the virulent pathogen, Pst DC3000 (Supplemental Figure 1B). In contrast, flg22, a pathogen-mimic peptide flagellin (Zipfel et al., 2004), did not discernibly affect SnRK2.8 transcription except for a slight induction at 48 h after treatment (Supplemental Figure 1C). Lipopolysaccharide and coronatine also did not discernibly affect SnRK2.8 transcription.
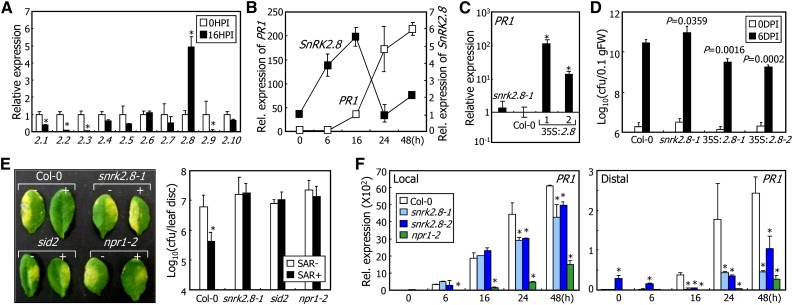
Figure 1.
SnRK2.8 Mediates SAR.
Four-week-old plants grown in soil were used for pathogen infection and gene expression analysis. Transcript levels were examined by RT-qPCR. Biological triplicates were averaged and statistically treated using Student’s t test (*P < 0.01). Bars indicate sd of the mean.
(A) Effects of pathogen infection on SnRK transcription. Col-0 plants were infected with Pst DC3000/avrRpt2 cells. Total RNA was extracted from distal leaves. A group of SnRK2 genes is represented on the x axis. HPI, hours postinfection.
(B) Transcription kinetics of SnRK2.8 and PR1 genes in distal leaves. The 4th and 5th leaves were pressure-infiltrated with Pst DC3000/avrRpt2 cells, and two upper leaves were harvested at each time point after infection.
(C) PR1 transcription in snrk2.8-1 mutant and 35S:2.8 transgenic plants. The 4th and 5th leaves of uninfected plants were used for total RNA extraction.
(D) Bacterial cell growth in snrk2.8-1 mutant and 35S:2.8 transgenic plants. Plants were spray-inoculated with Pst DC3000 cells and incubated for 6 d before counting bacterial cells. Three measurements were averaged and statistically treated using Student's t test. P values were annotated. Bars indicate sd of the mean. cfu, colony-forming units; DPI, days postinfection.
(E) SAR induction in snrk2.8-1 mutant. Halves of the 4th and 5th leaves were pressure-infiltrated with 10 mM MgCl2 (−) or Pst DC3000/avrRpt2 cells (+) and incubated for 2 d. Two upper leaves were infiltrated with Pst DC3000 cells and incubated for 3 d before taking photographs (left panel) and counting bacterial cells (right panel).
(F) PR1 transcription in the local and distal leaves of snrk2.8 mutants. Pathogen infection was conducted as described in (A). Infected leaves (local) and two upper leaves (distal) were used for gene expression analysis.
SnRK2.8 Mediates SAR
We examined the kinetics of SnRK2.8 transcription in distal tissues after primary infection. It was found that SnRK2.8 transcription was rapidly induced in distal leaves within 6 h following the local infection with Pst DC3000/avrRpt2 cells (Figure 1B). In contrast, systemic expression of PR1 initiated 16 h after local infection (Figure 1B). To further examine the temporal relationship between the induction of SnRK2.8 and PR1, we generated 35S:2.8 transgenic plants that overexpress SnRK2.8 driven by the CaMV 35S promoter (Supplemental Figure 1D). The SnRK2.8-overexpressing plants exhibited slightly enhanced growth (Supplemental Figure 1D), as has been reported previously (Shin et al., 2007). We found that PR1 transcription was markedly elevated in the 35S:2.8 plants, although it was not discernibly affected in the SnRK2.8-defective snrk2.8-1 mutant (Figure 1C), suggesting that SnRK2.8 acts upstream of PR1. On the basis of the elevation of PR expression, we tested whether the transgenic plants are resistant to pathogen infection. The SnRK2.8-overexpressing plants exhibited enhanced bacterial resistance, whereas snrk2.8-1 mutant displayed slightly reduced resistance in bacterial susceptibility compared with Col-0 plants (Figure 1D).
To investigate potential roles of SnRK2.8 in SAR, we examined the propagation of the virulent Pst DC3000 cells in the distal leaves of snrk2.8 mutants that were infected with the avirulent Pst DC3000/avrRpt2. Notably, SAR was not induced in the snrk2.8 mutants, while it was efficiently induced in Col-0 plants (Figure 1E; Supplemental Figure 1E). The reduced SAR phenotype of the snrk2.8 mutants was similar to that of the SA-deficient sid2 mutant and the NPR1-defective npr1-2 mutant, supporting the notion that SnRK2.8 plays a role in SAR, as has been reported for SA and NPR1 (Delaney et al., 1994; Vernooij et al., 1994; Cao et al., 1997; Fu and Dong, 2013).
Examination of PR1 transcription in SAR-induced plants revealed that PR1 induction was slightly reduced in the local leaves but largely diminished in the distal leaves of the snrk2.8 mutants (Figure 1F). In contrast, PR1 transcription was significantly reduced in both the local and distal leaves of npr1-2 mutants, as has been reported previously (Cao et al., 1997). We also examined whether SnRK2.8 is involved in effector-triggered immunity, which occurs in local tissues (Nomura et al., 2011), using the avirulent Pst DC3000/avrRpt2 cells. Bacterial resistance of the snrk2.8-1 mutant was not different from that of Col-0 plants (Supplemental Figure 1F). Together, these observations indicate that SnRK2.8 plays a role primarily in distal tissues.
SnRK2.8-Mediated Induction of SAR Depends on SA
Our data show that SnRK2.8 plays a role in inducing SAR. It is known that SA is essential for the induction of SAR (Delaney et al., 1994; Fu and Dong, 2013). Therefore, a question was whether SnRK2.8 is functionally associated with SA in inducing SAR.
We examined the effects of SA on SnRK2.8 transcription. It was found that SA only marginally affects SnRK2.8 transcription (Figure 2A). In addition, sid2 mutation did not alter SnRK2.8 transcription in distal leaves (Figure 2B), showing that SA is not the upstream signal for SnRK2.8 induction. It is known that SID2 expression is rapidly induced upon pathogen infection (Wildermuth et al., 2001). We observed that the SID2 gene is still induced by pathogen infection in the local leaves of snrk2.8-1 mutants (Supplemental Figure 2A), suggesting that SnRK2.8 does not affect SA biosynthesis.
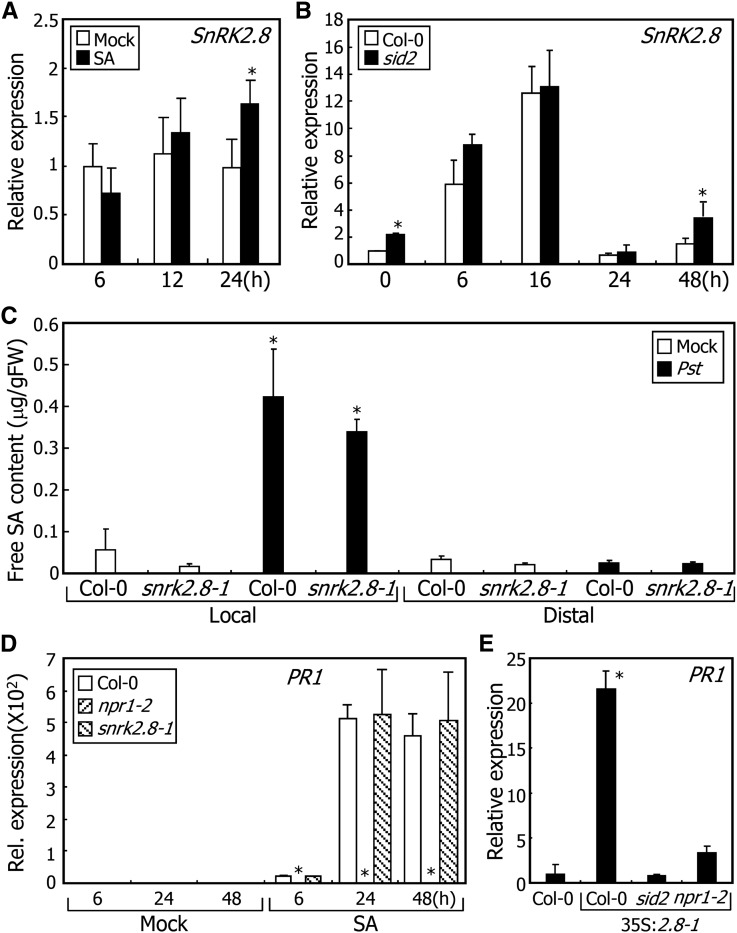
Figure 2.
SnRK2.8-Mediated Induction of SAR Requires SA.
In (A) and (D), 2-week-old plants grown on MS-agar plates were used for gene expression analysis. Transcript levels were examined by RT-qPCR. Biological triplicates were averaged and statistically treated (t test, *P < 0.01). Bars indicate sd.
(A) Effects of SA on SnRK2.8 transcription. Plants were treated with 0.5 mM SA for the indicated periods before harvesting whole-plant materials.
(B) Kinetics of SnRK2.8 transcription in distal leaves. The 4th and 5th leaves of 4-week-old plants grown in soil were pressure-infiltrated with Pst DC3000/avrRpt2 cells, and two upper leaves were harvested at the indicated time points for total RNA extraction.
