Abstract
Cell based assays are essential tools utilized by research labs in a wide range of fields, including cell biology, toxicology, and natural product discovery labs, however in some situations the need for cell based assays does not justify the costs of maintaining cell culture facilities and retaining skilled staff. The Kit-On-A-Lid-Assay (KOALA) technology enables accessible low-cost and pre-packageable microfluidic platforms that can be operated with minimal infrastructure or training. Here, we demonstrate and characterize high-density KOALA methods for high throughput applications, achieving an assay density comparable to that of a 384 well-plate and usability by hand with no liquid handling equipment. We show the potential for high-content screening and complex assays such as quantitative immunochemistry assays requiring multiple steps and reagents.
Keywords: Microfluidic, High-throughput, Cell-based, Assay, Low-cost
Introduction
Cell-based assays and in vitro models are essential tools for a wide range of biological and chemical applications such as cell biology, drug discovery, and toxicology.1 However, traditional assays, including microfluidic assays, suffer from significant limitations preventing more widespread use.2,3 Amongst these barriers to adoption are the cost of cells and reagents, the limited scalability of the assays due to the reliance on a serial (vs. parallel) operation process, and the requirement of both preparation and equipment – such as pipettes and centrifuges (Fig. 1). Perhaps more importantly, traditional assays require trained staff which limits accessibility of these assays to labs that don’t have cell culture expertise and/or facilities, such as natural product discovery labs, educational labs, engineering labs, or environmental research labs.4 The need for simplified assays has been highlighted by assays such as the SlipChip.5 Yet, there remains a dearth of technologies for cell-based assays that are low cost, simple to operate, and that integrate the ability to perform sequential and complex fluid replacements.
Figure 1.
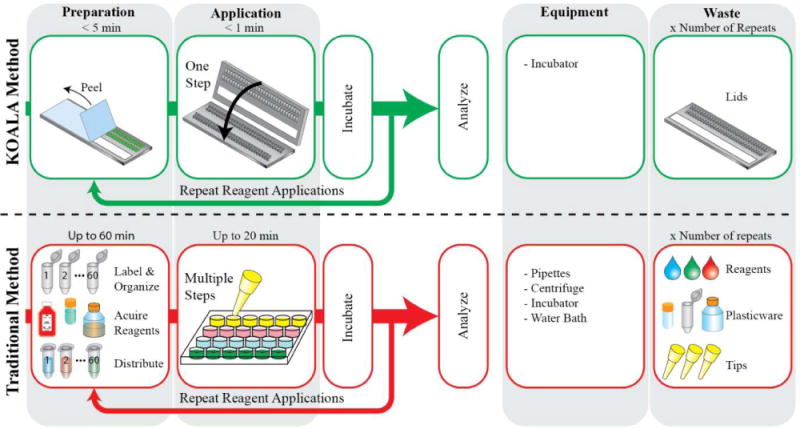
Schematic represents the workflow differences between a KOALA microfluidic platform (top) and a traditional assay (bottom). KOALA enables prepackaging of reagents into lids, parallel fluid applications, minimal demands for equipment, and minimal waste. Traditional assays often require significant preparation, a serial process for fluid application, several pieces of equipment, and large amounts of waste.
The Kit-On-A-Lid-Assay (KOALA) technology is a novel microscale platform that aims to reduce barriers to adoption of cell-based assays by enabling precise and advanced fluidic handling in a self-contained, user-friendly interface.6,7 KOALA integrates several features to simplify complex assays. First, pre-packaging eliminates the need to prepare reagents, label tubes, and distribute reagents to each assay, a time-consuming and often under-estimated part of traditional assays. Second, KOALA is a micro-to-micro platform, reducing waste by eliminating the need to dilute/prepare reagents for the assay via macro-scale tubes (as is currently done even for microscale platforms). Finally, the integrated fluid handling system of KOALA eliminates the need for fluid handling equipment, such as pipettes, mixers, and centrifuges. The KOALA fluid handling system utilizes a series of lids that are used in conjunction with a single base containing a set of microchannels. Different sets of lids are used for different assays. Lids are prepackaged with only the necessary volumes of pre-diluted reagents required for each step of the assay. Each step (i.e., fluid exchange step needed to apply a treatment or reagent on the cell culture) is then performed by removing a lid from the freezer, thawing it, peeling a protective layer covering micro-wells containing reagents, and “clicking” the lid onto the base (Fig. 2A).
Figure 2.
