Abstract
Staphylococcus aureus is an important nosocomial and community-acquired pathogen. Its genetic plasticity has facilitated the evolution of many virulent and drug-resistant strains, presenting a major and constantly changing clinical challenge. We sequenced the ≈2.8-Mbp genomes of two disease-causing S. aureus strains isolated from distinct clinical settings: a recent hospital-acquired representative of the epidemic methicillin-resistant S. aureus EMRSA-16 clone (MRSA252), a clinically important and globally prevalent lineage; and a representative of an invasive community-acquired methicillin-susceptible S. aureus clone (MSSA476). A comparative-genomics approach was used to explore the mechanisms of evolution of clinically important S. aureus genomes and to identify regions affecting virulence and drug resistance. The genome sequences of MRSA252 and MSSA476 have a well conserved core region but differ markedly in their accessory genetic elements. MRSA252 is the most genetically diverse S. aureus strain sequenced to date: ≈6% of the genome is novel compared with other published genomes, and it contains several unique genetic elements. MSSA476 is methicillin-susceptible, but it contains a novel Staphylococcal chromosomal cassette (SCC) mec-like element (designated SCC476), which is integrated at the same site on the chromosome as SCCmec elements in MRSA strains but encodes a putative fusidic acid resistance protein. The crucial role that accessory elements play in the rapid evolution of S. aureus is clearly illustrated by comparing the MSSA476 genome with that of an extremely closely related MRSA community-acquired strain; the differential distribution of large mobile elements carrying virulence and drug-resistance determinants may be responsible for the clinically important phenotypic differences in these strains.
The impact on human health of Staphylococcus aureus infections in community and hospital settings has lead to intensive investigation of this organism over recent years. It is the causative agent of a wide range of diseases, from carbuncles and food poisoning, through more serious device and wound-related infections, to life threatening conditions, such as bacteremia, necrotizing pneumonia, and endocarditis. S. aureus produces a plethora of virulence factors that facilitate attachment, colonization, cell-cell interactions, immune evasion, and tissue damage. The number of effective antibiotics has been reduced by the emergence of resistance to penicillin, methicillin, and, more recently, vancomycin (1, 2), a problem that has been compounded by the recent emergence of methicillin-resistant S. aureus (MRSA) carriage and disease in the community (3, 4).
The genomes of two recently isolated clinical S. aureus strains were sequenced: a hospital-acquired MRSA strain (MRSA252), representative of the highly successful epidemic EMRSA-16 clone, and a community-acquired methicillin-susceptible S. aureus (MSSA) strain (MSSA476). More complete genomes are now available for S. aureus than for any other bacterial species, thus providing a detailed insight into the evolutionary processes leading to strains of differing virulence and drug-resistance potential. Our comparative analyses used the previously published genomes of closely related hospital-acquired MRSA and vancomycin intermediately susceptible S. aureus strains (N315 and Mu50, respectively) (5), and a community-acquired MRSA strain (MW2) (6). Furthermore, extensive multilocus sequence typing (MLST) data have been generated for this species, allowing the comparison of the sequenced strains within the context of the whole S. aureus population (7). MRSA252 belongs to the clinically important EMRSA-16 clone that is responsible for half of all MRSA infections in the U.K. (8) and is one of the major MRSA clones found in the U.S. (USA200) (9). Its emergence in the last 10 years has been associated with a significant increase in the numbers of S. aureus infections in U.K. hospitals, and it is regarded as endemic in the majority of U.K. hospitals (8). MSSA476 was chosen because it caused severe invasive disease in an immunocompetent child in the community and belonged to a major clone associated with community-acquired disease that also contains the community-acquired MRSA strain MW2 (10).
In this study, we describe general features of the two new genomes and highlight the significance of mobile genetic elements for the acquisition of traits of clinical importance. We also present comparisons of very similar and very diverged genomes that reveal the patterns of genomic diversification over time and in particular illustrate the speed with which resistance genes and putative virulence genes move between strains by lateral gene transfer.
Methods
Bacterial Strains. Hospital-acquired, epidemic strain MRSA252. MRSA252 was isolated in 1997 from a 64-year-old female who acquired MRSA postoperatively. The organism was part of a miniature outbreak at an intensive treatment unit where all three of the patients who became infected died as a direct consequence of MRSA septicemia (septic shock or multiorgan failure subsequent to septic shock). MRSA252 is resistant to penicillin, ciprofloxacin, erythromycin, and methicillin and sensitive to fusidic acid, rifampicin, tetracycline, trimethoprim, gentamicin, and amikacin. The strain has been typed as belonging to the EMRSA-16 clone by pulsed-field gel electrophoresis and to sequence type (ST)36 by MLST (8, 10).
