Arabidopsis PPRD1 and -2, orthologs of human SRD5A3 (steroid 5α reductase type 3), encode polyprenol reductases responsible for conversion of polyprenol to dolichol.
Abstract
Dolichol is a required cofactor for protein glycosylation, the most common posttranslational modification modulating the stability and biological activity of proteins in all eukaryotic cells. We have identified and characterized two genes, PPRD1 and -2, which are orthologous to human SRD5A3 (steroid 5α reductase type 3) and encode polyprenol reductases responsible for conversion of polyprenol to dolichol in Arabidopsis thaliana. PPRD1 and -2 play dedicated roles in plant metabolism. PPRD2 is essential for plant viability; its deficiency results in aberrant development of the male gametophyte and sporophyte. Impaired protein glycosylation seems to be the major factor underlying these defects although disturbances in other cellular dolichol-dependent processes could also contribute. Shortage of dolichol in PPRD2-deficient cells is partially rescued by PPRD1 overexpression or by supplementation with dolichol. The latter has been discussed as a method to compensate for deficiency in protein glycosylation. Supplementation of the human diet with dolichol-enriched plant tissues could allow new therapeutic interventions in glycosylation disorders. This identification of PPRD1 and -2 elucidates the factors mediating the key step of the dolichol cycle in plant cells which makes manipulation of dolichol content in plant tissues feasible.
INTRODUCTION
Dolichol (Dol) is a member of the isoprenoids, which are functionally and structurally the most diverse group of natural products (Thulasiram et al., 2007). Dolichyl phosphate (Dol-P) as a cofactor is necessary for protein glycosylation, a ubiquitous posttranslational modification found in all domains of life (Pattison and Amtmann, 2009; Schwarz and Aebi, 2011). Glycosylation is crucial for protein functioning, since it modulates folding and quality control, and is a prerequisite for diverse biological recognition events (Liu and Howell, 2010; Moremen et al., 2012). The oligosaccharide precursor used for N-glycosylation of proteins is assembled on dolichyl diphosphate (Glc3Man9 GlcNAc2-PP-Dol), and the resulting tetradecasaccharide is cotranslationally transferred to an asparagine residue (Asn-X-Ser/Thr sequence) of the growing polypeptide. In addition, the activated monosaccharides Dol-P-Man and Dol-P-Glc are also utilized during protein N-glycosylation, O- and C-mannosylation, and glycosylphosphatidylinositol (GPI) anchor biosynthesis.
In line with the indispensable role of Dol in protein glycosylation, impaired Dol and/or Dol-P biosynthesis leads to disorders called Congenital Disorders of Glycosylation Type I (CDG-I), while disruption of the subsequent steps of the formation of Glc3Man9 GlcNAc2-PP-Dol together with peptide glycosylation and glycan maturation are collectively called CDG-II (Lefeber et al., 2011 and references therein).
Besides their role in protein glycosylation, polyisoprenoid alcohols, i.e., Dols and their α-unsaturated counterparts, polyprenols, are involved in cell adaptation to adverse environmental conditions (Bergamini, 2003; Bajda et al., 2009). Whether polyisoprenoids protect senescing tissues via their highly increased accumulation upon aging (summarized in Swiezewska and Danikiewicz, 2005) remains a matter of debate. Dol is also suggested to be involved in the intracellular trafficking of proteins (Sato et al., 1999; Belgareh-Touzé et al., 2003) and in macromolecular complex assembly (e.g., of glycan biosynthetic enzymes; Jones et al., 2009).
The biosynthesis of Dol comprises three steps: (1) formation of isopentenyl diphosphate and dimethylallyl diphosphate, five-carbon building blocks of isoprenoids biosynthesized in plant cells by a concomitant involvement of the mevalonate and the methylerythritol phosphate pathways; (2) formation of farnesyl diphosphate and its elongation via subsequent condensations of farnesyl diphosphate with isopentenyl diphosphate molecules (performed by cis-prenyltransferase [CPT]) leading to a mixture of homologous polyprenyl diphosphates with a species-dependent composition; and (3) hydrogenation of the double bond in the OH-terminal isoprene unit of polyprenol and/or polyprenyl diphosphate resulting in the formation of a corresponding mixture of Dols (Figure 1A). Among these, the last step in Dol biosynthesis was the least studied until the identification of members of the steroid α-reductase family, mammalian SRD5A3 and yeast Dfg10, as key enzymes responsible for polyprenol hydrogenation (Cantagrel et al., 2010). This was achieved by elucidation of the molecular basis of a rare Mendelian disease, demonstrating that mutations in the SRD5A3 or DFG10 gene cause increased polyprenol accumulation at the expense of Dol, leading to defective protein N-glycosylation. Moreover, a loss-of-function mutation the Srd5a3 gene is embryo-lethal in mouse (Cantagrel et al., 2010). In human, mutations in SRD5A3 lead to a neurological disease with developmental delay, ataxia, and early visual impairment with optic atrophy. In some cases, ichthyosiform dermatitis is reported with liver dysfunction and coagulation abnormalities (for summary, see Buczkowska et al. [2015] and references therein).
Figure 1.
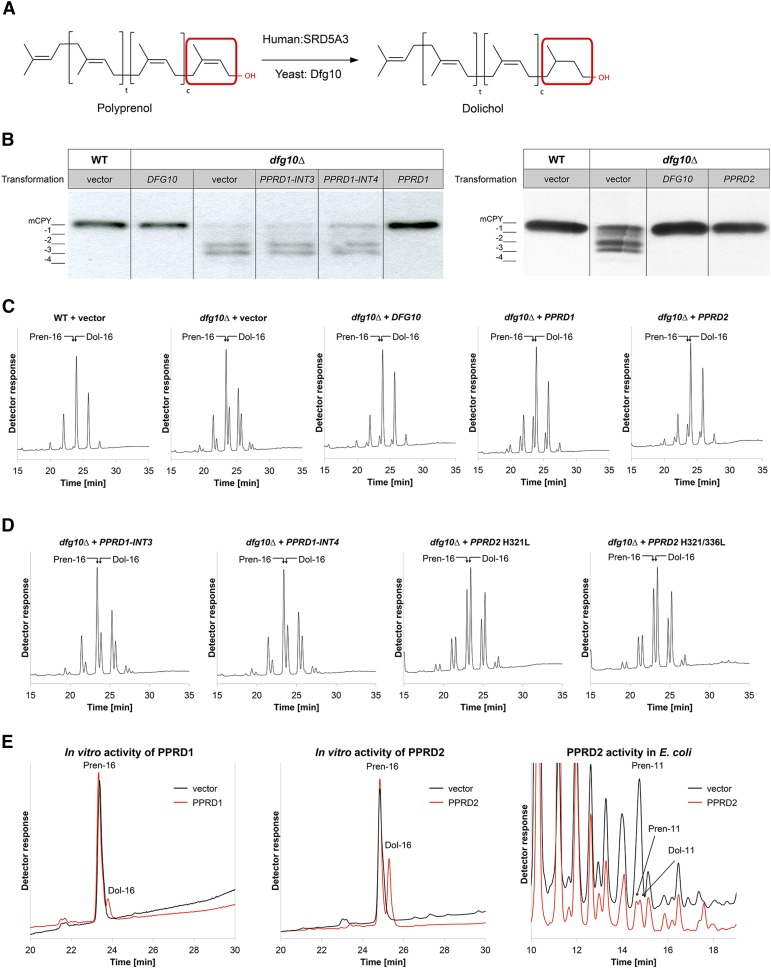
Analysis of Enzymatic Activity of PPRD1 and PPRD2 in Vivo and in Vitro.
(A) Reduction of polyprenol to Dol, schematic representation; SRD5A3 and Dfg10 are the human and yeast enzymes, respectively.
(B) and (C) Analysis of the glycosylation status of CPY (B) and polyisoprenoid profile (C) of the dfg10∆ yeast mutant transformed with either yeast DFG10 or Arabidopsis PPRD1, PPRD2, PPRD1-INT3, or PPRD1-INT4 constructs; dfg10∆ and wild-type transformants with empty vector were used as a negative and positive control, respectively. The CPY glycosylation and Pren:Dol ratio of dfg10∆ are rescued by DFG10, PPRD1, and PPRD2 expression. CPY (mature and hypoglycosylated forms) is detected by specific anti-CPY antibody. Positions of fully glycosylated CPY (mCPY) and the four hypoglycosylated forms (-1, -2, -3, and -4) are indicated.
(D) Polyisoprenoid profiles of dfg10∆ mutant transformed with PPRD1 variants: PPRD1-INT3, PPRD1-INT4, or mutants of PPRD2 carrying substitutions in conserved histidine residues (H321/336L). In contrast to PPRD1-INTs, which do not changed the lipid profile, expression of mutated constructs partially rescues the Pren:Dol ratio of the dfg10∆ mutant.
(E) Reduction of exogenous Pren-16 in vitro by recombinant PPRD1 or PPRD2 produced in E. coli. Extracted lipids were analyzed by HPLC/UV. Black traces represent negative control reactions (bacteria transformed with an empty vector), and red traces represent reactions with cell lysates from bacteria expressing PPRD1 or PPRD2. The rightmost chromatogram shows that Dol-11 is produced from endogenous Pren-11 in bacteria expressing PPRD2. Only relevant regions of chromatograms are shown with positions of external standards indicated. See also Supplemental Figure 1 and Supplemental Table 1.
Despite the fact that a considerable amount of Dol is accumulated in plant roots (Skorupińska-Tudek et al., 2003; Jozwiak et al., 2013), a plant polyprenol reductase has not been identified until now. In this study, two genes encoding polyprenol reductase (PPRD-1 and -2), orthologs of SRD5A3 and DFG10, were identified in Arabidopsis thaliana. The newly identified PPRDs were found to be involved in the regulation of plant growth and reproductive processes.
RESULTS
Two Genes Encode Functional Polyprenol Reductases in Arabidopsis
Three putative polyprenol reductases were found in the Arabidopsis proteome (gene loci At1G72590, At2G16530, and At3G43840) using BLASTP. The amino acid sequences of those putative Arabidopsis polyprenol reductases were similar to human (47 to 54% similarity and 28 to 30% identity) and yeast (39 to 47% similarity and 26 to 27% identity; Supplemental Table 1) orthologs and the predicted POLYPRENOL REDUCTASE1 (PPRD1) and PPRD2 polypeptides comprised 320 and 343 amino acids, respectively, while the putative PPRD3 contained 84 amino acids corresponding to the C-terminal part of PPRD1 and PPRD2. A multiple sequence alignment of PPRDs from various plant species showed the presence of at least eight highly conserved regions (Supplemental Figures 1 and 2). Membrane topology analysis (TMHMM server) predicted five transmembrane domains (TMDs) in PPRD1 and PPRD2 proteins, similar to the six TMDs in SRD5A3 (Cantagrel et al., 2010), while only one TMD was predicted in PPRD3 (Supplemental Figure 3).
To determine whether Arabidopsis PPRD1 and PPRD2 are true orthologs of yeast DFG10, functional complementation of the yeast dfg10 mutants (dfg10Δ and dfg10-100, a transposon insertion in DFG10 promoter) was performed with PPRD1 and PPRD2 coding sequences and with PPRD1 variants, PPRD1-INT3 or PPRD1-INT4 carrying intron 3 or intron 4, respectively (Supplemental Figure 4). Analysis of the N-glycosylation status of carboxypeptidase Y (CPY), which in the mature form contains four N-glycan chains, revealed that in contrast with the wild type (BY4741), hypoglycosylated (tri-, di-, and mono-) CPY variants were clearly detectable for dfg10 (Figure 1B). Transformation with PPRD1 and PPRD2, similarly to DFG10, fully rescued the CPY hypoglycosylation, in contrast with PPRD1-INT3 and PPRD1-INT4. Similar results were obtained for the both dfg10 mutants and only those for dfg10∆ are shown (Figure 1B). These results indicate that PPRD1 and -2 are functional orthologs of the yeast DFG10.
