The Medicago truncatula transcription factor NIN coordinates diverse processes during nodulation, restricting ENOD11 expression in the root epidermis and promoting CRE1 expression in the root cortex.
Abstract
Biological nitrogen fixation in legumes occurs in nodules that are initiated in the root cortex following Nod factor recognition at the root surface, and this requires coordination of diverse developmental programs in these different tissues. We show that while early Nod factor signaling associated with calcium oscillations is limited to the root surface, the resultant activation of Nodule Inception (NIN) in the root epidermis is sufficient to promote cytokinin signaling and nodule organogenesis in the inner root cortex. NIN or a product of its action must be associated with the transmission of a signal between the root surface and the cortical cells where nodule organogenesis is initiated. NIN appears to have distinct functions in the root epidermis and the root cortex. In the epidermis, NIN restricts the extent of Early Nodulin 11 (ENOD11) expression and does so through competitive inhibition of ERF Required for Nodulation (ERN1). In contrast, NIN is sufficient to promote the expression of the cytokinin receptor Cytokinin Response 1 (CRE1), which is restricted to the root cortex. Our work in Medicago truncatula highlights the complexity of NIN action and places NIN as a central player in the coordination of the symbiotic developmental programs occurring in differing tissues of the root that combined are necessary for a nitrogen-fixing symbiosis.
INTRODUCTION
The formation of a nitrogen-fixing nodule requires the initiation of two independent processes: nodule organogenesis in the root cortex and bacterial infection through infection threads that are initiated at the root epidermis (Oldroyd and Downie, 2008). While there is much evidence to show that these two processes can be genetically separated, it is clear that they must be coordinated both spatially and temporally in order to ensure that nodule organogenesis occurs below the site of bacterial infection. The root cortex in legumes consists of many cell layers, and in Medicago truncatula, the nodule primordium forms in the inner root cortex abutting the endodermis, with the earliest responses occurring in inner cortical and pericycle cells (Timmers et al., 1999; Xiao et al., 2014). Hence, coordination of bacterial infection with nodule organogenesis requires the integration of two different processes occurring multiple cell layers apart.
Many of the processes associated with nitrogen fixation are initiated in the plant following the perception of the bacterial-derived Nod factor (NF) signals (Dénarié et al., 1996; Oldroyd and Downie, 2008). This perception involves a suite of receptor-like kinases (Endre et al., 2002; Stracke et al., 2002; Madsen et al., 2003; Radutoiu et al., 2003; Arrighi et al., 2006), some of which bind NFs (Broghammer et al., 2012), and these in turn activate oscillations in nuclear-associated calcium levels (Ehrhardt et al., 1992; Sieberer et al., 2009) via a number of cation channels and calcium pumps located on the nuclear membranes (Wais et al., 2000; Charpentier et al., 2008; Capoen et al., 2011). A calcium and calmodulin-dependent protein kinase can decode these calcium oscillations (Lévy et al., 2004; Mitra et al., 2004; Miller et al., 2013) and phosphorylates the CYCLOPS transcription factor, promoting gene expression and the activation of nodulation (Yano et al., 2008; Singh et al., 2014). A suite of additional transcription factors act downstream or parallel to calcium and calmodulin-dependent protein kinase, including Nodule Inception (NIN), GRAS-type Nodulation Signaling Pathway (NSP1), NSP2, heterotrimeric CCAAT binding Nuclear Transcription Factor Y (NF-YA1), NF-YA2, ERF Required for Nodulation (ERN1), and ERN2 (Schauser et al., 1999; Kaló et al., 2005; Smit et al., 2005; Andriankaja et al., 2007; Marsh et al., 2007; Middleton et al., 2007; Soyano et al., 2013; Laloum et al., 2014). These transcriptional regulators act to coordinate the expression of nodulation-associated genes such as Early Nodulin 11 (ENOD11), a marker gene for NF-induced responses (Andriankaja et al., 2007).
The promotion of nodule organogenesis is associated with cytokinin signaling (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007), and the external application of cytokinin or an autoactivated cytokinin receptor (Lotus Histidine Kinase 1/M. truncatula Cytokinin Response 1 [CRE1]) is sufficient to induce nodule-like structures in the root cortex (Gonzalez-Rizzo et al., 2006; Murray et al., 2007; Tirichine et al., 2007; Heckmann et al., 2011; Plet et al., 2011). It was recently shown that NFs rapidly promote the accumulation of cytokinins within the zone of the root susceptible to rhizobial infection and that this cytokinin response accounts for a significant proportion of the gene induction changes associated with NF treatment (van Zeijl et al., 2015). NIN is essential for this cytokinin promotion of nodule organogenesis and is also required for the initiation of bacterial infection in the root epidermis (Schauser et al., 1999; Marsh et al., 2007; Madsen et al., 2010). During these processes, NIN has been shown to activate essential genes, such as Nodulation Pectate Lyase (NPL), which is required for bacterial infection (Xie et al., 2012), and NF-YA1 and NF-YA2, which are associated with cortical cell divisions (Soyano et al., 2013). NIN also activates the expression of a number of CLE peptides that act as root derived signals promoting autoregulation of nodulation (Soyano et al., 2014), a shoot-derived suppressive effect on the total levels of nodulation (Oka-Kira and Kawaguchi, 2006). This modality of action may explain NIN negative regulation of rhizobial infection (Yoro et al., 2014) and direct outputs of the NF signaling pathway: root hair deformation and gene expression measured by ENOD11 (Schauser et al., 1999; Marsh et al., 2007). Hence, NIN appears to play both an essential role in promoting multiple processes required for the onset of the nitrogen-fixing symbiosis in legumes as well as negatively regulating the degree of nodulation.
In order to better define the function of NIN in the activation of the diverse processes associated with the formation of a nitrogen-fixing nodule, we attempted to define some of the tissue-specific modes of action of the NIN protein. We show that NIN can act as a bifunctional transcription factor that can directly suppress the transcription of ENOD11 in the root epidermis and can promote the transcription of the cytokinin receptor CRE1 in the root cortex. Constitutive expression of NIN in either the root epidermis or the cortex can promote spontaneous nodule organogenesis, but differences in genetic dependencies point at different modalities of action in these different tissues. We propose that NIN activates the cortical program leading to nodule organogenesis but suppresses further induction of NF responses in the root epidermis.
RESULTS
Early Stages of Nod Factor Signaling Associated with Calcium Oscillations Mainly Involve Epidermal Cells
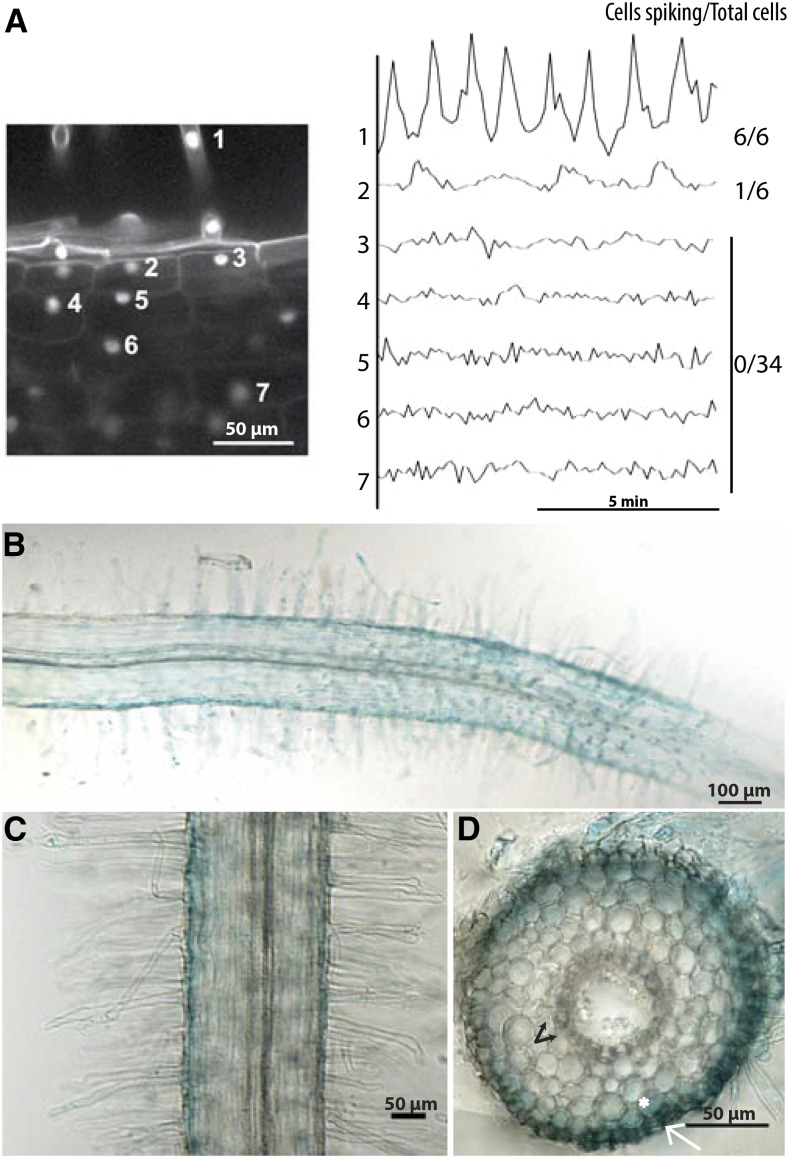
Considering that cell division in the root cortex occurs within 27 to 33 h after rhizobial inoculation (Timmers et al., 1999; Xiao et al., 2014), we wanted to see if NF-associated signaling could explain the activation of the cortical responses. NF signaling that is necessary for nodule organogenesis is associated with calcium oscillations (Ehrhardt et al., 1996; Wais et al., 2000), and using the nuclear-localized calcium reporter cameleon (Sieberer et al., 2009), it was shown that calcium oscillations are associated with the progression of rhizobial infection events in the root cortex (Sieberer et al., 2012). Using the nuclear-localized cameleon, we observed that external NF application led to calcium oscillations in the root epidermis, but no oscillations in the inner root cortex (Figure 1A) where nodule organogenesis is initiated. The outermost cortical cells do occasionally show a slight calcium response, but the frequency of the oscillations is reduced. Induction of NIN expression by external NF application occurs in the same cells where calcium spiking was observed: epidermal cells, with a slight induction in the outermost cortical cells, but no induction in the inner root cortex (Figures 1B to 1D). By 24 h after rhizobial treatment, gene expression changes are activated in the inner root cortex (for instance, see Held et al., 2014), and we conclude that these cortical responses cannot be a direct function of calcium oscillations, since these, along with NIN expression, are restricted to the outer root tissues during early Nod factor signaling.