Community-acquired, invasive strain MSSA476. MSSA476 was isolated in 1998 from a 9-year-old boy with community-acquired primary upper tibial osteomyelitis and bacteremia who had no predis-posing risk factors. Although resistant to penicillin and fusidic acid, the strain was susceptible to most commonly used antibiotics, including ciprofloxacin, rifampicin, tetracycline, trimethoprim, erythromycin, gentamicin, amikacin, and methicillin. After surgical intervention and treatment with i.v. flucloxacillin the patient made a full recovery. The strain has been assigned to ST1 by MLST (10).
Whole-Genome Sequencing. The two genome sequences were produced independently by separate teams within The Sanger Institute Pathogen Sequencing Unit to ensure that there was no cross contamination between the two projects. The sequence was assembled, finished, and annotated as described in ref. 11 by using artemis to collate data and facilitate annotation (12); detailed information is available in Supporting Methods, which is published on the PNAS web site.
Comparative Genomics. Comparison of the genome sequences was facilitated by using artemis comparison tool (K.R., unpublished data; see also www.sanger.ac.uk/Software/ACT) which enabled the visualization of blastn and tblastx comparisons between the genomes (13). Orthologous proteins were identified as reciprocal best matches by using fasta with subsequent manual curation. The genomes used for comparisons were for the S. aureus strains Mu50 (5), N315 (5), and MW2 (6).
Results
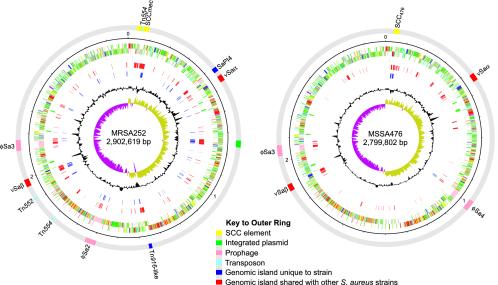
Features of the MRSA252 and MSSA476 Genomes. The MRSA252 and MSSA476 chromosomes are 2,902,619 bp and 2,799,802 bp in size and contain 2,671 and 2,565 predicted protein-coding sequences (CDSs) or genes, respectively. A summary of the general properties of the two genomes is presented in the supporting information. Comparisons of the chromosomes reveal they are collinear with each other and with those of the sequenced S. aureus strains N315, Mu50, and MW2 (5, 6). However, the conserved “core” genome is peppered with regions of small- and large-scale difference (Fig. 1).
Fig. 1.
Schematic circular diagrams of the MRSA252 and MSSA476 chromosomes. Where appropriate, categories are shown as pairs of concentric circles representing both coding strands. The outer colored segments on the gray outer ring represent genomic islands and horizontally acquired DNA (see figure for key). Inside the gray outer ring, the rings from outside to inside represent scale in Mbp, annotated CDS (colored according to predicted function), tRNA and rRNA (green), additional DNA compared to the other S. aureus strain described here (MSSA476 or MRSA252 where appropriate; red), additional DNA compared to other sequenced S. aureus strains [N315 (5), Mu50 (5), and MW2 (6); blue], percentage of G + C content, and G + C deviation (>0%, olive; <0%, purple). Color coding for CDSs is as follows: dark blue, pathogenicity/adaptation; black, energy metabolism; red, information transfer; dark green, surface-associated; cyan, degradation of large molecules; magenta, degradation of small molecules; yellow, central/intermediary metabolism; pale green, unknown; pale blue, regulators; orange, conserved hypothetical; brown, pseudogenes; pink, phage plus insertion sequence elements; gray, miscellaneous.
Small-scale variation, defined as genetic changes that affect an individual gene or small numbers of genes (<10 CDSs), includes the formation of pseudogenes by insertion sequence integration, point mutations, and variation in polymeric nucleotide repeats. Details of insertion sequence elements and pseudogenes for both strains is provided in Tables 2 and 3, which are published as supporting information on the PNAS web site. Large-scale variation (involving >10 CDSs) is associated with horizontally acquired DNA, often at loci that have been previously identified as mobile genetic elements or “genomic islands” (Fig. 1) (5, 6). Many of the genes contained in these regions are associated with virulence or drug resistance. A summary of the genomic islands of MRSA252 and MSSA476 and the important determinants they carry is given in Table 1.