(C) SA accumulation after pathogen infection. Col-0 plant and snrk2.8-1 mutant were infected with Pst DC3000/avrRpt2 cells as described in (B). Local and distal leaves were harvested 24 h after infection for SA extraction. Five measurements were averaged and statistically treated (t test, *P < 0.01).
(D) SA-mediated induction of PR1 transcription in snrk2.8-1 mutant. Plants were treated with 0.5 mM SA for the indicated periods.
(E) Effects of SnRK2.8 overexpression on PR1 transcription in sid2 and npr1-2 backgrounds. The 4th and 5th leaves of 4-week-old plants were harvested for gene expression analysis.
To further examine the functional relationship between SA and SnRK2.8 during SAR, we monitored changes in the contents of free SA in local and distal leaves following pathogen infection. It was found that free SA accumulates to similar levels in the local leaves of the infected Col-0 and snrk2.8-1 plants (Figure 2C). In contrast, free SA levels were not detectably elevated in the distal leaves of both the infected plants in our assay system (Figure 2C). Free SA accumulation was also unaltered in SnRK2.8-overexpressing plants (Supplemental Figure 2B), showing that SnRK2.8 is not associated with SA accumulation.
SA stimulates PR transcription in response to pathogen infection (Delaney et al., 1994). We found that SnRK2.8 overexpression induces PR1 expression (Figure 1C). We therefore investigated whether SnRK2.8 is functionally linked with SA in inducing PR transcription. It was observed that while SA-mediated induction of PR1 and PR5 disappears in npr1-2 mutants, it normally occurs in snrk2.8-1 mutants (Figure 2D; Supplemental Figure 2C). In addition, SA treatment induced pathogen resistance to similar degrees in Col-0 plants and snrk2.8-1 mutants (Supplemental Figure 2D), indicating that SA-mediated induction of PR genes does not require SnRK2.8. Notably, the high-level induction of PR1 observed in 35S:2.8 transgenic plants was significantly reduced in the sid2 and npr1-2 backgrounds (Figure 2E; Supplemental Figure 2E). Together, these observations indicate that although high-level induction of SA biosynthesis is not required for the SnRK2.8-mediated induction of PR genes, the SnRK2.8 function in inducing PR genes depends on SA.
SnRK2.8 Interacts with NPR1
SA accumulates only slightly in the distal leaves of infected plants (Nandi et al., 2004; Chanda et al., 2011; Kachroo and Robin, 2013; Gao et al., 2014). We observed that PR1 transcription was significantly reduced in the distal leaves of snrk2.8-1 mutants during SAR, raising the possibility that SnRK2.8 targets upstream regulators of SA signaling.
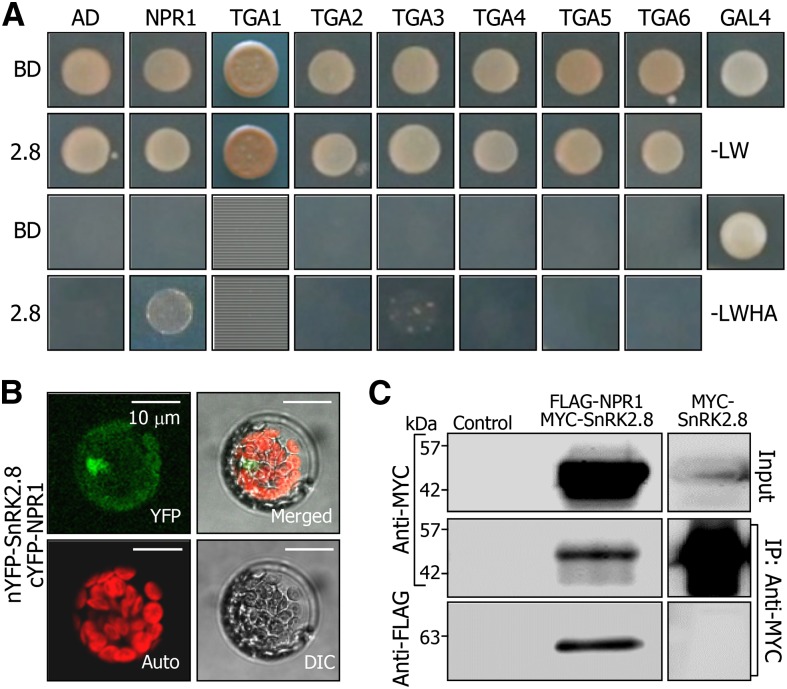
We examined whether SnRK2.8 directly regulates upstream regulators of plant immunity response, such as NPR1. NPR1, as a transcriptional coactivator, acts together with TGA transcription factors to regulate PR expression (Després et al., 2000, 2003). Yeast two-hybrid assays showed that SnRK2.8 interacted strongly with NPR1 among the upstream regulators tested (Figure 3A). It also interacted slightly with TGA3. The SnRK2.8-NPR1 interaction occurred in both the nucleus and the cytoplasm, as examined by bimolecular fluorescence complementation (BiFC) assay in Arabidopsis protoplasts (Figure 3B; Supplemental Figure 3). The in planta interaction of SnRK2.8 with NPR1 was also verified by coimmunoprecipitation (Co-IP) using tobacco (Nicotiana benthamiana) cells transiently coexpressing FLAG-NPR1 and MYC-SnRK2.8 fusion proteins (Figure 3C).
Figure 3.
SnRK2.8 Interacts with NPR1.
(A) Interaction of SnRK2.8 with NPR1 in yeast cells. Yeast cell growth on selective media lacking Leu, Trp, His, and Ade (-LWHA) represents positive interaction.
(B) BiFC. Partial YFP constructs were fused to SnRK2.8 or NPR1, and the fusions were coexpressed transiently in Arabidopsis protoplasts. YFP signals were visualized by DIC and fluorescence microscopy.
(C) Co-IP. MYC-SnRK2.8 and FLAG-NPR1 fusion constructs were coexpressed transiently in tobacco leaves. The input represents 10% of the protein extract. Control, protein extract without transient expression. IP, immunoprecipitation.
SnRK2.8 Phosphorylates NPR1
The SnRK2 family members are plant-specific S/T protein kinases that mediate plant development and environmental stress responses (Kulik et al., 2011). We found that SnRK2.8 interacts with NPR1. We therefore investigated whether SnRK2.8 phosphorylates NPR1. Recombinant proteins were prepared as maltose binding protein (MBP) fusions in Escherichia coli cells for in vitro phosphorylation assays. The results showed that SnRK2.8 phosphorylates NPR1 (Figure 4A; Supplemental Figure 4A). SnRK2.8 was autophosphorylated. SnRK2.6, which is a close homolog of SnRK2.8 (Boudsocq et al., 2004), was also autophosphorylated but did not phosphorylate NPR1 (Figure 4B). The observed reduction of SnRK2 autophosphorylation in the presence of nontarget proteins might be due to the inhibitory effects of excess nontarget proteins on the intermolecular interactions between kinase proteins (Kim et al., 2012).
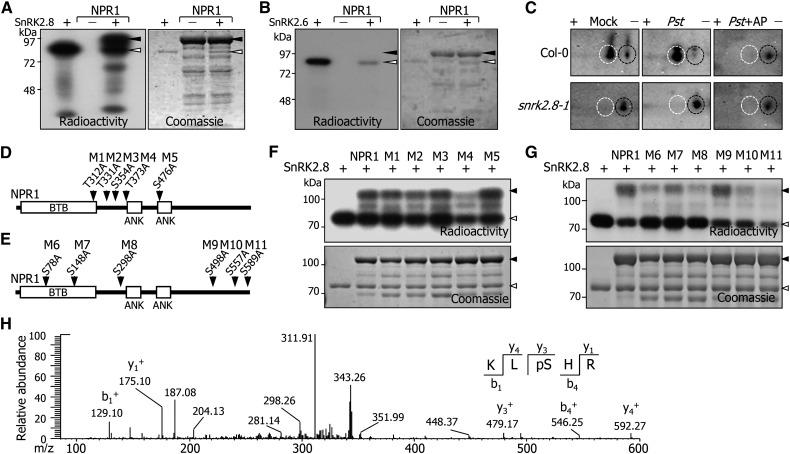
Figure 4.
SnRK2.8 Phosphorylates NPR1.
(A) In vitro phosphorylation assay. Recombinant SnRK2.8 and NPR1 proteins were prepared as MBP fusions in E. coli cells. MBP-SnRK2.8 (0.5 μg) and 5 μg of MBP-NPR1 were used. Black arrowheads indicate MBP-NPR1, and white arrowheads indicate MBP-SnRK2.8.
(B) SnRK2.6 does not phosphorylate NPR1. Phosphorylation assays in vitro using recombinant SnRK2.6 and NPR1 proteins were performed as described in (A). Black arrowheads indicate MBP-NPR1, and white arrowheads indicate MBP-SnRK2.6.
(C) In vivo phosphorylation assay by two-dimensional electrophoresis. A MYC-NPR1 fusion was overexpressed in Col-0 and snrk2.8-1 plants. The 4th and 5th leaves of 4-week-old plants grown in soil were pressure-infiltrated with Pst DC3000/avrRpt2 cells, and uninfiltrated leaves were harvested 24 h after infiltration. NPR1 proteins were detected by immunoblot assays using an anti-MYC antibody. Black circles indicate NPR1, and white circles indicate phosphorylated NPR1. AP, alkaline phosphatase; (+) and (−) indicate low and high pH, respectively.
(D) and (E) Protein structure of NPR1. M1 to M11 indicate the mutations of putative SnRK2.8 target residues. BTB, BTB/POZ core motif; ANK, ankyrin-repeat motif.