Figure 1.
NF Signaling Is Limited to the Root Epidermis.
(A) NF (10 nM) induced calcium oscillations, measured using the nuclear-targeted cameleon YC2.1, are mostly associated with epidermal cells, with a slight response in outer cortical cells. The vertical axis on the right shows the ratio of YFP to CFP (arbitrary units) for the cells indicated in the image on the left. No calcium oscillations are observed in inner cortical cells, even after long treatments (24 h).
(B) to (D) Nod factor (10 nM [B]; 100 pM [C] and [D]) induced NIN expression at 24 h after treatment, measured using the NIN promoter driving the expression of GUS. NIN expression is detected in epidermal cells (white arrow) and slightly in outer cortical cells (asterisk), but not in inner cortical cells (black arrow).
NIN Expression in the Root Epidermis Is Sufficient to Activate Cortical Cell Divisions
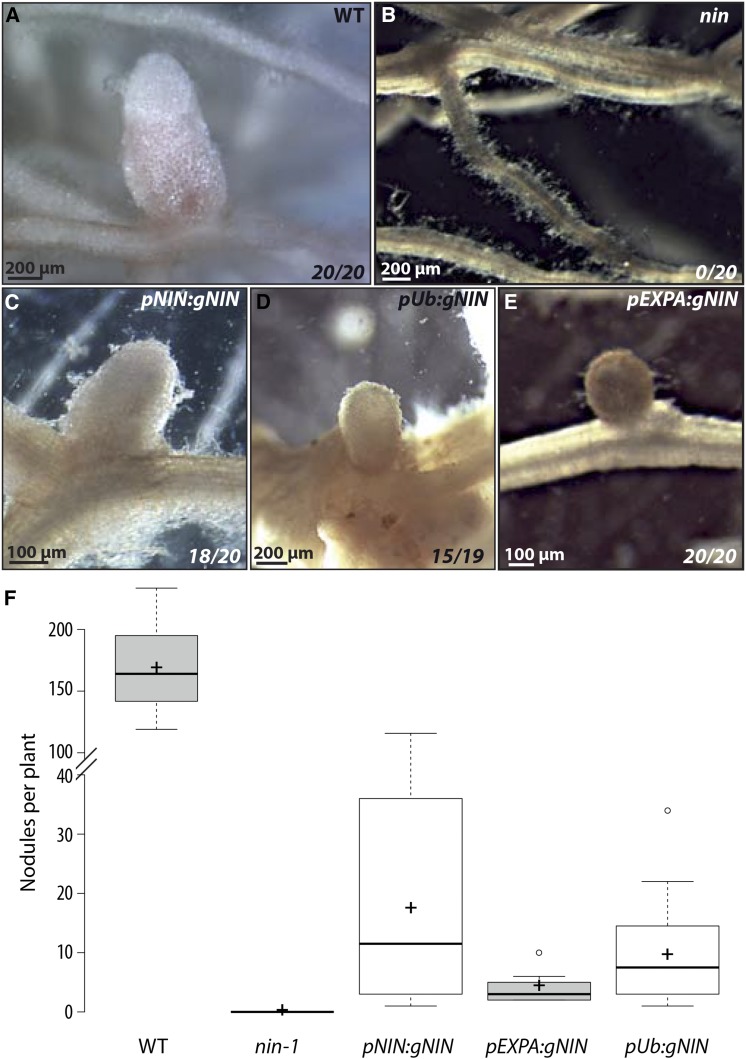
As NIN is initially expressed in the root epidermis (Figures 1B to 1D), we tested if this epidermal expression is sufficient to induce a cortical response. For this we used a root epidermal-specific promoter from M. truncatula, homologous to the Arabidopsis thaliana EXPANSIN A7 (EXPA) (Cho and Cosgrove, 2002; Kim et al., 2006). Expression from this promoter is restricted to the root epidermis of M. truncatula within the region of the root where nodulation responses occur, and this expression is not affected by NF treatment (Supplemental Figure 1). We assessed the ability of the genomic form of NIN expressed from the EXPA promoter to complement the nin-1 mutant. Complementation of nin-1 has proved challenging, with no promoters yet defined that allow full complementation of nin mutants leading to wild-type levels of N-fixing nodules. However, expression of NIN from its own promoter (used in the previous GUS assays; Figures 1B to 1D) and from the Lotus japonicus Ubiquitin promoter (Maekawa et al., 2008) led to some nodules in most nin-1 transgenic roots (Figure 2), although the nodules were less numerous than in wild-type plants transformed with the empty vector (Figure 2F), were generally smaller, and were presumably not fixing nitrogen (as indicated by white coloration; Figures 2A to 2D). This partial complementation with the native promoter and the L. japonicus Ubiquitin promoter has been previously observed (Soyano et al., 2014; Yoro et al., 2014), and while incomplete, it does enable the onset of a cortical response. NIN expressed from the EXPA promoter led to small nodule primordia in nin-1 transformed roots (Figure 2E), although these were observed only at late time points and at much lower frequency than nodulation in wild-type plants. As with the cases where NIN was expressed from the native and L. japonicas Ubiquitin promoters, nodules were white and therefore we presume not fixing nitrogen (Figure 2E). These results show that epidermal expression of NIN can induce a cortical program in response to Sinorhizobium meliloti, although at much lower efficiencies than when NIN is expressed in both the root epidermis and cortex, as in a wild-type plant.
Figure 2.
NIN Expression in the Root Epidermis Is Sufficient to Induce Nodule Formation.
(A) to (E) Wild-type (A) and nin-1 ([B] to [E]) M. truncatula roots were transformed with A. rhizogenes containing a control vector ([A] and [B]), pNIN:gNIN (C), pUb:gNIN (D), or pEXPA-gNIN (E) and inoculated with S. meliloti (50 d after inoculation). Numbers indicate the number of transformed plants showing nodules out of the total number of transformed plants.
(F) Nodules numbers at 50 DPI in wild-type (WT) and in nin-1 roots transformed with a control vector or pNIN:gNIN, pEXPA:gNIN, or pUb:gNIN. Only plants showing nodules are included in the analysis. Central lines show the medians, crosses show the averages, and the box delimits the 25th and 75th percentiles as determined by R. The whiskers extend 1.5 times the interquartile range from the 25th and 75th percentile, with outliers represented by dots.
The overexpression of NIN in the absence of rhizobia is sufficient to induce cortical cell divisions leading to spontaneous nodule-like structures (Soyano et al., 2013). To test if these cortical cell divisions are induced by NIN overexpression in the cortex and/or NIN overexpression in the epidermis, we placed NIN under the control of the EXPA promoter and the M. truncatula NRT1.3 promoter, which shows cortical-specific expression (Supplemental Figure 2A), and assessed for the spontaneous formation of nodule-like structures in the absence of S. meliloti. Spontaneous nodule-like structures were observed on most wild type plants overexpressing NIN from the L. japonicas Ubiquitin promoter (Supplemental Figure 2B; Table 1). When NIN was overexpressed only in the cortex (pNRT1.3:gNIN) of wild-type roots, approximately half of the plants showed spontaneous nodules (Supplemental Figure 2C; Table 1), and nodules were also observed in these lines when inoculated with S. meliloti (15/20 plants showed nodules, with an average of 2.5 nodules per plant). When NIN was overexpressed from the EXPA promoter in wild-type roots, we still observed spontaneous nodule-like structures, although the number of plants showing this response was reduced (Supplemental Figure 2D; Table 1), and the number of nodules was also reduced. RT-qPCR analysis showed that NIN expression levels from the EXPA promoter are higher than from the NRT1.3 promoter (Supplemental Figure 2E). These results show that NIN overexpression in wild-type roots in either the epidermis or the cortex is sufficient to induce cell divisions leading to nodule organogenesis. However, the scale of this response is reduced for epidermal expressed NIN, indicating that NIN function in the cortex is more effective when NIN is directly expressed there.
Table 1. Spontaneous Structures Induced by Tissue-Specific Expression of NIN in Different M. truncatula Mutants.
| pUb:gNIN | pNRT1.3:gNIN | pEXPA:gNIN | |
|---|---|---|---|
| A17 | +++ (18/21) | ++ (9/15) | + (6/48) |
| nin-1 | +++ (9/9) | +++ (10/12) | − (0/50) |
| cre1-1 | +++ (11/11) | +++ (4/5) | − (0/65) |
Numbers indicate number of plants showing spontaneous nodule-like structures per total number of plants. The (+) and (−) indicate the frequencies of plants showing nodules.
We were interested in the role of cytokinin signaling for NIN-induced spontaneous nodule-like structures when expressed in the root epidermis versus the root cortex. For this we expressed NIN from the EXPA and NRT1.3 promoters in the cre1-1 and nin-1 mutants. Constitutive expression of NIN from the L. japonicus Ubiquitin and cortical-specific expression of NIN from the NRT1.3 promoters led to spontaneous nodule-like structures in cre-1 and nin-1 mutants (Table 1), indicating that when NIN is expressed in the root cortex, it can promote nodulation independent of cytokinin signaling. In contrast when NIN was expressed from the EXPA promoter, we observed a dependence on CRE1 and NIN for activation of spontaneous nodule-like structures (Table 1). These results indicate that NIN epidermal expression requires cytokinin perception to activate nodule organogenesis, but cortical expression of NIN alone is sufficient for spontaneous cell divisions. This discrimination between the epidermal and cortical expression of NIN reveals mechanistic differences in their modes of induction of nodulation.