Table 1. Summary of the major genetic elements of the MRSA252 and MSSA476 genomes.
| MRSA252 | MSSA476 | |
|---|---|---|
| SCC | ||
| Type II SCCmec | mecA | — |
| SCC476 | — | far |
| Transposons | ||
| Tn554 | ermA (2), spc (2) | — |
| Tn552 | blaZ | — |
| Tn916-like | NI | — |
| Bacteriophage | ||
| ϕSa2 | NI | — |
| ϕSa3 | sea, sak | sea, sak, seg2, sek2 |
| ϕSa4 | — | NI |
| Genomic islands | ||
| νSaα | set (9), lpl (6)* | set (11), lpl (5) |
| νSaβ | hysA, spl (5), exotoxin (6)* | spl (4), lukDE, bsa |
| Pathogenicity islands | ||
| SaPl4 | NI | — |
| Plasmids | cadAC, arsBC (integrated) | blaZ, cadD (pSAS) |
| Insertion Sequences | ||
| IS1272 | 9 | — |
| IS431 | 2 | 1 |
| ISYZ | 5 | — |
| ISX | 6 | — |
| Others (remnants) | 8 | 4 |
The presence of genomic islands and other mobile regions is indicated by text that lists the genes involved in virulence or drug resistance; in the absence of genes proven to play a role in virulence, the presence of an element is indicated by neutral insertion (NI). Values in parentheses indicate the number of homologues in that region. For the insertion sequences, the number in each strain is given. The designation of genomic islands and prophage follows the convention previously described for S. aureus (6). —, not present. Asterisks indicate the presence of additional pseudogenes.
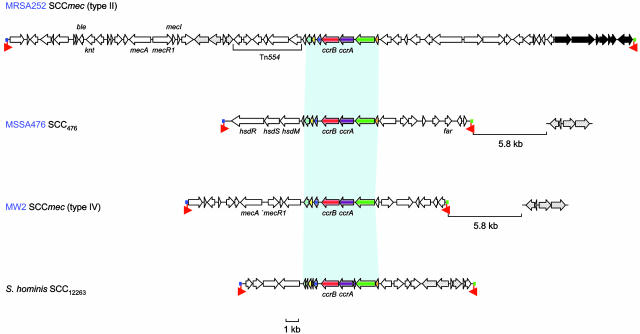
Genetic Elements Carrying Resistance Genes in MRSA252 and MSSA476. MSSA476 is resistant to penicillin and fusidic acid but susceptible to methicillin. Surprisingly MSSA476 contains a novel Staphylococcal chromosomal cassette (SCC)mec-like element (designated SCC476) integrated at the same site on the chromosome as SCCmec elements in MRSA strains (Fig. 1). The 22.8-kb SCC476 contains 18 CDSs and shares the same left and right boundaries (attL and attR, respectively) and similar inverted repeat sequences as SCCmec elements (Fig. 2) (14). However, unlike SCCmec elements, SCC476 does not carry the mecA gene (encoding penicillin-binding protein 2a) that confers resistance to methicillin, but it carries a CDS (SAS0043) encoding a protein similar (43.7% amino acid identity) to the plasmid-borne fusidic acid-resistance determinant Far1 (15). The only other apparent resistance protein contained within the MSSA476 genome is the pSAS1 plasmid encoded β-lactamase (pSAS19). This finding correlates well with the antibiotic resistance profile of MSSA476.
Fig. 2.
Comparison of SCC elements. Shown are the schematic diagrams of S. aureus type II SCCmec of MRSA252 (first schematic), non-mec SCC476 of MSSA476 (second schematic), the type IV SCCmec of MW2 (third schematic), and the non-mec SCC12263 from S. hominis (fourth schematic). Genes are marked in the direction of transcription as arrows. Genes with similarity in all elements are marked in the same color; genes with no similarity are white. Conserved regions in the SCC elements are joined by light blue shading. Genes within the S. hominis SCC12263 element that are similar to genes downstream of the SCC elements in the community-acquired strains MSSA476 and MW2 and within the MRSA252 SCCmec are shaded in gray. The black arrows in the MRSA252 SCCmec element indicate the additional CDS in comparison to the type II SCCmec elements of N315 and Mu50. The colored boxes at the end of the SCC elements mark the left and right attachment sites (blue, attL; green, attR). Inverted complementary repeats at the boundaries of the elements are marked by red arrows.