(F) and (G) In vitro phosphorylation assays on mutated NPR1 proteins. Phosphorylation assays in vitro using recombinant SnRK2.8 and mutated NPR1 proteins were performed as described in (A). Black and white arrowheads indicate NPR1 and SnRK2.8, respectively.
(H) LC-MS/MS spectrum of the peptide containing phosphorylated S589. Eight micrograms of recombinant MBP-NPR1 protein was phosphorylated by 4 μg of MBP-SnRK2.8, as described in (A). “b” and “y” indicate peptide fragment ions retaining charges at the N and C terminus, respectively. The subscript numbers indicate their positions in the identified peptide. The superscript “+” indicates singly protonated ions. The phosphorylated Ser is denoted as pS. LC-MS/MS was performed with three independent reactions, and only Ser-589 phosphorylation was detected.
We next examined the SnRK2.8-mediated phosphorylation of NPR1 in vivo by two-dimensional electrophoresis using tobacco leaves transiently coexpressing FLAG-NPR1 and MYC-SnRK2.8 fusions. It was found that an additional protein spot is detected in the protein samples extracted from tobacco leaves coexpressing FLAG-NPR1 and MYC-SnRK2.8 but not in the protein samples prepared from those expressing only FLAG-NPR1 (Supplemental Figure 4B), suggesting that SnRK2.8-mediated phosphorylation of NPR1 would occur in planta.
To further examine the potential phosphorylation of NPR1, we used transgenic Arabidopsis plants expressing MYC-NPR1 fusion in Col-0 or snrk2.8-1 background. Two-dimensional protein blots showed that NPR1 protein was phosphorylated in response to the induction of SAR (Figure 4C). In contrast, the phosphorylated NPR1 protein spots were not detected in the protein samples extracted from snrk2.8-1 mutant expressing the MYC-NPR1 fusion, indicating that NPR1 is phosphorylated in vivo by SnRK2.8 during SAR.
Basophilic kinases, such as SnRK2 members, preferentially phosphorylate Ser and Thr residues in the sequence context of K/RXXS/T, in which X represents any amino acid (Kim et al., 2012). We replaced the potential Ser and Thr residues of NPR1, which fit the consensus sequence context, with alanine (A), resulting in M1-M11 proteins (Figures 4D and 4E). Phosphorylation assays in vitro showed that SnRK2.8-mediated phosphorylation was reduced by ∼60 and 50% in the M4 and M11 proteins, respectively (Figures 4F and 4G), suggesting that Thr-373 and Ser-589 are major phosphorylation residues.
To further investigate the SnRK2.8-mediated phosphorylation of NPR1, we analyzed the reaction products of in vitro phosphorylation reactions by mass spectrometry. The mass spectrometry data showed that only Ser-589, which is located in the far C-terminal region of NPR1 (Figure 4E), is phosphorylated by SnRK2.8 (Figure 4H). It was unclear whether Thr-373 is phosphorylated by SnRK2.8 in the mass spectrometry assay because the peptide fragment containing Thr-373 was not detected in the assay (Supplemental Figure 4C).
It has been reported that the Ser-11 and Ser-15 residues of NPR1 are phosphorylated in SA-treated plants (Spoel et al., 2009). We found that a mutated NPR1 protein harboring S11A and S15A substitutions was still phosphorylated by SnRK2.8 (Supplemental Figure 4D), showing that the two Ser residues are not phosphorylated by SnRK2.8. Together, these observations indicate that SnRK2.8 phosphorylates primarily Ser-589 and possibly Thr-373 of NPR1.
It has been shown that Ser-154 and Thr-158 are important for the kinase activity of SnRK2.8 in phosphorylating its target NTL6 (Kim et al., 2012). We examined whether a mutated SnRK2.8 (mSnRK2.8) harboring S154A and T158A substitutions phosphorylates NPR1 or not. It was found that both autophosphorylation and NPR1 phosphorylation disappeared in the mSnRK2.8-mediated reactions (Supplemental Figure 4E), showing that the two residues are critical for SnRK2.8 activity. It is possible that Ser-154 and Thr-158 are the sites of autophosphorylation and autophosphorylation of SnRK2.8 is essential for its kinase activity, as has been shown previously with other plant kinases (Fujii and Zhu, 2009; M.H. Oh et al., 2010).
SnRK2.8 Does Not Affect the Monomerization Reaction of NPR1
A critical question was how SnRK2.8-mediated phosphorylation influences NPR1 function in plant immune responses. SA triggers NPR1 monomerization, which is required for its nuclear import (Mou et al., 2003). It has been reported that the oligomer-to-monomer reaction occurs in distal leaves, where SA level is slightly elevated during SAR (Mou et al., 2003).
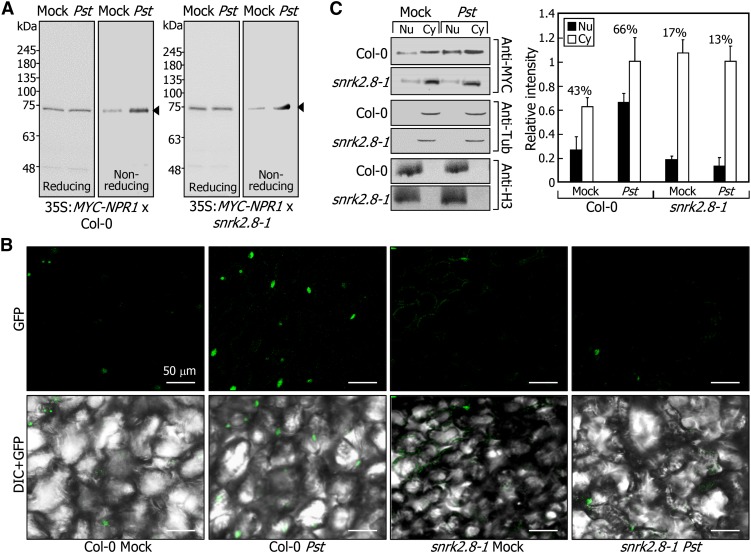
We therefore examined whether SnRK2.8 influences the NPR1 monomerization reaction using snrk2.8-1 mutants overexpressing a MYC-NPR1 fusion. The relative levels of NPR1 monomers were elevated equally in both Col-0 and snrk2.8-1 backgrounds after pathogen infection (Figure 5A), suggesting that SnRK2.8 does not affect the NPR1 monomerization reaction. We were not able to detect oligomeric forms of MYC-NPR1 proteins in our assays, possibly because the N-terminal region of NPR1 is buried internally in its oligomeric forms (Mou et al., 2003).
Figure 5.
SnRK2.8 Promotes the Nuclear Import of NPR1.
(A) SnRK2.8 does not affect the monomerization of NPR1. The MYC-NPR1 fusion was overexpressed driven by the CaMV 35S promoter in either Col-0 (left panel) or snrk2.8-1 (right panel). The 4th and 5th leaves of 4-week-old plants were pressure-infiltrated with Pst DC3000/avrRpt2 cells, and two upper leaves were harvested. Protein extracts were resuspended in loading buffer with or without β-mercaptoethanol (reducing or nonreducing, respectively). Arrowheads indicate NPR1 monomers.
(B) Nucleo-cytoplasmic distribution of NPR1 in the snrk2.8-1 mutant. The NPR1-GFP gene fusion was overexpressed driven by the CaMV 35S promoter in Col-0 and snrk2.8-1 plants. Plants were infected as described in (A). Distal leaves were visualized by DIC and fluorescence microscopy (left panel).
(C) Effects of pathogen infection on the nucleo-cytoplasmic distribution of NPR1. The MYC-NPR1 fusion was overexpressed in Col-0 or snrk2.8-1, as described above. Plants were infected as described in (A), and two upper leaves were harvested 24 h after infection for cell fractionation. NPR1 proteins were immunologically detected using an anti-MYC antibody (left panel). Antitubulin and anti-H3 antibodies were used for the detection of cytoplasmic and nuclear markers, respectively. Blots were quantitated using the ImageJ software (right panel). Band intensities of nuclear fractions were divided by those of cytoplasmic fractions to obtain relative values. Three blots were averaged and statistically treated (t test, *P < 0.01). Bars indicate sd. The relative intensity of pathogen-treated cytoplasmic fraction was set to 1. Percentage indicates the ratio of nuclear to cytoplasmic distribution. Nu, nuclear fraction; Cy, cytoplasmic fraction.
SnRK2.8 Is Required for the Nuclear Import of NPR1
It is notable that SnRK2.8 phosphorylates Ser-589 of NPR1, which is located within the nuclear localization sequence in the far C-terminal region (Kinkema et al., 2000). Therefore, we assessed whether SnRK2.8 regulates the nuclear import of NPR1 through protein phosphorylation. A gene encoding an NPR1-GFP fusion was overexpressed in Col-0 plants and snrk2.8-1 mutants, and the resultant transgenic plants were infected with Pst DC3000/avrRpt2 cells. After SAR induction, GFP signals were detected predominantly in the nuclei of distal leaf cells in the Col-0 background. In contrast, the GFP signals were relatively weaker in the nuclei of distal leaf cells in snrk2.8-1 background (Figure 5B). In local leaves, the nuclear localization patterns of NPR1 were similar in both Col-0 and snrk2.8 backgrounds (Supplemental Figure 5A). Together, these observations indicate that SnRK2.8 modulates the nuclear import of NPR1 in distal leaves during SAR.
