Abstract
Understanding the source of genetic variation in aging and using this variation to define the molecular mechanisms of healthy aging require deep and broad quantification of a host of physiological, morphological, and behavioral endpoints. The murine model is a powerful system in which to understand the relations across age-related phenotypes and to identify research models with variation in life span and health span. The Jackson Laboratory Nathan Shock Center of Excellence in the Basic Biology of Aging has performed broad characterization of aging in genetically diverse laboratory mice and has placed these data, along with data from several other major aging initiatives, into the interactive Mouse Phenome Database. The data may be accessed and analyzed by researchers interested in finding mouse models for specific aging processes, age-related health and disease states, and for genetic analysis of aging variation and trait covariation. We expect that by placing these data in the hands of the aging community that there will be (a) accelerated genetic analyses of aging processes, (b) discovery of genetic loci regulating life span, (c) identification of compelling correlations between life span and susceptibility for age-related disorders, and (d) discovery of concordant genomic loci influencing life span and aging phenotypes between mouse and humans.
Key Words: Database, Functional genomics, Mouse, Phenotype
The importance of animal models, and in particular the laboratory mouse, has been firmly established for basic and translational research. The mouse is especially powerful because it is a mammalian system with a relatively short life span and a deep repertoire of experimental resources for genetic and phenotypic characterization. Thousands of inbred and genetically modified strains are currently available and more are being created and phenotyped (1,2); new precision genetic reference and mapping populations have recently been developed (3–5); there is a deep reference genome and a growing number of fully sequenced mouse strains (6–9); 99% of its genes are shared with humans (10); commercial genotyping arrays are available (11); long-term housing is relatively economical; experimental conditions can be precisely controlled; and defined interventions can be performed that cannot be practically performed on humans.
The mouse has been the subject of studies on the basic biological mechanisms of aging, longevity, and health span for decades [for review, see (12)]. Early studies on renal aging by Shock and colleagues (13,14), hematological and physiological parameters (15–18), cognition and behavior (19–21), reproductive maturity (22,23), dietary restriction (24), and others have provided insights into the mechanisms and characteristics of mammalian aging. Several of these studies provided evidence of heritable variation in life span and aspects of health span in aging. Recent advances in genetic analysis (including the development of software and tools)—for the integration of broad-based phenotyping across panels of isogenic mice, for estimating genetic variation and covariation across large numbers of traits, for precisely manipulating the genomes of inbred mice of diverse backgrounds, and for precisely mapping the source of genetic variation in age-related phenotypes—motivated the broad characterization of inbred mouse strains across many measures of life span and health span in aging.
The Jackson Laboratory Nathan Shock Center of Excellence in the Basic Biology of Aging
The Jackson Laboratory Nathan Shock Center (JAX NSC) has extensively characterized more than 30 commonly used inbred strains of mice for life span and health span-related phenotypes (25), greatly increasing the genetic diversity of the mouse models available for aging research. This is the most comprehensive well-controlled life span study in mammals to date. The study includes both longitudinal and cross-sectional assessments. Longitudinal data include life span and a host of noninvasive multisystem physiological measures. The cross-sectional study includes invasive assessments, pathology, and the collection of tissues to evaluate apoptosis and chromosome stability. These data have been placed in a rich and interactive software environment, the Mouse Phenome Database [MPD; phenome.jax.org (26)], which serves as the authoritative source for JAX NSC data. All primary data and protocols are freely available for download or interactive analysis on the MPD website. Table 1 shows current JAX NSC strain surveys in MPD. Most studies have at least one associated publication, as shown.
Table 1.
