Abstract
A small and compact DNA cube with zeptoliter volume is constructed by means of a generalized DNA brick concept using short synthetic oligonucleotides with varying lengths. By mimicking design principles from the DNA origami technique, the DNA cube offers higher stability and assembly yields compared to other approaches. Its potential application as nanoscale fluorescent probe is demonstrated using super-resolution imaging.
Keywords: DNA nanostructures, DNA bricks, DNA-PAINT, super-resolution microscopy
Our ability to create large supramolecular structures from DNA with dimensions of tens of nm and larger has advanced considerably over the past few years. For many applications it would be desirable, however, to create well-defined, sequence-addressable small assemblies from DNA rather than ever larger structures. Small structures comparable in size with typical proteins or nanoparticles could penetrate biological barriers more easily, diffuse and react faster, and could be potentially produced in larger quantities more easily. Based on the previously developed “DNA brick” strategy, we here demonstrate the scaffold-free construction of a small compact cube from DNA with a side-length of only 10 nm. The cube offers a large number of sequence-addressable functionalization sites concentrated in a zeptoliter volume and can be used, e.g., as probe for nanoscale imaging applications.
Following the conceptualization of structural DNA nanotechnology more than three decades ago,[1] various approaches towards the realization of DNA-based nanostructures were developed. After the initial creation of DNA wireframe polyhedra,[2] DNA “tiles” – molecular structures composed of several DNA double helices connected by multiple DNA crossovers – were used to create extended, two-dimensional lattices.[3] Using the same DNA crossover scheme as in tile-assembly, the scaffolded DNA origami technique later succeeded in the creation of discrete supramolecular objects from DNA with almost arbitrary shape.[4] In DNA origami, a long single-stranded DNA molecule (usually the genome of phage M13mp18 with a length of approximately 7 kilobases, or a derivative thereof) is used as a scaffold, which is connected at multiple positions by approximately 200 DNA “staple strands”, folding the scaffold into the desired shape.[4a] The size of the resulting objects is determined by the scaffold length – using an M13mp18-based scaffold a wide range of geometries can be created, amongst others a flat (single-layered) structure of dimensions (90 nm × 70 nm × 2 nm)[4a] or three-dimensional cuboids with side lengths (20 nm × 20 nm × 30 nm).[5] Using much longer scaffolds (such as the λ phage genome) correspondingly larger structures may be created.[6]
A more recent, scaffold-free approach towards the construction of supramolecular structures with sizes comparable to DNA origami objects is based – similar to traditional tile assembly – on single-stranded tiles (SST) or DNA bricks.[7] These are short synthetic DNA strands with distinct sequence, each of them containing four specific domains, which bind to exactly four local neighbors and thus constitute a discrete two- or three-dimensional structure. The single-stranded tile and brick concepts are part of a larger design space using short oligonucleotides to build complex DNA structures. Desired DNA nanostructure features can be engineered by varying motif parameters such as domain length, symmetry, and crossover patterns.[8]
Surprisingly, the construction of compact DNA objects, which are much smaller in size than DNA origami or brick structures, is quite challenging, partially because of the strong electrostatic repulsion between close-packed DNA helices. More loosely packed or even hollow DNA structures such as Seeman’s original DNA cube,[2] DNA-based platonic bodies [9] or other wireframe structures [10] do not suffer from this problem. In compact objects, however, the repulsion has to be counteracted by attractive interactions mediated by DNA crosslinks (i.e., base-pairing interactions). Unavoidably, the crossover density is reduced for smaller structures, because strands located at their faces have a reduced number of neighbors (and consequently can establish less crossovers) – a fact which has a stronger impact on structures with smaller volumes and larger surfaces. As a consequence, for instance, the DNA brick motif previously failed in the assembly of a 4 × 4 helix cube with 32 base pairs in height.[7b]
The creation of small DNA structures using the alternative origami technique requires the use of much shorter scaffold strands than the M13mp18 genome. To this end, fragments of plasmids or viral genomes have been used as scaffolds. For instance, a zeptoliter box with dimensions of (18 × 18 × 24) nm3 was based on a truncated pUC plasmid with a length of 1983 nucleotides (nt).[11] A single-layered structure made from four parallel helices and with dimension 63 × 11 nm2 was based on a 756 nt long scaffold sequence produced by PCR amplification from longer DNA.[12] To date the smallest compact DNA nanostructures – triangles, cubes and other shapes – were made with a 704 nt short fragment of the M13 scaffold termed M1.3.[13] The production of very short scaffolds with a length of only a few 100 nts is demanding, however. Biochemical restrictions make specific truncation of long strands difficult, while synthetic DNA strands are rarely produced at lengths exceeding 200 nt.