To explore the biochemical effects underlying the yeast complementation, lipid profiles of dfg10 strains and the transformants mentioned above were analyzed using HPLC/UV. The two dfg10 mutants displayed a high polyprenol:Dol ratio in contrast to the wild-type strain, suggesting a block in the polyprenol reduction step and confirming the literature data (Cantagrel et al., 2010). Transformation of the dfg10∆ strain with DFG10, PPRD1, or PPRD2 almost completely rescued this chemotype; only traces of polyprenols were observed (Figure 1C). By contrast, transformation with PPRD1-INT3 or PPRD1-INT4 did not complement the Dol deficiency (Figure 1D); most likely truncated nonfunctional PPRD1 proteins devoid of 86 or 53 C-terminal amino acids were expressed in those cases (Supplemental Figure 4). Similar results were obtained for complementation of the dfg10-100 mutant (data not shown). Thus, PPRD1 and PPRD2 are functional orthologs of yeast polyprenol reductase.
The Catalytic Domain of PPRD Is Localized at the C Terminus
In silico analysis of the PPRD1 and -2 amino acid sequences led to the identification of the catalytic 3-oxo-5-α-steroid 4-dehydrogenase domain (PF02544) in the C-terminal part of both PPRDs (Supplemental Figure 3) similarly to SRD5A3 (Cantagrel et al., 2010). Again, PPRD3 seemed to be a truncated form of PPRD1 and -2 comprising solely this domain (87 and 61% identity with C-terminal fragments of PPRD2 and PPRD1, respectively; Supplemental Table 2). Thus, only PPRD1 and -2 were subjected to further analysis.
To confirm the location of the catalytic domain, directional mutagenesis was performed aiming at substitution of conserved histidines, which are suggested to take part in the reduction reaction (Wigley et al., 1994; Cantagrel et al., 2010). PPRD2 mutants with His-321 or His-336, corresponding to His-296 or His-309 in hSRD5A3, substituted by Leu were employed to complement the yeast dfg10∆ mutant (Supplemental Figure 3). For the H321L and double H321L H336L mutants, 47 and 45% of the wild-type reductase activity was recovered, respectively, supporting the C-terminal location of the catalytic domain. These results are also in line with the total loss of activity of the putative C-terminally truncated PPRD1 proteins encoded by PPRD1-INT3 and PPRD1-INT4 (Figure 1D). Nevertheless, these findings suggest that besides His-321 or His-336, other residues of PPRD2 might be involved in the reduction since H296G substitution in hSRD5A3 abolished its enzymatic activity (Cantagrel et al., 2010).
PPRD1 and PPRD2 Catalyze Reduction of Polyprenol to Dol in Vitro
To confirm the enzymatic activity of PPRDs as polyprenol reductases, an in vitro assay using lysates containing the recombinant proteins expressed in the bacterial system was performed. Escherichia coli, like most other bacteria, does not possess endogenous PPRD activity. The concentration of detergent, Triton X-100, used to solubilize exogenous Prenol-16 (Pren-16) required careful optimization to ensure the availability of the hydrophobic substrate to PPRD and simultaneously to avoid loss of its enzymatic activity. For both lysates from bacteria overexpressing either PPRD1 or PPRD2, a notable amount of Dol-16, the expected product of Pren-16 reduction (∼30 or 11% of the substrate for PPRD2 or PPRD1, respectively) was detected, while no Dol was formed in the presence of lysate from control bacteria (empty vector) (Figure 1E). Interestingly, in the strain expressing PPRD2 endogenous bacterial P-11 was also reduced to Dol-11 (Figure 1E).
Taken together, these results confirm that PPRD1 and PPRD2 encode polyprenol reductases, which upon expression in heterologous systems catalyze efficient reduction of prenol (Pren) to Dol both in vivo and in vitro.
Polyprenol Reductases Are Expressed in a Tissue-Specific Manner in Arabidopsis and Are Diversely Localized within the Plant Cell
Variable expression profiles of PPRDs were found in Arabidopsis organs (roots, leaves, stems, flowers, and pollen) during the plant life span. The expression of PPRD1 was consistently lower compared with that of PPRD2. Both genes were expressed in young seedlings (Figure 2A). In older plants, PPRD1 was expressed only in the roots and flowers, unlike PPRD2, which was expressed in all organs analyzed (Figure 2A). With age, expression of PPRD1 increased in the roots and decreased in the leaves, while expression of PPRD2 was fairly constant in the roots and decreased in the leaves. Interestingly, in pollen exclusively the PPRD2 transcript was detected (Figure 2A). Expression of PPRD1-INT3 and PPRD1-INT4, probably resulting from alternative splicing of PPRD1, was also observed in the leaves (Supplemental Figure 4).
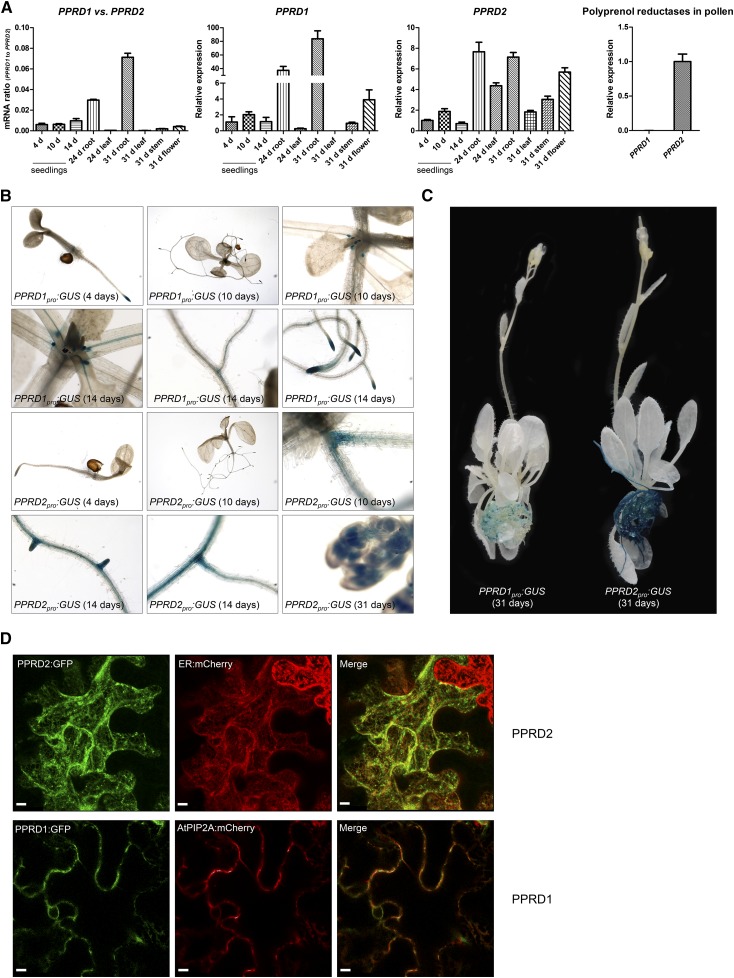
Figure 2.
Expression Pattern of Genes Encoding Polyprenol Reductases in Arabidopsis and Subcellular Localization of PPRDs.
(A) Expression of PPRDs in various organs of Arabidopsis was analyzed at different time points; the leftmost panel shows the ratio of PPRD1 versus PPRD2 transcript accumulation. PPRD2 expression is higher and more specific than PPRD1. PPRD1 transcript is virtually absent from Arabidopsis pollen. Values are the mean ± sd of three independent experiments.
(B) and (C) Histochemical localization of GUS expression driven by the PPRD1 or PPRD2 promoter in transgenic Arabidopsis was analyzed at various time points: 4, 10, and 14 d after germination (B) and in 31-d-old plants (C). In contrast to PPRD1, PPRD2 promoter drives broad expression in Arabidopsis tissues.
(D) Colocalization of GFP-tagged PPRD2 or PPRD1 (green) with ER and PM markers: ER:mCherry and PIP2A:mCherry (red). Representative pictures are presented. See also Supplemental Figure 5. Bar = 10 μm.
To characterize further the expression pattern of PPRD1 and PPRD2, a fusion of their promoter regions with the β-glucuronidase (GUS) reporter gene was employed. Substantial activity of the PPRD1 promoter (PPRD1pro:GUS) was detected mainly during the extensive development phase in tissues characterized by actively dividing cells (e.g., root apical meristem, root division and elongation zones, and leaf primordia) and restricted only to a small number of cells (Figure 2B). The promoter activity of PPRD2 (PPRD2pro:GUS) was induced in a time-dependent manner; the older the tissue, the higher the expression. It was virtually absent in 4-d-old seedlings and expressed along the whole root in 2-week-old and older plants, mostly in the junctions between lateral roots and the primary root, in the root division and elongation zones, and in root cap (Figures 2B and 2C). GUS staining was also visible in young stamen (Figure 2B).
Taken together, these results show that the expression of PPRDs is regulated temporally and in a tissue-specific manner, suggesting that the encoded enzymes might function in differentiation. The diverse patterns of expression of the two paralogs suggest divergent roles of these enzymes in the plant. In contrast to the more ubiquitously expressed PPRD2, PPRD1 seems to undergo induction in particular tissues/cells in response to the current metabolic requirements.
To compare the intracellular localization of polyprenol reductases, PPRD1:G3GFP and PPRD2:G3GFP constructs, encoding N-terminal fusions of the respective reductase with fluorescent protein, were transiently coexpressed with plant organelle markers (mCherry fusions) in Nicotiana benthamiana. The localization information for the ER:mCherry marker was provided by a well-established short targeting signal while plasma membrane (PM) labeling was based on the full-length coding region of At-PIP2A, a plasma membrane aquaporin (Nelson et al., 2007). Microscopy observations of transformed plants showed that PPRD2 was colocalized mostly with the marker of the endoplasmic reticulum (ER), while PPRD1 colocalized with that of the PM (Figure 2D; Supplemental Figure 5). Additionally, weak colocalization signals for PPRD2 with PM and PPRD1 with ER markers were also visible in all experiments (n = 4); moreover, weak colocalization signals for PPRD2 with Golgi:mCherry marker were observed in some (n = 2) experiments too (data not shown).