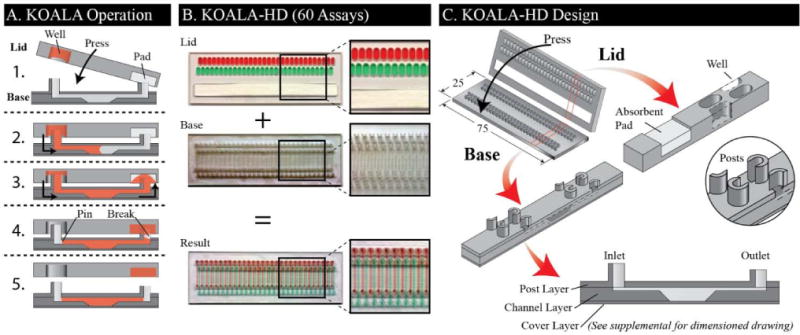
(A) KOALA operation entails pressing a lid, containing micro-wells with reagents and an absorbent pad, onto a base, containing microchannels. When the microfluidic post contacts the fluid, flow is initiated by capillary action. Once the channel is full, an absorbant pad sustains the flow until the well is empty. Once empty, the fluid pins at the entrance of the channel and an air bubble forms in the channel exit. This bubble breaks the fluidic connection between the channel and the absorbent pad – assuring that the channel is not emptied – and completes the fluid transfer. (B) An image of the KOALA-HD pre-filled lid and base, and the resultant base after application of the lid. (C) KOALA-HD compresses 60 assays into the foot print of a traditional glass slide. Lids house micro-wells and absorbent pads, while the bases consist of staggered microchannels. A post is located at both ends of the microchannel. A dimensioned drawing is available in the supplementary information.
Current KOALA methods have described the basic principles for exchanging fluid between a lid and a base. However, significant limitations remain for using these methods for high-density and robust screening applications6, including modes of cross-contamination between neighboring wells and channels, fluid replacement efficiency and overall fluid characterization, and validating the technology on for functional cancer cell assays. We address these limitations through multiple design enhancements that enable a high-density KOALA (KOALA-HD) format with throughput on par with 384 well plates in a platform that can be operated by hand without any fluid handling equipment (Fig. 2B). The following describes the geometrical parameters (i.e. feature characteristics, orientation, and dimensions) and fabrication processes that are necessary to enable KOALA-HD while preventing corss contamination. We present and characterize the washing efficiency, operation, and performance of the KOALA-HD platform for repeated and higher-density applications in terms of fluidic function and for facilitating cell-culture applications lasting more than 24 hours. Finally, we demonstrate the utility of KOALA-HD for high-throughput cell-based applications by performing a screening assay with complex immunohistochemistry (IHC) endpoints.
Materials and Methods
Device Fabrication and Preparation
The lids and bases are fabricated by CNC milling (PCNC 770, Tormach, Waunakee, WI) with standard microscope slide dimensions (25 mm × 75 mm). Bases are comprised of three layers: (1) a post layer, (2) a channel layer, and (3) a cover layer (Fig. 2C and S1). (1) The post layer is milled from 2 mm thick PS (#ST313200, Goodfellow, Coraopolis, PA) and encompasses an inlet and an outlet post for each channel. Both posts have a horseshoe like cross section with an outside diameter of 1 mm. The inlet post for each channel is 1.5 mm tall with a slot width of 0.8 mm and the outlet post is 1 mm tall with a slot width of 0.5 mm. Lengthwise the spacing of the posts matches that of the channels, however widthwise the posts and channels are staggered by 3 mm to facilitate close packing. (2) The channel layer is milled from 1.2 mm thick polystyrene (PS) (#ST313120, Goodfellow) and encompasses 60 microchannels spaced by 1 mm (from channel centers). Each channel is 0.5 mm wide, 2 mm long, and 0.3 mm deep. In the center of each channel is a micro-well that is 0.5 mm wide, 4 mm long, and 1.2 mm deep, with bevels at both ends to facilitate fluid flow. (3) The cover layer is 25 mm × 75 mm and is cut from 0.190 thick PS (#ST313190, Goodfellow). Prior to assembly, all three layers are treated with oxygen plasma (Femto, Thierry Corp., Royal Oak, MI) for 50s at 50 W. Layers are assembled by acetonitrile bonding.8 In brief, Acetonitrile (#271004, Sigma Aldrich, St. Louis, MO) is applied drop wise to one layer of the device, which is then promptly mated with a second layer. The excess acetonitrile is aspirated from the channels and the mated pair is gently pressed together on a hot plate preheated to 70° C. After 30 s the process is repeated for the third layer. After bonding, the device is plasma treated again with the same settings, and hydrophobic recovery is induced between the posts using a wiper.9
Lids are milled from 2 mm PS. Each lid contains a micro-well for each channel (i.e., 60 micro-wells) and a slot for absorbent pads. Each micro-well has an elliptical shape which is 1.5 mm wide, 4 mm long, and 2 mm deep, and is spaced to match the pattern of the input posts. The slot for the absorbent pad is 10 mm wide, 72 mm long, and 2 mm deep. The lids are assembled by first applying an optical adhesive cover (#04 729 757 001, Roche, Basel, Switzerland) to the back of the lid then placing two layers of absorbent pad (#CFSP223000, Millipore, Billerica, MA) in the designated slot. Both bases and lids are UV sterilized for 30 minutes, prior to cell culture, and lids are loaded with reagents prior to each application.