Six CDSs within SCC476 are conserved with respect to other types of SCCmec elements (Fig. 2) (16), including the two site-specific recombinases ccrA and ccrB, which were most similar to those found in type I SCCmec elements (17). However, the highest similarity of these proteins was to the ccrA and ccrB homologues of a Staphylococcus hominis non-mec SCC element (SCC12263) (18). At the left-hand end of the SCC476 element there are three CDSs that are similar to components of a type I restriction modification system: hsdM, hsdS, and hsdR.
The hospital-acquired MRSA252 is resistant to a larger number of antibiotics than MSSA476; as a corollary it contains more mobile element-encoded resistance determinants than the community-acquired strain. The genome contains a 58.8-kb SCCmec element at a site near the origin of replication (Fig. 1). The structure of the SCCmec element in MRSA252 corresponds to that of a type II SCCmec element (Fig. 2) and is very similar to those reported in the N315 and Mu50 genomes (14, 17). The SCCmec element carries an integrated copy of pUB110 with bleomycin- and kanamycin-resistance genes and a copy of Tn554 carrying erythromycin- and spectinomycin-resistance genes. The right-hand end of the MRSA252 SCCmec element contains six previously uncharacterized CDSs (three hypothetical proteins and three transposases) and a gene remnant. Immediately downstream of the SCCmec element is a 15.6-kb region encoding hypothetical proteins, proteins with similarity to modification and specificity components of a putative type-IV restriction enzyme system, a number of proteins with diverse predicted functions (SAR0087-SAR0103). In addition to the SCCmec element, there are two copies of a Tn554 transposon carrying erythromycin resistance; one is located in the type II SCCmec element (Fig. 2), and the other copy is inserted into a homologue of the DNA repair gene radC (SAR1733 and SAR1740). The MRSA252 chromosome also contains a Tn552 transposon that encodes the BlaI, BlaR, and BlaZ components of the inducible S. aureus β-lactamase. There is also an element integrated into the chromosome that contains similarity to the Tn916 transposon (Fig. 1); however, it does not appear to carry any obvious resistance determinants. An integrated plasmid confers resistance to the heavy metals arsenic and cadmium (Table 1). MSSA476 also carries resistance to heavy metals on a free plasmid, pSAS1.
Distinct from the characterized drug-resistance determinants associated with mobile genetic elements in MRSA252, there are several chromosomally encoded determinants, including a fluoroquinolone resistance protein (norA), SAR0748; a putative fosfomycin resistance protein (fosB), SAR2419; and teicoplanin-resistance-associated membrane proteins (tcaAB), (SAR2442 and SAR2441). There are also several other hypothetical proteins with similarity to families of proteins associated with drug resistance.
Genetic Elements Carrying Virulence Genes in MRSA252 and MSSA476. S. aureus pathogenicity islands (SaPI) are mobile genetic elements that often carry superantigen genes, such as toxic shock syndrome toxin and enterotoxins B and C, implicated in toxic shock and food poisoning. MRSA252 carries a SaPI-like element, SaPI4, that contains homologues of pathogenicity island proteins and displays synteny (conserved gene order) with the previously characterized pathogenicity islands SaPI1, SaPIbov, and “SaPI3” (19-21). SaPI4 has a integrase gene and insertion site downstream of the ribosomal protein gene rpsR but contains several hypothetical proteins with no similarity to characterized virulence genes.
The genomic islands νSaα and νSaβ have been identified in the genomes of all strains of S. aureus sequenced thus far (5, 6). The islands are found at the same loci and contain genes associated with pathogenicity. MRSA252 contains variants of these islands (Fig. 1 and Table 1), whereas those of MSSA476 match those of MW2 (6).
The transfer of toxin genes by lysogenic bacteriophage, or phage conversion, is an important mechanism in the evolution of virulent strains. The MRSA252 and MSSA476 genomes each contain two prophages (Fig. 1 and Table 1). Of the four prophages, three can be categorized into the previously described groups [φSa2 (252), φSa3 (252), and φSa3 (476)] (6). The fourth prophage is integrated at a locus in MSSA476 and has been designated φSa4 [φSa4 (476)]. This prophage has a mosaic structure, sharing regions of similarity with φSa2 (252), φSa2(MW2) (6), and φ12 (22) and does not carry any known virulence determinants. φSa4 integrase is most closely related to the integrase from phage L54a (23) and is integrated into the promoter region of the putative serine protease htrA. φSa2 (252) is a 46-kb prophage and shares similarity with φSa2(MW2), φPVL, and φSLT (6, 24, 25), but unlike these three, it does not contain the Panton-Valentine leukocidin toxin subunits or any other obvious virulence determinants. Α φSa3 family phage has been identified in all of the sequenced S. aureus genomes and in each case is integrated into the hlb (β-hemolysin) gene. These prophages typically harbor well characterized virulence factors, such as staphylokinase and pyrogenic toxin superantigen proteins. Both the φSa3 (252) and φSa3 (476) prophages contain staphylokinase (sak) and enterotoxin type A (sea) genes.