Rapid Activation of Cytokinin Signaling in the Root Cortex Is Dependent on NIN
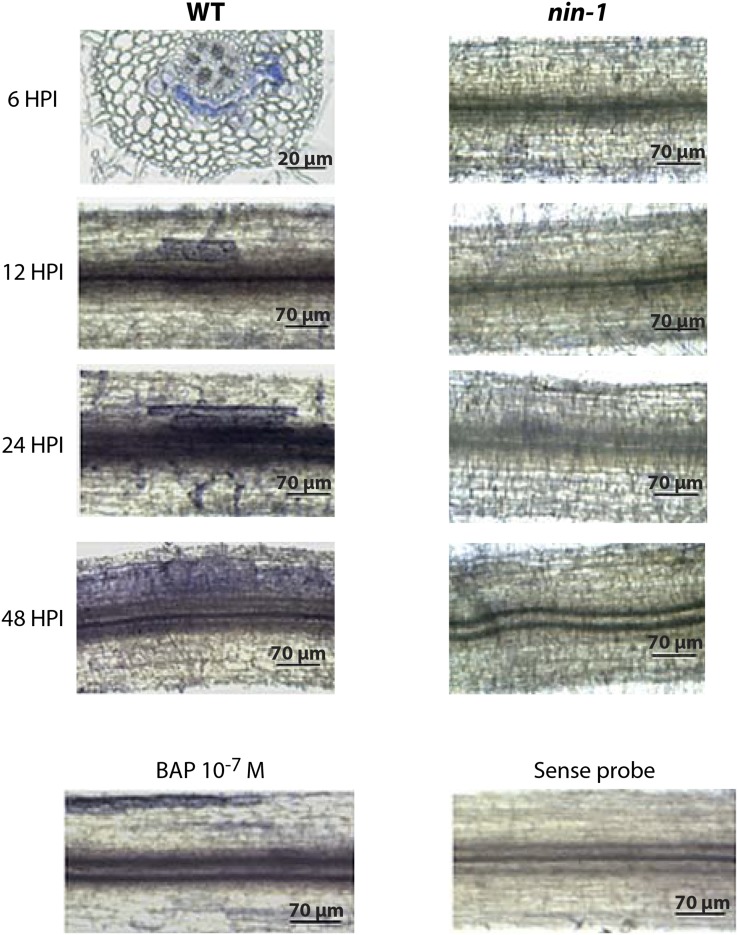
To further explore the role of NIN in cortical signaling processes, we checked by in situ hybridization Response Regulator 4 (RR4) mRNA localization in M. truncatula roots. To increase RR4 signal intensity, we used S. meliloti spot inoculation rather than NF application. Six hours after inoculation, RR4 was detected in the inner root cortical cells, and this is consistent with previous promoter-GUS analyses and with analysis of a cytokinin reporter (Plet et al., 2011; Held et al., 2014; van Zeijl et al., 2015). As time progressed, the region of RR4 expression expanded within the inner cortex, eventually filling a significant region of the entire root cortex 48 h after inoculation (Figure 3). This pattern of RR4 expression is consistent with a previous study using the TCS cytokinin reporter that revealed initial induction in the root cortex, with expansion from this region to fill a significant portion of the root undergoing rhizobial infection (Held et al., 2014). The induction of RR4 in the inner cortex requires NIN, even at the earliest time points, when NIN expression is restricted to the root epidermis. External application of the synthetic cytokinin benzylaminopurine (BAP) led to RR4 expression in the root epidermis and outer cortical cells, but not in the inner root cortex (Figure 3). NF induction of RR4 is dependent on NIN (Supplemental Table 1). Hence, it appears that early cytokinin signaling associated with nodulation is restricted to the inner root cortex and this is initiated at very early stages, within 6 h of rhizobial inoculation. At these early time points, no bacterial infection has yet been initiated (Timmers et al., 1999), and we propose that NF signaling will be restricted to the root epidermis.
Figure 3.
NIN-Dependent Expression of RR4 in Cortical Cell Layers.
Wild-type and nin-1 roots were spot inoculated with S. meliloti and harvested 6, 12, 24 and 48 h after inoculation (HPI) or treated with 10−7 M BAP for 6 h. Whole-mount in situ hybridization was conducted with the RR4 antisense probe (indicated by blue staining). No signal was detected with the sense probe. A transverse section is shown for wild-type plants at 6 h after inoculation, whereas only whole roots are shown for the other conditions.
NIN Binds to the CRE1 Promoter and Activates CRE1 Expression in the Cortex
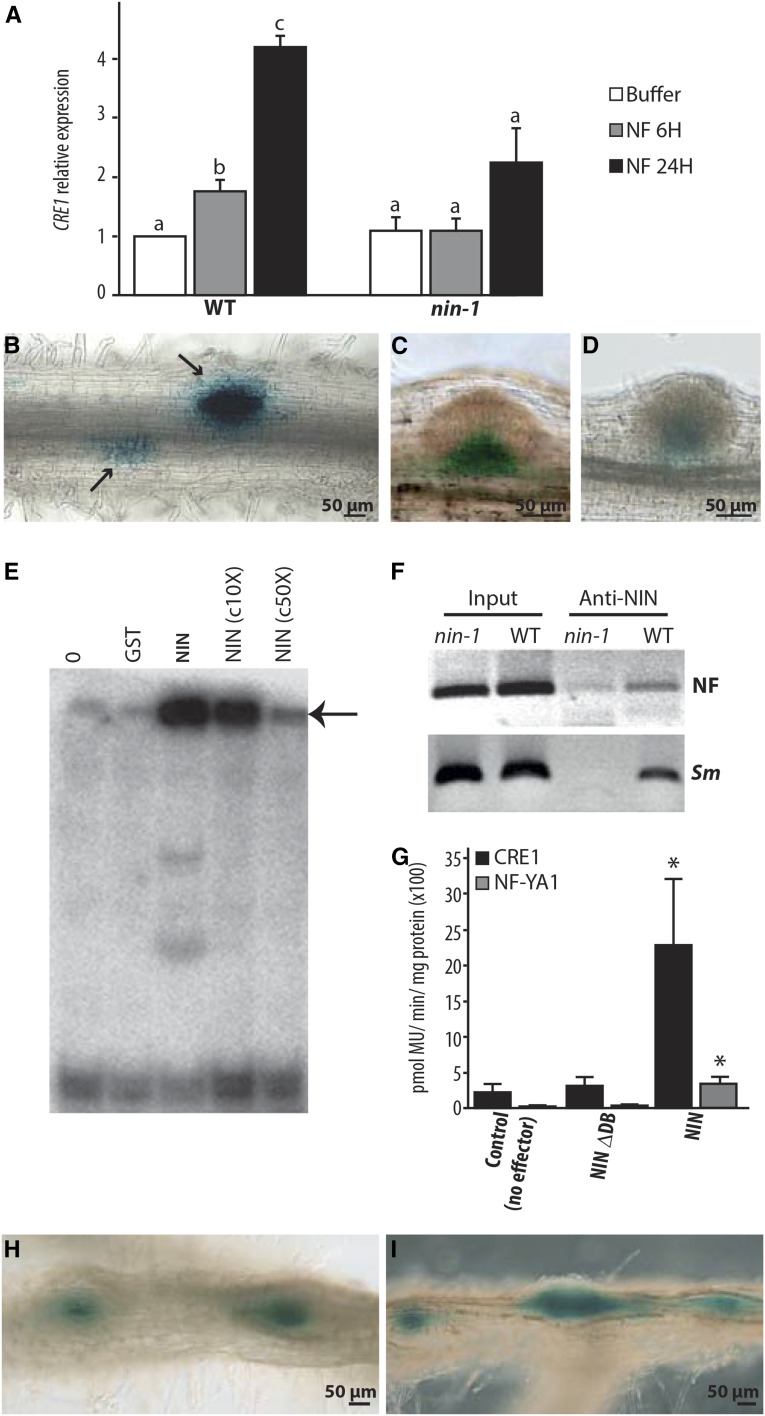
The cytokinin receptor CRE1 is essential for nodule organogenesis (Gonzalez-Rizzo et al., 2006; Plet et al., 2011), and it has been shown that this gene is upregulated during nodulation. CRE1 expression is induced by NF application, and this induction is dependent on NIN (Figure 4A). CRE1 expression has been shown to be associated with young nodule primordia (Lohar et al., 2006). We used a pCRE1:GUS construct (Lohar et al., 2006) to assess NIN-dependent CRE1 induction. The region of the CRE1 promoter that we used is sufficient to allow complementation of the cre1 mutant using a pCRE1:CRE1 construct (Supplemental Figure 3A). Strong GUS expression was detected in young dividing cortical cells, and no significant signal was detected in the root epidermis (Figures 4B and 4C; Supplemental Figures 3B, 3D, and 3E). This induction of CRE1 is dependent on NIN (Figure 4D; Supplemental Figure 3C).
Figure 4.
NIN Directly Regulates CRE1 Expression.
(A) CRE1 induction by NF requires NIN as evidenced by RT-qPCR analysis. CRE1 expression was normalized with ACTIN11. The bars represent the ratio relative to the buffer control. Error bars indicate sd for three biological replicates. Different letters indicate statistical differences as determined by pairwise t tests in each genotype (P < 0.05).
(B) to (D) CRE1 expression measured using the CRE1 promoter driving the expression of GUS in the wild type ([B] and [C]) and in the nin-1 mutant (D), inoculated with S. meliloti. In wild-type roots, CRE1 is specifically expressed in response to rhizobia in cortical cells (arrows), while nonsymbiotic expression of CRE-1 is observed in the base of lateral root primordia (C). In nin-1 (D), only the nonsymbiotic expression of CRE1 is found in the base of lateral root primordia.
(E) NIN binds a radiolabeled CRE1 promoter probe (−1487 to −971), causing its retardation, indicated with an arrow. c10X and c50X, unlabeled competitor DNA in 10- and 50-fold excess reduce this degree of binding. Lane 0, no proteins incubated with radiolabeled probe; lane GST, GST protein incubated with radiolabeled probe.
(F) In vivo association of NIN with the CRE1 promoter measured using ChIP. Wild-type and nin-1 roots were treated with 1 nM NF for 24 h or with S. meliloti (Sm).
