Abstract
Inherited colorectal cancer syndromes in humans exhibit regional specificity for tumor formation. By using mice with germline mutations in the adenomatous polyposis coli gene (Apc) and/or DNA mismatch repair genes, we have analyzed the genetic control of tumor regionality in the mouse small intestine. In C57BL/6 mice heterozygous for the Apc multiple intestinal neoplasia mutation (ApcMin), in which tumors are initiated by loss of heterozygosity by means of somatic recombination, tumors form preferentially in the distal region of the small intestine. By contrast, the formation of tumors initiated by allelic silencing on the AKR ApcMin genetic background is strongly skewed toward the ileocecal junction. A third tumor regionality is displayed by tumors that develop in MMR-deficient ApcMin/+ mice, in which mutation of the Apc gene is responsible for tumor initiation. Thus, tumor regionality in the small intestine of ApcMin/+ reflects the mechanism by which the wild-type allele of Apc is inactivated. We have reexamined the mechanism of Apc loss in tumors from Apc1638N/+ mice, in which tumors of the small intestine develop in a regional pattern overlapping that of mismatch repair-deficient mice. In contrast to previous reports, we find that tumors from Apc1638N/+ mice on a congenic C57BL/6 background maintain the wild-type allele of Apc. Our studies demonstrate a pathway-specific regionality for tumor development in mouse models for inherited intestinal cancer, an observation that is reminiscent of the regional preference for tumor development in the human colon. Perhaps, the power of mouse genetics and biology can be harnessed to identify genetic and other factors that contribute to tumor regionality.
The mammalian intestine is highly proliferative and is continually exposed to a genotoxic environment. As such, it is a common site of cancer; cancers of the gastrointestinal tract account for >80,000 deaths per year (1). Clearly, it is important to understand gastrointestinal cancer pathology at the molecular, cellular, and histologic levels.
In the United States, the most common gastrointestinal malignancy is colorectal cancer. The majority of colorectal cancer cases are sporadic, without a known hereditary component. Yet rare inherited forms of this disease exist and have been highly informative. Familial adenomatous polyposis (FAP; Online Mendelian Inheritance in Man no. 175100), or Gardner's syndrome, is an autosomal dominant disease caused by a germline mutation in the adenomatous polyposis coli (APC) tumor suppressor gene. Individuals with FAP can develop several thousand benign polyps throughout the colon. Although the benign lesions themselves are not dangerous, 5% of these adenomas will progress to malignancy (2). A second form of inherited colorectal cancer, hereditary nonpolyposis colorectal cancer (HNPCC; Online Mendelian Inheritance in Man no. 114500), results from mutations in DNA mismatch repair (MMR) genes (3). Individuals with HNPCC have an increased risk for cancer of the colon, endometrium, and ovary, among other organs (3). HNPCC results from mutation in any of a number of genes that function in the DNA MMR pathway, although mutations in MutS homolog 2 and MutL homolog 1 (MLH1) predominate (4, 5).
In these inherited cancers, it is notable that the tumors develop in distinct regions of the colon. Adenomas develop primarily in the descending colon of patients with FAP (6), whereas tumors from HNPCC patients arise predominantly in the ascending colon (7). Likewise, sporadic tumors with APC mutations tend to develop in the descending colon, and tumors with MMR mutations tend to develop in the ascending colon. What are the genetic and/or environmental factors that control this regional bias?
The laboratory mouse provides a setting in which to study the many genetic pathways that regulate intestinal tumorigenesis. Multiple intestinal neoplasia (Min) mice, a model of FAP, are heterozygous for ApcMin/+(Min), a germ-line truncating mutation at codon 850 of the Apc gene and develop multiple intestinal neoplasms (8). On the C57BL/6 (B6) genetic background, Min/+ mice develop dozens of adenomas throughout the intestinal tract, involving loss of heterozygosity (LOH) through somatic recombination (9, 10). By contrast, on the AKR genetic background, the LOH pathway is suppressed and tumors form predominantly through apparently epigenetic silencing of the wild-type Apc allele (11, 12). A knockout allele of Apc, Apc1638N (1638N), was constructed by gene targeting, selecting for the neomycin-resistance cassette carried in the targeting vector. This allele has a phenotype quite distinct from that of Min; on the B6 genetic background, 1638N/+ mice develop only one or two tumors in the small intestine (13). The mechanism of Apc loss in these tumors has been reported to involve LOH (14, 15)
Mice with mutations in MMR genes recapitulate many of the cellular and organismal phenotypes of humans with HNPCC (16-18). As observed with HNPCC patients, intestinal tumorigenesis in mice deficient for MMR proceeds primarily through mutational inactivation of Apc (16).