PPRD2 Knockout Plants Are Not Viable
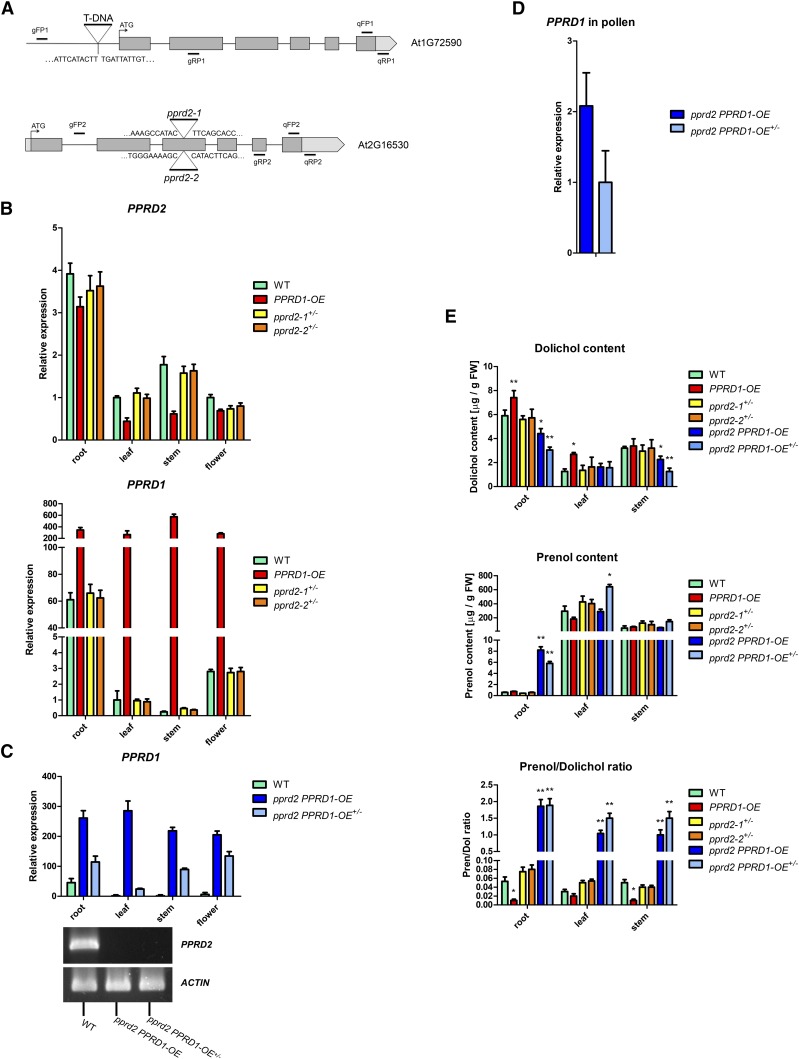
To analyze further the role of the polyprenol reductases, relevant PPRD T-DNA insertion lines were studied (Figure 3A). One line homozygous for PPRD1 insertion was available (GabiKat_575B02), but it turned out to contain much higher (up to 600-fold) levels of the PPRD1 transcript compared with wild-type plants in all organs (Figure 3B). Subsequent analysis revealed that in this line, the T-DNA insert was localized in the PPRD1 promoter region (Figure 3A). Such localization of the insert may actually lead to upregulation of gene expression. Indeed, in this line, Dol content was increased, reaching 213 and 126% of control in the leaves and roots, respectively. What is more, the increase of the dolichol content in the leaves seemed to occur at the expense of polyprenol; its content was decreased to 62% of the wild type, albeit no such phenomenon was observed in the roots (Figure 3E). Thus, the GabiKat_575B02 homozygous line will be further referred to as PPRD1-OE (overexpressing).
Figure 3.
Expression Pattern of Genes Encoding Polyprenol Reductases and Analysis of Polyisoprenoid Alcohols in Arabidopsis Lines.
(A) Schematic representation of PPRD1 and PPRD2. Gray and light-gray boxes represent exons and 5′- and 3′-untranslated regions; location of T-DNA insertion in PPRD1-OE, pprd2-1, and pprd2-2 is shown. Positions of primers used for PCR-based genotyping or qPCR are indicated. Sequences of the borders of T-DNA inserts are provided.
(B) to (D) Expression of PPRDs in Arabidopsis.
(B) Expression in various organs of 35-d-old Arabidopsis mutant plants normalized to the expression in leaves of same age wild-type plants. T-DNA PPRD gain-of-function (homozygous PPRD1-OE) and loss-of-function (heterozygous pprd2-1+/− and pprd2-2+/−) mutant lines were analyzed.
(C) and (D) Expression in various organs (C) and pollen (D) of pprd2 PPRD1-OE crosses (see Figure 4). Considerably increased expression of PPRD1 (upper panel) and totally abolished expression of PPRD2 (lower panel) are visible. Values are the mean ± sd of three independent experiments.
(E) Content of polyisoprenoid alcohols in leaves, stems, and roots of wild-type, PPRD1-OE, pprd2-1+/−, pprd2-2+/−, pprd2 PPRD1-OE, and pprd2 PPRD1-OE+/− plants; the rightmost panel shows the ratio of Pren versus Dol content. Considerably increased ratio (higher polyprenol content) is observed for pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines.
Values (±sd) represent mean of three, four, and five independent experiments for roots, stems, and leaves, respectively. Statistically significant differences compared with wild-type plants are indicated; *P < 0.05 and **P < 0.01. See also Supplemental Figure 4.
Two heterozygous T-DNA PPRD2 insertion lines, pprd2-1 (SALK_113221C) and pprd2-2 (SALK_006421) (Figure 3A), were used in this study since no plants homozygous for PPRD2 were found during genetic screens of pprd2-1 or pprd2-2 progeny (Table 1). In these lines, the expression levels of PPRD1 and PPRD2 were almost identical to those in wild-type plants (Figure 3B), as were the Dol and Pren contents (Figure 3E). Sequencing of the T-DNA insert confirmed its different location for pprd2-1 and pprd2-2 lines (Figure 3A).
Table 1. Segregation of pprd2 Alleles.
| Self-Pollination | |||||
|---|---|---|---|---|---|
| pprd2-1, n = 328 | pprd2-2, n = 196 | ||||
| Genotype |
Expected |
Observed |
Genotype |
Expected |
Observed |
| pprd2-1−/− | 82 (25%) | 0 | pprd2-2−/− | 49 (25%) | 0 |
| pprd2-1+/− | 164 (50%) | 158 (48%) | pprd2-2+/− | 98 (50%) | 96 (49%) |
| Wild type | 82 (25%) | 170 (52%) | Wild type | 49 (25%) | 100 (51%) |
| χ2 = 174.56*** | χ2 = 100.07*** | ||||
F1 progeny of self-pollinated pprd2-1 and pprd2-2 mutants. Analysis was performed by χ2 test against the H0 hypothesis that segregation is Mendelian. Asterisks indicate significant difference from Mendelian; P < 0.001.
To establish the reason for the inability to obtain homozygous pprd2 plants, heterozygous mutant lines, pprd2-1+/− or pprd2-2+/−, were self-pollinated and F1 progeny was genotyped. The lack of pprd2 homozygotes among almost 200 plants of each mutant line suggested that disruption of this gene was lethal (Table 1). Segregation analysis of heterozygous pprd2-1+/− and pprd2-2+/− plants and χ2 test (Table 1) confirmed the lethality of the pprd2−/− (homozygous) state, which could be caused by pollen sterility since these lines produced siliques with seed number and germination rate comparable to those of the wild type (Supplemental Figure 6 and Supplemental Table 3).
To gain a deeper insight into the pprd2 lethality, we performed reciprocal crosses of pprd2-1+/− and pprd2-2+/− with wild-type plants. For both the mutant alleles, pollination of the heterozygous stigma (♀) with wild-type pollen (♂) resulted in 50% of heterozygous and 50% wild-type plants in the offspring (Table 2). The reciprocal pollination of wild-type stigma (♀) with pollen from heterozygous plants (♂) produced only wild-type plants (Table 2). Therefore, the pprd2+/− plants are defective in the transmission of pprd2 alleles through the male gametophyte.
Table 2. Reciprocal Crosses of pprd2-1+/− and pprd2-2+/− with the Wild-Type Plants.
| Reciprocal Crosses | ||||
|---|---|---|---|---|
| Crosses | F1 Genotypes, n = 60 | |||
| Male |
Female |
Wild Type |
Heterozygote |
Homozygote |
| pprd2-1+/− | Wild type | 60 | 0 | 0 |
| Wild type | pprd2-1+/− | 30 | 30 | 0 |
| pprd2-2+/− | Wild type | 60 | 0 | 0 |
| Wild type | pprd2-2+/− | 31 | 29 | 0 |
Reciprocal crosses of pprd2-1+/− and pprd2-2+/− with the wild-type plants; 60 plants of each cross were genotyped.
To further elucidate the lethality of pprd2 disruption, pprd2-1+/− stamens were pollinated with PPRD1-OE to produce double heterozygote pprd2-1+/− PPRD1-OE+/− (F1) plants and in the next (F2) generation the double homozygote pprd2 PPRD1-OE was isolated. As expected, this mutant displayed no PPRD2 expression and an increased (at least 200-fold) level of PPRD1 mRNA (Figure 3C). This result suggests that overexpression of PPRD1 can rescue the pprd2 lethality since the pprd2 PPRD1-OE plants (double homozygote) displayed only mild morphological abnormalities (Figures 4A and 4B).
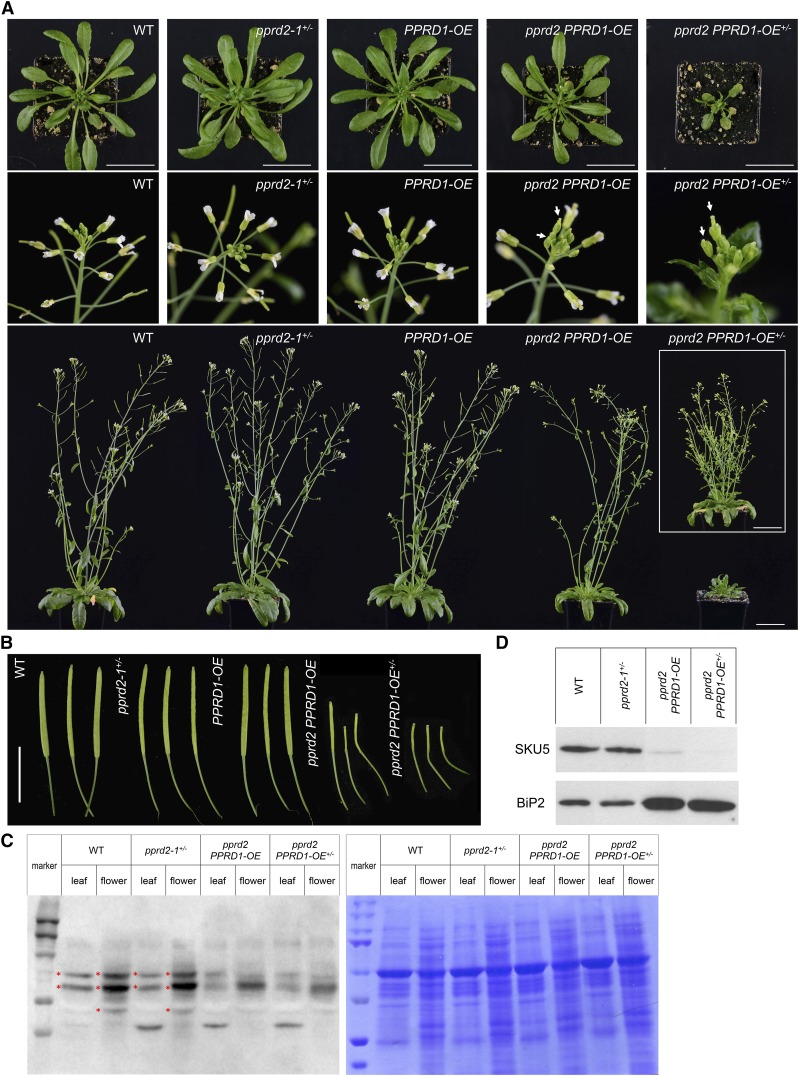
Figure 4.
Phenotypic Characteristics of PPRD1-OE and pprd2+/− Plants and Their Crosses.
(A) Rosettes of 5-week-old plants (upper panel), flowers (middle panel), and whole 7-week-old plants (lower panel) are presented. Arrows indicate delayed flowering of pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines. To illustrate that the pprd2 PPRD1-OE+/− plants show delayed rather than blocked development, the inset presents a 10-week-old plant. Bar = 5 cm.
(B) Siliques of 7-week-old (wild type, pprd2-1+/−, PPRD1-OE, and pprd2 PPRD1-OE) and 10-week-old (pprd2 PPRD1-OE+/−) plants. Bar = 1 cm.