The Jackson Laboratory Nathan Shock Center Projects in Mouse Phenome Database (MPD)
| Study | No. of Strains | Ages Tested | No. of Measurements | MPD Project | Ref |
|---|---|---|---|---|---|
| Longitudinal | |||||
| Life span | 31 | — | 1 | Yuan2 | (25) |
| IGF-1 and body weight | 33 | 6, 12, 18 | 6 | Yuan1 | (27,28) |
| Blood chemistry | 32 | 6, 12, 18 | 51 | Yuan3 | (29) |
| Hematology | 30 | 6, 12, 18, 24 | 96 | Peters4 | |
| Peripheral blood leukocytes | 32 | 6, 12, 18, 24 | 68 | Petkova1 | (30) |
| Cardiac conduction activity | 29 | 6, 12, 20 | 18 | Xing1 | (31) |
| Urine albumin and creatinine | 30 | 12, 18, 24 | 9 | Korstanje1 | (32) |
| Glomerular mesangial matrix expansion | 29 | 6, 12, 20 | 3 | Korstanje2 | (33) |
| Lymphocytic infiltration in kidneys | 23 | 20 | 2 | Hillebrands1 | (34) |
| Activity and food and water intake | 14 | 6 | 17 | Evsikova1 | |
| Grip strength and gait analysis | 32 | 6, 12, 18, 24 | 36 | Seburn2 | (35) |
| Gait analysis compliance | 31 | 6, 12, 18 | 3 | Xing2 | |
| Vaginal opening and sexual maturation | 33 | — | 1 | Yuan4 | (28,36) |
| Cross-sectional | |||||
| Bone mineral density and body composition | 32 | 6, 12, 20 | 33 | Ackert1 | |
| Chromosome instability and apoptosis | 30 | 6, 12, 20 | 18 | Mills1 | |
| Pulmonary adenomas | 28 | 20 | 2 | Berndt4 | (37) |
| Histopathology | 28 | — | — | Sundberg1 | (38) |
Mouse Phenome Database
The Mouse Phenome Project was launched in 2001 to complement mouse genome sequencing efforts by promoting new phenotyping initiatives under standardized conditions and collecting the data in a central public database. MPD houses a wealth of strain characteristics data to facilitate the use of the laboratory mouse. Data are acquired from investigators throughout the research community and integrated into a standardized platform supporting correlations, criteria-fit, and other analyses. MPD collects, annotates, integrates and maintains primary quantitative phenotype data representing a broad scope of endpoints and disease-related characteristics in naïve mice, aging mice, and those exposed to drugs, environmental agents, or other treatments. MPD provides a catalog of phenotypic assays and a common framework for data access and dissemination along with a suite of analysis tools to explore genetic variation and facilitate mouse research, including features to:
compare strains
compare results of diverse phenotypic assays
identify strains with significant sex differences for a phenotype of interest
identify outlier strains for a phenotype of interest
determine influence of environment on a phenotype of interest
find mouse models relevant to a research application
select optimal genetic backgrounds for new or existing mutations
identify phenotypes that are significantly correlated
identify gene expression probesets that correlate with a phenotype of interest
identify phenotypes that correlate with gene expression (via a gene of interest)
formulate and test hypotheses
discover phenotype–genotype relationships
validate and refine results from traditional mapping studies
benchmark experimental data using detailed protocols
Although originally developed to address a community need for a central repository of well-executed and documented inbred strain characterization data (39), MPD currently houses phenotype, gene expression, and/or genotype data for >1,330 strains of mice, including inbred, recombinant inbred (RI), F1 hybrid, chromosome substitution strains, and new genetic resource populations such as Collaborative Cross (CC) lines (3,4) and Diversity Outbred (DO) populations (5). Over 270 investigators from 14 countries have contributed data and are supported by 130+ funding agencies and research foundations worldwide, including all institutes of the NIH. There are now thousands of phenotypic measurements with associated protocols, including the life span and health span measures collected by JAX NSC.
Methods
Mice
All mice were obtained from The Jackson Laboratory. A list of strains tested is shown in Figure 1. Four strains are wild-derived and represent the major subspecies of laboratory mice: CAST/EiJ (Mus musculus castaneus), MOLF/EiJ (M. molossinus), PWD/PhJ (M. musculus), and WSB/EiJ (M. domesticus). The remaining classical strains were chosen because they were recommended by the Mouse Phenome Project based on genetic diversity and common use (http://phenome.jax.org/db/q?rtn=strains/searchstep2&reqpanel=mpd_priority&nosearch=1). Note that BTBR T+ tf/J (Jax 002282) is used as an abbreviation for BTBR T+ Itpr3 tf/J (official nomenclature). Mice received ad libitum access to autoclaved pelleted diet with 6% fat (Lab diet 5K52, PMI Nutritional International, Bentwood, MO) and ad libitum access to acidified water (pH 2.8–3.1). Mice of the same sex were housed 4–5 per pen (fighting mice were separated into single cages) in pressurized individually ventilated (PIV) polycarbonate cages measuring 31cm × 31cm × 214cm divided into two pens supplied with high efficiency particulate air (HEPA) filtered air (Thoren Caging Systems Inc., Hazleton, PA). Autoclaved white pine shavings (Crobb Box Co., Ellsworth, ME) were used as bedding. Animal room environmental parameters were as follows: 12:12 hour light-dark cycle, ~50% relative humidity, 21–23°C. Mice were kept in a barrier facility where room entry procedures required personnel to don caps, face masks, disposable gowns, shoe covers, and gloves. Mouse colonies in this facility were monitored four times a year for 15 viruses, 17 bacterial species, two Mycoplasma species, external and internal parasites, and Encephalitozoon cuniculi.