(G) Transactivation studies in N. benthamiana cells transiently transformed with the pCRE1:GUS or pNF-YA1:GUS reporters and respective effectors. The asterisk indicates a statistically significant difference (P < 0.05, Student’s t test) compared with the reference. NIN-∆DB, NIN carrying an internal deletion that removes the DNA binding domain.
(H) and (I) CRE1:GUS expression observed in wild-type roots expressing pEXPA:gNIN (H) or pNRT1.3:gNIN (I). GUS expression (in blue) is localized in spontaneous nodule-like structures.
Epidermally expressed NIN requires CRE1 for induction of nodulation-like structures, and one possible scenario is that CRE1 itself may be a target of NIN. In vitro binding studies using the CRE1 promoter revealed direct binding by the NIN C terminus, which contains the predicted DNA binding domain (Figure 4E). To determine the optimal DNA sequence to which NIN binds, we performed random binding site selection that revealed a consensus binding sequence of AAG(A/C)T (Supplemental Figure 4A), and this is comparable to informatic studies used to identify the NIN cis-element (Soyano et al., 2013). The CRE1 promoter contains multiple potential NIN binding cis elements (Supplemental Figure 3F), and using a native NIN antibody (Supplemental Figure 4B), we found that NIN associates with the CRE1 promoter in vivo following NF or S. meliloti treatment (Figure 4F). The relevance of this binding was revealed in transactivation studies in Nicotiana benthamiana, where we observed that expression of NIN alone is sufficient to activate the CRE1 promoter (Figure 4G) and this transactivation is much stronger than NIN induction of NF-YA1, a gene previously proposed to be a direct target of NIN in the root cortex (Soyano et al., 2013). To assess how CRE1 expression was affected by constitutive NIN we cotransformed pEXPA:gNIN or pNRT1.3:gNIN and pCRE1:GUS into wild-type M. truncatula roots. CRE1 was induced in the spontaneous nodule-like structures observed in the root cortex even when NIN was overexpressed only in the epidermis (Figures 4H and 4I). Very rarely, CRE1 expression was also detected in the root epidermis (Supplemental Figure 3G).
NIN Binds to the ENOD11 Promoter and Negatively Regulates Its Transcription
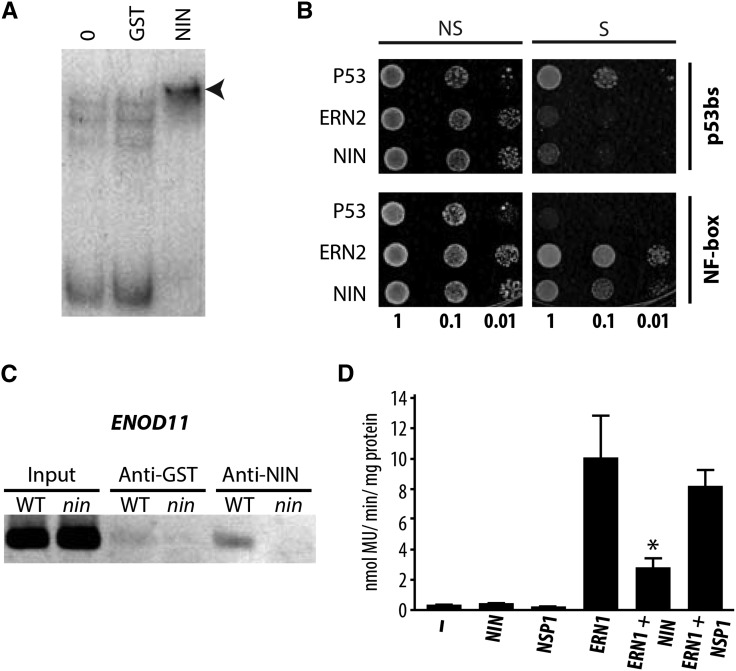
We previously observed that the spatial expression of ENOD11 in the root epidermis (Marsh et al., 2007) was greatly expanded in the nin-1 mutant, suggesting that NIN may negatively regulate ENOD11, in contrast to what we have observed for CRE1 and others have observed for NF-YA1/NF-YA2 (Soyano et al., 2013). Transcriptional profiling in the nin-1 mutant reveals that the majority of NIN-dependent changes in response to Nod factor at 24 h are genes that NIN suppresses (Supplemental Figure 5), implying that the negative regulation revealed by studies of ENOD11 is a significant function of NIN action. To investigate whether NIN could have a direct negative regulatory effect, we first showed that NIN can directly bind to the ENOD11 promoter (Figure 5A) and the mutation of the A1, A2, G3, or T5 nucleotides in the NIN binding site greatly reduced or abolished NIN binding (Supplemental Figure 4C). NF induction of the ENOD11 promoter is a function of the NF-responsive NF-box (Andriankaja et al., 2007), and using yeast one-hybrid analysis, we found that NIN binds to this region of the ENOD11 promoter (Figure 5B). In M. truncatula roots treated with S. meliloti, the NIN antiserum coprecipitated NIN and the ENOD11 promoter (Figure 5C).
Figure 5.
NIN Functions as a Negative Regulator That Binds the Nod Factor Box of the ENOD11 Promoter.
(A) NIN binds the ENOD11 promoter in vitro, evidenced by the retardation of migration (arrowhead) of the radiolabeled ENOD11 promoter probe (−1046 to +3, that contains the NF-box) following incubation with the DNA binding domain of NIN. Lane 0, no proteins incubated with radiolabeled probe; lane GST, GST protein incubated with the radiolabeled probe.
(B) The yeast YM4271 reporter strains carrying the NF-box in quadruplicate or the p53 cis-elements (p53bs) in triplicate were transformed with plasmids expressing the Gal4 Activating Domain (AD)-ERN2, AD-NIN, and the mouse AD-p53 factor that interacts with the p53 binding site. Growth of yeast in selective media (S) indicates specific DNA-protein interaction. OD 1, 0.1, and 0.01 are indicated; NS, nonselective medium; S, selective medium.
(C) ChIP of the ENOD11 promoter using anti-NIN antibodies in wild-type and nin-1 roots inoculated with S. meliloti indicating NIN binding to the ENOD11 promoter, within the region of the NF-box.
(D) Transactivation studies in N. benthamiana cells transiently transformed with the 4xNF-box:GUS reporter and respective HA-tagged transcription factors. The asterisk indicates a statistically significant difference (P < 0.05, Student’s t test) compared with ERN1 alone. Note that the GRAS transcription factor NSP1 does not significantly affect the ERN1-mediated transcription of the NF-box reporter.
The NIN binding site within the NF-box is only 2 bp away from a GCC-like motif essential for NF induction (Andriankaja et al., 2007). The GCC motif is recognized by the transcription factors ERN1 and ERN2, which are positive regulators of NF gene expression (Andriankaja et al., 2007; Middleton et al., 2007); thus, competitive binding to the NF-box is a possible mechanism for NIN suppression of ENOD11 expression. ERN1 is sufficient to activate ENOD11 in M. truncatula (Cerri et al., 2012), and in a heterologous system, it drives the expression of a synthetic promoter with four tandem copies of the NF-box (Andriankaja et al., 2007). To test for NIN interference of ERN1 action, we cotransformed N. benthamiana leaves with NIN, ERN1, NSP1 (used as a negative control), and the synthetic 4xNF-box promoter driving the expression of GUS. We found significant NIN suppression of ERN1 induction of the synthetic NF-responsive promoter (Figure 5D), revealing a possible mechanism for NIN suppression of epidermal ENOD11 expression.
DISCUSSION
It is becoming increasingly clear that NIN is a central regulator of nodulation. It plays essential roles in both the root epidermis and in the root cortex, where it is necessary for the initiation of bacterial infection and promotion of nodule organogenesis, respectively (Schauser et al., 1999; Marsh et al., 2007; Soyano et al., 2013). In addition, NIN also acts as a negative regulator, inhibiting additional NF responses after the initial activation of this signaling pathway in the root epidermis and promoting autoregulation of nodulation that limits the final number of nodules that form (Marsh et al., 2007; Soyano et al., 2014; Yoro et al., 2014).
External application of NF is only able to promote NIN symbiotic expression in the root epidermis and to a lesser extent in the outer root cortex, and this is consistent with the sites of calcium spiking. From this we infer that NF is unable to diffuse into the root tissue and that symbiosis signaling is cell autonomous in agreement with previous calcium studies (Miwa et al., 2006; Sieberer et al., 2009). It has been shown that root cortical cells have the capability to activate symbiotic calcium signaling, but this only occurs with the progression of infection threads (Sieberer et al., 2012), delivering bacteria and presumably NF into the root cortex. At the earliest stages of this symbiotic association, we propose that NF signal transduction is restricted to outer root tissues (epidermis and outer cortex), and this is consistent with the expression patterns of genes such as NIN, ENOD11, ERN1, and ERN2 (Schauser et al., 1999; Journet et al., 2001; Charron et al., 2004; Heckmann et al., 2011; Cerri et al., 2012). Interestingly, the upstream components of the symbiosis signaling pathway are only required in the root epidermis (Hayashi et al., 2014), implying that this epidermal induction of the pathway is sufficient to promote cortical processes associated with infection and nodule organogenesis.
The induction of NIN expression is a downstream response of NF signaling, being induced by phosphorylated CYCLOPS, a transcription factor that sits within a complex with the calcium decoder of symbiosis signaling (Yano et al., 2008; Horváth et al., 2011; Singh et al., 2014). NIN appears to act within a negative feedback loop that suppresses further outputs from NF signaling, within 24 h of initial signaling (Marsh et al., 2007; Yoro et al., 2014). The impact of this is a temporal restriction in the response to NF: In a nin mutant, pENOD11-GUS induction continues into the newly growing root zone, whereas in wild-type plants, the response remains restricted to the initial region of the root where the first response occurred (Marsh et al., 2007). NF induction of ENOD11 appears to be the function of the ERF transcription factors ERN1 and ERN2 (Andriankaja et al., 2007; Middleton et al., 2007), and we show that NIN can competitively inhibit ERN action for induction of the NF-box within the ENOD11 promoter. In addition to this effect on ENOD11 expression, nin mutants also show excessive root hair curling in response to NF that extends well beyond the initial responsive region of the root (Schauser et al., 1999). This temporal restriction to the root hair response is similar to that observed with ENOD11, and perhaps there are comparable modes of action between NIN restriction of gene expression and restriction of root hair curling.