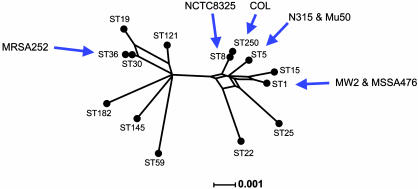
Diversity of S. aureus Strains. MLST is a technique widely used for typing S. aureus clinical isolates and has been used to show that the population structure of S. aureus is highly clonal (7). For a selection of S. aureus strains, including those whose genomes have been sequenced, the sequences of the seven housekeeping gene fragments used in MLST (sequences were obtained from the S. aureus MLST database, http://saureus.mlst.net) were joined end-to-end (concatenated) and a split decomposition graph was constructed (Fig. 3). MRSA252 is the most phylogenetically distinct of the strains sequenced thus far (ST36; Fig. 3). MSSA476, on the other hand, is much more closely related to the other sequenced strains (which are all relatively closely related in genotype; Fig. 3), and is indistinguishable by MLST from another sequenced community-acquired strain MW2 (both ST1) (6). Two other published genome strains, N315 and Mu50 (5), also have an identical MLST type: ST5. The remaining two unpublished but sequenced strains, NCTC8325 (www.genome.ou.edu/staph.html; ST8) and COL (www.tigr.org; ST250), are also very closely related to each other, differing at only one MLST locus.
Fig. 3.
Phylogenetic diversity of the sequenced S. aureus strains. The split decomposition tree was constructed by using concatenated sequences of the seven loci used in MLST for a selection of STs from the staphylococcal MLST database. The graph was constructed by using splitstree Version 3.1 (34). Distances were estimated by using hamming (uncorrected) distances. The positions of all seven strains for which complete genome sequences are available are shown.
Degree and Nature of Genetic Diversity Between MSSA476 and MW2. MW2 and MSSA476 are two community-acquired invasive strains isolated from distinct geographical locations (MW2 was isolated in the U.S. in 1998) (3) that are indistinguishable in genotype by MLST but differ in their susceptibility to methicillin. Global comparison reveals three large-scale differences that are the result of at least five separate horizontal gene transfer events. MSSA476 lacks the SCCmec type IV cassette, the SaPI3 pathogenicity island, and the bacteriophage φSa2(MW2) (6), but it contains SCC476 and bacteriophage φSa4 (476) (Table 1). These genomic differences are reflected in phenotypic differences between the strains; most notably MSSA476 is susceptible to methicillin and lacks three toxins, including the Panton-Valentine leukocidin toxin associated with necrotizing pneumonia (26). In contrast to these large-scale differences in gene content, the sequence of the core genome of MSSA476 and MW2 is extremely similar as predicted by MLST. An examination of all orthologous CDSs in these two genomes revealed only 285 single nucleotide polymorphisms within functional coding regions. A high proportion of these mutations (70%) were nonsynonymous, raising the possibility that they may have some adaptive significance. However, this observation can be more simply explained by assuming that these very recent mutations may be slightly deleterious, but purifying selection has not yet had time to remove them from the population (7).
Degree of Genetic Diversity Between the Phylogenetically Distinct MRSA252 and Other S. aureus Strains. Comparisons of the phylogenetically distinct genomes of MRSA252, MSSA476, and N315 by using blastn (score cut-off set at 100) were used to identify DNA regions that were unique to the individual strains. Encoded in these regions were 166 CDSs in MRSA252 (6.1% of the total number of CDS of MRSA252), compared with 109 CDSs (4.2%) in MSSA476 and 54 CDSs (2.1%) in N315. Many of the unique CDSs found in MRSA252 are found in small regions of inserted DNA (Fig. 1). Such regions have previously been called “genomic islets,” and have been observed in the genomes of other species of pathogenic bacteria (e.g., Escherichia coli O157 and Salmonella enterica serovar Typhi) (27, 28). Forty-one MRSA252-unique CDSs were found in these islets, representing ≈1.5% of the total CDSs (see Table 4, which is published as supporting information on the PNAS web site). Twenty-four CDSs are associated with islets of three CDSs or less. The other 17 are found in two regions of 7 and 10 CDS, the larger of which lies downstream of the type II SCCmec element. Analysis of the predicted functions of CDSs within these regions failed to identify any virulence factors. Many of the CDSs are predicted to have metabolic and transport functions and may therefore increase the metabolic repertoire of MRSA252.