Here we used mice with germ line mutations in Apc and Mlh1 to analyze the genetic factors that control tumor regionality in the mouse intestine. Our studies indicate that the location of tumors within the small intestine reflects the mechanism of tumor initiation. This finding is consistent with the observation that colonic tumors from humans with FAP develop with a distinct regional distribution different from that in individuals with HNPCC.
Materials and Methods
Mouse Husbandry and Genotyping. Animals were bred and housed at the McArdle Laboratory for Cancer Research and maintained on a Purina 5020 diet with 9% fat and 20% protein. Min/+, 1638N/+, and Mlh1 mice were genotyped as described in refs. 9 and 19-21. Mlh1-mutant animals were obtained originally from R. M. Liskay (Oregon Health Sciences University, Portland).
Analysis of Intestinal Tumors. Each animal was killed at 85-95 days of age, and its entire intestinal tract was removed, flushed with 1× PBS, and laid out on bibulous paper. Samples were fixed overnight in 10% neutral-buffered formalin and then transferred to 70% ethanol for long-term storage. Quantitative PCR for LOH analysis in tumors from Min/+ mice was performed as described (22).
LOH analysis of the 1638N allele of Apc was performed as follows: Intestinal tumors and adjacent normal tissue were dissected out of B6 1638N/+ mice and frozen in liquid nitrogen. DNA was then purified with the Tissue and Hair Extraction Kit from Promega by following the manufacturer's instructions.
To obtain allele ratios, 35 ng of DNA from each sample was amplified in a 12.25-μl PCR containing 10.2 mM Tris·HCl (pH 9.0 at 25°C); 51 mM KCl; 0.1% Triton X-100; 1.0 mM MgCl2; 200 μM concentrations (each) of dCTP, dGTP, dTTP, and dATP; 1.6 μM forward primer (1638N F-AGCTTTACACCAGGGGATGA); 0.8 μM Neo reverse primer (Neo R-ACCAAATTAAGGGCCAGCTC); 0.8 μM wild-type reverse primer (WT R-GATTTTTCCTCGCTGAGACAT); 1.0 unit Taq polymerase (Promega); and 0.033 μM [α-32P]dCTP (3,000 Ci/mmol) (NEN).
The forward primer is specific to a region of DNA upstream of the Neo insert. The Neo reverse primer is specific to a region in the Neo insert and produces a 249-bp product from the mutant allele. The wild-type reverse primer is specific to a wild-type sequence downstream of the forward primer and produces a 225-bp product from the wild-type allele.
PCR was performed in a Programmable Thermal Controller (MJ Research, Cambridge, MA) under the following conditions: 1 cycle at 94°C for 3 min followed by 30 cycles at 94°C for 15 s, 63°C for 2 min, and 72°C for 1 min 30 s followed by 1 cycle at 72°C for 10 min. Duplicate amplifications were done for each sample. Amplified samples were separated on a 6% polyacrylamide gel, dried on filter paper, and exposed for at least 12 h to a phosphor screen and scanned with a PhosphorImager (Molecular Dynamics). Quantitation of the bands was as described in ref. 22.
The Apc heterozygosity index (HetApc) was defined by the ratio of two ratios: the wild-type to mutant Apc band intensity ratio for tumor DNA normalized to that ratio for DNA from adjacent normal tissue, prepared and analyzed completely in parallel to the tumor DNA.
The regional distribution of tumors in the small intestine was assessed by counting tumors in each 2-cm section of the small intestine, beginning at the duodenum and ending at the ileocecal junction. Although the plots are normalized for intestinal length of each individual animal, there were no significant differences in absolute intestinal length between the genotypic classes.
Statistical Analysis. The regionality of intestinal tumors for a given genotypic class was assessed by generating the regional cumulative distribution function (CDF) for each group of animals. For a given group A, the CDF is defined as follows: FA(y) = the probability of a tumor being a distance y or less from the duodenum in set A. Letting FB be the CDF of another genotypic set, if FB(y) < FA(y), then we would expect a higher fraction of set A tumors to lie proximal to a point y than the corresponding fraction for B animals.