Impaired nuclear import of NPR1 in snrk2.8-1 mutant was also examined by cell fractionation assays. After primary infection with Pst DC3000/avrRpt2 cells, nuclear and cytoplasmic fractions of distal leaf cells were separately prepared from transgenic plants overexpressing the MYC-NPR1 gene fusion in either Col-0 or snrk2.8-1 background, and NPR1 protein was detected immunologically in each fraction. In mock-treated Col-0 plants, 43% of NPR1 was localized in the nucleus (Figure 5C). Notably, after SAR induction, the portion of nuclear NPR1 increased up to 66%. In contrast, 17% of NPR1 was localized in the nuclei of mock-treated snrk2.8-1 mutants, and the portion of nuclear NPR1 was not significantly affected by SAR induction, supporting the role of SnRK2.8 in inducing the nuclear import of NPR1 in distal tissues.
Our data showed that SnRK2.8 triggers the nuclear import of NPR1 by phosphorylating Ser-589 and Thr-373, entailing that mutated NPR1 proteins harboring S589A or T373A would be excluded from the nucleus. To examine this hypothesis, GFP fusions of the mutated NPR1 proteins and CFP-SnRK2.8 fusion were coexpressed driven by the CaMV 35S promoter in Arabidopsis protoplasts. It was found that NPR1 proteins having either one of the substitutions localized predominantly in the cytoplasm (Supplemental Figure 5B), further supporting the idea that the SnRK2.8-mediated phosphorylation of NPR1 is important for its nuclear import.
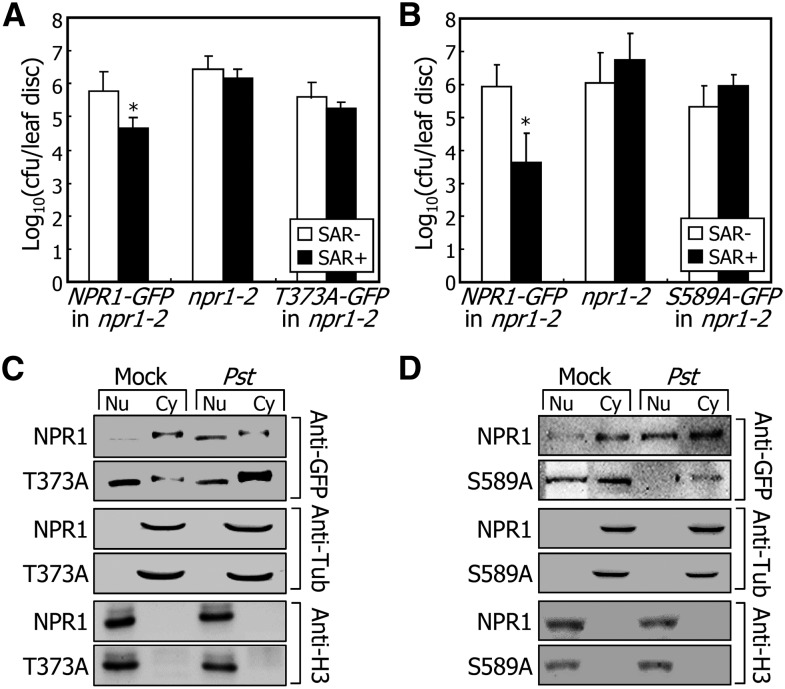
To verify that SnRK2.8 is required for the nuclear import of NPR1 in planta, we overexpressed the T373A-GFP and S589A-GFP fusions in the npr1-2 mutant. We examined whether the nuclear localization of the mutated NPR1 proteins is altered and SAR is affected in the transgenic plants. It was found that SAR was not induced in the npr1-2 mutant expressing the T373A-GFP or S589A-GFP proteins, while it was efficiently induced in the npr1-2 mutant expressing NPR1-GFP protein (Figures 6A and 6B). Notably, the SAR-induced nuclear import of NPR1 was disrupted by the Thr-373 or Ser-589 mutations (Figures 6C and 6D). These observations indicate that the SnRK2.8-mediated phosphorylation of NPR1 is critical for its nuclear import during SAR.
Figure 6.
SnRK2.8-Mediated Phosphorylation Is Important for the Nuclear Import of NPR1.
(A) and (B) SAR induction in npr1-2 mutant expressing T373A-GFP or S589A-GFP gene fusions. Four-week-old plants grown in soil were used for pathogen infection. Thr-373 (A) or Ser-589 (B) of NPR1 was mutated to alanine and overexpressed driven by the CaMV 35S promoter in npr1-2 mutant. Bacterial cell counting was performed as described in Figure 1E. Four measurements were averaged and statistically treated using Student's t test (*P < 0.01). Bars indicate sd.
(C) and (D) Nucleo-cytoplasmic distribution of T373A-GFP and S589A-GFP in pathogen-infected plants. Plants used were identical to those described in (A) and (B). The 35S:T373A-GFP (C) and 35S:S589A-GFP (D) transgenic plants were infected as described in Figure 5A. The α-tubulin and Η3 proteins were immunologically detected as described in Figure 5C. NPR1-GFP, T373A-GFP, and S589A-GFP proteins were detected using an anti-GFP antibody.
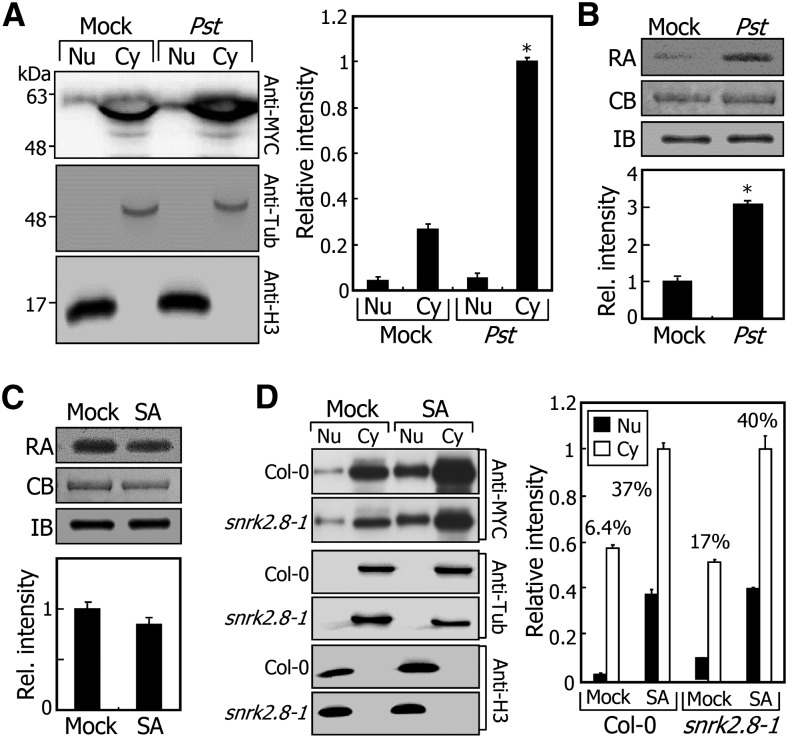
The role of SnRK2.8 in triggering the nuclear import of NPR1 raised a question as to the subcellular localization of SnRK2.8 during SAR. Col-0 plants overexpressing the MYC-SnRK2.8 gene fusion (35S:MYC-2.8) were infected with Pst DC3000/avrRpt2 cells to induce SAR, and nuclear and cytoplasmic fractions were prepared from the distal leaves for immunological detection of SnRK2.8 proteins. We found that SnRK2.8 exists in both the nucleus and cytoplasm but with a predominant distribution in the latter (Figure 7A). In addition, the nucleo-cytoplasmic distribution of SnRK2.8 did not alter during SAR, which is relevant to the notion that SnRK2.8 phosphorylates NPR1 in the cytoplasm.
Figure 7.
SnRK2.8-Mediated Nuclear Import of NPR1 Is Not Directly Linked with SA.
In (A) to (C), band intensities of three blots were averaged and statistically analyzed (t test, *P < 0.01). Bars indicate sd.
(A) Nucleo-cytoplasmic distribution of SnRK2.8 in pathogen-infected plants. A MYC-SnRK2.8 gene fusion was overexpressed driven by the CaMV 35S promoter in Col-0 plant. Plants were infected as described in Figure 5A. MYC-SnRK2.8, α-tubulin, and Η3 proteins were detected as described in Figure 5C.
(B) Elevation of SnRK2.8 activity after pathogen infection. Four-week-old 35S:MYC-SnRK2.8 transgenic plants grown in soil were infected with Pst DC3000/avrRpt2 cells. Two upper leaves were harvested 24 h after infection for immunoprecipitation of MYC-SnRK2.8 proteins. Five micrograms of recombinant MBP-NPR1 proteins was mixed with immunoprecipitated MYC-SnRK2.8 proteins and incubated as described in Figure 4A. MBP-NPR1 and MYC-SnRK2.8 proteins in the reaction mixtures were detected by Coomassie blue staining (CB) and immunoblot hybridization (IB), respectively (upper panel). Band intensities were quantitated using the ImageJ software (lower panel). Relative radioactivity (RA) to the amount of MYC-SnRK2.8 protein was displayed.
(C) Effects of SA on SnRK2.8 activity. Two-week-old 35S:MYC-SnRK2.8 transgenic plants grown on MS-agar plates were treated with 0.5 mM SA as described in Figure 2A. Whole plants were harvested 24 h after SA treatment for immunoprecipitation of MYC-SnRK2.8 proteins. Five micrograms of recombinant MBP-NPR1 proteins was mixed with immunoprecipitated MYC-SnRK2.8 proteins and incubated as described in Figure 4A. MBP-NPR1 and MYC-SnRK2.8 proteins were immunologically detected and quantitated as described in (B).
(D) Nucleo-cytoplasmic distribution of NPR1 in SA-treated plant cells. Two-week-old plants grown on MS-agar plates were treated with 0.5 mM SA for 24 h. Whole plants were used for cell fractionation. MYC-NPR1, α-tubulin, and Η3 proteins were immunologically detected as described in Figure 5C.
