Abstract
Electronic medical records are emerging as a major source of data for clinical and translational research studies, although phenotypes of interest need to be accurately defined first. This article provides an overview of how to develop a phenotype algorithm from electronic medical records, incorporating modern informatics and biostatistics methods.
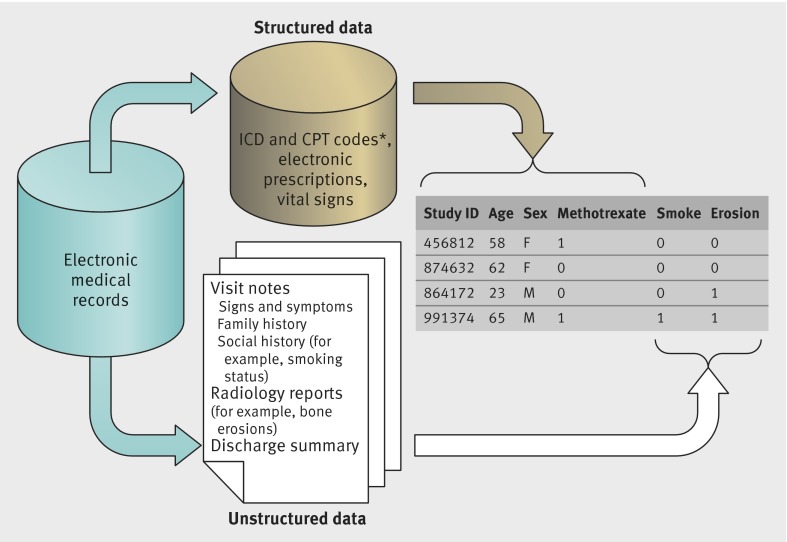
The increasing use of electronic medical records (EMR), driven mainly by efforts to improve the quality of patient care, have also launched a discipline of research using EMR data. In the past decade, methods and tools specifically used to conduct EMR research have allowed for sophisticated analyses including pharmacovigilance,1 genetic association,2 and pharmacogenetic studies.3 Phenotype algorithms using EMR data to classify patients with specific diseases and outcomes is a foundation of EMR research. Diagnoses or billing codes are typically used in these algorithms, and are examples of structured EMR data. These data are readily available and searchable (fig 1), but vary in accuracy. Recent work has focused on incorporating other informative EMR data to develop robust phenotype algorithms.
Fig 1 Overview of the two main types of EMR data, structured and unstructured, and how these data can be integrated for research studies. In this instance, the figure illustrates the development of a phenotype algorithm for rheumatoid arthritis. *Including ICD-9 (international classification of diseases, 9th revision) codes and CPT (current procedural terminology) codes
Beyond billing and diagnoses codes, advanced EMRs contain a variety of structured data such as electronic prescriptions and laboratory values. A substantial portion of clinical data is also embedded in unstructured data in the form of narrative text notes, either typed or dictated by physicians (fig 1). Extracting accurate information from narrative notes is a well known challenge to clinical researchers and is typically obtained through laborious medical record review. Natural language processing (NLP),4 a specialty of computer science and informatics, has greatly helped researchers extract clinical data from narrative notes in a high throughput manner. While cutting edge NLP technologies have been successfully applied to internet search engines and automatic speech recognition, they are only now being adapted with new methods for biomedical research.
Overall methods for EMR phenotype algorithms,5 including NLP algorithms, have been specified elsewhere.6 7 8 However, the implementation of these algorithms with a team of clinical domain experts, bioinformaticians or NLP experts, biostatisticians, EMR informaticians, and genomics researchers has only been analysed tangentially. The focus on this implementation process by a multidisciplinary team was an objective of the Informatics for Integrating Biology and the Bedside (i2b2) project, with the overarching goal to harness the output of the healthcare system for discovery research. As part of the i2b2 project, we applied one general approach to develop several phenotype algorithms: depression,9 diabetes mellitus (V Kumar, in preparation), inflammatory bowel disease (ulcerative colitis and Crohn’s disease),10 multiple sclerosis,11 and rheumatoid arthritis.12 This method was also successfully applied to EMR data at other institutions.13 In this article, we present a roadmap of the tools and methods used in our approach to develop EMR phenotype algorithms.