Fluid handling
Fluid exchanges are characterized by (i) measuring the duration, (ii) washing efficacy, and (iii) cross contamination during fluid replacements. Fluid replacements are initiated by mating the lid with the base, thereby inserting the series of posts into the micro-wells. (i) To assess time required for fluid replacement, the micro-wells are filled with 15 μL of DI water and the lid is clicked on the base. Time is measured from the point of initial fluid contact, between the post and the fluid, to the point at which the micro-well is emptied. (ii) Washing efficacy is assessed by measuring the depletion of a fluorescent dye after individual wash steps. Channels are pre-loaded with a fluorescent dye (Alexa Fluor 488, #D-22910, Life Technologies, Carlsbad, CA), then washed by applying lids containing 15 μL of DI water per micro-well. Channels are imaged after each wash is completed (IX-70, Olympus, Shinjuku, Tokyo, Japan) and fluorescence intensities are measured using ImageJ. The background autofluorescence, inherent to the PS, is measured, averaged (n = 8 locations), and subtracted from the channel fluorescence. After subtraction, all channels are normalized to the average of the starting fluorescence intensity, and averaged (n = 60 channels) (error bars represent one standard deviation). (iii) To assess cross contamination, alternating micro-wells are filled with 15 μL of either Alexa Fluor 488 or Texas Red (#D-3328, Life Technologies). After completing the fluid exchange, each channel is imaged for both dyes and fluorescence intensity is quantified using ImageJ. Autofluorescence noise is subtracted from each channel, for both wavelengths.
Cell Culture for IHC Assay
In this section, all fluid transfers, including cell seeding, are executed by filling each of the 60 micro-wells with 15μL of the respective reagent then mating the lid with the base. NMuMG cancerous epithelial cells are seeded in KOALA-HD channels at a surface density of 400 cells/mm2, and cultured at 37° C for a total of 72 hr. For the first 24 hours, cells are cultured in DMEM media with 4.5 g/L glucose (#10-017-CV, Corning Cellgro, Manassas, VA) supplemented with 10 μg/mL insulin and 10% fetal bovine serum (FBS). For the next 48 hours, the cells are cultured in the same media supplemented with a 0, 2, 20, or 200 pM concentration of TGF-β1 (#100-21, Pepro Tech, Rocky Hill, NJ). The media is replaced every 24 hours. Cells are washed with 1x phosphate buffered saline (PBS), fixed with 4% paraformaldehyde (#43368, Alfa Aesar, Ward Hill, MA) for 15 min, and permeabilized with 0.1% Triton X-100 (#807426, MP Biomedicals, Santa Ana, CA) for 30 min. Channels are washed with 1x PBS, then blocked with 1x PBS supplemented with 3% bovine serum albumin (PBS+BSA) for 30 min. Primary antibody solutions are prepared by separately diluting anti-β-catenin (rabbit) to 2 μg/mL, anti-E-Cadherin (mouse) (#610182, BD Biosciences, San Jose, CA) at 1 μg/mL, anti-FAK (rabbit) (#ab40794, Abcam, Cambridge, England, UK) to 2 μg/mL, anti-N-Cadherin (rabbit) (#ab18203, Abcam) to 6 μg/mL, or anti–Vimentin (mouse) at 1:50 into PBS+BSA supplemented with 0.1% Tween 20 (PBST+BSA) (#P1379, Sigma Aldrich). Primary antibody solutions are added to appropriate channels (with n = 3 for each concentration of TGF-B1) for 1.5 hours, then washed twice with PBS+BSA. Secondary antibody solutions are prepared by diluting both Alexa 488 Goat anti-mouse (#A-11029, Life Technologies) and Alexa 568 Goat anti-rabbit (#A-11036, Life Technologies) to 4 μg/mL into a single solution of PBST+BSA, which is applied to all channels for 1 hour. A Hoechst 33342 nuclear stain is applied to all channels for 15 minutes then washed twice with PBS+BSA. Bases are imaged on an inverted fluorescent microscope (Eclipse Ti, Nikon, Tokyo, Japan) in three wavelengths: 345, 488, and 572 nm. Channel images are taken at 4x magnification and stitched automatically (NIS-Elements Ar, Nikon), and high magnification images are taken at 10x magnification and overlaid using ImageJ. ImageJ is also used to measure the fluorescence intensity of the channels. Background fluorescence is taken from surrounding regions, averaged, and subtracted from the fluorescence values of the channels. Within each staining category (i.e., β-catenin, E-cadherin, etc.) the channels are normalized to the maximum intensity of their respective category, and averaged (n = 3). Error bars represent one standard deviation.