Discussion
Over the past 50 years, S. aureus has undergone incremental changes in genetic complement that have resulted in the emergence of strains that are antibiotic-resistant and that appear to be successful at transmission and causing disease in the hospital setting. More recently, MRSA strains appear to have become established within the community. MLST has demonstrated that although many different genetic lineages cause invasive disease in hospital and community settings, only a limited number of these have acquired SCCmec and become successful MRSA clones (29). Several hospital MRSA clones have become multidrug resistant, and reduced susceptibility to vancomycin, although rare, has been found in each of these (30). There is little doubt that if vancomycin resistance (vanA) genes become established in S. aureus, they would also spread into successful hospital MRSA lineages. A better understanding of the features that make MSSA and MRSA strains successful in both community and hospital settings is urgently needed. Sequencing of diverse S. aureus strains facilitates description of the full range of putative pathogenic determinants carried by this species and provides a basis for predicting rates of horizontal gene transfer. Such information may provide clues to the observed variability in pathogenesis of different strains and could inform future efforts to prevent and treat serious S. aureus disease. The strains chosen for sequencing during this project were phylogenetically similar to (MSSA476) and markedly divergent from (MRSA252) previously sequenced isolates (Fig. 3), thereby maximizing the amount of informative data generated.
One of the most significant findings of this study is that ≈6% of the MRSA252 genome is previously undescribed when compared with other published S. aureus genomes, including the other hospital-acquired MRSA strains. These additional genes fall into the accessory category. MRSA252 belongs to the clinically important EMRSA-16 clone that is responsible for half of all MRSA infections in the U.K. (8), and the recent detection of EMRSA-16 isolates in several other countries has highlighted the pandemic potential of this clone (9, 31). The genetic diversity observed was generated by numerous mechanisms involving the horizontal acquisition of mobile DNA, both on the large and small scale. For example, the MRSA252 genome contains examples of two previously uncharacterized genomic islands that appear to be horizontally transferred together with new allelic variants of previously characterized islands (6). Gene accretion has also been supplemented in MRSA252 by the acquisition of genomic islets. The presence of genes encoding putative metabolic functions in many of these islets may enhance the survival and growth of this strain in the hospital environment, thereby contributing to the success of EMRSA-16.
The distinctness of the MRSA252 genome is also likely to be a reflection of the choice of strains that have been sequenced thus far; MRSA252 is highly divergent from strains MSSA476, MW2, N315, Mu50, and from NCTC8325 and COL (whose genomes are not yet published), which all cluster together on a phylogenetic tree and represent only a fraction of the diversity within the species as a whole (7) (Fig. 3). This bias has practical implications for postgenomic analysis, for example, during the construction of microarrays that aim to include the full complement of accessory genes within the species S. aureus. We predict that many of the genes identified here will be found elsewhere once a more representative sample of the S. aureus population is sequenced and that additional genes will be found, particularly if the choice of strains takes account of strain phylogeny.
Global comparisons of the five published genomes demonstrated marked variation in the distribution of genomic islands, indicating that mobile DNA is exchanged readily in the S. aureus population. The potential for rapid evolution of strains via this mechanism is highlighted by the comparison of the two community-acquired strains, MSSA476 and MW2, which are very closely related. By comparing their genomes, it is possible to examine in detail the processes by which genetic variation arises during the short-term evolution of S. aureus and during the emergence of MRSA strains from MSSA strains; such a comparison also pinpoints regions of the genome that are evolving at an exceptional speed. There appear to have been at least five separate acquisition or loss events that distinguish these strains, leading to markedly different virulence factor and drug-resistance repertoires. Rapid spread of genetic elements is also suggested by their phylogenetic distribution, which does not correlate with the genetic relatedness inferred by MLST. This lack of correlation suggests that mobile elements facilitate the exchange of virulence factors and antibiotic resistance determinants between S. aureus lineages and may lead to rapid changes in the pathogenic potential or drug resistance of strains. It is worth noting that all but one of the antibiotic-resistance determinants that account for the antibiotic-resistance profile of MRSA252 are encoded on mobile genetic elements.