Summary points
Successful application of natural language processing (NLP) into a phenotype algorithm developed from electronic medical records (EMR) requires a multidisciplinary team—clinical investigator, biostatisticians, EMR informaticians, and NLP experts—working in close collaboration
In the Informatics for Integrating Biology and the Bedside study, NLP improved the sensitivity of all algorithms, classifying more patients with high accuracy than algorithms using only structured data
Despite other robust methods to develop EMR phenotype algorithms, the positive predictive value, along with the percentage of patients with the phenotype classified by the algorithm, are the best metrics for evaluating the performance of EMR phenotype algorithms, regardless of the method for development
Toolbox: basic components needed to create EMR phenotype algorithms
The research question
The first step in creating an EMR phenotype algorithm is defining the major research objectives and the ideal study design and population. For example, the initial objective of the rheumatoid arthritis study was to determine the genetic risk factors for the disorder. In genetic studies, a clean phenotype is needed to ensure adequate power to detect risk alleles associated with the disease. Thus, we aimed to develop a classification algorithm for rheumatoid arthritis that would identify a sufficient number of patients with a high positive predictive value (PPV>90%) for the disorder.
Research database of structured EMR data
EMRs were developed primarily for patient care; therefore, their data formats are typically not ideal for research studies. Loading and storing the data in a relational database14 enables investigators to perform queries to obtain preliminary data. For example, an investigator can query the dataset for the number of patients who received an electronic rofecoxib prescription and subsequently had a new code for myocardial infarction from the ICD-9 (international classification of diseases, 9th revision) within five years. At Partners Healthcare (where the i2b2 project was based), structured data included ICD-9 codes, current procedural terminology codes, electronic prescriptions, and laboratory tests, along with the dates of evaluation.
Natural language processing
NLP4 is a computational method for processing text to extract information using the rules of linguistics. When notes are processed, NLP breaks down sentences and phrases into words, and assigns each word a part of speech—for example, a noun or adjective. The NLP program then applies the rules of linguistics to interpret the possible meaning of the sentence. In creating EMR phenotypes, we relied on the NLP task that identified so-called concepts in narrative clinical text. A concept is a meaning; for example, the terms “atrial fibrillation(s)” and “auricular fibrillation(s)” are different ways of expressing the same concept.15
Incorporating data extracted by NLP into a phenotype algorithm has several advantages. First, NLP provides data that are not available in the structured data or where the accuracy of the structured data is low. For example, before 2012, no specific ICD-9 code existed for basal cell carcinoma, a common skin condition.16
Second, NLP can systematically link several terms to a concept. For example, smoking is an important risk factor for many chronic diseases, but most information on smoking status is in a patient’s narrative notes. Determining a patient’s smoking status can be challenging because it is described in multiple forms, such as “tobacco,” “pack-year,” or “cigarettes.”17 18 NLP differs from a “find” command because it can recognize that the terms “tobacco,” “pack-year,” and “cigarettes” are all related to the concept of smoking. This NLP task is made possible by databases that standardize health terminologies, define the terms, and relate terms to each other and to a concept.
Such databases include the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT), which organizes health terminologies into categories (such as body structure or clinical finding), and RxNorm, which links drug names to other drug names in major pharmacy and drug interaction databases (table 1). In RxNorm, simvastatin is linked to its brand name, Zocor, as well as drugs that form a combination pill with simvastatin (such as sitagliptin/simvastatin (Juvisync), niacin/simvastatin (Simcor), and ezetimibe/simvastatin (Vytorin).
Table 1.