In order to address the technological challenges for the production of small and compact DNA structures, we here develop a hybrid approach that expands the general concept of DNA bricks by borrowing design motifs from the DNA origami approach, and use this hybrid approach to create a compact DNA cube with a side length of only 10 nm (i.e., a volume of (10 × 10 × 10) nm3 = 10−24 m3 = 1 yocto cubic meter). As in the DNA brick/canvas strategy,[7b] the cube exclusively consists of short linear DNA oligonucleotides. Similar to the origami technique, however, several oligonucleotides are elongated and routed through a larger portion of the entire target structure compared to typical staple strands. The present approach therefore generalizes and extends the design space of the DNA brick concept. As one potential application of the compact cube structure, we demonstrate its use as multiply functionalized probe for nanoscale imaging.
Design considerations
In order to construct a cube with target dimensions of (10 × 10 × 10) nm3 we used an assembly strategy based on a generalized DNA brick approach. As described above, conventional tile or brick assembly previously failed to fold such a small DNA structure. The DNA origami technique in its usual form is not applicable, as a continuous or even circular routing of a single strand through the entire structure cannot be easily realized (cf. discussion below). In our hybrid assembly strategy, we instead used two sets of oligonucleotides of different lengths – one set (the longer oligos) constitutes the “X” strands, whereas a set of shorter Y strands serves as a connector set, stapling initially decoupled X layers together. All strands are short enough to be produced synthetically with high yield, which also allows the incorporation of arbitrary modifications.
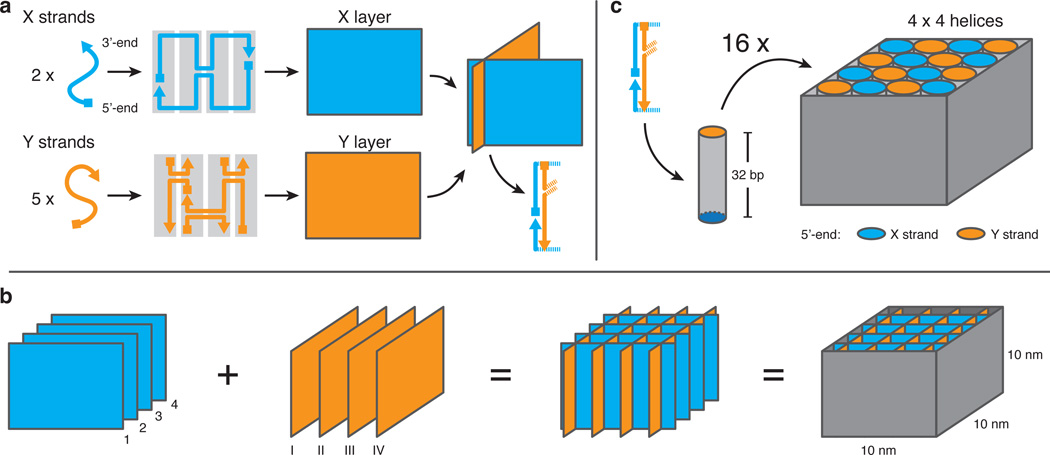
The basic assembly concept for the cube is shown in Figure 1a & b. The cube is constructed from four parallel DNA layers (numbered 1–4) defined by the X strands (blue), which are connected with each other by Y strands (orange) routed through layers I–IV perpendicular to the X layers (Figure 1b). X layers contain two X strands of 64 nt length each, while their routing through the layer allows for the establishment of either one or two classical four-way junctions (Supporting Figure S1).