Community- and hospital-acquired MRSA evolve from prevalent MSSA upon acquisition of SCCmec (29). Hospital MRSA contain one of four SCCmec types (I-IV), but the emergence of community-acquired MRSA is associated overwhelmingly with the acquisition of SCCmec IV (32), the smallest element and one that confers only resistance to β-lactams. Community MRSA are more diverse genetically than hospital MRSA (4) as a consequence of the increased frequency of acquisition of SCCmec IV compared with other SCCmec types (33). The sequenced community-acquired MRSA strain MW2 is assigned by MLST to ST1, which previously was identified as a cause of invasive disease in the community (3, 10). Surprisingly, MSSA476, an MSSA isolate assigned to ST1, also carries SCC476 but does not contain the mec gene. The presence of a putative fusidic acid-resistance gene in SCC476 highlights the potential of these elements to act as carriers for genes that provide a selective benefit in the absence of methicillin. The lack of overall similarity, coupled with the sequence divergence of conserved components (ccrA and ccrB), makes it unlikely that SCC476 is derived from SCCmec. Therefore, SCC476 represents a new type of SCC antibiotic-resistance carrying element (SCCfar) that is distinct from SCCmec, which indicates the potential spread of fusidic acid resistance by means of horizontal gene transfer. Our findings also raise the possibility that SCC elements are transferred readily through the S. aureus population (and perhaps other members of the genus) independently of the mec gene and that previously undetected SCC may be common within MSSA strains. Investigation into the prevalence and role of such elements in the spread of antibiotic resistance genes, putative virulence factors, and genes that enhance bacterial fitness is required. We propose that additional sequencing of further diverse clinical strains with known epidemic potential will contribute further to our understanding of S. aureus evolution and virulence.
Supplementary Material
Acknowledgments
We thank Keiichi Hiramatsu and Akio Oguchi for checking and confirming the sequence of the single-nucleotide polymorphisms in the MW2 assembly database. We also thank the Wellcome Trust Sanger Institute Core Sequencing and Informatics Groups. This work was supported by the Wellcome Trust through its Beowulf Genomics initiative. S.J.P. holds a Wellcome Trust Career Development Award in Clinical Tropical Medicine, and B.G.S. is a Wellcome Trust Principal Fellow. E.J.F. is supported by an Medical Research Council Career Development Award. M.C.E. is a Royal Society University Research Fellow.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MLST, multilocus sequence typing; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus; SCC, Staphylococcal chromosomal cassette; ST, sequence type; SaPI, Staphylococcus aureus pathogenicity islands.
Data deposition: The sequences and annotation of the MRSA252 and MSSA476 genomes, including the pSAS1 plasmid from MSSA476, have been deposited in the EMBL database (accession nos. BX571856 -BX571858).
References
- 1.Anonymous. (2002) Morbid. Mortal. Wkly. Rep. 51, 565-567. [Google Scholar]
- 2.Anonymous. (2002) Morbid. Mortal. Wkly. Rep. 51, 902. [Google Scholar]
- 3.Anonymous. (1999) Morbid. Mortal. Wkly. Rep. 48, 707-710. [Google Scholar]
- 4.Okuma, K., Iwakawa, K., Turnidge, J. D., Grubb, W. B., Bell, J. M., O'Brien, F. G., Coombs, G. W., Pearman, J. W., Tenover, F. C., Kapi, M., et al. (2002) J. Clin. Microbiol. 40, 4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuroda, M., Ohta, T., Uchiyama, I., Baba, T., Yuzawa, H., Kobayashi, I., Cui, L. Z., Oguchi, A., Aoki, K., Nagai, Y., et al. (2001) Lancet 357, 1225-1240. [DOI] [PubMed] [Google Scholar]
- 6.Baba, T., Takeuchi, F., Kuroda, M., Yuzawa, H., Aoki, K., Oguchi, A., Nagai, Y., Iwama, N., Asano, K., Naimi, T., et al. (2002) Lancet 359, 1819-1827. [DOI] [PubMed] [Google Scholar]
- 7.Feil, E. J., Cooper, J. E., Grundmann, H., Robinson, D. A., Enright, M. C., Berendt, T., Peacock, S. J., Smith, J. M., Murphy, M., Spratt, B. G., et al. (2003) J. Bacteriol. 185, 3307-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson, A. P., Aucken, H. M., Cavendish, S., Ganner, M., Wale, M. C. J., Warner, M., Livermore, D. M. & Cookson, B. D. (2001) J. Antimicrob. Chemother. 48, 143-144. [DOI] [PubMed] [Google Scholar]