Results and Discussion
KOALA-HD represents a novel class of high-throughput microfluidic platforms that can be operated quickly, reproducibly, and reliably with minimal demands for training or equipment. The KOALA-HD compresses 60 microfluidic assays into the 25 × 75 mm footprint of a traditional glass slide, enabling medium- to high-throughput applications by hand without the need for an automated liquid handler. The number of assays that can be performed in this footprint, i.e. assay density, is comparable to that of a 384 well plate. However, unlike assays performed in 384 well plates, fluid replacements for KOALA-HD are performed in all 60 assays simultaneously. The high density format of the channels in the base resulted in design challenges that needed to be resolved in order to prevent cross-contamination of reagents and support reliable operation of the device. These design challenges and solutions are discussed here.
Fluid exchanges in KOALA-HD
All fluid handling mechanisms necessary to operate KOALA are integrated into the KOALA lids and bases. Each lid is a one-time-use component comprised of 60 micro-wells, which houses reagents and an absorbent pad to drive fluid flow (Fig. 2C). Similarly, each base is comprised of 60 microfluidic channels; however, unlike the lids, the base is used for the duration of the assay. Each fluid replacement – performed by clicking a lid onto the base – is completed for all 60 channels within 15 seconds (See Supplemental Video). By measuring the average time required to empty each well, we find that the average flow rate through the channel is ~7 μL/sec. Standard microfluidic techniques to produce alternative channel designs or absorbent pad compositions can be used to achieve different flow rates for other applications if needed. When the lid is applied to the base fluid flows from the micro-well (of the lid), through the channel (of the base), and into the absorbent pad (of the lid), thus completing a fluid exchange. After which, the lid is disposed of and the base is incubated until the next fluid replacement. The novel design of the base leverages horseshoe shaped posts to enable fluid exchanges through several functionalities.6 The posts provide a method of connecting the base to the lid to prevent air bubbles from being trapped in the channels, while ensuring proper volumes during fluid exchanges. Posts are located at the channel inlet and outlet, which, when a lid is applied, make contact with the fluid in the micro-well and which the absorbent pad, respectively. Fluid flows through the channel initially driven by capillary forces in the channel and subsequently by wicking in the absorbent pad. When the micro-well is emptied, the fluid pins in the bottom of the inlet post, and the outlet post severs the fluid flow by allowing a bubble to form within the horseshoe, thus disconnecting the absorbent pad from the channel. This process completes the fluid exchange.
To make KOALA suitable for HD applications, however, several obstacles must be overcome including: (1) determine a configuration suitable for densification, (2) eliminate cross contamination, and (3) achieve complete and defined fluid exchanges in the new HD format.
To determine a setup that best supports the densification we test two designs, a format that uses individual posts as described above and a common rail configuration (Fig. S2). The lid for the common rail design comprises a single trench surrounded by half elliptical micro-wells while the base comprises a single rail with several protruding fins. When the lid is applied to the base, the fins provide the conduit for wicking fluid from the lid into the channel. We found that a design with individual posts for each channel is superior for two reasons. First, the separation between the inlet posts prevents cross contamination via Concus-Finn filaments. More explicitly, the corners between vertical and horizontal features act as a conduit for fluids, which leads to cross contamination. Second, the small diameter inlet and outlet posts prevent excessive displacement of fluid in the micro-wells, and support improved contact with the absorbent pad.