Figure 1. Design principle of the cube.
a) X strands (blue) and Y strands (orange) span across single-stranded layers comprising four parallel domains (light grey) which represent one double helix, respectively (the depicted routing through the layers is an example for one layer, other layers might be routed differently depending on the availability of potential crossover connections, see Supporting Figure S1). The single-stranded X and Y layers are arranged perpendicular to each other, establishing a duplex domain at their line of contact consisting of hybridized X and Y strands. b) The orthogonal assembly of parallel X and perpendicularly positioned Y layers forms a cubic structure at which each line of contact establishes a double helix like in a). Each X layer is connected to an adjacent X layer by four classic four-way junctions, with each junction being established by one of the Y layers, respectively. The target dimensions of the designed cube are (10 × 10 × 10) nm3. c) The total cube consists of 4×4 double helices with a height of 32 base pairs. The structure is formed by hybridization of a total of 28 oligonucleotides.
Y layers contain five shorter Y strands with an average length below 50 nt (Figure 1a). Each Y layer includes three classical four-way junctions, all oriented perpendicular to the X layers and each junction placed between two adjacent X layers, e.g. between 1–2, 2–3, and 3–4, respectively. As there is a set of four Y layers in total (I–IV) and each Y layer connects two neighboring X layers with one four-way junction, each X layer is connected to its next neighbors via four crossovers in total (one from each set of Y strands). The Y strands protruding from the top and bottom are extended with four thymine nucleotides to prevent stacking of multiple cubes (i.e. aggregation) on the blunt ends (Supporting Figure S1).
As a result, the cube consists of 4 × 4 parallel double helices, each helix with a height of 32 base pairs (bp) (Figure 1c). The overall size of the cube is then defined by the double stranded regions, which sum up to an overall length of 16 × 32 bp = 512 bp. Assuming an interhelical distance of up to 0.3 nm[4b] and a length of 3.5 nm per helical turn, the cube reaches theoretical dimensions of approximately (9 × 9 × 11) nm3. As the DNA strands pass through adjacent helices in an antiparallel manner, the 5’ ends of the oligonucleotides at the top and bottom of the cube are alternating and are thus arranged in a typical chessboard pattern (Figure 1c).
At this point we would like to briefly discuss why a circular routing of a single strand through the entire cube (as in the DNA origami technique) is not practical: With a height of only 32 bp, the number of potential strand crossovers between neighboring helices is limited. Ordinary square lattice-based origami structures have previously implemented crossovers to each of the neighbors of a helix every 32 bp (corresponding to an average distance between crossovers per helix of 16 bp in a 2D layer). By establishing a crossover, every helix involved is fragmented into shorter sequence domains. A reduction of the distance for crossovers below 32 bp would result in domain lengths that are not thermodynamically stable any longer. We, therefore, maintained a typical crossover distance of 32 bp for this cube and allowed a maximum of two crossovers per double helix over its full height. This restriction still allows for a cyclic routing through one layer of the structure (see Figure 1a). However, the orientation of the strands at the ends of a helix is such that they cannot cross over to a helix of the neighboring layer (which would require an additional turn by ± 90°). This could be enforced by either insertion of flexible linkers or additional base pairs, which both would cause a distortion and thus a deviation from the cubic target structure.
In order to illustrate this point more clearly, we also designed a similar cube with a circular scaffold routing using caDNAno (see Supporting Figure S2). It turns out that a circular design cannot provide a clean cubic shape as it is demonstrated in Figure 1, and further automatically results in unstable duplex domains with lengths below 4 bp. For this reason, a full or even cyclic routing of a single scaffold strand was discarded for our design.
The sequences of the eight X strands were created by first generating ten different sets of sequences by means of a random sequence generator.[7b] Each set was then analyzed for intra- and inter-oligonucleotide secondary structures using NUPACK.[14] The set showing the best orthogonality with the lowest degree of interactions between the strands was then chosen as the X set.