It was notable that the level of cytoplasmic SnRK2.8 was elevated by more than 3-fold during SAR, showing that its protein stability is enhanced by pathogen-triggered signals. To examine whether pathogen infection influences the kinase activity of SnRK2.8, we performed NPR1 phosphorylation assays using MYC-SnRK2.8 protein immunoprecipitated from the 35S:MYC-2.8 transgenic plants. We found that endogenous SnRK2.8 phosphorylates NPR1 and its kinase activity is elevated by ∼3-fold (Figure 7B). These observations indicate that NPR1 phosphorylation by SnRK2.8 is enhanced after pathogen infection. However, SA feeding did not influence the SnRK2.8 kinase activity (Figure 7C), further supporting the idea that the basal level of SA is sufficient for the activation of SnRK2.8.
SA triggers the nuclear import of NPR1 (Kinkema et al., 2000). We therefore asked whether SnRK2.8-mediated nuclear import of NPR1 is associated with SA. In mock-treated Col-0 plants, 6.4% of NPR1 was nuclear-localized, and the nuclear portion of NPR1 significantly increased up to 37% after SA treatment (Figure 7D). Notably, a similar nucleo-cytoplasmic distribution pattern of NPR1 was observed in snrk2.8-1 mutants, indicating that SA is not directly linked with the SnRK2.8-mediated nuclear import of NPR1.
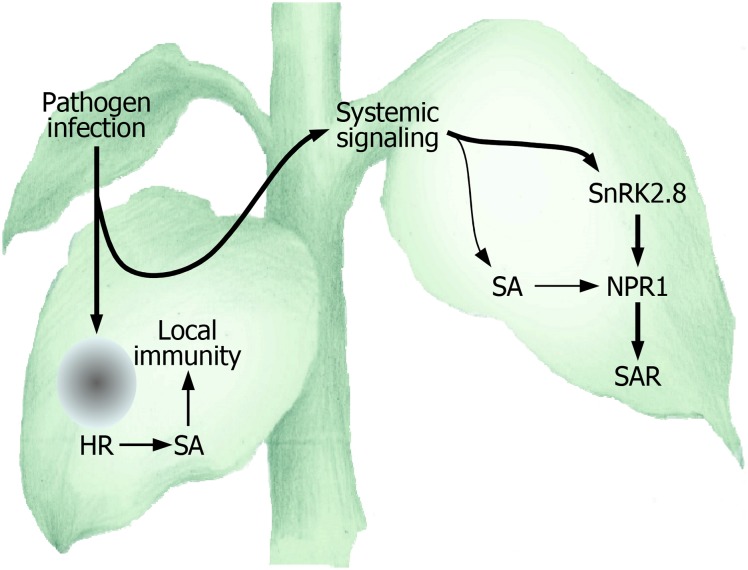
Altogether, our data demonstrate that SnRK2.8 phosphorylates NPR1 to facilitate its nuclear import (Figure 8). The SnRK2.8-mediated nuclear import of NPR1 depends on SA. A plausible working scenario would be that SA triggers the formation of NPR1 monomers, which are phosphorylated by SnRK2.8 for its nuclear import. It is notable that NPR1 is activated through a dual-step process during SAR, providing an answer to the long-standing question as to the highly efficient induction of SAR even though SA accumulates marginally in distal leaves.
Figure 8.
SnRK2.8 Is Required for NPR1 Activation during SAR.
SnRK2.8 modulates the nuclear import of NPR1 in inducing SAR. In this signaling network, the SnRK2.8-mediated NPR1 activation is integrated with SA signaling to ensure SAR to occur in distal tissues. The signaling steps examined or elucidated in this work are indicated by bold arrows.
DISCUSSION
Convergence of SA and SnRK2.8-Mediated Signals at NPR1
In this work, we showed that SAR-induced activation of NPR1 requires protein phosphorylation in addition to the monomerization reaction. While SA is essential for SAR development, it needs to be maintained at low levels in parts of the plant other than the site of hypersensitive response to avoid harmful effects, such as growth retardation (Rivas-San Vicente and Plasencia, 2011), DNA damage (Yan et al., 2013), and physiological disturbance (Scott et al., 2004). In this view, it is envisioned that SA serves as an initiating signal that links local infection with the induction of SAR in distal tissues; therefore, high levels of SA would not be necessarily required for the NPR1 activation. We propose that SA-mediated monomerization and SnRK2.8-mediated phosphorylation coordinately function to activate NPR1 in developing SAR in distal tissues. Our findings will contribute to the identification of long-distance signaling molecules other than SA during SAR development in the future.
It has been reported that Cys-82 and Cys-216 residues of NPR1 are important for its oligomerization and a mutated NPR1 protein having substitutions of the two Cys residues are constitutively nuclear-localized (Mou et al., 2003). PR genes are highly expressed in transgenic plants overexpressing the mutated NPR1 even without pathogen infection (Mou et al., 2003), signifying the nuclear import of NPR1 in the immune response. We observed that PR genes are highly expressed in SnRK2.8-overexpressing plants (Figure 1C). The elevated PR transcription in the transgenic plants would be due to the increased nuclear import of NPR1 monomers (Supplemental Figure 5B), since SA content is not altered in SnRK2.8-overexpressing plants (Supplemental Figure 2B). However, the elevation of PR transcription disappears in sid2 mutants, suggesting that a basal level of SA is necessary for the SnRK2.8-mediated activation of NPR1.
Do Upstream Signals Activate SnRK2.8 during SAR?
Our data show that SA is not the upstream regulator of SnRK2.8 function during SAR. A question is how pathogen infection in local tissues triggers SnRK2.8 activation in distal tissues. Plants produce a variety of mobile immune signals to induce systemic immunity after pathogen infection (Fu and Dong, 2013). The systemic signals would cause redox changes in distal tissues, which lead to the monomerization reaction of NPR1. It has been suggested that glycerol 3-phosphate, DEFECTIVE IN INDUCED RESISTANCE1, azelaic acid, pipecolic acid, and dehydroabietinal act as mobile immune signals in inducing SAR (Maldonado et al., 2002; Jung et al., 2009; Chanda et al., 2011; Chaturvedi et al., 2012; Návarová et al., 2012). It will be interesting to investigate the functional relationship between the systemic signals and NPR1 monomerization during SAR.
In this report, we propose that SnRK2.8 is required for the nuclear import of NPR1 monomer (Figures 5A and 5C). SA does not induce SnRK2.8 transcription and its protein activity. It has been reported that nitric oxide causes NPR1 translocation into the nucleus, while it does not trigger the monomerization reaction of NPR1 (Lindermayr et al., 2010), suggesting that nitric oxide is important for the nuclear import of NPR1 monomers. It will be interesting to examine whether nitric oxide is an upstream regulator of SnRK2.8 function.
Phosphorylation of NPR1 Is Required for Its Nuclear Import
NPR1 phosphorylation at several residues is important for its function (Spoel et al., 2009; Xie et al., 2010). SA triggers NPR1 phosphorylation at Ser-11 and Ser-15 (Spoel et al., 2009). When SA level is high, the phosphorylated NPR1 at these residues is targeted to the ubiquitin-dependent proteasome pathway (Spoel et al., 2009). It has been suggested clearing the phosphorylated NPR1 from the nucleus is required to reinitiate PR induction by adopting fresh NPR1 from the cytoplasm (Spoel et al., 2009). The C-terminal region of NPR1 is phosphorylated by PROTEIN KINASE SOS2-LIKE5, which is important for the induction of WRKY38 and 62 genes after pathogen infection (Xie et al., 2010). We found that SnRK2.8 phosphorylates Ser-589 and Thr-373 of NPR1 to facilitate its nuclear import (Figures 6C and 6D). It is now evident that multiple residues of NPR1 are phosphorylated by several different kinases to regulate distinct aspects of its function. It is tempting to examine how the multiple phosphorylation schemes are coordinated to modulate NPR1 function.
Two potential nuclear localization signals exist in the C-terminal region of NPR1 (Kinkema et al., 2000), each ranging from Lys-537 to Lys-554 and Lys-582 to Arg-593 (NLS1 and NLS2, respectively). It has been shown that NLS1 is important for the nuclear import of NPR1 at high concentrations of SA (Kinkema et al., 2000). Notably, one of the SnRK2.8-mediated phosphorylation sites, Ser-589, is located within NLS2. While SAR-induced activation of SnRK2.8 is independent of SA, the SnRK2.8-mediated activation of NPR1 depends on a basal or slightly elevated level of SA. On the basis of the NLS1-SA and NLS2-SnRK2.8 connections, it is suggested that while SA triggers the nuclear import of NPR1 via NLS1 at high levels of SA as observed in local tissues, SnRK2.8 directs the nuclear import of NPR1 via NLS2 in distal tissues, where SA level is low. The dual control of the NPR1 nuclear import would be an adaptation strategy to minimize unfavorable effects of high concentrations of SA on plant growth.
We found that T373A mutation disrupts the nuclear-cytoplasmic distribution of NPR1. The mutated NPR1 proteins having the T373A mutation are distributed in both the nucleus and cytoplasm, and their distribution pattern is not affected by SAR. Thus, it is likely that the nuclear import of NPR1, rather than its nuclear distribution, is important for developing SAR. Alternatively, its phosphorylation at Thr-373 may be necessary for the transcriptional regulatory activity of NPR1 in the nucleus. Investigation of the mutated NPR1 with TGA transcription factors and transcriptional activity assays of the mutated NPR1 would be helpful to clarify the molecular mechanisms by which protein phosphorylation modulates NPR1 function.