The CDFs for each group can be estimated by the empirical CDF: F̂A(y) = the proportion of tumors a distance y or less from the duodenum in animals of type A. Observing F̂B(y) < F̂A(y) gives evidence that type B animals are more likely to have tumors at locations distal to point y than type A animals. The significance of such a difference can be assessed by constructing confidence intervals for the CDFs.
The construction of confidence intervals is complicated because tumor locations within any one group may not be independently distributed. For example, if the locations within a single animal are clustered and more alike than between animals, then the standard approach for forming confidence intervals could give incorrect coverage probabilities by not accounting for between-animal variation. Therefore, we have constructed confidence intervals with a nonparametric approach that accounts for both within- and between-animal variation. We have used a version of the bootstrap (23), which, for each genotypic group, generates a sequence of bootstrap samples {F̂(1),..., F̂(B)} that can be used to generate confidence intervals for the CDF. For example, a 95% bootstrap confidence interval for F(y) has endpoints equal to the 2.5% and 97.5% quantiles of the bootstrap sample values {F̂(1)(y),..., F̂(B)(y)}. We have used a bootstrap method similar to that of Rao and Wu (24) for stratified survey data to account for the within- and between-group variability. The generation of a single bootstrap estimate of F, F̂*, proceeds as follows:
Letting m be the number of animals in the population and nj be the number of tumors in animal j,
Construct a set of numbers {i1,..., im} by sampling m numbers with replacement from {1,..., m};
For each sampled number ij, sample with replacement nij tumor locations from animal ij in the population.
The above process generates a bootstrap dataset of tumor locations from which the bootstrap estimate F̂* is calculated. Repeated application of the algorithm generates a sequence of bootstrap estimates {F̂(1),..., F̂(B)}. Note that the variability among the bootstrap estimates is a composite of between-animal variability (step 1 of the bootstrap) and within-animal variability (step 2 of the bootstrap).
The data in this study are nonstandard in that variability occurs at two stages of the sampling. Tumor location is variable both within an animal and between animals. This variation motivates the use of the hierarchical bootstrap procedure described above to form confidence intervals for the distributions of tumor location. Because the precision of our estimates depends on two sources of variation, the sizes of the confidence intervals depend on both the number of tumors for a given animal and the number of mice in each genotypic class. The former determines how well we can estimate the distribution function of a single mouse, whereas the latter determines how precisely we can estimate the mean CDF of all mice within a given class. The confidence intervals reflect the cumulative precision at both of these levels; this procedure has allowed us to directly compare the distribution of tumors throughout the intestinal tract between genotypic classes.
Results and Discussion
Generation and Characterization of AKR Mlh1 Congenic Animals. Our studies of tumor regionality have been seeded by studies comparing mouse models of FAP and HNPCC. We have generated and analyzed B6 animals with mutations in Apc and Mlh1. We found that loss of Mlh1 function increased the number of tumors in B6 Min/+ mice 3-fold (23). We have now generated Min/+; Mlh1-/- mice on AKR, a genetic background that carries multiple modifiers of the Min phenotype (25). We used marker-assisted selection to generate a congenic strain, starting with a null allele of Mlh1 carried on the B6 genetic background (20). At the N3 backcross generation, the AKR Mlh1 strain was >92% AKR across the genome, except for the Mlh1-bearing chromosome 9, which carried AKR alleles from the centromere to a breakpoint between 15 and 31 centimorgans. At the N6 backcross generation, AKR Mlh1+/- animals were crossed to AKR Min/+ congenic animals N14 (12). N7 AKR Min/+; Mlh1+/- progeny were then crossed to AKR Mlh1+/- littermates to generate N7F1 progeny that carried ApcMin and segregated for the null allele of Mlh1. As on the B6 background, AKR Min/+; Mlh1-/- arose at the expected Mendelian frequency (χ2 = 6.88, P > 0.2).