Useful web resources for EMR phenotype development*
| Resource | Description | URL |
|---|---|---|
| Dictionary of all concepts: maps concepts to unique concept identifiers | ||
| UMLS | Unified Medical Language System | www.nlm.nih.gov/research/umls |
| Biomedical terminology systems used by NLP: organizes concepts by categories such as drug class, signs and systems, and diagnoses | ||
| RxNorm | Normalized names for clinical drugs | www.nlm.nih.gov/research/umls/rxnorm |
| SNOMED CT | Systemized Nomenclature of Medicine-Clinical Terms | www.ihtsdo.org/snomed-ct |
| NLP open sources systems: parses notes to identify medical terms and maps terms to concepts | ||
| cTAKES | Apache clinical Text Analysis and Knowledge Extraction System |
http://ctakes.apache.org |
| HITex | Health Information Text Extraction | https://www.i2b2.org/software/projects/hitex/hitex_manual.html |
| Multicenter EMR projects applying NLP to EMR phenotype algorithms in clinical and translational studies | ||
| eMERGE | Electronic Medical Records and Genomics Network |
http://emerge.mc.vanderbilt.edu/ |
| i2b2 | Informatics for Integrating Biology and the Bedside project |
www.i2b2.org |
*This table lists examples of resources used by the i2b2 team for EMR phenotype development or mentioned in this article; it is not a comprehensive list.
Both SNOMED CT and RxNORM are part of the Unified Medical Language System,19 20 a resource linking standardized biomedical terms together into a concept. Each concept is assigned a unique concept identifier. For example, “atrial fibrillation(s)” and “auricular fibrillation(s)” are both defined under one unique concept identifier, C0004238. Similarly, all forms of simvastatin are represented by the identifier C0074554. As a result, terms expressed differently in clinical notes can link to one concept and one unique concept identifier.
An investigator studying simvastatin can use NLP (linked to RxNorm and the Unified Medical Language System) to process EMR narrative notes to identify patients on any formulation of simvastatin across different notes, patients, and EMR systems. For example, the variable of simvastatin (unique concept identifier C0074554) would have a value of 1 if there was a mention of simvastatin in the medical notes, or a value of zero if there was no mention. The equivalent task would require a manual search for each individual drug, generic name, and trade name using keywords terms. Although searches for individual terms are feasible for some concepts, if the study involved multiple diseases, drugs, and outcomes, the search terms needed could increase exponentially.
The i2b2 project used two open source, NLP software systems to extract concepts: the Health information Text Extraction system and the Apache clinical Text Analysis and Knowledge Extraction System21 (table 1).
Methods used to develop EMR phenotype algorithms
Creation of a sensitive data mart
The PPV, or accuracy, of an algorithm depends on the prevalence of the disease. The relationship between PPV and prevalence is shown in the following formula:
PPV=(sensitivity×prevalence)/[sensitivity×prevalence+(1−specificity)×(1−prevalence)]
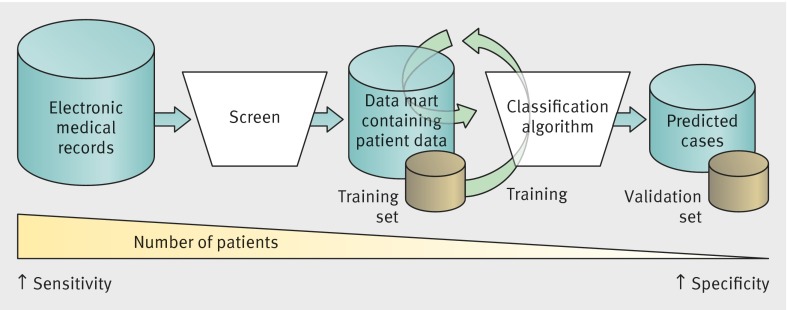
From large, population based epidemiological studies, the prevalence of four of the i2b2 study phenotypes, Crohn’s disease, multiple sclerosis, rheumatoid arthritis, and ulcerative colitis, was 1% or less in the general population in the United States.22 23 24 Developing an algorithm for these phenotypes using all patients in the EMR would substantially limit the PPV, owing to the phenotypes’ low expected prevalence in the EMR population. Therefore, as a first development step for all algorithms, we applied a screen selecting for patients with any data suggestive for the phenotype and excluded those with no evidence of the phenotype (fig 2). Patients with any data suggesting that they had the phenotype were included in a highly sensitive data mart. Clinical domain experts—who were the team physician scientists in the i2b2 study—determined the components of the screen. For example, a multiple sclerosis screen would include any patient with an ICD-9 code for “multiple sclerosis,” “encephalitis, myelitis, and encephalomyelitis,” and “other demyelinating disease of the central nervous system.”11