It has been reported that MODIFIER OF SNC1,7 (MOS7), a component of the nuclear pore complex, is required for the nuclear import of NPR1 in innate immunity and SAR (Cheng et al., 2009; Wiermer et al., 2010). In a MOS7-defective mutant (mos7-1), both PR expression and SAR are largely reduced (Cheng et al., 2009), which would be due to the impaired nuclear import of NPR1. Considering the phenotypic and physiological similarities between mos7-1 and snrk2.8 mutants, it will be worthy of investigating the biochemical linkage between SnRK2.8-mediated NPR1 phosphorylation and MOS7-mediated nuclear import of NPR1 during SAR.
Potential Roles of SnRK2.8 in Linking SAR with Metabolism
SnRK2 members are involved in abscisic acid (ABA)-mediated signaling pathways, including those regulating stomatal aperture, water stress, and seed germination (Fujita et al., 2009; Nakashima et al., 2009; Wege et al., 2014). The kinase activities of SnRK2.2, 2.3, 2.6, 2.7, and 2.8 are elevated by ABA treatments, while those of SnRK2.7 and SnRK2.8 are only slightly induced under the identical conditions (Boudsocq et al., 2004). ABA suppresses SAR and SAR inhibits ABA signaling (Yasuda et al., 2008). Since SnRK2.8 activates NPR1 to confer SAR (Figure 5C), it is not likely that SnRK2.8 mediates ABA-mediated suppression of SAR and vice versa. On the other hand, SnRK2.6 might be involved in ABA-mediated inhibition of SAR. Its activity is regulated by nitric oxide (Wang et al., 2015), which is one of the key molecules that induce SAR (Lindermayr et al., 2010). SnRK2.6 is also involved in PAMP-induced stomatal closure, which contributes to innate immunity (Melotto et al., 2006). It will be interesting to examine the functional relationship between SnRK2.6 and SAR.
Accumulating evidence supports the idea that the disease resistance response is intimately associated with plant metabolism (Berger et al., 2007; Bolton, 2009), although the underlying molecular mechanisms are largely unknown. SnRK2.8 also phosphorylates various proteins involved in plant metabolism. SnRK2.8-overexpressing plants exhibit enhanced shoot and root growth, suggesting a role of SnRK2.8 in metabolic processes (Shin et al., 2007). A screening for SnRK2.8 targets has identified more than seven distinct proteins (Shin et al., 2007). Most of them have been expected to be involved in diverse aspects of metabolism, and a 14-3-3 protein is related to the basal immune response (C.S. Oh et al., 2010). Our observations indicate that SnRK2.8 is not involved in basal immunity (Figure 1D), suggesting that SnRK2.8 is functionally distinct from that of 14-3-3 protein. It is currently unclear whether SnRK2.8 mediates the linkage between disease resistance response and metabolism.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana lines used were in Columbia (Col-0) background. Arabidopsis plants were grown either in soil or on 0.5× Murashige and Skoog-agar (MS-agar) plates under long days (LDs; 16 h light and 8 h dark) at 23°C with relative humidity of 60%. White light illumination (120 μmol photons m−2 s−1) was provided by fluorescent FLR40D/A tubes (Osram). The SnRK2.8-deficient snrk2.8-1 and snrk2.8-2 mutants (SALK-073395 and SALK-053423, respectively) were isolated from a T-DNA insertional mutant pool deposited in TAIR (Ohio State University, OH). The NPR1-deficient npr1-2 mutant, which has been described previously (Cao et al., 1997), was obtained from Xinnian Dong. Homozygotic mutant lines were obtained by selection for three or more generations and analysis of segregation ratios. Lack of gene expression in the mutants was confirmed by RT-PCR. The SnRK2.8 coding sequence (CDS) was overexpressed driven by the CaMV 35S promoter in Col-0 plants, resulting in 35S:2.8 transgenic plants. Multiple independent transgenic lines were obtained, and two representative lines were assayed in parallel. Agrobacterium tumefaciens-mediated Arabidopsis transformation was performed by a modified floral dip method (Zhang et al., 2006). Overexpression of transgenes in the transgenic plants was examined by RT-qPCR.
Gene Transcription Assay
RT-qPCR was employed to examine the relative levels of gene transcripts. Total RNA preparation, reverse transcription, and quantitative polymerase chain reactions were performed following the proposed rules to ensure reproducible measurements (Udvardi et al., 2008). RT-qPCR reactions were performed using 96-well blocks with an Applied Biosystems 7500 real-time PCR system using the SYBR Green I master mix in a volume of 20 μL. PCR primers are listed in Supplemental Table 1. The two-step thermal cycling profile employed was 15 s at 95°C for denaturation and 1 min at 60°C for annealing and polymerization. An eIF4A gene (At3g13920) was included in the reactions as internal control in order to normalize the variations in the amounts of cDNA used.
RT-qPCR reactions were performed in biological triplicates using total RNA samples extracted from three independent plant materials grown under identical conditions. The comparative ΔΔCT method was employed to evaluate relative quantities of each product amplified from the samples. The threshold cycle (CT) was automatically determined for each reaction using the default parameters of the system.
Preparation of Recombinant Proteins
SnRK2.8 and NPR1 CDSs were subcloned into the pMAL-c2X Escherichia coli expression vector (NEB) harboring an MBP-coding sequence. Recombinant MBP-SnRK2.8 and MBP-NPR1 proteins were synthesized in E. coli Rosetta2 (DE3) pLysS strain (Novagen). Harvested cells were suspended in MBP buffer (20 mM Tris-Cl, pH 7.4, 200 mM NaCl, 1 mM EDTA, 10 mM 2-mercaptoethanol, and 1 mM PMSF) supplemented with protease inhibitor cocktail (Sigma-Aldrich). Cell lysates were prepared by running three cycles of freezing and thawing followed by centrifugation. The fusion proteins were purified by affinity chromatography as described previously (Seo et al., 2011).
Yeast Two-Hybrid Assay
Yeast two-hybrid assays were employed to examine whether SnRK2.8 interacts with NPR1 using the BD Matchmaker system (Clontech). The pGADT7 vector was used for GAL4 activation domain fusion, and the pGBKT7 vector was used for GAL4 DNA binding domain fusion. PCR products of SnRK2.8 and NPR1 CDSs were digested with NdeI and EcoRI and subcloned into the pGBKT7 and pGADT7 vectors. The yeast strain AH109, which harbors chromosomally integrated reporter genes lacZ encoding β-galactosidase and HIS mediating histidine biosynthesis under the control of the GAL1 promoter, was used for transformation of the expression constructs. Transformation of AH109 cells was performed according to the manufacturer’s instructions. Colonies obtained were streaked on medium without His, Ade, Leu, and Trp.
BiFC Assay
BiFC assays were performed using Arabidopsis mesophyll protoplasts as described previously (Seo et al., 2011). SnRK2.8 CDS was fused in frame to the 3′ end of a DNA sequence encoding the N-terminal half of EYFP (nYFP) in the pSATN-cEYFP-C1 vector (E3082). NPR1 CDS was fused in frame to the 3′ end of a DNA sequence encoding the C-terminal half of EYFP (cYFP) in the pSATN-nEYFP-C1 vector (E3081). Cotransfection of the nYFP-SnRK2.8 and cYFP-NPR1 constructs into Arabidopsis protoplasts was performed by a polyethylene glycol-calcium transfection method (Yoo et al., 2007). Sixteen hours after cotransfection, the subcellular distribution of the fusion constructs was visualized by differential interference contrast (DIC) and fluorescence microscopy using a Zeiss LSM510 confocal microscope (Carl Zeiss). Reconstitution of YFP fluorescence was observed using a confocal microscope with the following YFP filter setup: excitation 515 nm, 458/514 dichroic, and emission 560- to 615-nm band-pass filter.
Cell Fractionation
A MYC-NPR1 fusion construct, in which a MYC-coding sequence was fused in frame to the 5′ end of NPR1 CDS, was overexpressed driven by the CaMV 35S promoter in Col-0 or snrk2.8-1 background. The 4th and 5th leaves of 4-week-old plants grown in soil under LDs were pressure-infiltrated with the avirulent Pst DC3000/avrRpt2 cells for 2 d. Two upper leaves were harvested for cell fractionation. Preparation of nuclear and cytoplasmic fractions was performed as described previously (Wang et al., 2011). Three percent of cytoplasmic fractions and 10% of nuclear fractions were used for immunoblot assays. The cellular fractions were analyzed on 10% SDS-PAGE and blotted onto an Immobilon-P membrane (Millipore). NPR1 proteins were immunologically detected using an anti-MYC antibody (Millipore). The α-tubulin protein, a cytoplasmic localization marker (Saslowsky et al., 2005), was detected using an antitubulin antibody (Sigma-Aldrich). The H3 protein, a nuclear localization marker (Feys et al., 2005), was detected using an anti-H3 antibody (Millipore). Blots were quantitated using the ImageJ software (http://rsbweb.nih.gov/ij/).
Pathogen Infection Assay and SA Treatment
Four-week-old plants grown in soil under LDs at 23°C were used for pathogen infection and gene expression assay. Bacterial cells of the avirulent Pst DC3000/avrRpt2 strain were cultured for 24 h at 28°C in Luria-Bertani medium supplemented with rifampicin (50 mg/L). A bacterial cell suspension was prepared at OD600 = 0.02 in 10 mM MgCl2 and pressure-infiltrated into the 4th and 5th rosette leaves at zeitgeber time 9. The inoculated plants were transferred to a growth chamber set at 23°C with relative humidity of 80% and further grown for 2 d under LDs. Two upper leaves were then infiltrated with the virulent Pst DC3000 cells and incubated for three additional days before taking photographs and counting bacterial cells, as previously described (Park et al., 2007).