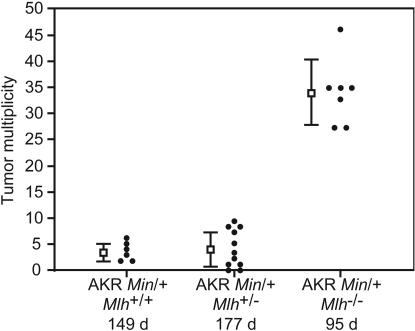
AKR Apc+/+; Mlh1+/- animals were phenotypically indistinguishable from normal AKR mice and frequently lived beyond 180 days of age. By contrast, AKR Apc+/+; Mlh1-/- animals rapidly developed thymic lymphomas and died by 100 days of age. Just as B6 Apc+/+; Mlh1-/- mice typically develop intestinal adenomas and skin papillomas only at ages >6 months, no intestinal or skin lesions were found in AKR Apc+/+; Mlh1-/- animals by 100 days of age. AKR Min/+; Mlh1-/- developed thymic lymphoma and generally died by 100 days of age. We found that AKR Min/+; Mlh1-/- mice have a significantly higher tumor multiplicity over the total intestinal tract than AKR Min/+; Mlh1+/+ or AKR Min/+; Mlh1+/- animals (Fig. 1). The Mlh1-deficient class was significantly different from the other two (P = 2.5 × 10-3 vs. AKR Min/+; Mlh1+/+, P = 4.6 × 10-4 vs. AKR Min/+; Mlh1+/- by Wilcoxon rank-sum analysis). The Mlh1+/- and Mlh1+/+ classes of AKR Min/+ animals did not differ significantly (P = 0.92 by Wilcoxon rank-sum analysis), demonstrating that Mlh1- is completely recessive, not haplo-insufficient.
Fig. 1.
Enhancement of tumor multiplicity in AKR Min/+;Mlh1-/- mice. Each dot represents the tumor multiplicity along the entire intestinal tract from a single animal. The square and error bars to the left of each distribution represent the mean ± SD. The tumor multiplicities for the classes are AKR Min/+;Mlh1+/+, 3.7 ± 1.6; AKR Min/+;Mlh1+/-, 4.0 ± 3.5; and AKR Min/+;Mlh1-/-, 34 ± 6.4. The average lifespan is denoted below each genotype.
Loss of Apc function in intestinal adenomas occurs by LOH through somatic recombination on the B6 Min/+ genetic background (22). When Mlh1 function is eliminated, however, a mutational pathway is initiated and many of the adenomas from B6 Min/+; Mlh1-/- carry a newly mutated form of Apc (26). On the AKR background, many of the tumors from Min/+ mice also maintain the wild-type Apc+ allele, apparently through silencing of Apc+ (12). We observed that all of the tumors from AKR Min/+; Mlh1-/- mice maintained the wild-type sequence at the Min site (12/12; K.M.H., unpublished data). By extension of the effect of Mlh1 deficiency on the B6 Min/+ background, the strong increase in tumor multiplicity generated by the Mlh1 deficiency on the AKR Min/+ background, compared with the AKR Apc+/+ background, implies that these tumors have incurred mutational hits to the wild-type allele.
From our analysis, AKR Min/+; Mlh1-/- appears to be a strain in which most tumors arise through a mutational pathway. This characteristic separates AKR Min/+; Mlh1-/- mice from B6 Min/+ and from AKR Min/+ mice, in which tumors arise through somatic recombination and apparent allelic silencing, respectively (Table 1). Overall, we noted that the distribution of tumors throughout the small intestine is consistent within any one strain but varies widely between strains.
Table 1. Data used to formulate regionality curves.
| Genotype | No. of mice | Tumor multiplicity, mean* | Predominant mechanism of loss of Apc function (%) | Source† |
|---|---|---|---|---|
| B6 Min/+ | 20 | 102.0 | LOH (≈100) | 22 |
| AKR Min/+ | 29 | 2.6 | Silencing (≈100) | 12 and this study |
| AKR Min/+; Mlh 1−/− | 5 | 31.0 | Mutation (≈90) | This study |
| B6 1638N/+ | 23 | 1.0 | Mutation (≈100) | This study |
Tumors from the entire small intestine only.
Reference for the initial characterization of the mechanism of loss of Apc function.
Tumor Regionality Is Regulated by the Mechanism of Apc Loss. To analyze the regionality phenomenon in more detail, we developed a statistical analysis that allows us to investigate differences in tumor locations by comparing the CDFs for the different genotypic groups: B6 Min/+, AKR Min/+, and AKR Min/+; Mlh1-/- (Fig. 2 A-D).