(C) and (D) Analysis of glycosylated proteins in wild-type, pprd2-1+/−, pprd2 PPRD1-OE, and pprd2 PPRD1-OE+/− plants. Protein extracts from leaves and flowers were separated by SDS-PAGE and blots were probed with Concanavalin A labeled with horseradish peroxidase (left image) (C). Gel stained with Coomassie Brilliant Blue is presented in right image. Marker lines contain PageRuler Prestained Protein Marker. Asterisks mark bands present in wild-type extracts but absent in pprd2 lines. Alternatively, blots were probed with anti-SKU5 or anti-BiP2 antibody (D). In pprd2 homozygotes, SKU5 protein is (nearly) absent, while the BiP2 level is elevated compared with the wild type. Equal amounts of protein were loaded in all lanes.
See also Supplemental Figures 6 and 7 and Supplemental Table 3.
By contrast, the pprd2 PPRD1-OE+/− (PPRD1-OE heterozygote) plants did show obvious growth defects, such as substantially delayed growth rate and flowering (Figures 4A and 4B), undulation of some rosette leaves (Supplemental Figure 6), a significantly increased number of auxiliary branches at maturity (first-order and higher order rosette branches and cauline branches), and no apical dominance (the length of the first-order branches was almost the same at different node positions) (Figure 4A). Both pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines displayed problems with flower development (small petals and short stamen), pollination, and silique maturation (short siliques with very few seeds, ∼30 and 10% of wild type for pprd2 PPRD1-OE and pprd2 PPRD1-OE+/−, respectively), and in fact the latter plants were almost fully sterile (Supplemental Figure 6). The differences in pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− phenotypes correlated well with the level of PPRD1 transcript, which was considerably lower in all analyzed organs (including pollen) of pprd2 PPRD1-OE+/− compared with pprd2 PPRD1-OE (Figures 3C and 3D). Moreover, Dol contents in the organs of pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines were considerably different, namely, for pprd2 PPRD1-OE plants, Dol was 70.2 and 74.8% of the control wild type, while for pprd2 PPRD1-OE+/−, Dol was 38.9 and 51.6% of control for stems and roots, respectively. Interestingly, Dol content was not changed in the leaves of these double mutant lines despite the increased transcript accumulation of PPRD1 in this tissue. Additionally, Pren had highly increased accumulation in leaves of pprd2 PPRD1-OE+/− (∼2-fold) and especially in roots of pprd2 PPRD1-OE (∼14-fold) and pprd2 PPRD1-OE+/− (∼10-fold of control) (Figure 3E). Consequently, in roots of mutant lines (homozygous for pprd2), the ratio of Pren versus Dol, calculated for the five most prevalent homologs Pren/Dol-14 to Pren/Dol-18, was increased from 0.05 for wild-type to ∼2 for mutants (Figure 3E). Interestingly, the qualitative profile of polyisoprenoids was not changed in these tissues. Thus, the shift of Pren:Dol ratio might indicate different mechanisms of regulation of PPRD enzymatic activity in leaves than in the other analyzed tissues.
Taken together, characterization of the pprd2 mutant lines points to defects of the male gametophyte as the reason for pprd2 lethality. Lack of PPRD2 is partially compensated for by overexpression of PPRD1, and this compensation is gene-dose dependent. Additionally, the impaired development of siliques of pprd2 PPRD1-OE+/− suggests some role of PPRD2 in female gametophyte function.
Lack of PPRD2 Results in Male Sterility Due to Disturbed Protein Glycosylation and Is Rescued by Exogenous Dol
Interestingly, the protein N-glycosylation status was clearly affected in the pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines compared with the wild type as shown by specific staining of total leaf and flower proteins (Figure 4C; Supplemental Figure 7). This was additionally confirmed by the fate of SKU5 protein, involved in regulation of root growth and cell expansion, which in its native form is both N-glycosylated and GPI-anchored (Sedbrook et al., 2002). SKU5 was absent from flowers of pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− plants, most probably due to proteolytic degradation of the unmodified polypeptide (Figure 4D) as earlier observed for pnt1 mutant devoid of GPI anchor biosynthesis (Gillmor et al., 2005). Similarly, mass spectrometry-based differential proteomics of wild-type and pprd2 PPRD1-OE+/− flower proteins revealed the absence of numerous proteins of vital physiological functions in pprd2 PPRD1-OE+/− plants (Supplemental Table 4).
Moreover, an elevated level of BiP2 protein, a marker of ER stress, was observed in both pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− lines (Figure 4D). Consistent with this observation, BiP2 expression was increased in flowers and especially leaves of pprd2 PPRD1-OE+/− plants, and, interestingly, the level of BiP2 appeared to be negatively related with the level of PPRD1 mRNA in individual plants (Supplemental Figure 6).
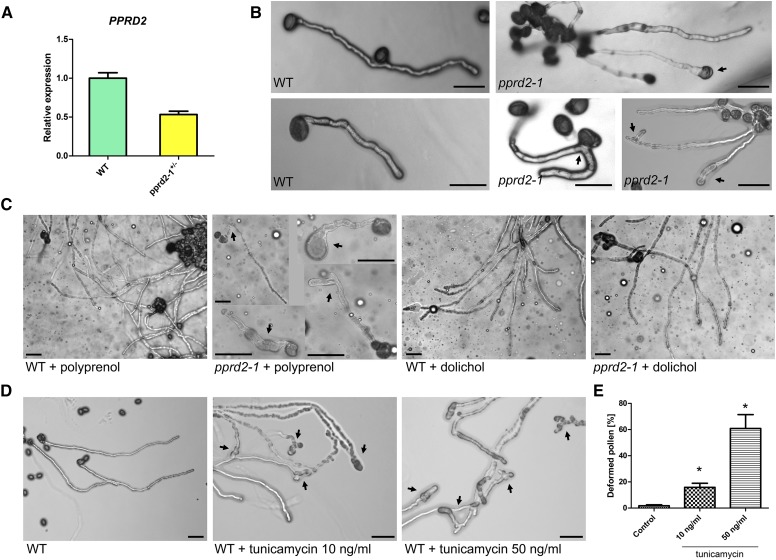
The development of male reproductive organs was monitored with Alexander staining; viable pollen grains are stained purple and dead ones green (Lalanne et al., 2004). Both wild-type and pprd2-1+/− anthers were full of purple pollen grains, indicating that pprd2-1+/− pollen was viable (Supplemental Figure 8). In parallel, in vitro pollen germination on solid medium revealed that ∼43% of germinated pprd2-1+/− pollen grains displayed defects in tube growth and shape: swelling, shortening, and branching (Figure 5B; Supplemental Table 5). By contrast, <4% of wild-type pollen produced abnormal pollen tubes (Figure 5B; Supplemental Table 5). These phenotypic differences were in accordance with the 50% decrease of PPRD2 mRNA level in the pollen of heterozygous pprd2-1+/− plants compared with the wild type (Figure 5A). Aniline blue staining of wild-type pistils hand-pollinated with pollen of wild-type, pprd2+/−, or pprd2 PPRD1-OE plants revealed inhibition of mutant pollen tube growth compared with the wild type, with more short tubes in the transgenic lines and fewer tubes reaching the bottom of the transmitting tract (Supplemental Figure 8).
Figure 5.
Development of Pollen of the pprd2-1+/− Line: Effect of Supplementation with Dol.
(A) Expression of PPRD2 in wild-type and pprd2-1+/− pollen. Values are the means ± sd of three independent experiments.
(B) to (D) Germination of pollen grains from wild-type and heterozygous pprd2-1+/− plants on regular medium (B), medium supplemented with polyprenol or Dol (C), or with tunicamycin ([D]; final concentration 10 or 50 ng/mL). Tube deformations (arrows), significantly more frequent for pprd2-1+/− than wild-type plants, are rescued by Dol but not Pren. Tunicamycin significantly increases number of deformed pollen tubes. Bar = 100 μm.
(E) Quantification of the effect of tunicamycin on pollen tube development. Number of pollen tubes viewed is 164, 203, and 241 for control, 10 and 50 ng/mL tunicamycin, respectively (Supplemental Table 5). Means ± sd are shown. Statistically significant differences from the wild type are indicated: *P < 0.05
See also Supplemental Figure 8 and Supplemental Table 5.
To confirm that the shortage of functional PPRD was the reason for the pollen tube malformations, a rescue experiment was performed. Since Dol is the end product of the enzymatic activity of PPRD, the solid medium used for pollen germination was supplemented with Dol or, in a control experiment, with polyprenol. Wild-type pollen displayed no abnormalities upon germination on either medium (Figure 5C). The supplementation of the growth medium with Dol resulted in an almost 100% rescue of the pprd2-1+/− pollen germination defect; the pollen tubes were of regular length and no malformations could be detected (Figure 5C). By contrast, the supplementation with Pren did not improve the pollen tube growth, confirming that specifically Dol, not just any polyprenol, was lacking in the pprd2-1+/− plants (Figure 5C; Supplemental Table 5).
To verify the mechanism of the effect of Dol shortage on pollen germination, wild-type pollen grains were germinated on solid medium supplemented with tunicamycin, an inhibitor of N-acetylglucosamine transferase (an essential enzyme of protein glycosylation) and inducer of ER stress. The morphology of tunicamycin-treated wild-type pollen tubes was almost identical to that of pprd2+/− pollen germinated on regular medium; swollen, shortened, and branched tubes were noted (∼20 or 60% of total germinating pollen for 10 or 50 ng/mL tunicamycin, respectively; Figures 5D and 5E; Supplemental Table 5). These pollen germination experiments strongly support the concept that the male sterility of pprd2 plants is caused by shortage of Dol and consequent defects in protein glycosylation.
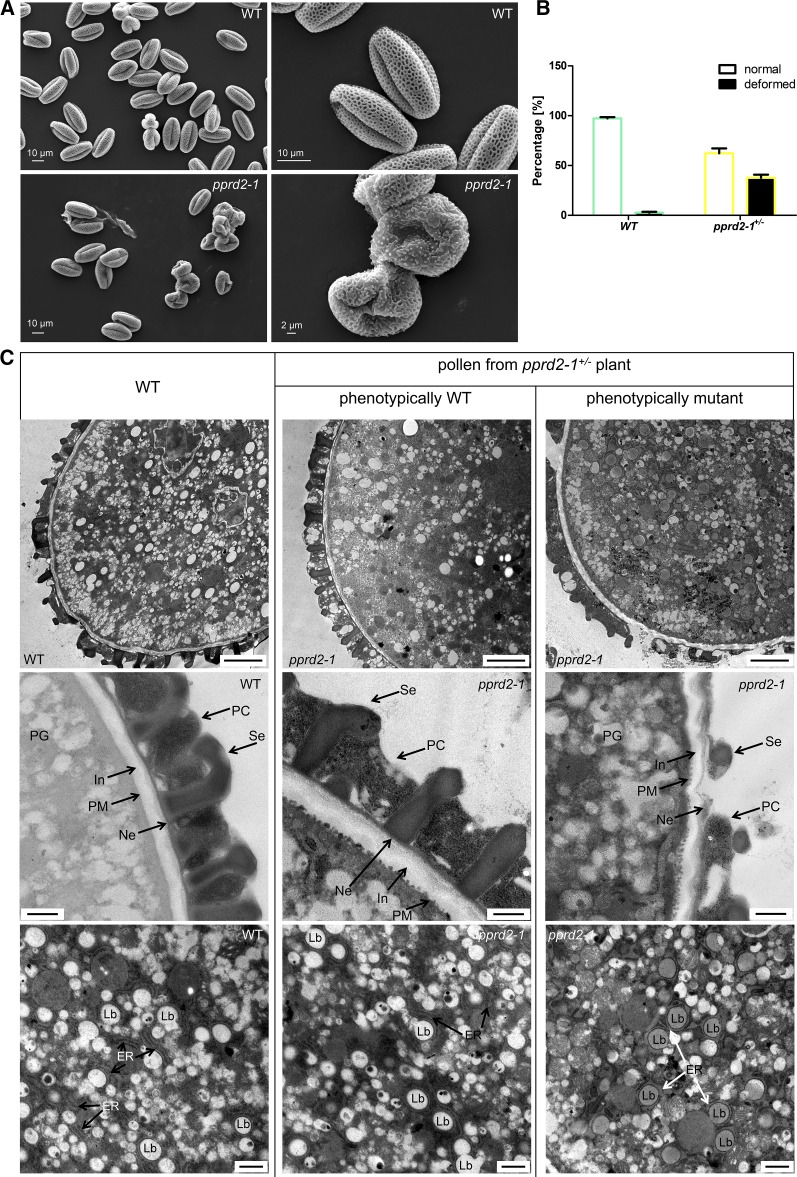
Additionally, scanning electron microscopy examination revealed abnormalities in the exine structure of pprd2-1+/− pollen grain (Figure 6A). Consistently with the pollen germination results, ∼38% of the observed grains were deformed (collapsed, shrunken, and wrinkled) in contrast to wild-type pollen with only 3% deformed grains (Figure 6B). Upon transmission electron microscopy (TEM) analysis, the pprd2-1+/− pollen grains showed an abnormal cell wall of variable thickness, and some of the grains also had underdeveloped exine, uneven intine, and slightly malformed ER structures. Furthermore, the electron density of lipid bodies was changed, suggesting their modified composition (Figure 6C). In contrast, wild-type pollen showed a uniformly structured cell wall with clearly visible exine (T-shaped baculae and tecta) and regular intine structures and undisturbed cellular membranes.