In the root epidermis, NIN is also necessary for the onset of bacterial infection (Fournier et al., 2015), and this is consistent with its requirement for NPL induction, a gene associated with the formation of infection threads (Xie et al., 2012). NIN is also strongly associated with the initiation of nodule organogenesis in the root cortex (Schauser et al., 1999; Plet et al., 2011). We have shown that cortical expression of NIN is sufficient to autoactivate nodulation, and this occurs independently of the nodulation-associated cytokinin receptor CRE1. It has already been shown that cytokinin signaling promotes NIN induction in the root cortex, and in the absence of this cortical promotion of NIN, no nodule organogenesis occurs (Gonzalez-Rizzo et al., 2006; Heckmann et al., 2011; Plet et al., 2011). Hence, the predominant effect of symbiotic-associated cytokinin signaling in the root cortex is the promotion of NIN expression, and once induced in these cells, NIN can activate nodule organogenesis, in part through the promotion of NF-YA1 and NF-YA2 expression (Soyano et al., 2013).
We have shown that the cytokinin response occurring in the root cortex is induced very rapidly upon rhizobial inoculation. We observed RR4 expression in inner root cortical cells within 6 h of rhizobial inoculation, and this is consistent with previous studies (Plet et al., 2011; Held et al., 2014; van Zeijl et al., 2015). At this time point, NF signaling, associated with calcium oscillations, is restricted to the root epidermis and NIN induction is also restricted to the root epidermis. Despite this, the cytokinin responses in the root cortex are dependent on NIN. We found that the expression of NIN in the root epidermis was sufficient to promote some nodule organogenesis in the root cortex with or without the addition of rhizobia. There must be a mobile signal that moves between the epidermis and the root cortex (Hayashi et al., 2014; Held et al., 2014; van Zeijl et al., 2015), and this signal must either be NIN itself or a product of NIN action. Despite much effort, we have been unable to detect full-length NIN-GFP fusions in M. truncatula roots; therefore, it is impossible to state whether NIN is mobile. It was recently proposed that cytokinin itself may act as the mobile signal, since its production is enhanced in the root cortex very rapidly after the perception of NF (van Zeijl et al., 2015). However, Held et al. (2014) only observed cytokinin signaling in the epidermis after the initiation of cell divisions in the cortex. If cytokinin were the mobile signal, then one would expect to see evidence for cytokinin at first in the epidermis and secondarily in the root cortex. We believe that this points at an alternative mobile signal being the more likely explanation.
Interestingly, the promotion of nodule organogenesis by epidermal-expressed NIN required CRE1 and the wild-type NIN. The best explanation for this is a dependence on NIN expression in cortical cells, acting as a positive feedback mechanism, to promote nodule organogenesis, and this is consistent with previous work (Plet et al., 2011; Soyano et al., 2013). One inconsistency in our results is the fact that the nin-1 mutant complementation with pEXPA-NIN, with secondary treatment with S. meliloti, did result in a few small nodules. Perhaps the S. meliloti infection promotes the movement of NIN or a product of NIN action, from the epidermis to the cortex, allowing a few small nodules to form. The dependence on CRE1 suggests that epidermal expression of NIN is sufficient to activate cytokinin signaling in the root cortex and this in turn induces NIN expression in the root cortex that promotes nodule organogenesis. We found that NIN is necessary and sufficient to activate the expression of CRE1 that is induced in the root cortex at early stages in this interaction (Lohar et al., 2006). One exciting possibility is that NIN acts as a mobile signal moving from the root epidermis into the root cortex where it activates cytokinin signaling through the promotion of CRE1 expression. However, we cannot discriminate this hypothesis from alternative explanations; for instance, NIN induction in the root cortex could lead to the induction of CRE1 as a positive feedback mechanism. Furthermore, the absence of spontaneous nodule induction in nin-1 by expression of pEXPA:NIN could be interpreted as disproving that NIN acts as the mobile signal. We viewed this result differently: that cortical expression of NIN is required to sustain nodule organogenesis. Clearly, further work is necessary to define the nature of the elusive mobile signal.
In an attempt to rationalize our findings, we propose a model for NIN action (Figure 6). Initial induction of NIN in the root epidermis following perception of NF promotes the early stages required for subsequent bacterial infection (Madsen et al., 2010; Xie et al., 2012; Fournier et al., 2015). Later on, NIN suppresses further NF-associated signaling (Marsh et al., 2007), and this is likely the result of NIN induction of CLE peptides (Soyano et al., 2014) and the direct action of NIN negatively regulating promoters controlled by ERN1 and ERN2 (Andriankaja et al., 2007). Expression of NIN in the epidermis is also sufficient to promote cytokinin signaling in the root cortex; however, the mechanism by which this occurs remains unclear. Cytokinin signaling promotes the expression of NIN in the root cortex (Gonzalez-Rizzo et al., 2006; Plet et al., 2011), and when sufficient levels of NIN accumulate in cortical cells, nodule organogenesis is activated. Hence, it would appear that NIN expression in the root cortex is both necessary and sufficient for nodule organogenesis. NIN expression in the root epidermis can promote nodule organogenesis, but only via the intermediaries, cytokinin signaling in the root cortex, and an as yet undefined mobile signal linking the epidermal responses to those in the cortex. NIN induction of CRE1 expression could be associated with the early establishment of the nodule program, or it could act within a positive feedback loop within the root cortex cementing the commitment to nodule production. It is interesting to note that even though NIN alone is sufficient to promote CRE1 induction in a heterologous system, when NIN was expressed throughout the root tissue, CRE1 induction was only observed in punctate regions of the root cortex associated with nodule organogenesis. This points to a more complex modality of CRE1 induction that transactivation studies suggest.
Figure 6.
A Model of NIN Action.
(1) NIN expression is induced in epidermal cells in response to NF. Within the epidermis, NIN promotes NPL expression (Xie et al., 2012) and represses ENOD11 expression by competing with ERN1. NIN promotes movement of a signal from the epidermis to the cortex that initiates cytokinin signaling in the cortex. (2) Cytokinin signaling promotes NIN expression in cortical cells. (3) NIN and cytokinin signaling act within a positive feedback loop, with NIN promoting CRE1 expression and cytokinin signaling promoting NIN expression, expanding the degree of nodulation responses within the root cortex. (4) High levels of NIN in the cortex drive nodule organogenesis through promotion of NF-YA1 and NF-YA2 (Soyano et al., 2013). In the cortex, NIN activates the expression of CLE peptides (Soyano et al., 2014) implicated in the negative regulation of nodulation. Cells expressing NIN are shown in blue. Full lines indicate direct targets of transcription factors. Dotted arrows indicate indirect targets.
There is one area of disagreement between the model stated above and evidence from the literature. The L. japonicus nena and symrk-14 mutants both show nodulation, but in the apparent absence of epidermal induction of NIN (Groth et al., 2010; Kosuta et al., 2011), suggesting that nodule organogenesis in the root cortex can be activated in the absence of epidermal NIN expression, and clearly inconsistent with the model we have proposed. This inconsistency may reflect differences between determinate and indeterminate nodulation processes in L. japonicus and M. truncatula, or it may reflect a more important inconsistency that highlights a current gap in our knowledge.
Our work and the recent work of others have started to differentiate the diverse and complex roles of NIN during nodulation. This protein appears to be the central regulator of this response, activating both bacterial infection and nodule organogenesis, but also regulating the degree of the response to the nitrogen-fixing bacteria and ultimately the numbers of nodules that arise. For such a complex functionality, it is important to discriminate the tissue-specific modes of action of this protein, and our work has begun to differentiate the roles of NIN in the different tissues of the root. It is clear that the epidermal and cortical responses must be tightly coordinated and NIN is clearly involved in this coordination. But whether it is NIN alone or a product of NIN action remains to be shown.
METHODS
NIN, CRE1, and EXPA-Promoter GUS Analyses
To generate the pNIN:GUS construct, a 2.18-kb fragment was amplified from the CR936325.2 genomic BAC sequence (primers in Supplemental Table 2). The NIN promoter was subcloned into the pGEM-T vector (Promega) and then inserted into the pKWGFS7 vector (Karimi et al., 2002) using the Gateway cloning system (Invitrogen). The pCRE1:GUS construct was provided by K. Vandenbosch (Lohar et al., 2006), and the pEXPA:GFP-GUS construct is described in this article (see below). Agrobacterium rhizogenes (ARqua1) was used for plant transformation of the fusion constructs in Medicago truncatula roots (Boisson-Dernier et al., 2001). One-month-old transformed roots were inoculated with S. meliloti RCR2011 pXLGD4 (GMI6526) (OD 0.02) or treated with NF (10 nM and/or 100 pM) or liquid buffered nodulation medium (BNM) (Ehrhardt et al., 1992). Histochemical GUS staining was performed essentially as described previously (Boisson-Dernier et al., 2001). Root sections (100- to 150-µm thick) were prepared in 4% agarose with a vibrating microtome (Vibratome 1000 plus) and observed with a Leica DMR microscope.
Calcium Spiking Analysis
After stratification at 4°C for 4 d, seeds were germinated overnight at room temperature. Seedlings were grown on BNM plates for 24 h before treatment with NFs. Seedling roots were fixed into a small chamber made on a cover glass using vacuum grease. The chamber was filled with 500 mL of BNM buffer, and the roots were treated with 10 nM NF. For the time-lapse confocal scanning images, the fluorescence was measured with a Zeiss LSM 510 Meta confocal scanning microscope equipped with a 40× EC Plan-Neofluor 1.3 oil immersion objective (Zeiss). The nuclear-targeted cameleon calcium sensor (Sieberer et al., 2009) was excited with the argon ion 458-nm laser and imaged using emission filters 476 to 486 nm for CFP and 529 to 540 nm for YFP. Images were acquired at 5-s intervals, and the scanning resolution was 512 × 512 pixels.