Fig 2 Overview of methods used to develop EMR phenotype algorithms
Algorithm variables
For each phenotype algorithm, the clinical domain experts created a comprehensive list of potential variables and terms (known as a customized dictionary). For rheumatoid arthritis, this list included “rheumatoid arthritis,” “bone erosions,” “synovitis,” “rheumatoid factor positivity,” and first line treatments such as methotrexate. The list was converted to available structured data including ICD-9 and current procedural terminology codes, electronic prescriptions, and laboratory tests. We also identified potential negative predictors, such as phenotypes with similar clinical presentations: for example, a negative predictor for ulcerative colitis was Crohn’s disease and vice versa.10 The clinical domain experts and NLP experts then mapped the list of terms to the concepts and unique concept identifiers using the Unified Medical Language System. In the i2b2 studies, we used NLP to process all clinical text notes including progress notes, discharge summaries, radiology reports, and pathology reports. NLP transformed the narrative data into data that could be readily analyzed—such as the number of times “bone erosions” was mentioned in the radiology reports. The final dataset for analysis included structured data (patient study identifier, number of ICD-9 codes for rheumatoid arthritis, and number of electronic prescriptions for methotrexate), alongside the narrative data (such as smoking status (yes v no) and number of mentions of “bone erosions”; fig 1).
The accuracy of each variable to define the phenotype was not as important as how the variables together in the algorithm could predict the phenotype. For all phenotypes, both the ICD-9 code and NLP concept for the phenotype were among the top five most predictive variables in the algorithm, despite low accuracy (PPV=20%12) for some ICD-9 codes. Thus, although we reviewed sentences labeled by NLP as containing a concept to assess whether the correct concept was identified, we did not systematically validate each potential variable for the algorithm. For example, we reviewed 100 sentences labeled by NLP as discussing cardiac catheterization to ensure that the sentence was not instead describing other forms of catheterization, such as urinary catheterization. After creating a comprehensive list of candidate variables, we relied on these data to inform which variables were most predictive for the phenotype.
Training set
We created a training set by selecting patients from the data mart at random (fig 2). The size of the training set was determined by the number of candidate variables and the prevalence of the phenotype. A phenotype with a substantial number of candidate variables and lower prevalence would need a large training set to achieve a robust classification algorithm. The clinical domain experts reviewed medical records of all patients in the training set and classified each patient as having the phenotype or not based on expert opinion and, when available, validated classification criteria from their respective clinical societies (for example, the American College of Rheumatology and the European League Against Rheumatism Classification Criteria for rheumatoid arthritis25).
Developing the classification algorithm
We identified the predictive variables for the algorithm and their weights using the adaptive LASSO penalized logistic regression26 method. The final classification algorithm (fig 2) was a logistic regression model, which assigned each patient a probability of having the phenotype based on their values for each variable. A hypothetical classification algorithm for phenotype A (PA) would be as follows:
Logit (probability of PA) = intercept − 0.16(sex) + 0.73 log(1+(NLP PA)) + 0.88 log(1+(ICD-9 PA)) + 0.63(NLP treatment) + ...
In the algorithm above, the input from the phenotype A data mart included a patient’s sex (1=female, 0=male), number of mentions of the NLP concept phenotype A (NLP PA) from the narrative notes, number of ICD-9 codes for phenotype A (ICD-9 PA), and whether a treatment for phenotype A was mentioned in any of the narrative notes (NLP treatment; 1=yes, 0=no). The end result of applying the algorithm to the phenotype A data mart was a calculated probability for phenotype A ranging between 0 and 1.0 for each patient.