Results
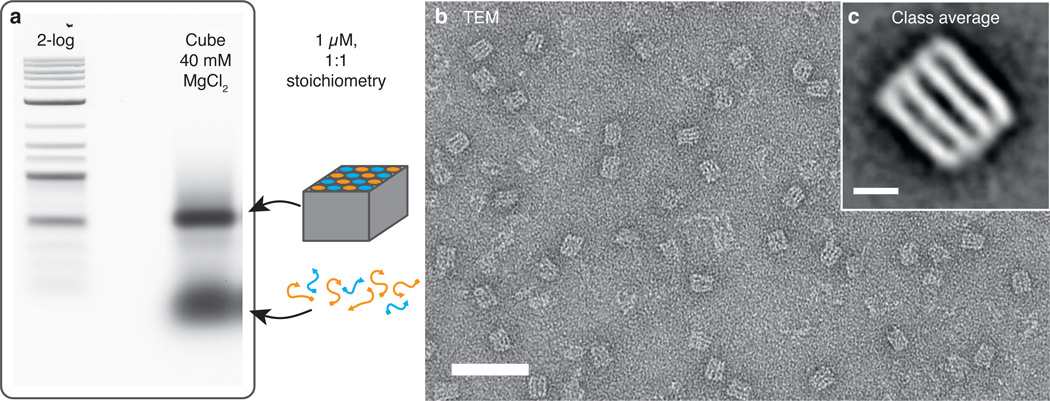
The successful folding of structures was verified using gel electrophoresis (Figure 2a). A clear band separated from unfolded strands identified the compact cubes, which could subsequently be extracted from the gel. Gel-purified DNA structures were further characterized using transmission electron microscopy (TEM). Although the structures were quite small, their compact and dense design conveyed excellent image contrast in TEM, which also simplified their detection and selection for class averaging (Figure 2b and Supporting Figure S4). The TEM images confirm the proper formation of the targeted cube structure. The class average image shown in Figure 2c impressively illustrates the design principle of the cube and its rigidity by directly visualizing the four parallel dsDNA layers schematically shown in Figure 1b. One of the major benefits of small and compact DNA structures such as the cube is the possibility to concentrate a well-defined number of chemical functionalizations in a tiny volume. To demonstrate a potential application for this capability, we utilized the cube as a multi-functionalized probe for fluorescence imaging.
Figure 2. Verification of folding.
a) Cubes are folded in 1:1 stoichiometry and with 40 mM MgCl2 (magnesium screening in Supporting Figure S3). Successful folding of cube structures is proven by means of gel electrophoresis, resulting in a clear band separated from unfolded or excess strands. b) TEM images of folded and gel purified cube structures. Length scale: 50 nm. c) A class average image of the cube highlights the four parallel layers building the basic shape. Because the cubes tend to lie on its sides, a differentiation between X and Y layers is not possible, consequently the class average is based on a mixture of X and Y layers standing perpendicular on the image plane. Length scale: 5 nm.
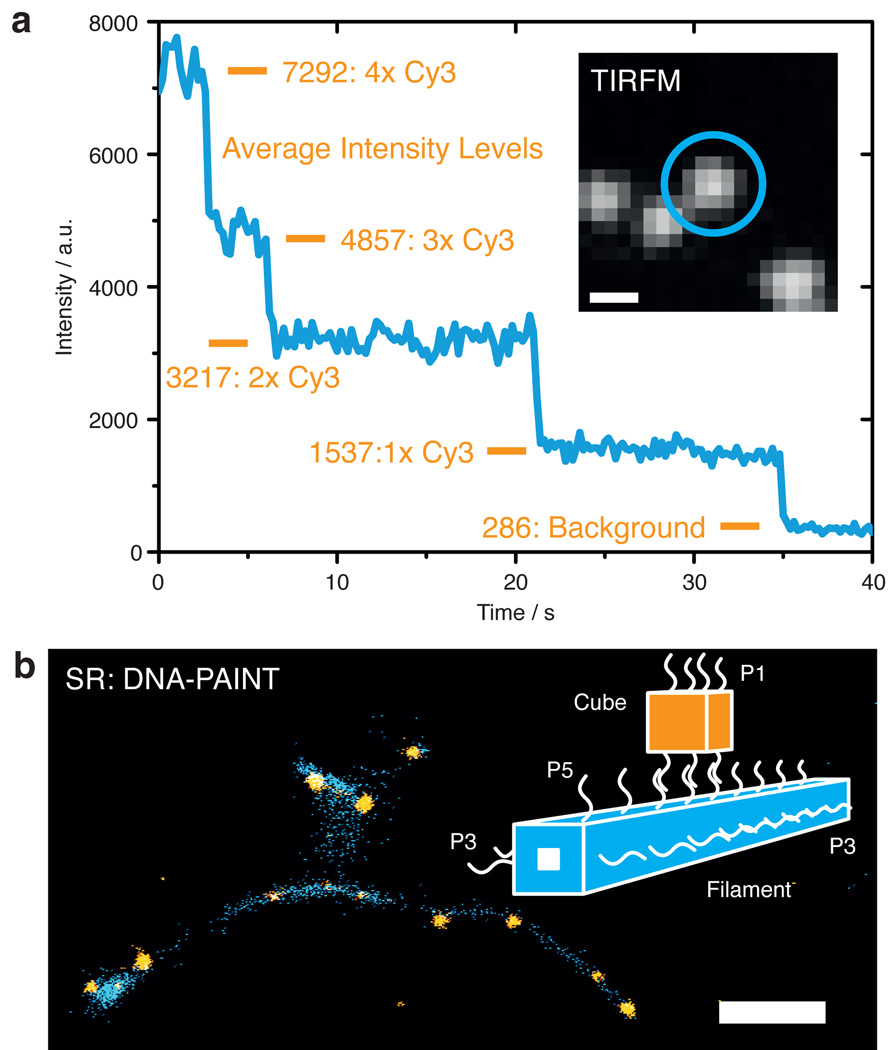
For direct single molecule fluorescence imaging, the DNA cubes were first modified at both helical faces to enable labeling and surface immobilization. Four Y strands (positions 2-III, 3-II, 3-IV, and 4-III in Figure 1b) were each extended at their 3’-ends (at the top of the cube) with a common 21 nt long sequence domain S2, and fluorescently labeled oligonucleotides with the complementary sequence S2* were hybridized to the cube after folding and purification. The cube was further biotinylated at positions 1-IV, 2-I, 3-IV, and 4-I (Figure 1b) at its bottom side to facilitate binding to a glass slide coated with BSA-biotin and streptavidin (cf. Experimental Section). In the inset of Figure 3a, a typical diffraction limited image of single cube structures is shown, which were obtained on a total internal reflection fluorescence microscope (TIRFM). The proper functionalization of the cube with exactly four Cy3 fluorophores is qualitatively demonstrated by the four bleaching steps visible in the fluorescence time trace shown in Figure 3 (see also Supporting Figure S5).