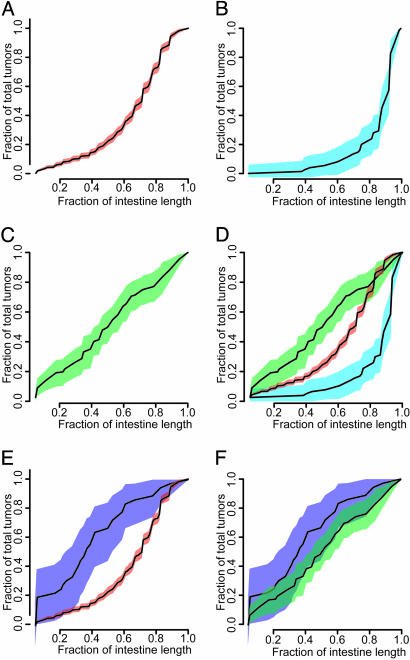
Fig. 2.
Tumor regionality versus genetic background and Apc allele. The CDF is shown for each genotypic class, with the 95% confidence intervals flanking the CDF in color. The number of animals and tumors represented by each curve are collated in Table 1. (A)B6 Min/+.(B) AKR Min/+.(C) AKR Min/+;Mlh1-/-. (D)B6 Min/+ vs. AKR Min/+ and AKR Min/+;Mlh1-/-.(E)B6 1638N/+ vs. B6 Min/+. (F) B6 1638N/+ vs. AKR Min/+;Mlh1-/-. CDFs in which the 95% confidence intervals do not overlap are considered significantly different. The CDF for B6 1638N/+ does not overlap with that of B6 Min/+, indicating that loss of Apc function in tumors from B6 1638N/+ mice does not occur by LOH. By contrast, the CDF from B6 1638N/+ overlaps that of AKR Min/+;Mlh1-/-, a strain for which Apc+ is inactivated by mutation.
We have found that tumors do not develop uniformly throughout the intestine of Min/+ animals. More than 75% of tumors from B6 Min/+ mice developed in the distal half of the small intestine (Fig. 2 A). If tumors were uniformly distributed throughout the small intestine, they would partition evenly between the proximal and distal regions, with the regionality curve approaching a straight line. Even more strikingly, in AKR Min/+ mice, 75% of the tumors developed in the distal 20% of the small intestine (Fig. 2B). The change in regionality seen in AKR Min/+ mice is also not simply a result of a reduction in tumor multiplicity compared with B6 because B6 Min/+ mice carrying the Robertsonian translocation Rb(7.18)9Lub have a reduced tumor multiplicity but a regionality that is not significantly different from that of B6 Min/+ (10). Regionality is also unchanged by homozygosity for the resistance allele of Mom1 (modifier of Min 1) in Min/+ mice (R.B.H., unpublished data).
The regionality of tumors from AKR Min/+; Mlh1-/- gave evidence for a uniform distribution, given that the CDF for this class is almost linear (Fig. 2C). This CDF indicates that tumors develop randomly throughout the small intestine of AKR Min/+; Mlh1-/- mice. The regionality of tumors in these mice also is consistent with a mutational mechanism of tumor initiation; indeed, tumors from wild-type B6 mice treated with N-ethyl-N-nitrosourea develop with similar regionality.
Our study of tumor regionality has yielded distinct distributions for the three genetic backgrounds that we have analyzed. These three regionality distributions are classified as significantly different from one another in the regions for which the 95% confidence intervals of the curves do not overlap (Fig. 2D). We note that these three genetic backgrounds differ in the mechanisms by which intestinal tumors are initiated. Thus, the regionality of tumors within the small intestine appears to reflect the mechanism by which Apc function is lost.
Analysis of B6 1638N/+ Mice. To investigate further the hypothesis that tumor regionality reflects the mechanism of Apc loss, we have studied regionality in B6 1638N/+ mice. To our surprise, the CDF for 1638N/+ did not overlap with B6 Min/+ (Fig. 2E) but, instead, overlapped with AKR Min/+; Mlh1-/- (Fig. 2F). This result seemed to contradict the hypothesis, because the initiation of intestinal tumors in 1638N/+ mice has been reported predominantly to involve LOH (14, 15).
To investigate this apparent contradiction, we reanalyzed the status of the Apc locus in tumors from B6 1638N/+ mice by using a newly developed quantitative LOH assay. Contrary to previous reports, we find that tumors from B6 1638N/+ mice maintain the wild-type allele of Apc (Fig. 3). Given that the CDF for 1638N/+ mice overlaps with that of AKR Min/+; Mlh1-/- mice, we believe that adenomas from these mice are preferentially initiated by mutation of Apc.