For spray inoculation, 4-week-old plants grown in soil were sprayed with Pst DC3000 cell suspensions of OD600 = 0.4 in 10 mM MgCl2 containing 0.05% Silwet L-77. The sprayed plants were incubated for 6 d in growth chamber set at 23°C under LDs before counting bacterial cells.
For SA treatments, 2-week-old plants grown on MS-agar plates were sprayed with 0.5 mM SA and incubated for appropriate time periods before harvesting whole plant materials.
Co-IP Assay
The Co-IP assays were performed using Nicotiana benthamiana cells as described previously (Song et al., 2012). The MYC-SnRK2.8 and FLAG-NPR1 fusion sequences were subcloned into the pBA002 and pEarleyGate202 vectors (ABRC), respectively, and the expression constructs were transiently coexpressed driven by the CaMV 35S promoter in the tobacco leaves. Plant materials were ground in liquid nitrogen, and proteins were extracted in Co-IP buffer (50 mM Tris-Cl, pH 7.4, 500 mM NaCl, 10% glycerol, 5 mM EDTA, 1% Triton X-100, and 1% Nonidet P-40) containing protease inhibitor cocktail (Sigma-Aldrich). Ten percent of the extract was used as input control. An anti-MYC antibody (Sigma-Aldrich) was added to the extract and incubated for 2 h. Protein A agarose beads were then added and further incubated for 2 h. The beads were washed five times with Co-IP buffer lacking protease inhibitor cocktail. To elute bound proteins, 50 μL of 2× SDS-PAGE loading buffer (100 mM Tris-Cl, pH 6.8, 4% SDS, 0.2% bromophenol blue, 20% glycerol, and 200 mM DTT) was added and boiled for 5 min. Twenty percent of the eluted proteins was used as immunoprecipitation control. Anti-FLAG (Sigma-Aldrich) and anti-MYC (Millipore) antibodies were used for the immunological detection of FLAG-NPR1 and MYC-SnRK2.8 fusion proteins, respectively.
In Vitro Phosphorylation Assay
In vitro phosphorylation assays were performed as described previously (Kim et al., 2012). Five micrograms of purified recombinant NPR1 proteins and 1.5 μg of recombinant SnRK2.8 proteins were added to reaction buffer (20 mM HEPES, pH 7.4, 10 mM MgCl2, 1 mM Na3VO4, 2 mM DTT, 0.5 mM PMSF, 2 mM EDTA, and 200 nM dATP) containing 1 μCi of [γ-32P]ATP. The reaction mixture was incubated at 30°C for 30 min, and the reaction was terminated by adding 4 μL of 6× SDS-PAGE loading buffer. The mixture was boiled for 5 min and loaded onto 10% SDS-PAGE gel. The gel was stained with Coomassie Brilliant Blue R 250 and dried under vacuum for the detection of radioactive signals.
Kinase Activity Assay
A MYC-SnRK2.8 gene fusion, in which a MYC-coding sequence was fused in frame to the 5′ end of the SnRK2.8 CDS, was overexpressed driven by the CaMV 35S promoter in Col-0 plants. The 4th and 5th leaves of 4-week-old plants grown in soil under LDs were pressure-infiltrated with Pst DC3000/avrRpt2 cells for 2 d. Two upper leaves were harvested for kinase activity assays. Approximately 0.2 g of plant materials were ground in liquid nitrogen, and total proteins were extracted in immunoprecipitation buffer (50 mM Tris-Cl, pH 7.4, 500 mM NaCl, 10% glycerol, 5 mM EDTA, 1% Triton X-100, and 1% Nonidet P-40) containing protease inhibitor cocktail (Sigma-Aldrich). An anti-MYC antibody (Millipore) was added to the protein extract and incubated for 3 h to immunoprecipitate MYC-SnRK2.8 proteins. Protein G agarose beads were then added and further incubated for 2 h. The beads were washed three times with immunoprecipitation buffer without protease inhibitor cocktail. The beads and 5 μg of recombinant MBP-NPR1 protein were subjected to in vitro phosphorylation assay.
Mass Spectrometry
Eight micrograms of recombinant NPR1 protein was phosphorylated in vitro by 4 μg of SnRK2.8 protein. The reaction mixture was separated by SDS-PAGE, and NPR1 protein was eluted from the gel. The eluted NPR1 protein was digested with trypsin, and the resultant peptides were subjected to nano-liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis using an LTQ-Orbitrap (Thermo Fisher Scientific). Collision-induced dissociation method was employed for the fragmentation. Mass spectra were analyzed using the database search program Sequest (http://fields.scripps.edu/sequest/).
Measurement of SA Content
Four-week-old plants grown in soil were infected with Pst DC3000/avrRpt2 cells. The four upper leaves were harvested 24 h after the infection for SA measurement. Plant materials of 150 mg were ground in liquid nitrogen and resuspended in 1 mL of ethylacetate for centrifugation at 16,000g for 20 min at 4°C. The supernatant was transferred to a new tube and evaporated using a vacuum concentrator at 30°C for 1 h. The dried samples were resuspended in 100 μL of 70% methanol, and 50 μL was subjected to LC-MS/MS analysis.
In Vivo Phosphorylation Assay
Two-dimensional electrophoresis was employed for in vivo phosphorylation assays, as described previously (Kim et al., 2012). A MYC-NPR1 fusion was overexpressed driven by the CaMV 35S promoter in Col-0 and snrk2.8-1 plants, and the transgenic plants were grown for 4 weeks in soil. The 4th and 5th leaves were pressure-infiltrated with Pst DC3000/avrRpt2 cell suspensions, and uninfiltrated leaves were harvested 24 h after infiltration. Plant materials of 200 mg were ground in liquid nitrogen and resuspended in 1 mL of extraction buffer (50 mM Tris-Cl, pH 8.4, 100 mM NaCl, 1 mM MgCl2, 1 mM DTT, and 1 mM PMSF) supplemented with protease inhibitor cocktail (Sigma-Aldrich). For alkaline phosphatase treatment, 300 units of calf intestinal alkaline phosphatase (Takara) were added to the mixture and incubated at 37°C for 1 h. After the reaction, 3 mL of lysis buffer (8 M urea, 2% CHAPS, 100 mM DTT, 0.2% Bio-lytes [Bio-Rad], and 1 mM PMSF) was added to stop the reaction. Proteins were then precipitated by trichloroacetic acid-acetone method (Damerval et al., 1986), and 500 μg of total proteins was loaded onto an immobilized pH gradient gel strip (7 cm, pH 3 to 10) (Bio-Rad). After focusing, immunological detection of NPR1 proteins was performed using an anti-MYC antibody (Millipore).
For two-dimensional electrophoresis using tobacco plants, the MYC-SnRK2.8 and FLAG-NPR1 fusions were transiently coexpressed driven by the CaMV 35S promoter in N. benthamiana cells. Approximately 200 μg of total proteins was used for the isoelectric focusing and immunoblot as described above. An anti-FLAG antibody (Sigma-Aldrich) was used for the detection of FLAG-NPR1 proteins.
Statistical Analysis
The statistical significance between two means of measurements were determined using Student’s t test with P values < 0.01.
Accession Numbers
Sequence data from this article can be obtained from the Arabidopsis Genome Initiative databases under the following accession numbers: eIF4A (At3g13920), SnRK2.1 (At5g08590), SnRK2.2 (At3g50500), SnRK2.3 (At5g66880), SnRK2.4 (At1g10940), SnRK2.5 (At5g63650), SnRK2.6 (At4g33950), SnRK2.7 (At4g40010), SnRK2.8 (At1g78290), SnRK2.9 (At2g23030), SnRK2.10 (At1g60940), PR1 (At2g14610), PR5 (At1g75040), SID2 (At1g74710), NPR1 (At1g64280), TGA1 (At5g65210), TGA2 (At5g06950), TGA3 (At1g22070), TGA4 (At5g10030), TGA5 (At5g06960), and TGA6 (At3g12250).
Supplemental Data
Supplemental Figure 1. SnRK2.8 Is Required for Systemic Acquired Resistance.
Supplemental Figure 2. Functional Relationship between SnRK2.8 and Salicylic Acid.
Supplemental Figure 3. Control Reactions in Bimolecular Fluorescence Complementation.
Supplemental Figure 4. Biochemical Characterization of NPR1 Phosphorylation by SnRK2.8.
Supplemental Figure 5. SnRK2.8-Mediated Phosphorylation Is Important for Nuclear Import of NPR1.
Supplemental Table 1. Primers Used.
Supplementary Material
Acknowledgments
We thank Xinnian Dong for providing npr1-1 and npr1-2 mutants and Ohkmae K. Park for providing the virulent Pst DC3000 and the avirulent Pst DC3000/avrRpt2 cell lines. This work was supported by the Leaping Research (NRF-2015R1A2A1A05001636) and Global Research Lab (NRF-2012K1A1A2055546) programs provided by the National Research Foundation of Korea and the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center PJ0111532015) provided by the Rural Development Administration of Korea.