Figure 3. Fluorescence and super-resolution imaging.
a) Cubes immobilized on a surface and labeled with four Cy3 dyes show clear bleaching steps on a TIRFM setup. The inset shows a diffraction limited image of single cube structures. Length scale: 400 nm. b) Super-resolution image reconstruction of a DNA nanostructure-based complex (shown here as simplified illustration, highlighting the different binding domains P#). Cube structures (orange) hybridize to surface immobilized DNA origami filaments (blue) and are imaged using DNA-PAINT. Length scale: 350 nm.
We also attached DNA cube structures to extended supramolecular filaments made from DNA,[15] and imaged the resulting complexes using the super-resolution technique DNA-PAINT (Figure 3b).[16] The DNA filaments were created by polymerization of a hollow DNA origami building block described previously in Ref.[15] For DNA-PAINT imaging, the filaments were modified with 9nt long DNA-PAINT docking domains (P3) at their sides. Transient binding of ‘imager’ strands P3* (fluorescently labeled with Cy3b and sequence-complementary to P3) allowed for the localization-based reconstruction of a super-resolved image.
DNA filaments were used as supramolecular ‘targets’, on which DNA cubes – approximately 20 times smaller in volume than a filament monomer – could attach. In order to facilitate binding of the DNA cubes, the top side of the DNA origami filaments was extended with a double-row of 9 nt long extensions (domain P5). DNA cubes were in turn extended on their bottom side with eight 20 nt long ‘binding’ domains, to which up to eight ‘linker’ strands could hybridize. Linker strands carried an additional P5* domain at their end, which could further establish a connection to the DNA filament through hybridization with the P5 domains. DNA-PAINT imaging of the DNA cubes was achieved by extending top Y positions 2-III, 3-II, 3-IV, and 4-III (as above) with DNA-PAINT domains P1 to which ATTO 655 labeled P1* imager strands could bind. In Figure 3b, a super-resolved image is shown, in which single DNA cube structures (orange) are bound to DNA origami filaments (blue). The DNA-PAINT image shows the clear colocalization of both structures with negligible nonspecific binding of the cubes to the surface (see also Supporting Figure S6).