To eliminate cross contamination, hydrophilicity between the posts and fluid displacement in the micro-wells is decreased. After plasma treatment, we implement a novel approach, described by Guckenberger et al.9 to recover surface hydrophobicity between the posts, thus mitigating hydrophilicity based cross contamination. Inadequate contact between the outlet posts and the absorbent pad lead to cross contamination via rapid displacement of the fluid in the micro-wells. This issue is circumvented by ensuring proper connection between the outlet posts and the absorbent pad prior to inserting the inlet posts into the micro-wells. In the future, a small reusable device for repeatably aligning and mating the lid and base (i.e., a jig) will be designed to facilitate this connection and prevent the possibility of user-error.
To ensure complete and defined fluid exchanges for each channel, we leverage the designs of the micro-wells, the posts, and the absorbent pads. The volume allotted for each fluid exchange is dictated by the volume of the micro-well. However, maximizing use of the fluid in the micro-well, and thus minimizing dead volume, depends on the orientation of the inlet post and the size of the absorbent pad. We found that to minimize dead volume, the open side of the horseshoe should be directed towards the end of the micro-well (Fig. S3). In doing so, the dead volume of each micro-well is reduced to <3 μL. Further, to assure that all the micro-wells are emptied entirely, the absorbent pad has to be sufficiently large to assure that it does not saturate during the fluid exchange. Put together, these solutions make KOALA-HD fluid exchanges repeatable and robust.
Evaluating KOALA-HD Function
In order to make a highly functional platform to be used for toxicology and cell-based studies, we assess the platform for its ability to: (1) ensure proper fluid replacement in the channels, (2) ensure no cross contamination between adjacent channels, a difficultly particularly compounded by higher throughput, and (3) perform several fluid exchanges without failure.
We quantify the depletion of a fluorescent dye after washing steps to assess the efficiency of fluid replacements (Fig. 3A). To fulfill this, a lid filled with fluorescent dye is applied to the channels, followed by two washing lids. We quantify the fluorescent depletion after each wash and normalize it to the starting intensity. We found that one washing lid is capable of removing ~60% of the initial fluid, and a second washing lid reduced the dye concentration by over 98% (n = 60), which, based on the volumes of the channels and fluid exchanges used here, is comparable to washing of traditional microfluidic channels.10
Figure 3.
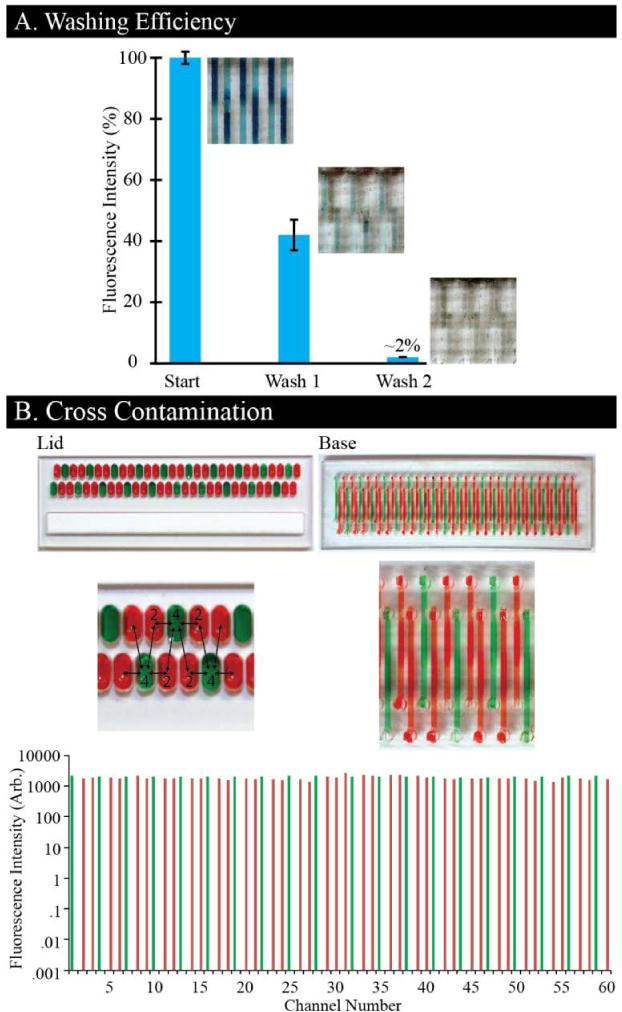
(A) Washing efficiency of lids is quantified by measuring a decrease in fluorescent intensity after each lid (n = 60). (B) Cross contamination is assessed by filling a lid with two different fluorescent dyes (depicted by food coloring), and applying that lid to a base. Fluorescent intensities for both red and green are measured and represented for each channel. No cross contamination is evident.