AUTHOR CONTRIBUTIONS
C.-M.P. conceived and designed the experiments. C.-M.P. prepared the manuscript with the contributions of H.-J.L. H.-J.L. and Y.-J.P. examined the SAR of snrk2.8 mutants and performed assays on gene expression, protein phosphorylation, coimmunoprecipitation, and protein localization. P.J.S. performed yeast two-hybrid screening and BiFC assay. J.-H.K. generated the vector constructs and provided scientific discussions on the manuscript. H.-J.S. and S.-G.K. measured SA content.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
PAMP-triggered immunity
- SAR
systemic acquired resistance
- SA
salicylic acid
- BiFC
bimolecular fluorescence complementation
- ABA
abscisic acid
- MS-agar
Murashige and Skoog-agar
- LD
long day
- CDS
coding sequence
- DIC
differential interference contrast
- Co-IP
coimmunoprecipitation
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
References
- Berger S., Sinha A.K., Roitsch T. (2007). Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J. Exp. Bot. 58: 4019–4026. [DOI] [PubMed] [Google Scholar]
- Block A., Alfano J.R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14: 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton M.D. (2009). Primary metabolism and plant defense--fuel for the fire. Mol. Plant Microbe Interact. 22: 487–497. [DOI] [PubMed] [Google Scholar]
- Boudsocq M., Barbier-Brygoo H., Laurière C. (2004). Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279: 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boyle P., Le Su E., Rochon A., Shearer H.L., Murmu J., Chu J.Y., Fobert P.R., Després C. (2009). The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. Plant Cell 21: 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron R.K., Paiva N.L., Lamb C.J., Dixon R.A. (1999). Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol. Mol. Plant Pathol. 55: 121–130. [Google Scholar]
- Cao H., Glazebrook J., Clarke J.D., Volko S., Dong X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63. [DOI] [PubMed] [Google Scholar]
- Chanda B., Xia Y., Mandal M.K., Yu K., Sekine K.T., Gao Q.M., Selote D., Hu Y., Stromberg A., Navarre D., Kachroo A., Kachroo P. (2011). Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43: 421–427. [DOI] [PubMed] [Google Scholar]
- Chaturvedi R., Venables B., Petros R.A., Nalam V., Li M., Wang X., Takemoto L.J., Shah J. (2012). An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71: 161–172. [DOI] [PubMed] [Google Scholar]
- Cheng Y.T., Germain H., Wiermer M., Bi D., Xu F., García A.V., Wirthmueller L., Després C., Parker J.E., Zhang Y., Li X. (2009). Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21: 2503–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C., de Vienne D., Zivy M., Thiellement H. (1986). Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis 7: 52–54. [Google Scholar]
- Delaney T.P., Uknes S., Vernooij B., Friedrich L., Weymann K., Negrotto D., Gaffney T., Gut-Rella M., Kessmann H., Ward E., Ryals J. (1994). A central role of salicylic acid in plant disease resistance. Science 266: 1247–1250. [DOI] [PubMed] [Google Scholar]
- Després C., Chubak C., Rochon A., Clark R., Bethune T., Desveaux D., Fobert P.R. (2003). The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell 15: 2181–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C., DeLong C., Glaze S., Liu E., Fobert P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12: 279–290. [PMC free article] [PubMed] [Google Scholar]
- Fan W., Dong X. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14: 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys B.J., Wiermer M., Bhat R.A., Moisan L.J., Medina-Escobar N., Neu C., Cabral A., Parker J.E. (2005). Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17: 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z.Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64: 839–863. [DOI] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. (2009). An autophosphorylation site of the protein kinase SOS2 is important for salt tolerance in Arabidopsis. Mol. Plant 2: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50: 2123–2132. [DOI] [PubMed] [Google Scholar]
- Gao Q.M., Kachroo A., Kachroo P. (2014). Chemical inducers of systemic immunity in plants. J. Exp. Bot. 65: 1849–1855. [DOI] [PubMed] [Google Scholar]
- Jung H.W., Tschaplinski T.J., Wang L., Glazebrook J., Greenberg J.T. (2009). Priming in systemic plant immunity. Science 324: 89–91. [DOI] [PubMed] [Google Scholar]
- Kachroo A., Robin G.P. (2013). Systemic signaling during plant defense. Curr. Opin. Plant Biol. 16: 527–533. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Park M.J., Seo P.J., Song J.S., Kim H.J., Park C.M. (2012). Controlled nuclear import of the transcription factor NTL6 reveals a cytoplasmic role of SnRK2.8 in the drought-stress response. Biochem. J. 448: 353–363. [DOI] [PubMed] [Google Scholar]
- Kinkema M., Fan W., Dong X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulik A., Wawer I., Krzywińska E., Bucholc M., Dobrowolska G. (2011). SnRK2 protein kinases--key regulators of plant response to abiotic stresses. OMICS 15: 859–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ehrhardt D.W., Rhee S.Y. (2006). Systematic analysis of Arabidopsis organelles and a protein localization database for facilitating fluorescent tagging of full-length Arabidopsis proteins. Plant Physiol. 141: 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C., Sell S., Müller B., Leister D., Durner J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado A.M., Doerner P., Dixon R.A., Lamb C.J., Cameron R.K. (2002). A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403. [DOI] [PubMed] [Google Scholar]
- Melotto M., Underwood W., Koczan J., Nomura K., He S.Y. (2006). Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980. [DOI] [PubMed] [Google Scholar]
- Mou Z., Fan W., Dong X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944. [DOI] [PubMed] [Google Scholar]
- Mudgett M.B., Staskawicz B.J. (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol. 32: 927–941. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50: 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nandi A., Welti R., Shah J. (2004). The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16: 465–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Návarová H., Bernsdorff F., Döring A.C., Zeier J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Mecey C., Lee Y.N., Imboden L.A., Chang J.H., He S.Y. (2011). Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh C.S., Pedley K.F., Martin G.B. (2010). Tomato 14-3-3 protein 7 positively regulates immunity-associated programmed cell death by enhancing protein abundance and signaling ability of MAPKKK α. Plant Cell 22: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M.H., Wang X., Wu X., Zhao Y., Clouse S.D., Huber S.C. (2010). Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc. Natl. Acad. Sci. USA 107: 17827–17832. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Park J.E., Park J.Y., Kim Y.S., Staswick P.E., Jeon J., Yun J., Kim S.Y., Kim J., Lee Y.H., Park C.M. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282: 10036–10046. [DOI] [PubMed] [Google Scholar]
- Rivas-San Vicente M., Plasencia J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62: 3321–3338. [DOI] [PubMed] [Google Scholar]
- Saslowsky D.E., Warek U., Winkel B.S.J. (2005). Nuclear localization of flavonoid enzymes in Arabidopsis. J. Biol. Chem. 280: 23735–23740. [DOI] [PubMed] [Google Scholar]
- Scott I.M., Clarke S.M., Wood J.E., Mur L.A. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135: 1040–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Park J.Y., Kim S.Y., Jeon J., Lee Y.H., Kim J., Park C.M. (2010). Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 61: 661–671. [DOI] [PubMed] [Google Scholar]
- Seo P.J., Kim M.J., Ryu J.Y., Jeong E.Y., Park C.M. (2011). Two splice variants of the IDD14 transcription factor competitively form nonfunctional heterodimers which may regulate starch metabolism. Nat. Commun. 2: 303. [DOI] [PubMed] [Google Scholar]
- Shin R., Alvarez S., Burch A.Y., Jez J.M., Schachtman D.P. (2007). Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. USA 104: 6460–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Smith R.W., To B.J., Millar A.J., Imaizumi T. (2012). FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336: 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S.H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., Zuo J., Dong X. (2008). Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvardi M.K., Czechowski T., Scheible W.R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20: 1736–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij B., Friedrich L., Morse A., Reist R., Kolditz-Jawhar R., Ward E., Uknes S., Kessmann H., Ryals J. (1994). Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Du Y., Hou Y.J., Zhao Y., Hsu C.C., Yuan F., Zhu X., Tao W.A., Song C.P., Zhu J.K. (2015). Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 112: 613–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Ye R., Xin Y., Fang X., Li C., Shi H., Zhou X., Qi Y. (2011). An importin β protein negatively regulates microRNA activity in Arabidopsis. Plant Cell 23: 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S., De Angeli A., Droillard M.J., Kroniewicz L., Merlot S., Cornu D., Gambale F., Martinoia E., Barbier-Brygoo H., Thomine S., Leonhardt N., Filleur S. (2014). Phosphorylation of the vacuolar anion exchanger AtCLCa is required for the stomatal response to abscisic acid. Sci. Signal. 7: ra65. [DOI] [PubMed] [Google Scholar]
- Whalen M.C., Innes R.W., Bent A.F., Staskawicz B.J. (1991). Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermer M., Germain H., Cheng Y.T., García A.V., Parker J.E., Li X. (2010). Nucleoporin MOS7/Nup88 contributes to plant immunity and nuclear accumulation of defense regulators. Nucleus 1: 332–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth M.C., Dewdney J., Wu G., Ausubel F.M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565. [DOI] [PubMed] [Google Scholar]
- Xie C., Zhou X., Deng X., Guo Y. (2010). PKS5, a SNF1-related kinase, interacts with and phosphorylates NPR1, and modulates expression of WRKY38 and WRKY62. J. Genet. Genomics 37: 359–369. [DOI] [PubMed] [Google Scholar]
- Yan S., Wang W., Marqués J., Mohan R., Saleh A., Durrant W.E., Song J., Dong X. (2013). Salicylic acid activates DNA damage responses to potentiate plant immunity. Mol. Cell 52: 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M., Ishikawa A., Jikumaru Y., Seki M., Umezawa T., Asami T., Maruyama-Nakashita A., Kudo T., Shinozaki K., Yoshida S., Nakashita H. (2008). Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis. Plant Cell 20: 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang X., Henriques R., Lin S.S., Niu Q.W., Chua N.H. (2006). Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 1: 641–646. [DOI] [PubMed] [Google Scholar]
- Zipfel C. (2009). Early molecular events in PAMP-triggered immunity. Curr. Opin. Plant Biol. 12: 414–420. [DOI] [PubMed] [Google Scholar]
- Zipfel C., Robatzek S., Navarro L., Oakeley E.J., Jones J.D., Felix G., Boller T. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.