Electrophoretic Mobility Shift Assay
The cDNA region coding for the DNA binding domain (residues 582 to 700) of NIN was amplified by PCR with gene-specific primers (Supplemental Table 2). The amplified products were gel-purified and inserted into a pBAD-DEST49 vector using a Gateway cloning system (Invitrogen). The recombinant proteins were produced in Escherichia coli and purified using ProBond resins (Invitrogen). The amount of purified proteins was estimated by the Bradford method using a protein assay kit (Bio-Rad). Oligonucleotide probes were labeled with [γ-32P]ATP using T4 nucleotide kinase (Invitrogen) or [α-32P]dTTP using Klenow fragment (Invitrogen). The binding reactions were performed in 20 μL binding buffer (25 mM HEPES-KOH, pH 8.0, 50 mM KCl, 1mM DTT, 0.05% Triton X-100, and 15% glycerol). After 20 min incubation at room temperature, the reactions were resolved by 6 to 8% native polyacrylamide gels with 0.5× TBE buffer.
RT-qPCR
Total RNA was extracted from roots using the RNeasy kits (Qiagen) following the manufacturer's protocol. RNAs were treated with Turbo DNA-Free (Ambion), and 1 μg RNA was reverse transcribed using Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Real-time RT-qPCR was performed using SYBR Green to monitor double-stranded DNA synthesis. Reactions were performed in a 96-well plate using the SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). Primers used were as follows: ACTIN11 (Gonzalez-Rizzo et al., 2006), CRE1 (Gonzalez-Rizzo et al., 2006), NIN (Gonzalez-Rizzo et al., 2006), and EF1 (Vernié et al., 2008).
Random Binding Site Selection
The random binding site selection was performed by mixing 100 ng of the double-stranded random oligonucleotides (420 possible combinations) with 200 ng of the recombinant proteins attached to 40 μL of ProBond resins (Invitrogen). The supernatant was removed, and the pellet was resuspended in 50 μL of water, boiled for 3 min, and centrifuged rapidly. The supernatant (5 µL) was used as the template for a PCR. The PCR product was purified from 2% agarose gel. The procedure was repeated 10 times. After the last PCR amplification step, the PCR products were ligated to a T/A cloning vector (Promega), and individual selected clones were sequenced.
Yeast One-Hybrid Assay
NIN (582 to 933 amino acids; 1059 bp) was amplified by PCR from M. truncatula cDNA with gene-specific primers (Supplemental Table 2). The amplified product was gel-purified and inserted into the Gateway donor vector pDONR207 (Invitrogen) by BP recombination reaction, sequenced, and then recombined into the Gateway destination vector pGADT7 by LR reaction, creating AD-NIN (582-933). YM4721 reporter strains carrying tetramer NF-box (Andriankaja et al., 2007) and trimer p53 cis-sequences (p53bs; Matchmaker one-hybrid system; Clontech) were transformed with plasmids expressing AD-ERN2 (Andriankaja et al., 2007), AD-NIN (582 to 933 amino acids), and the mouse AD-p53 factor (Matchmaker one-hybrid system) that interacts with the p53 binding site. Serial dilutions of transformed yeast cells were spotted onto nonselective SD Leu- (L-) or selective SD Leu-His- conditions on media supplemented with 5 mM 3-amino-1,2,4-triazole and incubated at 28°C for 3 d.
Chromatin Immunoprecipitation
About 5 g of fresh root tissues from 10-d-old seedlings were used for chromatin immunoprecipitation (ChIP) with the anti-NIN antibodies. The anti-NIN polyclonal antibodies were raised in rabbit by Eurogentec (http://www.eurogentec.be) against a NIN peptide (CRQHGITRWPSRK). The activity of the anti-NIN antibody was tested by immunoprecipitation in the wild type and nin-1. Nuclei were isolated as described previously (Delaney et al., 2006). Purified nuclei were fixed with 1% formaldehyde for 20 min immediately after extraction. The immunoprecipitation of purified chromatin was performed using a ChIP assay kit (Upstate) according to the manufacturer’s instructions. The ChIP products were used to detect the ENOD11 or CRE1 promoter by PCR analyses (primers in Supplemental Table 2). PCR products were resolved on 2% agarose gel.
Transient Expression in Nicotiana benthamiana Leaves and Fluorimetric GUS Assay
Full-length NIN (3130 bp) and coding sequence (CDS) (2802 bp) were amplified by PCR from M. truncatula genomic DNA and cDNA with gene-specific primers (Supplemental Table 2). The amplified products were gel-purified and inserted into the Gateway donor vector pDONR207 (Invitrogen) by BP recombination reaction, creating pENTRY gNIN and pENTRY NIN CDS. pENTRY NIN CDS was used to create a deleted version of NIN CDS without the DNA binding domain (deletion from 802 to 1495 bp, corresponding to a protein of 285 amino acids). Both pENTRYs were then used for recombination into the Gateway destination vector PAM-PAT35S-3xHA-GTW (Andriankaja et al., 2007) by LR reaction. Cultures of Agrobacterium tumefaciens strains GV3101 or GV3103 harboring the reporters: tetramer NF-box:GUS (Andriankaja et al., 2007), pCRE1:GUS (Lohar et al., 2006), or pNF-YA1:GUS (Laporte et al., 2014) combined or not with the effector constructs 35S-3xHA-ERN1 (Cerri et al., 2012), 35S-3xHA-NIN (1 to 933 amino acids), 35S-3xHA-NIN∆DB (285 amino acids), or 35S-3xHA-NSP1 (Cerri et al., 2012), were infiltrated into leaves of N. benthamiana plants as described previously (Andriankaja et al., 2007). Nine to twelve leaf discs were collected 36 h after inoculation in three independent replicates, frozen in liquid nitrogen, and used for total protein extraction in 1× GUS buffer (50 mM sodium phosphate, pH 7.5, 10 mM 2-mercaptoethanol, 10 mM Na2EDTA, 0.1% Triton X-100, 0.1% sodium lauryl-sarcosine). GUS activities were measured fluorimetrically using 1 µg of total protein extract as described previously (Andriankaja et al., 2007).
Plasmid Constructions for Tissue-Specific Expression and Complementation Assays
Publically available Arabidopsis thaliana root cell-type-specific gene expression data (Birnbaum et al., 2003; Nawy et al., 2005; Gifford et al., 2008) were analyzed to identify the most highly cell-type-specific Arabidopsis genes. From this, the putative M. truncatula orthologs of the top hits were computed by comparing Arabidopsis TAIR10 and M. truncatula Mt3.5 protein sequences in a reciprocal best BLAST hit analysis using a custom BLASTp script. The script first identifies the best BLASTp hit for protein(a-x) of species(a) in the proteome of species(b), then uses this protein sequence (protein b-x) as the basis for a BLASTp search in species(a). If this best hit returns protein a-x (i.e., the protein started with), then a reciprocal best BLAST hit has been found and a predicted orthology between the two proteins/genes is assigned. The reciprocal best BLAST hit of the Arabidopsis At3g21670 gene (nitrate transporter NTP3) was identified to be the M. truncatula gene Medtr5g085850, the dual-affinity nitrate transporter NRT1.3 (Morère-Le Paven et al., 2011). pEXPA (401 bp before ATG, DQ899790; Kim et al., 2006) and pNRT1.3 (858 bp before ATG, Medtr5g085850.1) were amplified by PCR from M. truncatula genomic DNA with gene-specific primers (Supplemental Table 2). The amplified products were gel-purified and inserted into the Gateway donor vector pDONR207 (Invitrogen) by BP recombination reactions, sequenced, and then recombined into the Gateway destination vectors pKGWFS7,0 (Karimi et al., 2002) and pBGWFS7 (Karimi et al., 2002) by LR reactions, creating pEXPA:GFP-GUS and pNRT1:GFP. pEXPA, pNRT1.3, and pLjUb (553 bp from ATG; Maekawa et al., 2008) were cloned by restriction into pK7WG2-R (Ding et al., 2008) between HindIII and SpeI sites (replacing 35S promoter), creating pK7WG2-R-pEXPA, pK7WG2-R-pNRT1.3, and pK7WG2-R-pUb. pENTRY gNIN was used for recombination into the Gateway destination vectors pK7WG2-R-pEXPA, pK7WG2-R-pNRT1.3, and pK7WG2-R-pUb to create pEXPA:gNIN, pNRT1.3:gNIN, and pUb:gNIN by LR reactions. pNIN (2180 bp; this article) and pCRE1 (2490 bp; Lohar et al., 2006) were cloned by restriction into pK7WG2-R (Ding et al., 2008) between HindIII and SpeI sites (replacing 35S promoter), creating pK7WG2-R-pNIN and pK7WG2-R-pCRE1. CRE1 CDS (3012 bp) was amplified by PCR from M. truncatula cDNA with gene-specific primers. The amplified product was gel-purified and inserted into the Gateway donor vector pDONR207 (Invitrogen) by BP recombination reaction, creating pENTRY CRE1. pENTRY gNIN (this article) and pENTRY CRE1 were used for recombination into the Gateway destination vectors pK7WG2-R-pNIN and pK7WG2-R-pCRE1 by LR reaction to create pNIN:gNIN and pCRE1:CRE1, respectively.