Discussion and Conclusion
With the 10 nm DNA cube presented in this work, we have generated one of the smallest compact DNA nanostructures to date. In contrast to small wireframes or larger compact DNA structures, folding of this cube requires relatively long annealing ramps, but their yield (at approximately 27 %, Supporting Figure S7) outperforms previous approaches. We have also found that the introduction of single-stranded extensions protruding perpendicularly from the helical axes – serving as docking positions for further functionalization – will reduce the folding yield. This indicates that in such cases the base-pairing interactions within the cube cannot compensate the additional electrostatic repulsion of the staple extensions and their excluded volume interactions any longer. We believe, however, that folding yields and assembly speeds of small compact structures could be further improved by optimized annealing protocols and buffers. Additional ligation of the staples or covalent crosslinking [17] of the structures could also increase their stability. More detailed future studies of the internal structure of such compact DNA structures performed by X-ray diffraction or cryo-EM [18] could be extremely valuable in this context.
The compact DNA cube offers a large number of sites for chemical functionalization concentrated in a zeptoliter volume. In contrast to DNA origami structures, which provide a similar functionalization density, the DNA cube can be constructed from a much smaller number of synthetic oligonucleotides. The assembly of this small and compact nanostructure – by generalizing the DNA brick concept – can be an advantage in many applications, for instance when fast diffusion or the penetration of biological barriers is required. The assembly approach introduced here is based on parallel oligonucleotide layers, which are interconnected in perpendicular direction, with two adjacent X layers forming the basic structural unit. In principle, the same approach could be used for the modular assembly of larger structures by simple repetition of this basic unit. As demonstrated in the present work, DNA cubes should be particularly useful as multifunctional labeling agents with potential applications in single molecule and super-resolution imaging.
Experimental Section
Assembly and folding
DNA strands were synthesized by Integrated DNA Technology. Unpurified X and Y strands were mixed in 1:1 stoichiometry (final concentration of 1µM per strand) in folding buffer (1× TAE buffer supplemented with 40 mM MgCl2). Modified strands (with DNA-PAINT domains, binding domains, etc.) were added with 2:1 excess, ‘linker’ strands with 4:1 excess. The strand mixture was annealed in a PCR thermo cycler by incubating the sample at 70 °C for 5 minutes followed by a cooling step to 65 °C and a further annealing ramp from 65 – 37 °C over 42 hours. After folding samples were held at room temperature and stored at −20 °C. Cy3 labeled S2* strands were added after folding and purification with 5:1 excess and incubated for 2 hours at room temperature. DNA origami filaments were prepared according to Jungmann et al.[15] (additional modifications are listed in the Supporting Tables S8).
Gel electrophoresis
Characterization of folding yield and purification was performed with gel electrophoresis. Samples were subjected to a 2 % agarose gel at 70 V for 2 hours (in 0.5 TBE, 10 mM MgCl2, gel prepared with 0.0001 % (v/v) SYBR® Safe) in an ice water bath. For purification target bands were excised and either placed into a Freeze ‘N Squeeze column (Bio-Rad Laboratories, Inc) and centrifuged at 7000 g for 5 minutes or the piece of gel was placed between to layers of PARAFILM, squeezed and extracted by pipetting the accumulated liquid drop at the boundary of the two PARAFILM layers.