Fig. 3.
LOH analysis in intestinal tumors. (A) HetApc values for adenomas in B6 Min/+, AKR Min/+, and congenic B6 1638N/+ animals. HetApc was defined by the ratio of two ratios: the wild-type to mutant Apc band intensity ratio for tumor DNA normalized to that ratio for DNA from adjacent normal tissue prepared and analyzed completely in parallel to the tumor DNA. The values of HetApc for tumors in B6 Min/+ animals (orange bars) track with the previously published Gaussian distribution (white bars; mean 0.32 ± 0.17). The HetApc values for tumors in AKR Min/+ and B6 1638N/+ animals (cyan and purple bars, respectively) differ strongly from those of the B6 Min/+ LOH distribution. Instead, they lie in the range previously categorized as involving maintenance with silencing of the Apc+ allele (12). (B) Resolution of allele-specific PCR products. Allele-specific primer pairs were designed as described in Materials and Methods, giving PCR products for the 1638N mutant allele (249 bp) and the wild-type allele (225 bp) that were resolved on 6% polyacrylamide gels and quantitated with a PhosphorImager. Adenomas and adjacent normal tissue were dissected and frozen in liquid nitrogen. DNA was extracted, purified, and used as template for the production of dCTP-labeled PCR products. The measured HetApc values for tumors 1 and 2 were 0.84 and 0.77, respectively.
Consistent with our finding that tumors from 1638N/+ mice maintain Apc+, we found that the regionality of 1638N/+; Mlh1-/- mice was not different from that of 1638N/+; Mlh1+/+ mice (data not shown). This result supports the idea that initiation of a mutator phenotype by loss of Mlh1 function enhances, but does not change, the fundamental mechanism whereby Apc function is lost in B6 1638N/+ mice.
The Basis for Regionality. What underlies the regional preference for tumor formation? The simplest hypothesis states that the regionality reflects the distribution of the cellular substrate or the genetic or epigenetic event that eliminates Apc function. In contrast to this “direct hypothesis,” an indirect hypothesis states that the regionality is a function of the somatic Apc genotype, which in turn varies with the distinct mechanism for loss of Apc function.
The direct hypothesis states that the development of tumors of a given class is simply a reflection of the regional incidence of these events. Tumors that initiate through LOH (i.e., those from B6 Min/+ mice) would develop preferentially in the distal half of the small intestine because recombinational LOH occurs preferentially in this region. Further, allelic silencing would occur preferentially in the distal 20% of the small intestine to give rise to the regionality seen in AKR Min/+ mice. By contrast, tumors from MMR-deficient animals would develop uniformly throughout the tissue because MMR functions uniformly throughout the entire small intestine to protect the genome from mutation. A specific version of the direct hypothesis states that the progenitor cell types that are prone to the different pathways are regionally heterogeneous. These direct hypotheses can be investigated further if assays can be developed for the salient progenitor cell type and the absolute frequency of each of the initiating events in vivo.
The indirect hypothesis states that particular somatic Apc genotypes cause neoplastic growth in a region-specific manner. In this hypothesis, the formation of a particular genotypic class would be regionally uniform. Cells that have undergone LOH by somatic recombination and thus express two Min alleles (Min/Min), would develop into tumors primarily in the distal half of the small intestine. By contrast, cells in which Apc has been silenced, and thus express only a single Min allele (Min/0), would lead to tumors only in the region of the small intestine near the cecum. Finally, loss of MMR would lead to a wide variety of mutant Apc alleles and would generate tumors throughout the intestinal tract.
We can envisage particular biological facets to this indirect model. The ability of different alleles to induce or support neoplastic growth may be linked to states of cellular differentiation: Differences between the morphologically distinct regions of the small intestine would be associated with differences in cellular differentiation. Whereas the proximal small intestine is composed largely of absorptive enterocytes, the distal small intestine and colon contain a larger proportion of goblet cells (K.M.H., unpublished data). Bjerknes and Cheng (27) formulated a model for cellular differentiation that posits distinct differentiative pathways for enterocytes and goblet cells. In B6 Min/+ mice, tumors develop by LOH through somatic recombination, primarily in the distal small intestine. In this indirect model, the enterocytic differentiation pathway that dominates the proximal small intestine would provide relative resistance to tumor formation or maintenance for Min/Min somatic recombinants but not for Min/Apcmut precursors generated by somatic mutation, whereas goblet cell precursors would be more sensitive to the Min/Min state.