Plant Material Used for Complementation and Spontaneous Nodulation Assays
Seeds of M. truncatula cv Jemalong A17, nin1-1 (Marsh et al., 2007), and cre1-1 (Plet et al., 2011) were surface-sterilized and placed on inverted agar plates in the dark for 3 d at 8°C and 1 d at 20°C. Seedlings were transformed with A. rhizogenes strain ARqua1 carrying the appropriate binary vector using standard protocols (Boisson-Dernier et al., 2001). Three weeks after transformation, plantlets were screened for positive DsRed expression and transferred to a 1:1 mix of sterilized Terragreen/sand (Oil-Dri Company). For complementation assays, plantlets were inoculated with Sinorhizobium meliloti RCR2011 pXLGD4 (GMI6526) (OD 0.02) and scored 50 d later. Lac Z staining was done as described by Vernié et al. (2008). For spontaneous nodulation assays, plantlets were kept under sterile conditions in small glasshouses, watered alternatively with sterile water and liquid BNM, and scored 11 weeks later (15 weeks old). Control vector used in all A. rhizogenes assays corresponds to pK7WG2-R. An average of 20 independent transformed plants was scored per construct, per phenotype. These transformed plants were generated from at least two independent transformation experiments. For RT-qPCR analyses on NIN tissue-specific expression, plantlets were transferred on BNM and 100 nM aminoethoxyvinylglycine with pouch paper for a week. Plantlets were then harvested and let in liquid BNM for 1, 6, 24, and 48 h before to be frozen. Three independent replicates were done per time points with at least five plants per time point. The 12 samples were analyzed in RT-qPCR. For cotransformation, roots were cotransformed with pCRE1:GUS and pEXPA:gNIN/pNRT1.3:gNIN using a mixed suspension (D0 1) of A. rhizogenes strains harboring respective constructs. Composite plants grown on 25 mg/mL kanamycin/Fahraeus medium were selected on the basis of the fluorescent DSRED marker, comprised in the T-DNA of the pEXPA:gNIN/pNRT1.3:gNIN vectors. DSRED+ transgenic roots were harvested and used for histochemical (blue) staining for GUS activity as described by Cerri et al. (2012), before stereomicroscope (Leica Microsystems) and light microscope (Axioplan 2 Imaging; Carl Zeiss) observations. Thirty cotransformed roots were analyzed for both combinations.
Whole-Mount in Situ Hybridization
The general protocol is derived from Drea et al. (2005). For RNA probe preparation, a 470-bp fragment derived from the 3′ end of the Mt-RR4 cDNA sequence was amplified and inserted into the pGEM-T (Promega) plasmid (primers in Supplemental Table 2). SP6 and T7 sites of the pGEM-T easy vector were used to make antisense and sense riboprobes. Probes were prepared by in vitro transcription. PCR was performed as follow: 94°C for 3 min, then 30 cycles of 94°C for 45 s, 63°C for 45 s, and 72°C for 1.5 min, with a final extension reaction at 72°C for 6 min. Two microliters of all products were checked on agarose gels. In vitro transcription, using 2 μL of the PCR product in 10-μL reaction mix, was performed for 2 h at 37°C in the presence of digoxigenin-UTP nucleotides (0.35 mM). The remaining DNA was removed by the mixed solution of 75 μL 1× MS (10 mM Tris-HCl, pH 7.5, 10mM MgCl2 buffer, and 50 mM NaCl), 2 μL tRNA (100 μg/μL), and 1 μL DNase (RNase-free). Samples were centrifuged, resuspended, and incubated at 37°C for 10 min. Hydrolysis was performed in 100 mM carbonate buffer, pH 10.2, at 60°C for 40 min, and the products were precipitated with 10% acetic acid, 3 M sodium acetate, and 3 volumes of ice-cold ethanol 2H (or overnight) at −20°C. The product was pelleted by centrifugation at 4000 rpm for 30 min, and the pellets were resuspended in 30 μL of TE buffer. The probe was diluted into water by 1:100. Probe label incorporation was checked by dot-blotting (Drea et al., 2005) on 1 μL of the diluted probe. For hybridization, 2 μL of probe, 2 μL of water, and 4 μL of deionized formamide were mixed together and heated at 80°C for 2 min. Followed by a few-minute cool down on ice, 32 μL of hybridization solution (Drea et al., 2005) was added. This mix solution was stored at –20°C until use.
For tissue fixation, hybridization, and washes, wild-type and nin-1 plants were grown on BNM for 5 d. The susceptible region of the roots, corresponding to NF-induced ENOD11 expression, was chosen for the spot inoculation with 10−7 M BAP, BNM, or S. meliloti 1021 at OD 0.1. The spot inoculation regions were taken 6, 12, 24, and 48 h after inoculation for the whole-mount in situ hybridization with antisense and sense RR4 probes. M. truncatula roots were softened by pretreatment with acetone for 30 s, covered with paraformaldehyde (4% w/v) in PBS (10× PBS is 1.3 M NaCl, 0.07 M Na2HPO4, and 0.03 M NaH2PO4, pH 6.5 to 7.0) for 15 min, then transferred to 50-mL falcon tubes with paraformaldehyde (4%) in PBS and a vacuum applied for 2 min. New 4% paraformaldehyde was exchanged and the samples were left overnight at 4°C. Before the in situ hybridization, the root tissues were cleared and dehydrated through an ethanol series as follows 25% ethanol for 2 h, 50% ethanol for 2 h, 75% ethanol for 2 h, 100% ethanol for 2 h, 75% ethanol for 2 h, and 75% stored overnight at 4°C. Prior to hybridization, the prepared root samples were treated with 25% ethanol for 2 h at room temperature and separated into 25-well square plastic plates, followed by a pre-in situ washing step as follows: 2× PBS for 30 min, 1× proteinase K (400 µg/mL) for 10 min at 37°C, 1× glycine for 10 min, 2× BS for 15 min, 1× acetic anhydride for 30 min, 1× PBS for 15 min, and 2× PBS + Tween 20 0.1% for 15 min. The probe in the hybridization solution was used to cover the root samples with overnight incubation at 50°C. The overnight incubated samples were taken out the next day for the stringent washing step as follows: 2× SSC/50% formamide, 0.1% Tween 20 (20× SSC is 3 M NaCl and 0.3 M trisodium citrate) for 15 min at 50°C, then in the same solution for 60 min at 50°C and 15 min at 50°C, 2× SSC, 0.1% Tween 20 for 15 min at 50°C, 0.2× SSC, 0.1% Tween 20 for 15 min at 50°C, 1× TBS, and 0.1% Tween 20 for 10 min at room temperature three times. Afterwards, the washed samples were labeled by antidigoxigenin as follows: 1× TBS, 0.5% (w/v) blocking reagent (Roche), 0.1% Tween 20 for 30 min at room temperature twice, 1× TBS, 1% BSA (w/v), 0.1% Tween 20 for 30 min at room temperature, 1× TBS, 1% BSA (w/v), 0.1% Tween 20, with 1:3000 dilution of antidigoxigenin-alkaline phosphatase for 30 min at room temperature, and stored at 4°C overnight. The antibody-labeled samples were carried through a series of six washing steps of 20 min in 1× TBS and 0.1% Tween 20 with a final step of 20 min in alkaline phosphatase buffer (100 mM Tris-HCl, 100 mM NaCl, and 50 mM MgCl2, pH 9.5). Then, the color reaction was developed in alkaline phosphatase buffer containing nitroblue tetrazolium (0.15 mg/mL) and 5-bromo-4-chloro-3-indolyl phosphate-p-toluidine salt (0.075 mg/mL) at room temperature for generally 4 h (or longer depending on probe quality). Water was used to stop the reaction, followed by sequential washings in 70, 95, 100, 95, and 70% ethanol to clear the background. The samples were stored at 4°C in water or 65% ethanol. Transverse sections (10 µm thick) were conducted after the whole-mount in situ hybridization.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Mt4.0v1 databases under the following accession numbers: NIN (Medtr5g099060), CRE1 (Medtr8g106150), NSP1 (AJ972478), ERN1 (EU038802), ERN2 (EU038803), EXPA (DQ899790), NRT1.3 (GU966590), and RR4 (Medtr5g036480).
Supplemental Data
Supplemental Figure 1. pEXPA expression is restricted to epidermal cells of control or Nod factor-treated M. truncatula roots.
Supplemental Figure 2. Tissue-specific expression of NIN in epidermal and cortical root tissues.
Supplemental Figure 3. CRE1 expression is associated with cortical cell divisions in response to S. meliloti.
Supplemental Figure 4. NIN binds the NF-box.
Supplemental Figure 5. A subset of Nod factor (100 pM) induced gene expression dependent on NIN.
Supplemental Table 1. Nod factors induce RR4 expression in wild-type roots but not in the nin-1 mutant roots.
Supplemental Table 2. Primers used in this work.
Supplementary Material
Acknowledgments
We thank David Barker for providing the nupYC2.1 construct, Wim Dejonghe for helping with pEXPA:GFP-GUS studies, and Sibylle Hirsch and Christian Rogers for providing Affymetrix data on nin-1. This work was supported by the European Union as a Marie Curie IEF to T.V. (255467) and the BBSRC through BB/J001872/1. We thank Allan Downie and Jeremy Murray for critical reading of the manuscript.
AUTHOR CONTRIBUTIONS
T.V., J.K., F.d.C.-N., and G.E.D.O. conceived the experiments. J.K., J.S., L.F., T.V., and D.G. conducted the experiments. T.V., J.K., and G.E.D.O. wrote the article.
Glossary
- NF
Nod factor
- BAP
benzylaminopurine
- BNM
buffered nodulation medium
- ChIP
chromatin immunoprecipitation
- CDS
coding sequence
Footnotes
Articles can be viewed without a subscription.