Transmission electron microscopy
For imaging, 3.5 µL of agarose-gel-purified sample was adsorbed for 4 minutes onto glow-discharged, carbon-coated TEM grids. The grids were then stained for 1 minute using a 2% aqueous uranyl formate solution containing 25 mM NaOH. Imaging was performed using a JEOL JEM-1400 operated at 80 kV. Class averages were obtained by EMAN2, a boxer routine-based image processing suite for single particle reconstruction.[19]
Fluorescence and super-resolution microscopy
Fluorescence imaging was carried out on an inverted Nikon Eclipse Ti microscope (Nikon Instruments) applying an objective-type TIRF configuration with an oil-immersion objective (CFI Apo TIRF 100×, numerical aperture (NA) 1.49, oil). For imaging an additional 1.5× magnification was used to obtain a final magnification of ~150-fold, corresponding to a pixel size of 107 nm. Two lasers were used for excitation: 561 nm (200 mW nominal, Coherent Sapphire) and 647 nm (300 mW nominal, MBP Communications). Fluorescence light was imaged on an electron-multiplying charge-coupled device (EMCCD) camera (iXon X3 DU-897, Andor Technologies) (for a detailed description see Jungmann et al.[15]). Cy3 labeled cubes were imaged at high EM gain (≈ 300), very low laser power and long integration times (200 ms, 2000 frames) to prevent photo bleaching. Cube-filament complexes were imaged with DNA-PAINT at low EM gain (≈ 50), higher laser power and shorter integration times (100 ms, 10000 frames). Super-resolution DNA-PAINT images were reconstructed using spot-finding and 2D-Gaussian fitting algorithms programmed in LabVIEW.[16]
Fluorescence and super-resolution imaging
Samples were imaged using self-built flow chambers made of an objective slide and a coverslip (# 1.5), assembled by means of double-adhesive tape. First, 20 µL of biotin-labeled bovine-albumin-serum (1 mg/mL, dissolved in buffer A: 1× TAE, 100 mM NaCl, 0.05 % Tween-20) was flown into the chamber and incubated for 2 minutes, followed by a washing step (40 µL buffer A) and further incubation with streptavidin (0.5 mg/ml, dissolved in buffer A) for 2 minutes. For imaging of Cy3 labeled cubes the flow chamber was washed (40 µL buffer B: 1× TAE, 10 mM MgCl2, 0.05 % Tween-20), cubes were added to the chamber (20 µL at approx. 200 pM) and incubated for 2 minutes. After a further washing step the chamber was sealed with two-component adhesive and was ready for imaging. For the cube-filament complex the initial preparation with BSA-biotin and streptavidin was the same. After washing (40 µL of buffer B) filaments (with P5 and P3) were carefully flown into the chamber and incubated for 5 minutes. After further washing a mixture of cubes (including the domains P1 and P5*), ATTO 655 imager (P1*) and Cy3b imager (P3*) in buffer B were added to the flow chamber (20 nM, respectively) and sealed without further washing.
Supplementary Material
Acknowledgements
We gratefully acknowledge financial support by the Deutsche Forschungsgemeinschaft (SFB 1032 TPA2 and cluster of excellence Nanosystems Initiative Munich (NIM)) to F.C.S. and ONR Grants N000141410610 and N000141310593 and NIH Transformative Research Award (1R01EB018659) to P.Y. M.B.S. would like to thank the TUM Graduate School and the NIM Graduate Program.
Footnotes
Supporting Information is available from the Wiley Online Library.
Contributor Information
Dr. Max B. Scheible, Physics Department and ZNN/WSI, Technische Universität München, Am Coulombwall 4a, 85748 Garching, Germany Nanosystems Initiative Munich, Schellingstr. 4, 80799 München, Germany.
Luvena L. Ong, Wyss Institute for Biologically Inspired Engineering, Harvard University, 3 Blackfan Cir, Boston, MA 02115, USA
Johannes B. Woehrstein, Wyss Institute for Biologically Inspired Engineering, Harvard University, 3 Blackfan Cir, Boston, MA 02115, USA Max Planck Institute of Biochemistry and LMU, Am Klopferspitz 18, 82152 Martinsried, Germany.
Dr. Ralf Jungmann, Max Planck Institute of Biochemistry and LMU, Am Klopferspitz 18, 82152 Martinsried, Germany
Dr. Peng Yin, Wyss Institute for Biologically Inspired Engineering, Harvard University, 3 Blackfan Cir, Boston, MA 02115, USA
Dr. Friedrich C. Simmel, Email: simmel@tum.de, Physics Department and ZNN/WSI, Technische Universität München, Am Coulombwall 4a, 85748 Garching, Germany; Nanosystems Initiative Munich, Schellingstr. 4, 80799 München, Germany.