An interesting conundrum is raised by our observation that the 1638N allele differs from the Min allele: The wild-type Apc allele is maintained in the 1638N/+ adenoma. Fodde and his colleagues (28) report that the 1638N allele is transcribed only weakly and suggest that it represents the null status of the Apc locus. An alternative explanation is that the NeoR insertion of the 1638N allele exerts a long-range position effect (29, 30), inactivating not only Apc but also a neighboring gene that is important for adenomagenesis under the conditions in which our experiments have been performed.
We have used a suite of mouse models to study an important but poorly characterized facet of inherited intestinal cancer: tumor regionality. We have found that the regional distribution of intestinal tumors reflects the major mechanism by which Apc function is lost. Further genetic and molecular analysis will lead to the identification of specific loci and ultimately the cellular and molecular networks that mediate the regionality of intestinal neoplasms in both mice and humans.
Acknowledgments
We thank Bryan Biehl for help with mouse maintenance and genotyping, Jane Weeks and Harlene Edwards for skilled preparation of histological samples, Linda Clipson for critical contributions to the formulation of this report, and Drs. Norman Drinkwater, Asit Parikh, Ilse Riegel, and Alexandra Shedlovsky for critical feedback. The exchange of the targeted allele 1638N was facilitated by Drs. Winfried Edelmann and Raju Kucherlapati under the aegis of the Mouse Models for Human Cancer Consortium. This work was supported by National Cancer Institute Grants P30-CA07075 (to the McArdle Laboratory) and R01-CA58085, R37-CA63677, and U01-CA84227 (to W.F.D.). K.M.H. was supported by National Institutes of Health Predoctoral Training Grant 5T32GM07133. This is publication No. 3620 from the Laboratory of Genetics, University of Wisconsin.
Abbreviations: APC, adenomatous polyposis coli; 1638N/+, Apc1638N/+; CDF, cumulative distribution function; FAP, familial adenomatous polyposis; HetApc, Apc heterozygosity index; HNPCC, hereditary nonpolyposis colorectal cancer; LOH, loss of heterozygosity; Min, multiple intestinal neoplasia; Min/+, ApcMin/+; MLH1, MutL homolog 1; MMR, mismatch repair.
References
- 1.American Cancer Society (2004) Cancer Facts & Figures 2004 (Am. Cancer Soc., Atlanta).
- 2.Giardiello, F. M. (1995) in Gastrointestinal Cancers: Biology, Diagnosis, and Therapy, ed. Rustgi, A. K. (Lippincott-Raven, Philadelphia).
- 3.Lynch, H. T. & Lynch, J. F. (2000) Semin. Surg. Oncol. 18, 305-313. [DOI] [PubMed] [Google Scholar]
- 4.Fishel, R., Lescoe, M. K., Rao, M. R. S., Copeland, N. G., Jenkins, N. A., Garber, J., Kane, M. & Kolodner, R. (1993) Cell 75, 1027-1038. [DOI] [PubMed] [Google Scholar]
- 5.Bronner, C. E., Baker, S. M., Morrison, P. T., Warren, G., Smith, L. G., Lescoe, M. K., Kane, M., Earabino, C., Lipford, J., Lindblom, A., et al. (1994) Nature 368, 258-260. [DOI] [PubMed] [Google Scholar]
- 6.Weber, W., Foeppl, M., Schlossberg, D. & Locher, K. (1990) Anticancer Res. 10, 543-546. [PubMed] [Google Scholar]
- 7.Lynch, H. T., Smyrk, T. C., Watson, P., Lanspa, S. J., Lynch, J. F., Lynch, P. M., Cavalieri, R. J. & Boland, C. R. (1993) Gastroenterology 104, 1535-1549. [DOI] [PubMed] [Google Scholar]
- 8.Shoemaker, A. R., Gould, K. A., Luongo, C., Moser, A. R. & Dove, W. F. (1997) Biochim. Biophys. Acta 1332, F25-F48. [DOI] [PubMed] [Google Scholar]