Cross contamination is assessed by quantifying fluorescence in each microchannel after applying a lid pre-filled with two fluorescent dyes (Fig. 3B). The lid is strategically filled such that each micro-well containing green dye is directly adjacent to four micro-wells with red dye (illustrated with food coloring), enabling detection of cross contamination from both horizontally and diagonally adjacent micro-wells. This lid is applied to a base, and each channel in the base is quantified for both red and green fluorescence (Fig. 3B, bottom). The results reveal no cross contamination between channels based on no increase in fluorescence intensity between channels (n = 1), which is confirmed by observing no fluidic connection between the adjacent posts or micro-wells. Notably, these data are collected using high-surface tension fluid (water), but for low surface tension fluids (e.g. oil, high-surfactant reagents) the lid and base may be inverted. This result sets the stage for using the KOALA-HD in toxicological screens or pharmaceutical identification of natural products, including readouts such as protein expression.
Finally, we assesses the ability to operate a fluidic assay without failure by applying a series of lids and verifying complete flow in each channel. A series of 20 lids are pre-filled with dyed water and applied to a single base. For comparison, about 15 fluid replacements are typically required for a traditional immunochemistry staining. Each lid is filled with alternating colors to make channel failure obvious. All 20 fluid exchanges are completed without a single channel failure.
Channel design for cell culture
To ensure suitability for cell culture, KOALA-HD is designed to: (1) allow precise readouts of cellular functions (proliferation, protein localization, etc.), (2) contain enough media to maintain cell viability throughout the incubation period, and (3) be negligibly impacted by evaporation. The channels are designed with a lowered region in the center that serves three purposes. First, the decreased plastic thickness enables better cellular readouts (e.g., higher magnification microscopy, less autofluorescence from the plastic). Second, the increased channel size reduces the shear stress by slowing fluid velocity. Third, the lowered region increased the volume and thus increases the reservoir of nutrients that are available for the cells. With this channel design, we are able to culture cells for 24 hours without media exchanges.11,12 However, for more highly-active or sensitive cells more frequent fluid replacements may be necessary. Evaporation is mitigated using previously described and characterized methods.13,14
IHC assays
We demonstrate the ability to perform high density IHC assays and complex staining processes reliably and without the need to prepare antibody dilutions or source specific reagents. We chose an IHC assay based on their ubiquity in screening applications and for the multitude of steps necessary to complete the assay (i.e., cell-culture, fixation, permeabalization, staining, washes, etc.). We use a relevant cancer specific assay where NMuMG epithelial cells are treated with various dosages of TGF-β1, inducing an epithelial-to-mesenchymal transition (EMT).15 Markers of this transition include: down regulation and localization of β-catenin and E-cadherin to cell-cell contact areas16, and increased expression of FAK17, N-cadherin18, and Vimentin.19 Such complex readouts require cell culture, treatment with a drug, and IHC staining. We demonstrate cell culture in KOALA-HD by culturing NMuMG cells in 60 channels for 72 hours. The process requires 15 sequential fluidic steps, all performed using the KOALA platform (i.e. cell seeding, media replacements, TGF-β1 dose treatments, fixing solution, washing buffer, primary antibody staining, and secondary antibody staining). The platform is able to perform each fluidic step in all channels simultaneously without failure or cross contamination. Each fluidic exchange requires about 15 seconds with a simple mating of the required lid with the base containing cells.
Cells are treated in four groups of 15 channels with varying concentrations of TGF-β1. After which, cells are fixed, permeabalized, stained with a nuclear stain, and stained in triplicate for β-catenin, E-cadherin, FAK, N-cadherin, and Vimentin (Fig. 4A). We found that β-catenin decreases and N-cadherin increases with increasing levels of TGF-β1, as expected. We did not observe any decrease of E-cadherin for increasing levels of TGF-β1, but we did observe localization to the cell-cell contact areas. FAK decreases with increasing levels of TGF-β1, which is inverted from typical results but has been observed in NMuMG cells expressing D119A-β3 integrin.17 Finally, prevalence of Vimentin, a mesenchymal marker, is evident in cells treated with TGF-β1 indicating a transition to a mesenchymal phenotype. Selected channels treated with the highest concentration of TGF-β1 (i.e., 200 pM) are imaged at high magnification (Fig. 4B). These images demonstrate localization of β-catenin and E-cadherin to the cell-cell contact areas. A noticeable feature in some of the images is the striated pattern formed by the cells arranging themselves along fabrication artifacts. These are the result of machining marks left by the CNC machine during the channel fabrication and can be removed by utilizing other fabrication methods such as injection molding. All channels are stained with both the Alexa 488 and Alexa 568 dyes to detect non-specific binding. Importantly, we demonstrate that KOALA-HD can – in the footprint of a glass slide – cover a broad range of treatments and readouts, providing high content microscopy images at a high throughput scale, without a need for automation or trained personnel. In future applications, this platform could be imaged with a scanner for higher throughput analysis.12,20
Figure 4.