Patients were classified as having phenotype A or not if their probability was above or below a threshold level, respectively. Unlike a Boolean approach (such as ≥1 ICD-9 code + treatment), this type of algorithm allows the investigator to adjust the threshold based on the scientific question. For the genetic study in rheumatoid arthritis, we found that a specificity of 95% (probability threshold ≥0.53) provided more power to detect an association with potential risk alleles (odds ratio 1.2) than a specificity of 97% (probability threshold ≥0.71).27 The improved power of the algorithm using a lower specificity threshold was driven largely by the classification of additional patients (from n=3585 to n=4575), with similarly high accuracy. Investigators interested in a pharmacovigilance study could consider setting a lower specificity threshold at 90% to capture additional patients.
Validation
We created a validation set comprising all patients classified with the phenotype, mixed with an additional 50% of random patients from the data mart. The clinical domain experts reviewed the records of all patients in the validation set using the same criteria to define the phenotype in the training set. Reviewers were blinded to the algorithm classification results. The performance of the algorithm was estimated using the validation set.
Other considerations
Use of NLP in phenotype classification algorithms
Incorporation of NLP improved the performance of all the algorithms studied in the i2b2 project. This improvement can be illustrated by the validation results for the algorithms for Crohn’s disease, multiple sclerosis, rheumatoid arthritis, and ulcerative colitis. For each phenotype, we compared the performance of a structured and NLP data algorithm with algorithms using only structured data or only data derived using NLP (table 2). For Crohn’s disease, multiple sclerosis, and ulcerative colitis, we achieved high accuracy (PPV≥94%) algorithms using structured data alone. NLP improved all algorithms using structured data by increasing the sensitivity while either maintaining or improving the accuracy, because NLP added independent predictive variables to the algorithm. For Crohn’s disease, the top two predictors for the phenotype were the number of ICD-9 codes followed by the number of NLP mentions. Therefore, although the structured and NLP variables described the same concept, the information they provided was not the same and were independently predictive of Crohn’s disease.
Table 2.
Comparison of performance algorithms using different types of data to classify phenotypes*
| Phenotype and performance characteristic | Performance algorithm | ||
|---|---|---|---|
| Structured data only | NLP data only | Structured and NLP data | |
| Inflammatory bowel disease10 | |||
| Crohn’s disease | |||
| Sensitivity (%) | 64 | 64 | 72 |
| PPV (%) | 98 | 98 | 98 |
| Ulcerative colitis | |||
| Sensitivity (%) | 60 | 68 | 73 |
| PPV (%) | 97 | 97 | 97 |
| Multiple sclerosis11 | |||
| Sensitivity (%) | 68 | 68 | 78 |
| PPV (%) | 94 | 94 | 95 |
| Rheumatoid arthritis12 | |||
| Sensitivity (%) | 51 | 56 | 63 |
| PPV (%) | 88 | 89 | 94 |
*Specificity cut-off for all phenotypes was set at 97%.