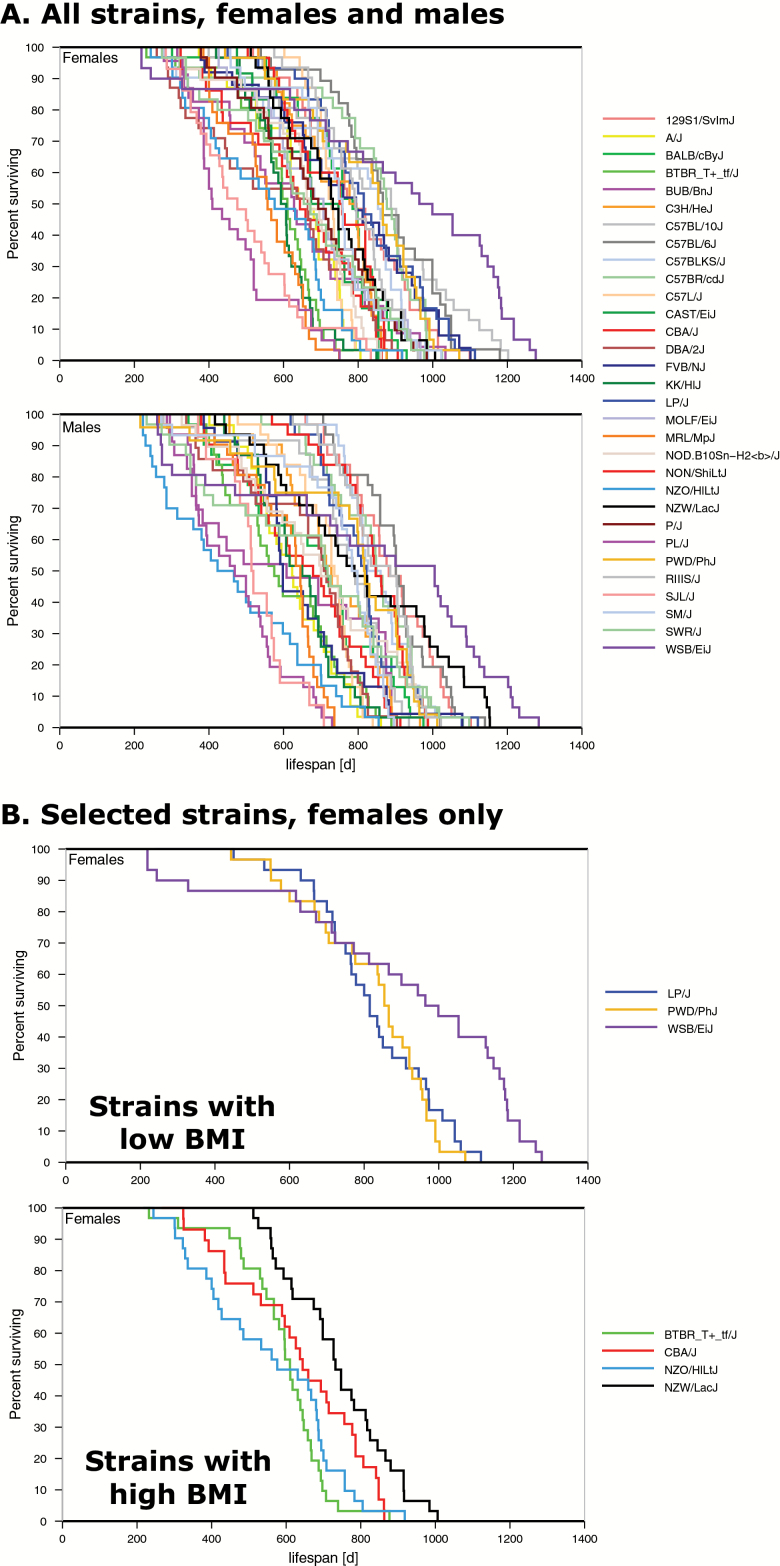
Figure 1.