Figure 6.
Pollen of pprd2-1+/− Line: Microscopy Analysis.
(A) Pollen grains from wild-type and pprd2-1+/− plants visualized by a scanning electron microscope.
(B) Quantification of deformed pollen grains under a scanning electron microscope. A significantly higher (by χ2 test) proportion of deformed grains is observed for pprd2-1+/− compared with the wild type. Number of pollen grains analyzed is 1208 and 1424 for the wild type and pprd2-1+/−, respectively. Means ± sd are shown.
(C) TEM images of morphology and ultrastructure of pollen from pprd2-1+/− plants. For phenotypic comparison, wild-type versus pprd2-1 pollen grains were derived from the same pprd2-1+/− mutant plant. Top row, general morphology of pollen grains; bars = 1 μm. Cell wall, intine, and exine (middle row; bars = 500 nm) and lipid bodies and ER (bottom row; bars = 250 nm) of pprd2-1+/− mutant are altered. Representative pictures are presented. In, intine; Ne, nexine; Se, sexine; PC, pollen coat; PG, pollen grain; LB, lipid body.
See also Supplemental Figure 8 and Supplemental Table 5.
To summarize, mutations in the PPRD2 gene lead to defects in pollen germination and development, and this explains the lack of functional pollen in pprd2 mutants and the inability to obtain a homozygous pprd2 sporophyte.
DISCUSSION
Identification and Functional Characterization of Arabidopsis Polyprenol Reductases
Given the presence of Dols in plants, we undertook a search for PPRDs and identified two genes potentially encoding PPRD: PPRD1 and PPRD2 . Enzymatic activity of the corresponding PPRDs was confirmed both in vivo and in vitro. The catalytic domain was localized at their C termini, similarly to hSRD5A3; in contrast to the human enzyme, however, the Arabidopsis PPRDs apparently had more than one catalytically active histidine residue.
Further studies are required to characterize PPRDs biochemically; however, the reduction of bacterial Pren-11 observed in this report together with the complex Dol mixture (Dol-7 to -35) present in vivo (Jozwiak et al., 2013; Surmacz et al., 2014) might suggest their broad substrate specificity. The function of the alternatively spliced PPRD1 with persisting intron 3 or intron 4 remains unclear. Alternative splicing of PPRD1 pre-mRNA might allow the plant to increase its adaptive potential in response to developmental and environmental cues, as suggested for other mRNA splice variants (Reddy et al., 2013). For instance, it might act as a gene expression regulatory mechanism since alternatively spliced PPRD1-INT3 and -INT4 give rise to enzymatically inactive and/or unstable proteins.
Polyprenol Reductase: A Hitherto Uncharacterized Component of Plant Cell Metabolism
Dol has been shown to participate in several vital cellular processes apart from protein glycosylation, e.g., aging and adaptation to adverse environmental conditions. Overexpression of PPRD1 protects plants against environmental factors and ER stress caused by tunicamycin and, in line with this, expression of PPRDs (especially PPRD2) is enhanced upon stress (Supplemental Figure 9). Most probably an elevated Dol level compensates, at least partially, for the tunicamycin-induced disturbances in protein glycosylation, as clearly shown for pollen. Similarly, increased Dol-P-Man biosynthesis results in an increased resistance of DPMS1-OE plants to tunicamycin (Jadid et al., 2011). It seems worth mentioning that neither of the mutant lines characterized in this study, pprd2-1+/− and pprd2-2+/−, showed hypersensitivity to ammonium salts (mentioned earlier; Jadid et al., 2011). This discrepancy could result from differences in the experimental conditions used.
The enhancement of transcript accumulation of PPRDs, especially PPRD2, with plant age is in line with the increased Dol accumulation observed in senescing plant and mammalian tissues (summarized in Jones et al., 2009). Additionally, the tissue-specific expression pattern of PPRDs fits the Dol versus polyprenol accumulation observed in the roots and leaves, respectively, even though the reason for this dichotomy remains obscure.
The differences between PPRD1 and -2 expression patterns suggest that they have specific roles in diverse organs. PPRD2 mutants lack functional pollen, as clearly shown by the results of reciprocal crosses and microscopy observations of pollen germination in vivo and in vitro. The defects due to PPRD2 deficiency could be rescued by PPRD1 overexpression only partially and in a gene-dose-dependent manner. Thus, the development of sporophyte of pprd2 PPRD1-OE+/− (heterozygous for PPRD1-OE) was more severely affected than that of pprd2 PPRD1-OE, and similarly, seed formation, although considerably compromised in both lines, was substantially more efficient in the latter. This gene-dose effect is also mirrored at the molecular level, upon transcriptomic and lipid analysis of both lines. The partial compensation for PPRD2 deficit by PPRD1 suggests that these enzymes are not fully redundant. This seems understandable in light of their different cellular locations, of mainly the ER and the plasma membrane for PPRD2 and PPRD1, respectively, as suggested by the colocalization experiment. This result corroborates well with the in silico prediction (http://suba.plantenergy.uwa.edu.au/). Arabidopsis organelle proteome screening (Dunkley et al., 2006) has suggested that PPRD2 localizes to the ER, similarly to hSRD5A3 (Sagami et al., 1993; Cantagrel et al., 2010). The five TMDs predicted in the PPRDs makes ER membrane localization plausible. It seems likely, however, that besides these main cellular sites of residence, PPRDs might also undergo relocalization toward other cellular compartments. Dol biosynthetic machinery is involved in protein sorting and intracellular vesicular transport (Sato et al., 1999; Belgareh-Touzé et al., 2003), and Dol overproduction resulting from PPDR expression might result in the enhancement of these processes. The ER localization of PPRD2 is in line with the intracellular organization of Dol biosynthesis; formation of polyprenyl diphosphate, the substrate of PPRDs, is completed in ER (Skorupinska-Tudek et al., 2008). Moreover, Dol kinase is also localized in the ER of mammalian cells (Shridas and Waechter, 2006), and Dol-P thus produced is easily accessible to the ER-resident saccharide transferases biosynthesizing Dol-P-linked (oligo)saccharides (Supplemental Figure 10).
In summary, PPRD2 plays a crucial role in sporophyte growth and is critical for normal development of the male gametophyte, suggesting that protein glycosylation is required for reproductive processes.
Polyprenol Reductase: A Regulator of Pollen Development
The molecular mechanism linking the Dol cycle with biosynthesis of the pollen grain surface layer awaits clarification. Pollen wall comprises intine and exine (Quilichini et al., 2015). The intine is the innermost layer made up of cellulose and pectin. It maintains the structural integrity of pollen grains, as Arabidopsis plants with mutations in primary cell wall cellulose synthases (CESAs) produce collapsed or malformed pollen grains with aberrant pollen walls that lack or have uneven intine layer (Persson et al., 2007). Interestingly, the morphology of pollen grains of pprd2 resembles that of mutants in genes encoding catalytic subunits of CESA (cesa1-1 and cesa3-1). Moreover, the unevenly deposited cell wall of pollen grain, especially the intine layer, seems a common feature of pprd2-1 and mutants in cellulose synthase-encoding genes (compared with Figure 3F in Persson et al., 2007 and Figure 6 of this article). CESA requires N-glycosylated (Kang et al., 2008) KORRIGAN (KOR1) protein for activity (Mansoori et al., 2014), a fact that might explain the involvement of PPRD2 in intine formation.
The exine surrounding the intine is a complex structure made of sporopollenin (i.e., covalently cross-linked phenolics and polyhydroxylated aliphatics containing most probably also fatty alcohols, since fatty acid reductase encoded by MALE STERILITY2 is required for exine formation). Sporopollenin provides the rigid and sculptured framework of the exine, which serves to encapsulate and protect the contents of spores/pollen, and to assist in stigmatic capture. Components of the exine are biosynthesized by the tapetal cells and deposited on the surface of developing microspores (Quilichini et al., 2015 and references therein). Recent identification of the involvement of GPI-anchored nonspecific lipid transfer proteins in the biosynthesis or deposition of sporopollenin (Edstam et al., 2013) might suggest the role of Dol, a compulsory cofactor of the biosynthesis of GPI-anchored proteins, in sporopollenin biosynthesis. Despite the observations described above, genes encoding proteins of the Dol cycle, e.g., CPT, DOK, and PPRD, escaped identification within a large-scale genetic screen aimed at detection of those involved in pollen exine production (Suzuki et al., 2008; Dobritsa et al., 2011).
Although neither Dol nor Pren have ever been identified as components of pollen grain surface layers, this possibility has been raised based on the expression of the cis-prenyltransferase-encoding gene LLA66 (accession number DQ911525) of Lilium longiflorum in the microspores and the anther cell wall. Expression and activity of Ll-CPT correlated with tapetal growth and disintegration (Liu et al., 2011). Interestingly, the genes encoding close Arabidopsis homologs of Ll-CPT, namely, CPT4 and CPT5 (Surmacz and Swiezewska, 2011), are expressed mainly in inflorescence, pollen, and flower (Genevestigator). Their lipid products, polyprenyl diphosphates of not yet identified chain length, might possibly serve as substrates for PPRD2.
Besides its putative role as component of pollen grain surface layer(s), Dol might also regulate the function of tapetum and/or other pollen grain surface layers via the biosynthesis of particular glycosylated and/or GPI-anchored proteins. So far, such a role might be predicted based on the presence of several potential N-glycosylation sites identified in the sequence of polygalacturonase inhibitory protein (Bc-MF19) expressed exclusively in the tapetal cells and microspores during anther development in Chinese cabbage (Brassica campestris) Bc-MF19 (Huang et al., 2011). Even more interestingly, a gene encoding SKU5-SIMILAR18 (AT1G75790) protein possessing four potential N-glycosylation sites is coexpressed (TAIR) with ACYL-COA SYNTHETASE5, which is critical for pollen development and sporopollenin biosynthesis (Quilichini et al., 2015). Moreover, Dol deficiency might disturb the function of COBL10, a GPI-anchored protein that is a component of pollen tube internal machinery critical for normal pollen tube growth and directional sensing in the female transmitting tract (Li et al., 2013). In line with this, an essential role of Dol kinase (DOK) in pollen development and the pollen tube reception pathway (Lindner et al., 2015) has been described recently.
Taken together, these observations might explain the intine and exine malformations observed in the pollen grains of Dol-deficient pprd2 mutants.