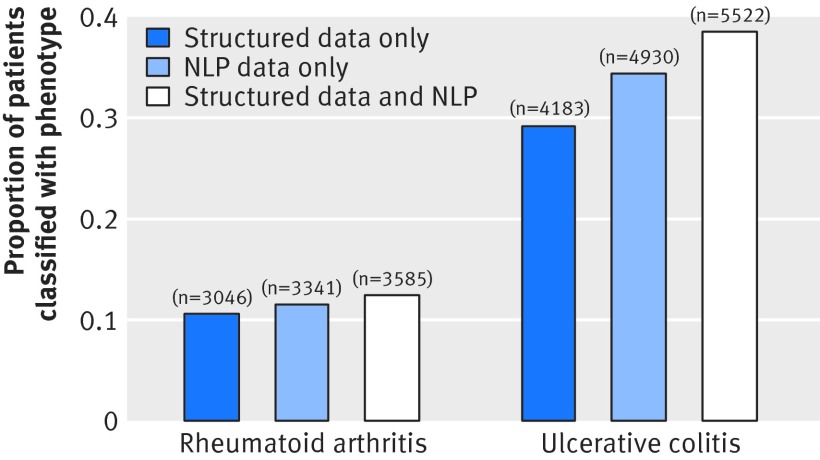
In absolute numbers, the addition of NLP increased the size of the ulcerative colitis cohort from 4183 to 5522 patients (fig 3) and improved the power for subsequent association studies.28 29 The addition of NLP to structured data in rheumatoid arthritis substantially improved the accuracy of the algorithm from 88% to 94% and increased the number of patients classified with the disorder from 3046 to 3585. But why did the addition of NLP data not improve accuracy in the algorithms for Crohn’s disease, multiple sclerosis, and ulcerative colitis? The accuracy of the structured data might explain some of this difference. Among a ulcerative colitis training set of 600 patients with at least one ICD-9 code for the disorder, 378 (PPV=64%) were confirmed to have the disorder.10 Among a rheumatoid arthritis training set of 500 patients, only 96 (PPV=19%) were confirmed to have the disorder,12 limiting the accuracy of an algorithm with structured data. In our experience, NLP had a greater impact on improving algorithms for phenotypes with a low prevalence and low accuracy for the phenotype ICD-9 code.
Fig 3 Proportion of patients in data marts for rheumatoid arthritis (n=28 982) and ulcerative colitis (n=14 335) who have been classified to have the phenotype. Numbers over each bar=EMR cohort size
The main limitation of using NLP in an EMR phenotype algorithm was the time and resources needed to identify and extract the variables for the algorithms. Such resources would be affected by the number of notes, number of variables in the customized dictionary, and NLP systems used for processing. For projects with many potential algorithm variables, mapping the clinical terms to NLP concepts was rate limiting, even with the use of available tools that assist with mapping. Several groups are now developing tools to accelerate the tuning process, in which terms are mapped accurately to NLP concepts. This process requires first tuning these tools on larger texts of medical knowledge, or using an automated tuning process that adapts or learns from mapping corrections made by the clinical domain experts.21 30 31 32 Finally, we note that the general phenotyping method presented in this article were successfully applied to a range of defined diseases and conditions but has not been extensively tested on outcomes such as drug response or adverse events.
EMR platform for clinical and translational studies
A unique aspect of EMR based cohorts is the ability to link clinical data with a biorepository, integrating clinical and genomic data in an EMR research platform for translational studies.33 With the appropriate infrastructure, EMR phenotype cohorts can be assembled in a relatively short period of time (12-18 months) compared with the years needed to recruit patients for prospective cohort studies, particularly for uncommon diseases. An EMR research platform containing linked data provides opportunities to conduct both hypothesis testing and generating studies. Hypothesis testing includes traditional, clinical, and genetic association studies. In a method unique to EMR research, the Phenome Wide Association Study allows for hypothesis generating studies and can be used as a screen to test for the association between genes or biomarkers and all phenotypes in the EMR.34 Moreover, the ability to apply EMR phenotype algorithms across institutions allows for the use of one phenotype for collaborative multicenter studies highlighted by the Electronic Medical Records and Genomics network35 as well as projects from our group.13
FAQs for clinical investigators
Can any institution with an EMR develop phenotype algorithms?
Any institution with an advanced EMR database—that includes data such as billing codes, electronic prescriptions, laboratory values, and narrative text notes—has the potential to develop phenotype algorithms. However, tapping into these data requires programmers with expertise in transforming the data into a useable format for research, such as a relational database structure. Such data reformatting requires an infrastructure that can support a research copy of the EMR, secure servers, terabytes of hardware space, and programmers who can manage and extract the data.
How would an investigator assemble a team to develop an EMR phenotype algorithm?