Survival curves for 31 inbred strains of mice. (A) All strains tested, both sexes. Percent survival is shown on the y-axis and life span on the x-axis in days. (B) Selected strains, females only, are shown as an example to illustrate the selection of strains of interest for plotting survival curves in Mouse Phenome Database. In this example, we chose strains with low body mass index (BMI) versus high BMI at 12 mo of age (see Survival Curves section), plotted them separately and downloaded each. We show them here in the same figure for comparison purposes. Strains are color-coded; some colors were altered for clarity. Survival data are from Yuan2; BMI data were accessed through the Ackert1 study (data not shown). To generate these figures: (A) From the homepage, select “Phenotype,” select “Longevity.” From the Yuan2 study select “Survival curve comparison,” and use the pulldown menu to select strain set = “all strains,” click on “Go.” (B) From landing page for (A), hit the back button, select strains where females at 12 mo of age are low for BMI (view this measurement in a new window through a search on “BMI”), generate the Kaplan–Meier plot by clicking on “Go”; repeat this process by selecting strains that are high for BMI. “eps” was selected as the “Result type”; images were downloaded for publication-quality figures.
Physiological Testing
Mice were evaluated by multiple clinical assays at three or four ages, usually in 6-month increments. Body composition, bone mineral density (Ackert1; Table 1), and heart function (Xing1) were determined. Neuromuscular function was assessed by forelimb grip strength and automated gait analysis (Seburn2, Xing2); kidney function by blood urea nitrogen (Yuan3), urine albumin and creatinine levels (Korstanje1), glomerular mesangial matrix expansion (Korstanje2), and tertiary lymphoid organ formation (Hillebrands1); liver function by alanine aminotransferase and bilirubin levels (Yuan3); and immunological function by fluorescence-activated cell sorting (Petkova1). Each evaluation included a complete hematological screen (Peters4) and routine blood clinical chemistries (Yuan3). Additionally, levels of hormones thought to be involved in the basic mechanisms of aging, such as insulin-like growth factor 1 (IGF-1) (Yuan1) and thyroxin (Yuan3) were measured. Micronuclei and apoptosis tests were carried out by spectral karyotyping to determine aging-associated DNA damage accumulation (Mills1). A reproductive study evaluated the age of sexual maturity in females (Yuan4). Necropsies were performed, and tissues and organs (including tumors) were preserved for histopathology (Sundberg1, Berndt4). Details of JAX NSC procedures for each project can be found on the MPD website (http://phenome.jax.org/db/q?rtn=projects/titlesbycat&req=LShock) and through associated publications listed in Table 1.
Life Span Monitoring
Mice were inspected at least once daily for the life span study (Yuan2). Moribund mice were sacrificed if severely ill and judged that they would not survive another 48 hours. A mouse was considered severely moribund if it exhibited more than one of the following signs: (a) inability to eat or drink, (b) abnormally low body temperature, (c) severe lethargy (reluctance to move when gently prodded with forceps), (d) severe balance or gait disturbance, (e) rapid weight loss for a week or more, or (f) an ulcerated or bleeding tumor. The age at which a moribund mouse was sacrificed was taken as the best available estimate of its natural life span. Mice dying abnormally, such as those experiencing severe bites, were censored for analysis.
Results and Discussion
MPD is a versatile data resource on the genetics of health metrics in aging mice. Over 350 JAX NSC measurements (Table 1) have been submitted to MPD, annotated with public ontologies and integrated into the larger MPD framework. JAX NSC measurements (primary and summary data) are publicly available for online analysis or downloading for custom analyses. Analysis tools used for this article are available through MPD, in addition to many other tools and features not shown. Examples demonstrate how one might use JAX NSC data to assess different research questions. Figures were generated from the MPD website as screenshots or downloadable, publication-ready images (except where noted). Users can save MPD graphics as png, eps, pdf, or svg.