- 9.McDougal, L. K., Steward, C. D., Killgore, G. E., Chaitram, J. M., McAllister, S. K. & Tenover, F. C. (2003) J. Clin. Microbiol. 41, 5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enright, M. C., Day, N. P. J., Davies, C. E., Peacock, S. J. & Spratt, B. G. (2000) J. Clin. Microbiol. 38, 1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkhill, J., Achtman, M., James, K. D., Bentley, S. D., Churcher, C., Klee, S. R., Morelli, G., Basham, D., Brown, D., Chillingworth, T., et al. (2000) Nature 404, 502-506. [DOI] [PubMed] [Google Scholar]
- 12.Rutherford, K., Parkhill, J., Crook, J., Horsnell, T., Rice, P., Rajandream, M. A. & Barrell, B. (2000) Bioinformatics 16, 944-945. [DOI] [PubMed] [Google Scholar]
- 13.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. (1990) J. Mol. Biol. 215, 403-410. [DOI] [PubMed] [Google Scholar]
- 14.Katayama, Y., Ito, T. & Hiramatsu, K. (2000) Antimicrob. Agents Chemother. 44, 1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Brien, F. G., Price, C., Grubb, W. B. & Gustafson, J. E. (2002) J. Antimicrob. Chemother. 50, 313-321. [DOI] [PubMed] [Google Scholar]
- 16.Hiramatsu, K., Cui, L., Kuroda, M. & Ito, T. (2001) Trends Microbiol. 9, 486-493. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., Katayama, Y., Asada, K., Mori, N., Tsutsumimoto, K., Tiensasitorn, C. & Hiramatsu, K. (2001) Antimicrob. Agents Chemother. 45, 1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama, Y., Takeuchi, F., Ito, T., Ma, X. X., Ui-Mizutani, Y., Kobayashi, I. & Hiramatsu, K. (2003) J. Bacteriol. 185, 2711-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindsay, J. A., Ruzin, A., Ross, H. F., Kurepina, N. & Novick, R. P. (1998) Mol. Microbiol. 29, 527-543. [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald, J. R., Monday, S. R., Foster, T. J., Bohach, G. A., Hartigan, P. J., Meaney, W. J. & Smyth, C. J. (2001) J. Bacteriol. 183, 63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yarwood, J. M., McCormick, J. K., Paustian, M. L., Orwin, P. M., Kapur, V. & Schlievert, P. M. (2002) J. Biol. Chem. 277, 13138-13147. [DOI] [PubMed] [Google Scholar]
- 22.Iandolo, J. J., Worrell, V., Groicher, K. H., Qian, Y. D., Tian, R. Y., Kenton, S., Dorman, A., Ji, H. G., Lin, S. P., Loh, P., et al. (2002) Gene 289, 109-118. [DOI] [PubMed] [Google Scholar]
- 23.Ye, Z. H. & Lee, C. Y. (1989) J. Bacteriol. 171, 4146-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaneko, J., Kimura, T., Narita, S., Tomita, T. & Kamio, Y. (1998) Gene 215, 57-67. [DOI] [PubMed] [Google Scholar]
- 25.Narita, S., Kaneko, J., Chiba, J., Piemont, Y., Jarraud, S., Etienne, J. & Kamio, Y. (2001) Gene 268, 195-206. [DOI] [PubMed] [Google Scholar]
- 26.Dufour, P., Gillet, Y., Bes, M., Lina, G., Vandenesch, F., Floret, D., Etienne, J. & Richet, H. (2002) Clin. Infect. Dis. 35, 819-824. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., Plunkett, G., Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529-533. [DOI] [PubMed] [Google Scholar]
- 28.Parkhill, J., Dougan, G., James, K. D., Thomson, N. R., Pickard, D., Wain, J., Churcher, C., Mungall, K. L., Bentley, S. D., Holden, M. T. G., et al. (2001) Nature 413, 848-852. [DOI] [PubMed] [Google Scholar]
- 29.Enright, M. C., Robinson, D. A., Randle, G., Feil, E. J., Grundmann, H. & Spratt, B. G. (2002) Proc. Natl. Acad. Sci. USA 99, 7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howe, R. A., Monk, A., Wootton, M., Walsh, T. R. & Enright, M. C. (2004) Emerg. Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 31.Chung, M., Dickinson, G., de Lencastre, H. & Tomasz, A. (2004) J. Clin. Microbiol. 42, 542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma, X. X., Ito, T., Tiensasitorn, C., Jamklang, M., Chongtrakool, P., Boyle-Vavra, S., Daum, R. S. & Hiramatsu, K. (2002) Antimicrob. Agents Chemother. 46, 1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, D. A. & Enright, M. C. (2003) Antimicrob. Agents Chemother. 47, 3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huson, D. H. (1998) Bioinformatics 14, 68-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.