Polyprenol Reductase: A Component of the Protein Glycosylation Machinery
Since Dol is an obligatory cofactor of protein glycosylation, mutations resulting in Dol deficiency should eventually lead to phenotypic aberrations observed for diverse mutants in this pathway. Indeed, developmental defects in male and female gametophytes leading to sterility similar to those observed for PPRD2 disruption have been reported for several null mutants, both upstream and downstream of Dol (Supplemental Table 6), e.g., for DOK deficiency (Kanehara et al., 2015).
For the pprd2 homozygous mutants, consistently with their Dol deficiency, defects in N-glycosylation were accompanied by degradation of some proteins, e.g., SKU5. Similarly, in Dol-P-Man synthase null mutants, reporter N-glycosylated and GPI-anchored proteins were affected (Jadid et al., 2011). The distinct phenotypes observed for various protein glycosylation-related mutants (Supplemental Table 6) are presumably caused by different degrees of hypoglycosylation of particular proteins.
Interestingly, in contrast to the mutations affecting the mevalonate pathway, those compromising the second isoprenoid-generating route in plants, the methylerythritol phosphate pathway, have not been shown to result in plant sterility.
The diverse clinical manifestations of hSRD5A3 mutations do not include impaired fertility. Moreover, the excess of polyprenols accumulated in SRD5A3 patients’ cells is considered toxic due to the polyprenol versus Dol competition during polyisoprenoidPP-tetradecasaccharide assembly (Gründahl et al., 2012). In Arabidopsis leaves, the content of polyprenols (Pren-10 dominating) highly exceeds that of Dols (Dol-16 dominating) (Gawarecka and Swiezewska, 2014), but the considerable difference of their chain length suggests that the substrate specificity of Dol kinase and/or saccharyl transferases protects these cells against the potential toxicity of Pren excess. Moreover, sequestration of Pren in plastids might also serve as a salvage mechanism.
In conclusion, the PPRD2 polyprenol reductase is essential for the development of gametophytes and the sporophyte, and the pprd2 knockout mutation is lethal due to male sterility. Defective protein glycosylation seems the major reason for these malfunctions, although disturbances in other cellular Dol-dependent processes could also contribute. Additionally, involvement of PPRDs in other pathways besides the Dol cycle, e.g., reduction of other vital cellular substrates, cannot be ruled out.
Supplementation of polyprenol reductase-deficient cells with Dol has been discussed as a method to compensate for deficiency in protein glycosylation, and, from a longer perspective, supplementation of the human diet with Dol-enriched plant tissues could allow new therapeutic interventions for glycosylation disorders. Complete elucidation of the Dol cycle in plant cells, via the current identification of PPRD1 and -2, renders manipulation of Dol content in plant tissues theoretically feasible.
The experimental model developed in this article, revealing beneficial effects of supplementation with exogenous Dol on PPRD2-deficient pollen tube development, might be suitable to test new therapeutic strategies of SRD5A3-CDG polyprenol reductase deficiency therapy since no such model is currently available.
Finally, an alternative albeit still not recognized pathway leading to Dol has been postulated in yeast and human to account for the residual Dol and partially retained protein glycosylation found in DFG10 and SDR5A3 and null mutants (Cantagrel et al., 2010; Gründahl et al., 2012). Whether a similar, SRD5A3/PPRD-independent Dol-producing pathway functions in plants awaits clarification (Supplemental Figure 10).
METHODS
Plants and Plant Growth Conditions
Arabidopsis thaliana ecotype Col-0 (wild-type control) as well as mutant lines pprd2-1 and pprd2-2 (SALK_113221C and SALK_006421, respectively) and PPRD1-OE (GK_575B02) were from the Nottingham Arabidopsis Stock Center.
Progeny of pprd2-1, pprd2-2, and PPRD1-OE lines were genotyped and PPRD1-OE homozygous line was isolated. pprd2 mutants were maintained as heterozygous segregating lines due to lethality of the homozygote.
In pprd2-2, an additional T-DNA insertion on the fifth chromosome was found, and after segregation, plants with single insertion located exclusively in PPRD2 coding sequence (CDS) on chromosome 2 (At2G16530 locus) were obtained. For the pprd2 PPRD1-OE and pprd2 PPRD1-OE+/− plants, PPRD1-OE was crossed to pprd2-1+/− and the F2 generation of the progeny was screened by PCR for homozygosity. F3 plants homozygous for both traits (pprd2 PPRD1-OE) or heterozygous for PPRD1-OE (pprd2 PPRD1-OE+/−) were used. PPRD1pro:GUS or PPRD2pro:GUS expressing lines were constructed by Agrobacterium tumefaciens transformation of wild-type Arabidopsis Col-0 with gateway binary vector pGWB633 vector carrying GUS under native PPRD1 or PPRD2 promoter (Nakamura et al., 2010); sequences of the primers are listed in Supplemental Table 7. The fourth homozygous generation after transformation was used for experiments.
Plants for segregation analysis, phenotyping, and pollen collection were grown in a greenhouse under a long-day (16 h light) photoperiod at 21/18°C. Plants for RNA extraction, RT-PCR, lipid analysis, and GUS histochemical analysis were grown in hydroponic culture in Gilbert medium in a growth chamber (AR-66L CLF Plant Climatics). Seedlings for GUS histochemical analysis were grown on plates with half-strength Murashige and Skoog (MS) medium supplemented with vitamins and 1% sucrose and solidified with 1.2% agar. To test the effect of stressors on seed germination, standard 0.5× MS growth medium supplemented as above and with an appropriate stressor, sorbitol (final concentration 300 mM) or NaCl (150 mM), was used. To test the effect of tunicamycin on plant development, 4-d-old seedlings grown on plates with standard 0.5× MS medium were transferred onto plates supplemented with tunicamycin (final concentration 200 ng/mL) and grown for 10 d.
Pollen for RNA extraction and gene expression analysis was collected as described earlier (Becker et al., 2003). Genotyping was performed with specific primers designed with the aid of T-primer design tool and LBb1.3 primer (Supplemental Table 7).
Cloning of Arabidopsis Genes Encoding Polyprenol Reductase, PPRD1 and PPRD2
Putative genes encoding polyprenol reductases were identified in the Arabidopsis genomic sequence by searching with human gene encoding polyprenol reductase hSRD5A3. Total RNA from Arabidopsis roots was isolated and purified using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. The mRNA was transcribed to cDNA, and PPRD1 and PPRD2 CDSs were amplified by PCR using specific primers (Supplemental Table 7). DNA fragments with CACC sequence at the 5′-end were cloned into the pENTR vector according to manufacturer’s instructions (pENTR D-TOPO; Invitrogen).
Heterologous Expression of PPRD1 and -2 and Analysis of CPY Glycosylation in Saccharomyces cerevisiae Transformants
PPRD1 and PPRD2 CDSs were recombined from pENTR to the yeast expression vector pYES-DEST52 using the LR Clonase enzyme mix according to manufacturer’s instructions (Gateway LR Clonase II Enzyme mix; Invitrogen).
Yeast S. cerevisiae strains (BY4741, L5366, and mutant strains dfg10∆ and dfg10-100 generated in the BY4741 and L5366 genetic background, respectively; Cantagrel et al., 2010) were grown on liquid minimal medium (0.67% Yeast Nitrogen Base, 2% galactose, and appropriate amino acids without uracil) to stationary phase.
Protein extraction was done using postalkaline extraction (Kushnirov, 2000).
Proteins were separated on 12% SDS-PAGE and transferred for 1 h to nitrocellulose membrane. The blots were incubated in a primary antibody solution (anti-CPY mouse antibody; Invitrogen) at a dilution of 1:2000 overnight in the cold room with agitation. They were then washed three times in PBS-T and incubated with secondary antibody (anti-mouse IgG horseradish peroxidase-conjugated; Sigma-Aldrich) diluted to 1:1000 for 1.5 h at room temperature with agitation. The blots were washed as above and developed for 1 min with ECL detection reagent (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific) according to the manufacturer’s instructions.
Transformation with empty vector or native yeast DFG10 was used as negative or positive control, respectively.
Site-Directed Mutagenesis of PPRD2
Mutations in PPRD2 CDS were introduced by PCR site-directed mutagenesis using specific primers with modified nucleotide sequence (Supplemental Table 7).
Polyisoprenoid Profiling of Yeast Strains
Yeast strains were cultured in minimal media and harvested in log phase (OD600 1). Cultures (100 mL, 48 h) were centrifuged for 10 min at 1500g (Allegra; Beckman). Supernatant was removed by decantation; the pellet was resuspended in 5 mL hydrolyzing mixture (0.5M KOH in 60% ethanol) and placed in hot water bath (85°C) for 1 h. After cooling down, 15 mL water and 5 mL hexane were added. Lipids were extracted five times with 5-mL portions of hexane. Pooled organic fractions were evaporated under stream of nitrogen and dissolved in 200 μL propan-2-ol. Polyisoprenoid alcohols were analyzed as described previously (Jozwiak et al., 2013). Polyprenol and Dol standards were from the Collection of Polyprenols (Institute of Biochemistry and Biophysics, Polish Academy of Sciences [IBB PAS]).
Heterologous Expression of PPRD1 and PPRD2 in E. coli
Plasmids harboring PPRD1 or PPRD2 were prepared by cloning appropriate nucleotide sequences into pGEX-4T-1 vector (GE Healthcare Life Sciences) cut blunt-ended with SmaI enzyme. The resulting vectors produced N-terminal GST fusions of PPRD1 or PPRD2 protein.
Escherichia coli BL21 strain was transformed with the above plasmids and grown on Luria-Bertani plates with ampicillin (100 μg/mL); empty pGEX-4T-1 vector was used as a negative control. Overnight bacterial culture (250 μL) was transferred to fresh medium (25 mL) and grown to OD600 of 1.0. Expression of PPRD1 and -2 was induced by the addition of 25 μL 0.1 M IPTG, followed by incubation for 4 h.
For protein purification and mass spectrometry analysis, 20 mL bacterial cell culture was harvested by centrifugation, resuspended in lysis buffer (50 mM Tris, pH 8.0, 10% glycerol, 0.1% Triton X-100, 100 μg/mL lysozyme, 1 mM PMSF, and 2 mM MgCl2), and incubated for 30 min on ice and sonicated for 5 × 1 min. Crude lysate was used for enzymatic assay. Protein bands corresponding to PPRDs were excised from SDS-PAGE gel and analyzed by mass spectrometry.
In Vitro Reduction of Polyprenol by PPRD1 or PPRD2
The assay was performed following the procedures described previously (Sagami et al., 1993; Cantagrel et al., 2010) with modifications. The reaction mixture consisted of 50 mM Tris-HCl (pH 8.0), 1 mM DTT, 50 mM KF, 20% glycerol, 2 mM MgCl2, 0.1% Triton X-100, and 3.3 μg Pren-16 (C80) in 5 μL ethanol. After 25 min sonication in a water bath, the reaction was initiated with the addition of 10 mM NADPH and 700 μg crude cell-extract proteins to give a final volume of 1250 μL. After incubation for 18 h at 24°C, the samples were supplemented with 1 mL chloroform:methanol (2:1; v/v), and lipids were extracted and analyzed by HPLC/UV as described previously (Jozwiak et al., 2013). Polyprenol and Dol standards were from the Collection of Polyprenols.
Cloning of Promoters of PPRD1 and -2 and Construction of Lines Expressing PPRD1pro:GUS and PPRD2pro:GUS
To generate reporter constructs, the promoter regions of PPRD1 (–1103 bp upstream of ATG) and PPRD2 (–1041 bp upstream of ATG) were amplified from genomic DNA and cloned into the Gateway entry vector pENTR (Invitrogen). The promoter sequences were then subcloned into the binary plant transformation vector pGWB633, resulting in reporter constructs. The reporter constructs were used to transform Agrobacterium.