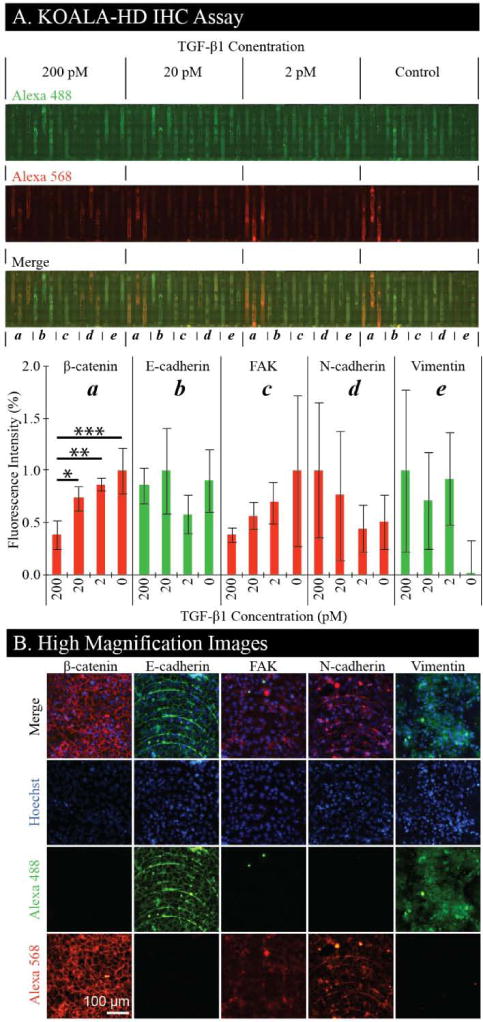
(A) NMuMG cells are cultured in KOALA-HD channels and treated in 15 channel segments with varying concentrations of TGF-β1. Within each segment, channels are stained with primary/secondary antibodies for (a) β-catenin/Alexa-568, (b) E-cadherin/Alexa-488, (c) FAK/Alexa-568, (d) N-cadherin/Alexa-568, and (e) Vimentin/Alexa-488 (Hoechst not shown). The bar graph demonstrates average fluorescence intensities for each of the treatment conditions and antibody stains (n = 3). Error bars represent standard deviation. Analysis of Variance (ANOVA) indicated significant differences within B-catenin data (p<0.01). P-values are indicated for protected Fisher’s Least Significant Difference (LSD) test as follows: * p≤0.05, ** p≤0.01, *** p≤0.001. (B) High magnification images of each antibody stain taken from channels treated with 200 pM concentration of TGF-β1. Each channel is imaged for Hoechst, Alexa 488, and Alexa 568.
Conclusion
The KOALA-HD platform is a microscale fluidic handling technology that simplifies cell-based assays such that anyone can perform them. The only operation is to place a lid onto a base. KOALA-HD enables: reagent prepackaging and cryopreservation – acting to reduce preparation time and waste, lower demands on infrastructure or trained personnel – expanding access of cell-based assays to more labs, and enable high-throughput microfluidic applications through the scalability offered by parallelized fluidic handling procedures. We demonstrate how feature dimensions and geometry can be altered to accommodate close packing of microfluidic channels. In addition, we characterize the robustness simultaneously performing 60 assays within the footprint of one microscope slide. We demonstrate that the KOALA-HD would be suitable for toxicology, environmental pollutant, and natural product research by showing functional and advanced cellular readouts in the form of protein expression and morphology. However, due to the simplistic nature and device modularity it is possible that KOALA-HD could be expanded into other areas, including patient screening in clinics7,21, water quality tests, or even food safety testing.
Supplementary Material
Acknowledgments
The authors thank Dr. Joshua Lang for use of his microscope.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH R01 EB010039.
Footnotes
Declaration of Confliction Interests
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: All authors hold equity in Salus Discovery LLC, which has licensed technology described in this manuscript.