References
- 1.Seeman NC. J Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen JH, Seeman NC. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 3.a Fu T, Seeman NC. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]; b Seeman NC. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]; c Winfree E, Liu F, Wenzler LA, Seeman NC. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]; d Hansen MN, Zhang AM, Rangnekar A, Bompiani KM, Carter JD, Gothelf KV, LaBean TH. J Am. Chem. Soc. 2010;132:14481–14486. doi: 10.1021/ja104456p. [DOI] [PubMed] [Google Scholar]; e Rangnekar A, Gothelf KV, LaBean TH. Nanotechnology. 2011;22:235601. doi: 10.1088/0957-4484/22/23/235601. [DOI] [PubMed] [Google Scholar]; f Liu H, He Y, Ribbe AE, Mao C. Biomacromolecules. 2005;6:2943–2945. doi: 10.1021/bm050632j. [DOI] [PubMed] [Google Scholar]; g Rothemund PWK, Fygenson DK, Winfree E. J Am. Chem. Soc. 2004;126:16344–16352. doi: 10.1021/ja044319l. [DOI] [PubMed] [Google Scholar]
- 4.a Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]; b Douglas SM, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Ke Y, Douglas SM, Liu M, Sharma J, Liu Y, Shih WM, Yan H. J Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Dietz H, Douglas SM, Shih WM. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl E, Martin TG, Praetorius F, Dietz H. Angew. Chem. Int. Ed. 2014;53:12735–12740. doi: 10.1002/anie.201405991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchi AN, Saaem I, Vogen BN, Brown S, LaBean TH. Nano Lett. 2014;14:5740–5747. doi: 10.1021/nl502626s. [DOI] [PubMed] [Google Scholar]
- 7.a Wei B, Dai M, Yin P. Nature. 2012;485:623–626. doi: 10.1038/nature11075. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ke Y, Ong LL, Shih WM, Yin P. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Yin P, Hariadi R, Sahu S, Choi H, Park S, Labean T, Reif J. Science. 2008;321:824–826. doi: 10.1126/science.1157312. [DOI] [PubMed] [Google Scholar]
- 8.Wei B, Dai M, Myhrvold C, Ke Y, Jungmann R, Yin P. J Am. Chem. Soc. 2013;135:18080–18088. doi: 10.1021/ja4062294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a Goodman RP, Schaap IAT, Tardin CF, Erben CM, Berry RM, Schmidt CF, Turberfield AJ. Science. 2005;310:1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]; b He Y, Ye T, Su M, Zhang C, Ribbe AE, Jiang W, Mao C. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 10.Simmel SS, Nickels PC, Liedl T. Acc. Chem. Res. 2014;47:1691–1699. doi: 10.1021/ar400319n. [DOI] [PubMed] [Google Scholar]
- 11.Zadegan RM, Jepsen MDE, Thomsen KE, Okholm AH, Schaffert DH, Andersen ES, Birkedal V, Kjems J. ACS nano. 2012;6:10050–10053. doi: 10.1021/nn303767b. [DOI] [PubMed] [Google Scholar]
- 12.Pound E, Ashton JR, Becerril HA, Woolley AT. Nano Lett. 2009;9:4302–4305. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]
- 13.Said H, Schüller VJ, Eber FJ, Wege C, Liedl T, Richert C. Nanoscale. 2013;5:284–290. doi: 10.1039/c2nr32393a. [DOI] [PubMed] [Google Scholar]
- 14.Dirks RM, Pierce NA. J Comput Chem. 2004;25:1295–1304. doi: 10.1002/jcc.20057. [DOI] [PubMed] [Google Scholar]
- 15.Jungmann R, Avendaño MS, Woehrstein JB, Dai M, Shih WM, Yin P. Nat Meth. 2014 doi: 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jungmann R, Steinhauer C, Scheible MB, Kuzyk A, Tinnefeld P, Simmel FC. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- 17.Tomás-Gamasa M, Serdjukow S, Su M, Müller M, Carell T. Angew. Chem. Int. Ed. 2014;54:796–800. doi: 10.1002/anie.201407854. [DOI] [PubMed] [Google Scholar]
- 18.Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, Pedersen JS, Birkedal V, Besenbacher F, Gothelf KV, Kjems J. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 19.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. J Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.