The Agrobacterium transformants were used to generate lines expressing PPRD1pro:GUS and PPRD2pro:GUS using floral dip method as described previously (Bent, 2006).
Histochemical Analysis of GUS Activity
Histochemical localization of GUS in several independent transgenic lines harboring the PPRD1pro:GUS or PPRD2pro:GUS constructs was performed as described by Jefferson et al. (1987) with some modifications. Sample tissues were infiltrated with the reaction buffer [50 mM Na2HPO4-NaH2PO4, pH 7.0, 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6] containing 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) as substrate under vacuum, and incubated at 37°C overnight. Plant pigments were extracted with 80% ethanol, and the histochemical staining was analyzed under a binocular microscope (SMZ1500; Nikon).
Subcellular Localization Analysis of PPRD1 and PPRD2
Agrobacterium-mediated transient expression in Nicotiana benthamiana was employed. PPRD1 and PPRD2 were cloned into the pGWB451 Gateway binary vector (Nakagawa et al., 2007). cd3-959, cd-967, and cd3-1007 vectors were used as ER, Golgi, and PM markers, respectively (Nelson et al., 2007). Recombinant plasmid (50 ng) was introduced into Agrobacterium strain GV3101 using the freeze/thaw method (Weigel and Glazebrook, 2006).
For infiltration, recombinant strains were grown overnight at 28°C with agitation in Luria-Bertani medium supplemented with spectinomycin (30 μg/mL) or kanamycin (30 μg/mL), respectively. Cells were pelleted at 3300g for 3 min at 20°C, resuspended in infiltration medium containing 10 mM MES (pH 5.6) and 10 mM MgCl2 supplemented with 100 μM acetosyringone, and diluted to OD600 of 0.4. After incubation at room temperature for 3 h, cells were infiltrated by injection into the bottom side of the third or fourth leaf of 6-week-old N. benthamiana plants with a needless syringe. After 24 to 48 h, leaf discs of the infiltrated areas were observed under a confocal microscope.
Quantitative PCR Analysis of Expression of Genes Encoding Polyprenol Reductase and BiP2
Total RNA was isolated and purified using the RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. First-strand cDNA synthesis was performed with 2 μg of RNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific) and oligo(dT) primers according to the manufacturer’s procedure.
PPRD1, PPRD2, or BiP2 expression analysis was performed in a total volume of 20 μL Luminaris Color HiGreen high ROX qPCR Master Mix (Thermo Scientific) using gene-specific primers in a StepOnePlus Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions.
The cycle threshold (Ct) was used to determine the relative expression level of a given gene using the 2−ΔΔCt method. The relative expression level of PPRD1, PPRD2, and BiP2 was normalized against ACTIN2. Statistical analysis of the qPCR data was performed using one-way ANOVA with Tukey’s post test.
Analysis of Protein Glycosylation
Proteins were isolated according to Guillaumot et al. (2009). Protein extracts (30 µg/line) were separated by SDS-PAGE (10%), and two methods, using either Concanavalin A or Emerald 300, were subsequently employed to detect glycosylated proteins.
For the Concanavalin A method, the SDS-PAGE gels prepared in parallel were either stained with Coomassie Brilliant Blue or transferred to membrane as described above; the membrane was probed for 17 h with Concanavalin A labeled with horseradish peroxidase (1 μg/mL; Sigma-Aldrich), washed with PBS for 50 min, and developed for 5 s with ECL reagent (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific) according to the manufacturer’s instructions. Detection was performed with Molecular Imager ChemiDoc XRS+ with ImageLab software (Bio-Rad).
For the Emerald 300 method, SDS-PAGE gels prepared in parallel were stained with either Coomassie Brilliant Blue or the Pro-Q Emerald 300 Glycoprotein Gel and Blot Stain Kit (Invitrogen) according to the manufacturer’s instructions.
Immunodetection of SKU5 and BiP2 by Immunoblotting
Proteins (30 µg/line) separated on 10% SDS-PAGE were transferred to membrane as described before and probed for 12 h with either anti-SKU5 (1:1000; provided by J.C. Sedbrook, Illinois State University) or anti-BiP2 (1:2000; Agrisera) antibody (catalog number AS09 481). They were then washed three times in PBS-T and incubated with secondary antibody (anti-rabbit IgG horseradish peroxidase-conjugated; Sigma-Aldrich) diluted to 1:1000 for 1 h at room temperature with agitation. The blots were washed and developed for 5 min with ECL detection reagent (SuperSignal West Pico Chemiluminescent Substrate; Thermo Scientific) according to the manufacturer’s instructions.
Pollen in Vitro Germination
In vitro pollen germination and growth were performed on solid medium for 16 h at 22°C according to Boavida and McCormick (2007). For the chemical complementation experiment, Pren-16 or Dol mix (Dol-16 to Dol-21) (Collection of Polyprenols) was added to the hot pollen germination medium at a final concentration of 10 mM.
Aniline Blue Staining of Pollen Tubes Growing in the Pistil
Hand-pollinated Arabidopsis pistils were collected 12 h after pollination and placed in Carnoy’s solution (60% ethanol, 30% chloroform, and 10% acetic acid). After ∼3 h, the fixative was changed to 70% ethanol and left for 10 min at room temperature. After that, the same treatment was performed using 50% ethanol, 30% ethanol, and water. The specimens were moved into alkaline solution (1 M NaOH) and left covered overnight at room temperature. Pistils were washed with water for 10 min and then stained with 0.1% aniline blue in 50 mM K3PO4 (pH 11) for 2 h in darkness. The morphology of pollen tube growth in the pistil was observed under an Eclipse E800 microscope (Nikon Instruments).
Staining of Pollen Anthers with Alexander Stain
Staining of pollen anthers with Alexander stain and microscopy observations were performed as described earlier (Gutkowska et al., 2015).
Microscopy Observations
The morphology of pollen germinating in vitro was observed under an Eclipse E800 microscope (Nikon Instruments) equipped with a CCD camera (Hamamatsu). Image acquisition was performed with Lucia software (Laboratory Imaging). At least 10 visual fields of material derived from six different plants obtained from three independent plant cultivations were used for scoring in each case. The morphology of flowers and siliques was studied under a Nikon D7000 camera equipped with a Nikon AF-S 60 f/2.8 G ED micro lens.
Subcellular localization of PPRD1 and PPRD2 was observed under a C1 confocal microscope (Nikon Instruments). Images were recorded with the EZ C1 image acquisition software (Nikon Instruments) and processed with EZ-C1 Viewer v.3.6 (Nikon Instruments).
For TEM observations, flowers (green buds) were fixed in 2.5% glutaraldehyde in 100 mM cacodylate buffer (pH 7.2) overnight, rinsed once in the buffer, and postfixed in 1% osmium tetroxide overnight. Samples were rinsed, dehydrated in a graded ethanol series (30, 50, 75, and 100%), and finally embedded in epoxy resin. Ultrathin sections were cut with a diamond knife on an MTX ultramicrotome (RMC Boeckeler Instruments). Specimens were examined using a LEO 912AB transmission electron microscope (Carl Zeiss). Grains derived from the same pprd2-1+/− mutant plant were used to compare the phenotypically wild-type versus pprd2-1 pollen. For scanning electron microscopy observations, pollen was spilled directly on microscope tables, coated with a thin layer of gold, and examined using a LEO 1430VP scanning electron microscope (Carl Zeiss).
Extraction of Lipids from Arabidopsis Tissues
Plant material (∼10g of fresh mass, collected from three plants) was supplemented with an internal standard (Prenol-19, 10 μL, 1 μg/μL; Collection of Polyprenols) and 20 mL chloroform:methanol mixture (1:1, v/v) was added. The tissue was homogenized with an Ultra-Turrax apparatus (IKA Labortechnik). After dispersion, the mixture was agitated for 24 h at room temperature. The homogenate was filtered under reduced pressure, and the remaining tissue was reextracted with 20 mL chloroform:methanol (2:1, v/v) and then 20 mL chloroform. The filtrates were pooled and evaporated under reduced pressure. Crude lipid extract was hydrolyzed, purified on a silica gel column, and analyzed by HPLC-UV as described previously (Jozwiak et al., 2013). Each lipid extraction was performed in triplicate.
Bioinformatics
In silico analyses of Arabidopsis polyprenol reductases were performed using BLAST P (comparison of protein sequences and identification of homologs), ClustalW (multiple sequence alignment; Supplemental Data Set 1), MEGA 6 (phylogenetic tree representation), TMHMM Server v. 2.0 (topology prediction and prediction of membrane-spanning segments), and the Pfam database (http://pfam.xfam.org/; Wellcome Trust Sanger Institute, Cambridge, UK; identification of steroid 5α-reductase domains). PPRD protein sequences were aligned using a Gonnet matrix by ClustalW (Larkin et al., 2007), with an open gap penalty of 10 and an extend gap penalty of 0.1 in pairwise alignments, an extend gap penalty of 0.2 in the multiple alignment, and a delay divergent setting of 30%. Phylogenetic relationships among the PPRDs were reconstructed using a neighbor-joining method by MEGA 6 (Tamura et al., 2013) with the Poisson amino acid substitution model. Two thousand bootstrap replicates were performed in each analysis to obtain the confidence support.
Accession Numbers
Sequence data from this article can be found in TAIR under the following accession numbers: PPRD1 (At1g72590), PPRD2 (At2g16530), BiP2 (At5g42020), and ACTIN-2 (At3g18780). SRD5A3 is in GenBank under NM_024592.4.
Supplemental Data
Supplemental Figure 1. Alignment of Amino Acid Sequences of Proteins with Steroid 5α-Reductase Domain from Human (hSRD5A3), Yeast Saccharomyces cerevisiae (Dfg10p), and 60 Plant Species.
Supplemental Figure 2. Evolutionary Tree of Proteins with Steroid 5α-Reductase Domain.
Supplemental Figure 3. Models of PPRDs.
Supplemental Figure 4. Schematic Representation and Expression of PPRDs.
Supplemental Figure 5. Subcellular localization of PPRD2 and PPRD1.
Supplemental Figure 6. Phenotypes of Wild-Type, pprd2-1+/−, PPRD1-OE, pprd2 PPRD1-OE, and pprd2 PPRD1-OE+/− Plants.
Supplemental Figure 7. Analysis of Glycosylated Proteins in Wild-Type, pprd2-1+/−, pprd2 PPRD1-OE, and pprd2 PPRD1-OE+/− Plants.
Supplemental Figure 8. Alexander Staining of Mature Anthers of Wild-Type and pprd2-1+/− Plants and Aniline Blue Staining of Pollen Tubes in the Pistil.
Supplemental Figure 9. Effect of At-PPRD1 Overexpression on Plant Tolerance to Stress.
Supplemental Figure 10. Dol Cycle in Plant Cells.
Supplemental Table 1. Comparison of Amino Acid Sequences of Putative Arabidopsis PPRDs with Human and Yeast Polyprenol Reductases Based on the Alignment.
Supplemental Table 2. Localization of Predicted 3-oxo-5-α-Steroid 4-Dehydrogenase Domain in Arabidopsis Polyprenol Reductases.
Supplemental Table 3. Seed Germination of pprd2-1+/− and pprd2-2+/− Plants on Solid Medium.
Supplemental Table 4. Mass Spectrometry-Based Differential Proteomics of Wild-Type and pprd2 PPRD1-OE+/− Plants.
Supplemental Table 5. Effect of Polyprenol, Dol, and Tunicamycin on Pollen Germination.
Supplemental Table 6. Phenotypes of Arabidopsis Mutants in Genes Encoding Elements of Isoprenoid Biosynthesis or Protein Glycosylation Pathways.
Supplemental Table 7. Primers Used for Genotyping, Cloning, Mutagenesis, and Expression Studies.
Supplemental Data Set 1. Text File of the Alignment Used for the Phylogenetic Analysis Shown in Supplemental Figure 2.