References
- Andriankaja A., Boisson-Dernier A., Frances L., Sauviac L., Jauneau A., Barker D.G., de Carvalho-Niebel F. (2007). AP2-ERF transcription factors mediate Nod factor dependent Mt ENOD11 activation in root hairs via a novel cis-regulatory motif. Plant Cell 19: 2866–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi J.F., et al. (2006). The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D.G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14: 695–700. [DOI] [PubMed] [Google Scholar]
- Broghammer A., et al. (2012). Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA 109: 13859–13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capoen W., Sun J., Wysham D., Otegui M.S., Venkateshwaran M., Hirsch S., Miwa H., Downie J.A., Morris R.J., Ané J.M., Oldroyd G.E. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 108: 14348–14353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerri M.R., Frances L., Laloum T., Auriac M.C., Niebel A., Oldroyd G.E., Barker D.G., Fournier J., de Carvalho-Niebel F. (2012). Medicago truncatula ERN transcription factors: regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 160: 2155–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. (2008). Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 20: 3467–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron D., Pingret J.L., Chabaud M., Journet E.P., Barker D.G. (2004). Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 136: 3582–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.T., Cosgrove D.J. (2002). Regulation of root hair initiation and expansin gene expression in Arabidopsis. Plant Cell 14: 3237–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney K.J., Xu R., Zhang J., Li Q.Q., Yun K.Y., Falcone D.L., Hunt A.G. (2006). Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 140: 1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dénarié J., Debellé F., Promé J.-C. (1996). Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu. Rev. Biochem. 65: 503–535. [DOI] [PubMed] [Google Scholar]
- Ding Y., Kalo P., Yendrek C., Sun J., Liang Y., Marsh J.F., Harris J.M., Oldroyd G.E. (2008). Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula. Plant Cell 20: 2681–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drea S., Corsar J., Crawford B., Shaw P., Dolan L., Doonan J.H. (2005). A streamlined method for systematic, high resolution in situ analysis of mRNA distribution in plants. Plant Methods 1: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt D.W., Atkinson E.M., Long S.R. (1992). Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science 256: 998–1000. [DOI] [PubMed] [Google Scholar]
- Ehrhardt D.W., Wais R., Long S.R. (1996). Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell 85: 673–681. [DOI] [PubMed] [Google Scholar]
- Endre G., Kereszt A., Kevei Z., Mihacea S., Kaló P., Kiss G.B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966. [DOI] [PubMed] [Google Scholar]
- Fournier J., Teillet A., Chabaud M., Ivanov S., Genre A., Limpens E., de Carvalho-Niebel F., Barker D.G. (2015). Remodeling of the infection chamber before infection thread formation reveals a two-step mechanism for rhizobial entry into the host legume root hair. Plant Physiol. 167: 1233–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rizzo S., Crespi M., Frugier F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18: 2680–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth M., Takeda N., Perry J., Uchida H., Dräxl S., Brachmann A., Sato S., Tabata S., Kawaguchi M., Wang T.L., Parniske M. (2010). NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 22: 2509–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Shimoda Y., Sato S., Tabata S., Imaizumi-Anraku H., Hayashi M. (2014). Rhizobial infection does not require cortical expression of upstream common symbiosis genes responsible for the induction of Ca2+ spiking. Plant J. 77: 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann A.B., Sandal N., Bek A.S., Madsen L.H., Jurkiewicz A., Nielsen M.W., Tirichine L., Stougaard J. (2011). Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 24: 1385–1395. [DOI] [PubMed] [Google Scholar]
- Held M., Hou H., Miri M., Huynh C., Ross L., Hossain M.S., Sato S., Tabata S., Perry J., Wang T.L., Szczyglowski K. (2014). Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell 26: 678–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth B., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24: 1345–1358. [DOI] [PubMed] [Google Scholar]
- Journet E.-P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. (2001). Medicago truncatula ENOD11: a novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 14: 737–748. [DOI] [PubMed] [Google Scholar]
- Kaló P., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308: 1786–1789. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kim D.W., Lee S.H., Choi S.B., Won S.K., Heo Y.K., Cho M., Park Y.I., Cho H.T. (2006). Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell 18: 2958–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuta S., Held M., Hossain M.S., Morieri G., Macgillivary A., Johansen C., Antolín-Llovera M., Parniske M., Oldroyd G.E.D., Downie A.J., Karas B., Szczyglowski K. (2011). Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 67: 929–940. [DOI] [PubMed] [Google Scholar]
- Laloum T., Baudin M., Frances L., Lepage A., Billault-Penneteau B., Cerri M.R., Ariel F., Jardinaud M.F., Gamas P., de Carvalho-Niebel F., Niebel A. (2014). Two CCAAT-box-binding transcription factors redundantly regulate early steps of the legume-rhizobia endosymbiosis. Plant J. 79: 757–768. [DOI] [PubMed] [Google Scholar]
- Laporte P., Lepage A., Fournier J., Catrice O., Moreau S., Jardinaud M.-F., Mun J.-H., Larrainzar E., Cook D.R., Gamas P., Niebel A. (2014). The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 65: 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy J., et al. (2004). A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303: 1361–1364. [DOI] [PubMed] [Google Scholar]
- Lohar D.P., Sharopova N., Endre G., Peñuela S., Samac D., Town C., Silverstein K.A., VandenBosch K.A. (2006). Transcript analysis of early nodulation events in Medicago truncatula. Plant Physiol. 140: 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640. [DOI] [PubMed] [Google Scholar]
- Madsen L.H., Tirichine L., Jurkiewicz A., Sullivan J.T., Heckmann A.B., Bek A.S., Ronson C.W., James E.K., Stougaard J. (2010). The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat. Commun. 1: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kusakabe M., Shimoda Y., Sato S., Tabata S., Murooka Y., Hayashi M. (2008). Polyubiquitin promoter-based binary vectors for overexpression and gene silencing in Lotus japonicus. Mol. Plant Microbe Interact. 21: 375–382. [DOI] [PubMed] [Google Scholar]
- Marsh J.F., Rakocevic A., Mitra R.M., Brocard L., Sun J., Eschstruth A., Long S.R., Schultze M., Ratet P., Oldroyd G.E. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton P.H., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19: 1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.B., Pratap A., Miyahara A., Zhou L., Bornemann S., Morris R.J., Oldroyd G.E. (2013). Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25: 5053–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R.M., Gleason C.A., Edwards A., Hadfield J., Downie J.A., Oldroyd G.E., Long S.R. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: Gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. USA 101: 4701–4705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H., Sun J., Oldroyd G.E., Downie J.A. (2006). Analysis of calcium spiking using a cameleon calcium sensor reveals that nodulation gene expression is regulated by calcium spike number and the developmental status of the cell. Plant J. 48: 883–894. [DOI] [PubMed] [Google Scholar]
- Morère-Le Paven M.C., Viau L., Hamon A., Vandecasteele C., Pellizzaro A., Bourdin C., Laffont C., Lapied B., Lepetit M., Frugier F., Legros C., Limami A.M. (2011). Characterization of a dual-affinity nitrate transporter MtNRT1.3 in the model legume Medicago truncatula. J. Exp. Bot. 62: 5595–5605. [DOI] [PubMed] [Google Scholar]
- Murray J.D., Karas B.J., Sato S., Tabata S., Amyot L., Szczyglowski K. (2007). A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315: 101–104. [DOI] [PubMed] [Google Scholar]
- Nawy T., Lee J.Y., Colinas J., Wang J.Y., Thongrod S.C., Malamy J.E., Birnbaum K., Benfey P.N. (2005). Transcriptional profile of the Arabidopsis root quiescent center. Plant Cell 17: 1908–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka-Kira E., Kawaguchi M. (2006). Long-distance signaling to control root nodule number. Curr. Opin. Plant Biol. 9: 496–502. [DOI] [PubMed] [Google Scholar]
- Oldroyd G.E., Downie J.A. (2008). Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59: 519–546. [DOI] [PubMed] [Google Scholar]
- Plet J., Wasson A., Ariel F., Le Signor C., Baker D., Mathesius U., Crespi M., Frugier F. (2011). MtCRE1-dependent cytokinin signaling integrates bacterial and plant cues to coordinate symbiotic nodule organogenesis in Medicago truncatula. Plant J. 65: 622–633. [DOI] [PubMed] [Google Scholar]
- Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592. [DOI] [PubMed] [Google Scholar]
- Schauser L., Roussis A., Stiller J., Stougaard J. (1999). A plant regulator controlling development of symbiotic root nodules. Nature 402: 191–195. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Fournier J., Timmers A.C., Barker D.G. (2012). A switch in Ca2+ spiking signature is concomitant with endosymbiotic microbe entry into cortical root cells of Medicago truncatula. Plant J. 69: 822–830. [DOI] [PubMed] [Google Scholar]
- Sieberer B.J., Chabaud M., Timmers A.C., Monin A., Fournier J., Barker D.G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S., Katzer K., Lambert J., Cerri M., Parniske M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15: 139–152. [DOI] [PubMed] [Google Scholar]
- Smit P., Raedts J., Portyanko V., Debellé F., Gough C., Bisseling T., Geurts R. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308: 1789–1791. [DOI] [PubMed] [Google Scholar]
- Soyano T., Hirakawa H., Sato S., Hayashi M., Kawaguchi M. (2014). Nodule Inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. USA 111: 14607–14612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyano T., Kouchi H., Hirota A., Hayashi M. (2013). Nodule inception directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9: e1003352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke S., Kistner C., Yoshida S., Mulder L., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J., Szczyglowski K., Parniske M. (2002). A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 417: 959–962. [DOI] [PubMed] [Google Scholar]
- Timmers A.C., Auriac M.C., Truchet G. (1999). Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628. [DOI] [PubMed] [Google Scholar]
- Tirichine L., Sandal N., Madsen L.H., Radutoiu S., Albrektsen A.S., Sato S., Asamizu E., Tabata S., Stougaard J. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315: 104–107. [DOI] [PubMed] [Google Scholar]
- van Zeijl A., Op den Camp R.H., Deinum E.E., Charnikhova T., Franssen H., Op den Camp H.J., Bouwmeester H., Kohlen W., Bisseling T., Geurts R. (2015). Rhizobium lipo-chitooligosaccharide signaling triggers accumulation of cytokinins in Medicago truncatula roots. Mol. Plant 8: 1213–1226. [DOI] [PubMed] [Google Scholar]
- Vernié T., Moreau S., de Billy F., Plet J., Combier J.P., Rogers C., Oldroyd G., Frugier F., Niebel A., Gamas P. (2008). EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20: 2696–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wais R.J., Galera C., Oldroyd G., Catoira R., Penmetsa R.V., Cook D., Gough C., Denarié J., Long S.R. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. USA 97: 13407–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T.T., Schilderink S., Moling S., Deinum E.E., Kondorosi E., Franssen H., Kulikova O., Niebel A., Bisseling T. (2014). Fate map of Medicago truncatula root nodules. Development 141: 3517–3528. [DOI] [PubMed] [Google Scholar]
- Xie F., Murray J.D., Kim J., Heckmann A.B., Edwards A., Oldroyd G.E., Downie J.A. (2012). Legume pectate lyase required for root infection by rhizobia. Proc. Natl. Acad. Sci. USA 109: 633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K., et al. (2008). CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 105: 20540–20545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoro E., Suzaki T., Toyokura K., Miyazawa H., Fukaki H., Kawaguchi M. (2014). A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 165: 747–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.