- 9.Haigis, K. M., Caya, J. G., Reichelderfer, M. & Dove, W. F. (2002) Proc. Natl. Acad. Sci. USA 99, 8927-8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haigis, K. M. & Dove, W. F. (2003) Nat. Genet. 33, 33-39. [DOI] [PubMed] [Google Scholar]
- 11.Moser, A. R., Pitot, H. C. & Dove, W. F. (1990) Science 247, 322-324. [DOI] [PubMed] [Google Scholar]
- 12.Shoemaker, A. R., Moser, A. R., Midgley, C. A., Clipson, L., Newton, M. A. & Dove, W. F. (1998) Proc. Natl. Acad. Sci. USA 95, 10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fodde, R., Edelmann, W., Yang, K., van Leeuwen, C., Carlson, C., Renault, B., Breukel, C., Alt, E., Lipkin, M., Khan, P. M., et al. (1994) Proc. Natl. Acad. Sci. USA 91, 8969-8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smits, R., Kartheuser, A., Jagmohan-Changur, S., Leblanc, V., Breukel, C., de Vries, A., van Kranen, H., van Krieken, J. H., Williamson, S., Edelmann, W., et al. (1997) Carcinogenesis 18, 321-327. [DOI] [PubMed] [Google Scholar]
- 15.Kuraguchi, M., Edelmann, W., Yang, K., Lipkin, M., Kucherlapati, R. & Brown, A. M. (2000) Oncogene 19, 5755-5763. [DOI] [PubMed] [Google Scholar]
- 16.Prolla, T. A., Baker, S. M., Harris, A. C., Tsao, J. L., Yao, X., Bronner, C. E., Zheng, B., Gordon, M., Reneker, J., Arnheim, N., et al. (1998) Nat. Genet. 18, 276-279. [DOI] [PubMed] [Google Scholar]
- 17.Edelmann, W., Yang, K., Kuraguchi, M., Heyer, J., Lia, M., Kneitz, B., Fan, K., Brown, A. M., Lipkin, M. & Kucherlapati, R. (1999) Cancer Res. 59, 1301-1307. [PubMed] [Google Scholar]
- 18.de Wind, N., Dekker, M., Berns, A., Radman, M. & te Riele, H. (1995) Cell 82, 321-330. [DOI] [PubMed] [Google Scholar]
- 19.Su, L. K., Kinzler, K. W., Vogelstein, B., Preisinger, A. C., Moser, A. R., Luongo, C., Gould, K. A. & Dove, W. F. (1992) Science 256, 668-670. [DOI] [PubMed] [Google Scholar]
- 20.Baker, S. M., Plug, A. W., Prolla, T. A., Bronner, C. E., Harris, A. C., Yao, X., Christie, D. M., Monell, C., Arnheim, N., Bradley, A., et al. (1996) Nat. Genet. 13, 336-342. [DOI] [PubMed] [Google Scholar]
- 21.Gould, K. A., Dietrich, W. F., Borenstein, N., Lander, E. S. & Dove, W. F. (1996) Genetics 144, 1769-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luongo, C., Moser, A. R., Gledhill, S. & Dove, W. F. (1994) Cancer Res. 54, 5947-5952. [PubMed] [Google Scholar]
- 23.Efron, B. (1979) Ann. Stat. 7, 1-26. [Google Scholar]
- 24.Rao, J. N. K. & Wu, C.-F. J. (1988) J. Am. Stat. Assoc. 83, 231-241. [Google Scholar]
- 25.Dietrich, W. F., Lander, E. S., Smith, J. S., Moser, A. R., Gould, K. A., Luongo, C., Borenstein, N. & Dove, W. F. (1993) Cell 75, 631-639. [DOI] [PubMed] [Google Scholar]
- 26.Shoemaker, A. R., Haigis, K. M., Baker, S. M., Dudley, S., Liskay, R. M. & Dove, W. F. (2000) Oncogene 19, 2774-2779. [DOI] [PubMed] [Google Scholar]
- 27.Bjerknes, M. & Cheng, H. (1999) Gastroenterology 116, 7-14. [DOI] [PubMed] [Google Scholar]
- 28.Fodde, R., Smits, R., Hofland, N., Kielman, M. & Khan, P. M. (1999) Cytogenet. Cell Genet. 86, 105-111. [DOI] [PubMed] [Google Scholar]
- 29.Herault, Y., Rassoulzadegan, M., Cuzin, F. & Duboule, D. (1998) Nat. Genet. 20, 381-384. [DOI] [PubMed] [Google Scholar]
- 30.Bunting, M., Bernstein, K. E., Greer, J. M., Capecchi, M. R. & Thomas, K. R. (1999) Genes Dev. 13, 1524-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]