Despite many advances in the development of tools to mine EMR data commodity (for example, NLP software for clinical researchers), carrying out these studies presently requires a specialized team. The core team members include a biostatistician, clinical researcher, EMR informatician, and NLP expert. With the growth of the NLP field and its applications to biomedical research, most large academic medical centers have NLP experts on staff. An often missed but essential member of the team is the EMR informatician, who can understand the particularities of healthcare system data, such as differences in the way diagnostic results are reported by various clinics and where the data are stored.
In the i2b2 project, team meetings with all members present (especially at the start of the project) were the most effective way to work through multidisciplinary questions and discuss key concepts from our respective specialties. For example, a simple request can take a few steps, such as extracting data for white blood cell counts. Although the EMR informaticians at our institution know where to obtain the data, they would need to know from the clinical investigators which of the 46 types of laboratory data pertaining to white blood cell counts, grouped in two ways, were the correct fields to extract from the database. In another example, our NLP team presented a smoking module and used what they considered “precision” and “recall” to describe the performance of the algorithms. After some discussion, the clinical investigators and biostatisticians learned that NLP “precision” and “recall” is the same as PPV and sensitivity, respectively.
We thank the i2b2 team members integral to the development of our EMR algorithms, including Andrew Cagan, programmer, Research Computing, Partners Healthcare; Su-Chun Cheng, senior research scientist, Harvard School of Public Health; Sergey Goryachev, software developer, Ariadne Labs; Vishesh Kumar, postdoctoral fellow, Massachusetts General Hospital; and Robert Plenge, vice president, Merck Laboratories (Boston, MA, USA).
Contributors: All authors participated in the conception and design of the article, worked on the drafting of the article and revised it critically for important intellectual content, and approved the final version to be published.
Funding: This study was funded by grants from the US National Institutes of Health (U54LM008748, AR 060257, K08 K23 DK097142), and the Harold and Duval Bowen Fund.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf (available on request from the corresponding author) and declare: support from US National Institutes of Health and the Harold and Duval Bowen Fund for the submitted work; KPL is supported by the National Institutes of Health and the Harold and Duval Bowen Fund; GKS is on the Advisory Board of Wired Informatics, which provides services and products for clinical NLP applications; no other relationships or activities that could appear to have influenced the submitted work.
Provenance and peer review: Not commissioned; externally peer reviewed.
Cite this as: BMJ 2015;350:h1885
References
- 1.Brownstein JS, Murphy SN, Goldfine AB, Grant RW, Sordo M, Gainer V, et al. Rapid identification of myocardial infarction risk associated with diabetes medications using electronic medical records. Diabetes Care 2010;33:526-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liao KP, Diogo D, Cui J, Cai T, Okada Y, Gainer VS, et al. Association between low density lipoprotein and rheumatoid arthritis genetic factors with low density lipoprotein levels in rheumatoid arthritis and non-rheumatoid arthritis controls. Ann Rheum Dis 2013;73:1170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramirez AH, Shi Y, Schildcrout JS, Delaney JT, Xu H, Oetjens MT, et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics 2012;13:407-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jurafsky D, Martin JH. Speech and language processing : an introduction to natural language processing, computational linguistics, and speech recognition. 2nd ed. Pearson Prentice Hall, 2009.