Supplementary Material
Acknowledgments
Financial support provided by the National Science Centre of Poland (Grant UMO-2012/06/M/NZ3/00155) is kindly acknowledged. We thank Vincent Cantagrel (Université René Descartes, Paris 5) for dfg10∆ and dfg10-100 yeast mutants, John C. Sedbrook (Illinois State University) for anti-SKU5 antibody, Ruslan Yatusevich (IBB PAS) for pGWB633 vector and for help with promoter analysis, and Przemyslaw Surowiecki (IBB PAS) for help with analysis of subcellular localization of PPRDs.
AUTHOR CONTRIBUTIONS
A.J., M.G., and E.S. designed the research. A.J., K.G., L.S., A.B., M.L., and J.N. performed the research. A.J., M.G., and E.S. analyzed data and wrote the article.
Glossary
- Dol
dolichol
- Dol-P
dolichyl phosphate
- GPI
glycosylphosphatidylinositol
- TMD
transmembrane domain
- CPY
carboxypeptidase Y
- PM
plasma membrane
- ER
endoplasmic reticulum
- Pren
prenol
- TEM
transmission electron microscopy
- MS
Murashige and Skoog
- CDS
coding sequence
- IBB PAS
Institute of Biochemistry and Biophysics, Polish Academy of Sciences
Footnotes
Articles can be viewed online without a subscription.
References
- Bajda A., et al. (2009). Role of polyisoprenoids in tobacco resistance against biotic stresses. Physiol. Plant. 135: 351–364. [DOI] [PubMed] [Google Scholar]
- Becker J.D., Boavida L.C., Carneiro J., Haury M., Feijó J.A. (2003). Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol. 133: 713–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh-Touzé N., Corral-Debrinski M., Launhardt H., Galan J.M., Munder T., Le Panse S., Haguenauer-Tsapis R. (2003). Yeast functional analysis: identification of two essential genes involved in ER to Golgi trafficking. Traffic 4: 607–617. [DOI] [PubMed] [Google Scholar]
- Bent A. (2006). Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 343: 87–103. [DOI] [PubMed] [Google Scholar]
- Bergamini E. (2003). Dolichol: an essential part in the antioxidant machinery of cell membranes? Biogerontology 4: 337–339. [DOI] [PubMed] [Google Scholar]
- Boavida L.C., McCormick S. (2007). Temperature as a determinant factor for increased and reproducible in vitro pollen germination in Arabidopsis thaliana. Plant J. 52: 570–582. [DOI] [PubMed] [Google Scholar]
- Buczkowska A., Swiezewska E., Lefeber D.J. (2015). Genetic defects in dolichol metabolism. J. Inherit. Metab. Dis. 38: 157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantagrel V., et al. (2010). SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 142: 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobritsa A.A., Geanconteri A., Shrestha J., Carlson A., Kooyers N., Coerper D., Urbanczyk-Wochniak E., Bench B.J., Sumner L.W., Swanson R., Preuss D. (2011). A large-scale genetic screen in Arabidopsis to identify genes involved in pollen exine production. Plant Physiol. 157: 947–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkley T.P.J., et al. (2006). Mapping the Arabidopsis organelle proteome. Proc. Natl. Acad. Sci. USA 103: 6518–6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstam M.M., Blomqvist K., Eklöf A., Wennergren U., Edqvist J. (2013). Coexpression patterns indicate that GPI-anchored non-specific lipid transfer proteins are involved in accumulation of cuticular wax, suberin and sporopollenin. Plant Mol. Biol. 83: 625–649. [DOI] [PubMed] [Google Scholar]
- Gawarecka K., Swiezewska E. (2014). Analysis of plant polyisoprenoids. Methods Mol. Biol. 1153: 135–147. [DOI] [PubMed] [Google Scholar]
- Gillmor C.S., Lukowitz W., Brininstool G., Sedbrook J.C., Hamann T., Poindexter P., Somerville C. (2005). Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell 17: 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründahl J.E., et al. (2012). Life with too much polyprenol: polyprenol reductase deficiency. Mol. Genet. Metab. 105: 642–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumot D., Guillon S., Déplanque T., Vanhee C., Gumy C., Masquelier D., Morsomme P., Batoko H. (2009). The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum-Golgi-localized membrane protein. Plant J. 60: 242–256. [DOI] [PubMed] [Google Scholar]
- Gutkowska M., Wnuk M., Nowakowska J., Lichocka M., Stronkowski M.M., Swiezewska E. (2015). Rab geranylgeranyl transferase β subunit is essential for male fertility and tip growth in Arabidopsis. J. Exp. Bot. 66: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Liu Y., Yu X., Xiang X., Cao J. (2011). A polygalacturonase inhibitory protein gene (BcMF19) expressed during pollen development in Chinese cabbage-pak-choi. Mol. Biol. Rep. 38: 545–552. [DOI] [PubMed] [Google Scholar]
- Jadid N., Mialoundama A.S., Heintz D., Ayoub D., Erhardt M., Mutterer J., Meyer D., Alioua A., Van Dorsselaer A., Rahier A., Camara B., Bouvier F. (2011). DOLICHOL PHOSPHATE MANNOSE SYNTHASE1 mediates the biogenesis of isoprenyl-linked glycans and influences development, stress response, and ammonium hypersensitivity in Arabidopsis. Plant Cell 23: 1985–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.B., Rosenberg J.N., Betenbaugh M.J., Krag S.S. (2009). Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim. Biophys. Acta 1790: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwiak A., Ples M., Skorupinska-Tudek K., Kania M., Dydak M., Danikiewicz W., Swiezewska E. (2013). Sugar availability modulates polyisoprenoid and phytosterol profiles in Arabidopsis thaliana hairy root culture. Biochim. Biophys. Acta 1831: 438–447. [DOI] [PubMed] [Google Scholar]
- Kanehara K., Cho Y., Lin Y.-C., Chen C.-E., Yu C.-Y., Nakamura Y. (2015). Arabidopsis DOK1 encodes a functional dolichol kinase involved in reproduction. Plant J. 81: 292–303. [DOI] [PubMed] [Google Scholar]
- Kang J.S., et al. (2008). Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105: 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- Lalanne E., Honys D., Johnson A., Borner G.H.H., Lilley K.S., Dupree P., Grossniklaus U., Twell D. (2004). SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. Plant Cell 16: 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- Lefeber D.J., et al. (2011). Autosomal recessive dilated cardiomyopathy due to DOLK mutations results from abnormal dystroglycan O-mannosylation. PLoS Genet. 7: e1002427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ge F.R., Xu M., Zhao X.Y., Huang G.Q., Zhou L.Z., Wang J.G., Kombrink A., McCormick S., Zhang X.S., Zhang Y. (2013). Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J. 74: 486–497. [DOI] [PubMed] [Google Scholar]
- Lindner H., Kessler S.A., Müller L.M., Shimosato-Asano H., Boisson-Dernier A., Grossniklaus U. (2015). TURAN and EVAN mediate pollen tube reception in Arabidopsis synergids through protein glycosylation. PLoS Biol. 13: e1002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Howell S.H. (2010). Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. Plant Cell 22: 2930–2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.C., Wang B.J., Huang J.K., Wang C.S. (2011). Expression, localization and function of a cis-prenyltransferase in the tapetum and microspores of lily anthers. Plant Cell Physiol. 52: 1487–1500. [DOI] [PubMed] [Google Scholar]
- Mansoori N., Timmers J., Desprez T., Kamei C.L., Dees D.C., Vincken J.P., Visser R.G., Höfte H., Vernhettes S., Trindade L.M. (2014). KORRIGAN1 interacts specifically with integral components of the cellulose synthase machinery. PLoS One 9: e112387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moremen K.W., Tiemeyer M., Nairn A.V. (2012). Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13: 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Mano S., Tanaka Y., Ohnishi M., Nakamori C., Araki M., Niwa T., Nishimura M., Kaminaka H., Nakagawa T., Sato Y., Ishiguro S. (2010). Gateway binary vectors with the bialaphos resistance gene, bar, as a selection marker for plant transformation. Biosci. Biotechnol. Biochem. 74: 1315–1319. [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136. [DOI] [PubMed] [Google Scholar]
- Pattison R.J., Amtmann A. (2009). N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14: 92–99. [DOI] [PubMed] [Google Scholar]
- Persson S., Paredez A., Carroll A., Palsdottir H., Doblin M., Poindexter P., Khitrov N., Auer M., Somerville C.R. (2007). Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 15566–15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilichini T.D., Grienenberger E., Douglas C.J. (2015). The biosynthesis, composition and assembly of the outer pollen wall: A tough case to crack. Phytochemistry 113: 170–182. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Marquez Y., Kalyna M., Barta A. (2013). Complexity of the alternative splicing landscape in plants. Plant Cell 25: 3657–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagami H., Kurisaki A., Ogura K. (1993). Formation of dolichol from dehydrodolichol is catalyzed by NADPH-dependent reductase localized in microsomes of rat liver. J. Biol. Chem. 268: 10109–10113. [PubMed] [Google Scholar]
- Sato M., Sato K., Nishikawa S., Hirata A., Kato J., Nakano A. (1999). The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz F., Aebi M. (2011). Mechanisms and principles of N-linked protein glycosylation. Curr. Opin. Struct. Biol. 21: 576–582. [DOI] [PubMed] [Google Scholar]
- Sedbrook J.C., Carroll K.L., Hung K.F., Masson P.H., Somerville C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol-anchored glycoprotein involved in directional root growth. Plant Cell 14: 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shridas P., Waechter C.J. (2006). Human dolichol kinase, a polytopic endoplasmic reticulum membrane protein with a cytoplasmically oriented CTP-binding site. J. Biol. Chem. 281: 31696–31704. [DOI] [PubMed] [Google Scholar]
- Skorupińska-Tudek K., Bieńkowski T., Olszowska O., Furmanowa M., Chojnacki T., Danikiewicz W., Swiezewska E. (2003). Divergent pattern of polyisoprenoid alcohols in the tissues of Coluria geoides: a new electrospray ionization MS approach. Lipids 38: 981–990. [DOI] [PubMed] [Google Scholar]
- Skorupinska-Tudek K., et al. (2008). Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of dolichols in plants. J. Biol. Chem. 283: 21024–21035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmacz L., Plochocka D., Kania M., Danikiewicz W., Swiezewska E. (2014). cis-Prenyltransferase atCPT6 produces a family of very short-chain polyisoprenoids in planta. Biochim. Biophys. Acta 1841: 240–250. [DOI] [PubMed] [Google Scholar]
- Surmacz L., Swiezewska E. (2011). Polyisoprenoids - Secondary metabolites or physiologically important superlipids? Biochem. Biophys. Res. Commun. 407: 627–632. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Masaoka K., Nishi M., Nakamura K., Ishiguro S. (2008). Identification of kaonashi mutants showing abnormal pollen exine structure in Arabidopsis thaliana. Plant Cell Physiol. 49: 1465–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewska E., Danikiewicz W. (2005). Polyisoprenoids: structure, biosynthesis and function. Prog. Lipid Res. 44: 235–258. [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiram H.V., Erickson H.K., Poulter C.D. (2007). Chimeras of two isoprenoid synthases catalyze all four coupling reactions in isoprenoid biosynthesis. Science 316: 73–76. [DOI] [PubMed] [Google Scholar]
- Weigel, D., and Glazebrook, J. (2006). In planta transformation of Arabidopsis. CSH Protoc. 2006: pdb.prot4668. [DOI] [PubMed]
- Wigley W.C., Prihoda J.S., Mowszowicz I., Mendonca B.B., New M.I., Wilson J.D., Russell D.W. (1994). Natural mutagenesis study of the human steroid 5 alpha-reductase 2 isozyme. Biochemistry 33: 1265–1270. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.