References
- 1.Bhadriraju K, Chen CS. Engineering cellular microenvironments to improve cell-based drug testing. Drug Discovery Today. 2002;7(11):612–620. doi: 10.1016/s1359-6446(02)02273-0. [DOI] [PubMed] [Google Scholar]
- 2.Barbulovic-Nad I, Yang H, Park PS, Wheeler AR, et al. Digital microfluidics for cell-based assays. Lab on a Chip. 2008;8(4):519–526. doi: 10.1039/b717759c. [DOI] [PubMed] [Google Scholar]
- 3.Michelini E, Cevenini L, Mezzanotte L, et al. Cell-based assays: fuelling drug discovery. Anal Bioanal Chem. 2010;398(1):227–238. doi: 10.1007/s00216-010-3933-z. [DOI] [PubMed] [Google Scholar]
- 4.Ewer K, Deeks J, Alvarez L, et al. Comparison of T-cell-based assay with tuberculin skin test for diagnosis of Mycobacterium tuberculosis infection in a school tuberculosis outbreak. The Lancet. 2003;361(9364):1168–1173. doi: 10.1016/S0140-6736(03)12950-9. [DOI] [PubMed] [Google Scholar]
- 5.Du W, Li L, Nichols KP, et al. SlipChip. Lab on a Chip. 2009;9(16):2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berthier E, Guckenberger DJ, Cavnar P, et al. Kit-On-A-Lid-Assays for accessible self-contained cell assays. Lab on a Chip. 2013;13(3):424–431. doi: 10.1039/c2lc41019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sackmann EK, Berthier E, Young EW, et al. Microfluidic kit-on-a-lid: a versatile platform for neutrophil chemotaxis assays. Blood. 2012;120(14):e45–53. doi: 10.1182/blood-2012-03-416453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou P, Young L, Chen Z. Weak solvent based chip lamination and characterization of on-chip valve and pump. Biomed Microdevices. 2010;12(5):821–32. doi: 10.1007/s10544-010-9436-z. [DOI] [PubMed] [Google Scholar]
- 9.Guckenberger DJ, Berthier E, Young EWK, et al. Induced hydrophobic recovery of oxygen plasma-treated surfaces. Lab on a Chip. 2012;12(13):2317–2321. doi: 10.1039/c2lc21052e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warrick J, Meyvantsson I, Ju J, et al. High-throughput microfluidics: improved sample treatment and washing over standard wells. Lab on a Chip. 2007;7(3):316–321. doi: 10.1039/b613350a. [DOI] [PubMed] [Google Scholar]
- 11.Su X, Theberge AB, January, et al. Effect of Microculture on Cell Metabolism and Biochemistry: Do Cells Get Stressed in Microchannels? Anal Chem. 2013;85(3):1562–1570. doi: 10.1021/ac3027228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paguirigan AL, Beebe DJ. From the cellular perspective: exploring differences in the cellular baseline in macroscale and microfluidic cultures. Integrative Biology. 2009;1(2):182–195. doi: 10.1039/b814565b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berthier E, Warrick J, et al. Managing evaporation for more robust microscale assays Part 1. Volume loss in high throughput assays. Lab on a Chip. 2008;8(6):852–859. doi: 10.1039/b717422e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Regehr KJ, Domenech M, Koepsel JT, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab on a Chip. 2009;9(15):2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavadil J, Bottinger EP. TGF-[beta] and epithelial-to-mesenchymal transitions. Oncogene. 2005;24(37):5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 16.Piek E, Moustakas A, Kurisaki A, et al. TGF-(beta) type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. Journal of Cell Science. 1999;112(24):4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- 17.Wendt M, Schiemann W. Therapeutic targeting of the focal adhesion complex prevents oncogenic TGF-beta signaling and metastasis. Breast Cancer Research. 2009;11(5):R68. doi: 10.1186/bcr2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhowmick NA, Ghiassi M, Bakin A, Aakre M, et al. Transforming Growth Factor-β1 Mediates Epithelial to Mesenchymal Transdifferentiation through a RhoA-dependent Mechanism. Molecular Biology of the Cell. 2001;12(1):27–36. doi: 10.1091/mbc.12.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vincent T, Neve EPA, Johnson JR, et al. SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-[beta] mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11(8):943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puccinelli JP, Su X, Beebe DJ. Automated High-Throughput Microchannel Assays for Cell Biology: Operational Optimization and Characterization. Journal of the Association for Laboratory Automation. 2010;15(1):25–32. doi: 10.1016/j.jala.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sackmann EK-H, Berthier E, Schwantes EA, et al. Characterizing asthma from a drop of blood using neutrophil chemotaxis. Proceedings of the National Academy of Sciences. 2014 doi: 10.1073/pnas.1324043111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