- 5.Rasmussen LV, Thompson WK, Pacheco JA, Kho AN, Carrell DS, Pathak J, et al. Design patterns for the development of electronic health record-driven phenotype extraction algorithms. J Biomed Inform 2014;51:280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrell DS, Halgrim S, Tran DT, Buist DS, Chubak J, Chapman WW, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol 2013;179:749-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murff HJ, FitzHenry F, Matheny ME, Gentry N, Kotter KL, Crimin K, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA 2011;306:848-55. [DOI] [PubMed] [Google Scholar]
- 8.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC Med Inform Decis Mak 2006;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perlis RH, Iosifescu DV, Castro VM, Murphy SN, Gainer VS, Minnier J, et al. Using electronic medical records to enable large-scale studies in psychiatry: treatment resistant depression as a model. Psychol Med 2011;42:41-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ananthakrishnan AN, Cai T, Savova G, Cheng SC, Chen P, Perez RG, et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm Bowel Dis 2013;19:1411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Z, Secor E, Chibnik LB, Bove RM, Cheng S, Chitnis T, et al. Modeling disease severity in multiple sclerosis using electronic health records. PLoS One 2013;8:e78927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao KP, Cai T, Gainer V, Goryachev S, Zeng-treitler Q, Raychaudhuri S, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res (Hoboken) 2010;62:1120-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll RJ, Thompson WK, Eyler AE, Mandelin AM, Cai T, Zink RM, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nalichowski R, Keogh D, Chueh HC, Murphy SN. Calculating the benefits of a research patient data repository. AMIA Annu Symp Proc 2006:1044. [PMC free article] [PubMed]
- 15.Pradhan S, Elhadad N, South BR, Martinez D, Christensen L, Vogel A, et al. Evaluating the state of the art in disorder recognition and normalization of the clinical narrative. J Am Med Inform Assoc 2015;22:143-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endicott M. New ICD-9-CM diagnosis codes for FY 2012. J AHIMA 2012;82:60-2;quiz 63. [PubMed] [Google Scholar]
- 17.McCormick PJ, Elhadad N, Stetson PD. Use of semantic features to classify patient smoking status. AMIA Annu Symp Proc 2008:450-4. [PMC free article] [PubMed]
- 18.Savova GK, Ogren PV, Duffy PH, Buntrock JD, Chute CG. Mayo clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.US National Library of Medicine. Unified medical language system terminology services. 2014. https://uts.nlm.nih.gov/home.html.
- 20.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao KP, Karlson EW. Classification and epidemiology of rheumatoid arthritis. In: Hochberg MC, Silman AJ, Smolen JS, Weinblatt ME, Weisman MH, eds. Rheumatology. 5th ed. Mosby Elsevier, 2011:823-4.
- 23.Kappelman MD, Rifas-Shiman SL, Kleinman K, Ollendorf D, Bousvaros A, Grand RJ, et al. The prevalence and geographic distribution of Crohn’s disease and ulcerative colitis in the United States. Clin Gastroenterol Hepatol 2007;5:1424-9. [DOI] [PubMed] [Google Scholar]
- 24.Simpson S Jr, Blizzard L, Otahal P, Van der Mei I, Taylor B. Latitude is significantly associated with the prevalence of multiple sclerosis: a meta-analysis. J Neurol Neurosurg Psychiatry 2011;82:1132-41. [DOI] [PubMed] [Google Scholar]
- 25.Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 Rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Arthritis Rheum 2010;62:2569-81. [DOI] [PubMed] [Google Scholar]
- 26.Zou H. The adaptive Lasso and its oracle properties. J Am Stat Assoc 2006;101:1418-29. [Google Scholar]
- 27.Kurreeman F, Liao K, Chibnik L, Hickey B, Stahl E, Gainer V, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ananthakrishnan AN, Cagan A, Gainer VS, Cai T, Cheng SC, Savova G, et al. Normalization of plasma 25-hydroxy vitamin D is associated with reduced risk of surgery in Crohn’s disease. Inflamm Bowel Dis 2013;19:1921-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ananthakrishnan AN, Cagan A, Gainer VS, Cheng SC, Cai T, Scoville E, et al. Thromboprophylaxis is associated with reduced post-hospitalization venous thromboembolic events in patients with inflammatory bowel diseases. Clin Gastroenterol Hepatol 2014;12:1905-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010;17:229-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garla VN, Brandt C. Knowledge-based biomedical word sense disambiguation: an evaluation and application to clinical document classification. J Am Med Inform Assoc 2013;20:882-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leaman R, Islamaj Dogan R, Lu Z. DNorm: disease name normalization with pairwise learning to rank. Bioinformatics 2013;29:2909-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy S, Churchill S, Bry L, Chueh H, Weiss S, Lazarus R, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res 2009;19:1675-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med 2011;3:79re1. [DOI] [PMC free article] [PubMed] [Google Scholar]