Survival Curves
To compare longevity among strains, survival curves (Kaplan–Meier) can be generated from life span data as shown in Figure 1A. As illustrated, median life span and qualitative aspects of the survival curves vary dramatically among inbred strains. WSB/EiJ is the longest-lived strain for both males and females while BUB/BnJ is one of the most short-lived for both sexes. MPD provides a convenient option to narrow results by choosing one or more strains of interest as shown in Figure 1B. Users may select MPD Priority Strains, CC founder strains, or other strain set; or users can individually select strains from a list of check boxes. In our example (Figure 1B), we have generated survival curves for females from several inbred strains with low body mass index (BMI) and strains with high BMI at 12 months of age (data accessed from Ackert1).
Analyzing Single Measurements
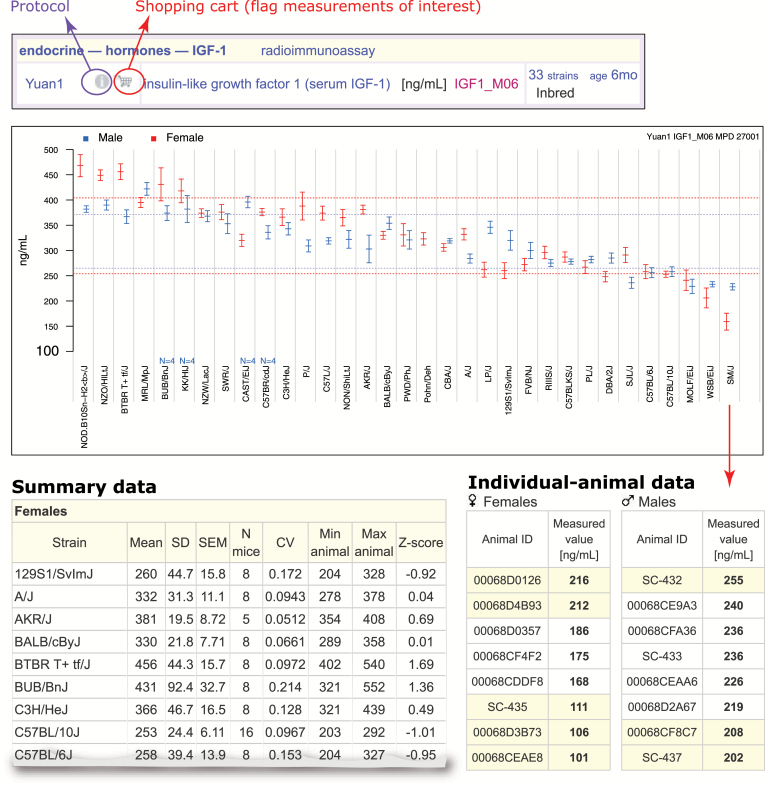
For each phenotypic measurement, a strain plot is provided along with summary statistics and individual animal data, as shown in Figure 2 for IGF-1 for 33 strains at 6 months of age (Yuan1). A wide range of phenotypic values is reflective of the genetic diversity of this panel of inbred strains. Through visualization of this data, it is easy to see that many strains exhibit no sex differences (e.g., NZW/LacJ, PWD/PhJ) while others show significant sex differences (e.g., LP/J, SM/J). A table of summary data can be found below the plot on the MPD website (a small section is shown in Figure 2, lower left panel). Users can also access individual animal data for any strain by clicking on relevant regions of the plot. Quantitative data may be downloaded or copy/pasted into other desktop applications.
Figure 2.
Measurement plot and underlying data for IGF-1. The upper panel (Mouse Phenome Database screenshot) identifies the measurement and its attributes (project (Yuan1), methodology used (radioimmunoassay), variable name (in magenta), number of strains tested (33), strain type (inbred) and age (6 mo). The “i” (info) icon (circled in purple) to the right of “Yuan1” takes users to a detailed protocol. The “shopping cart” icon (circled in red) allows users to select (flag) this measurement for use in more advanced analyses (described in Figs. 4, 5). The middle panel shows the plot with strains along the x-axis and IGF-1 levels on the y-axis, males in blue, females in red. The dotted lines indicate ±1 SD of the overall means across all strains for males and females, separately. Each strain mean is shown with 1 SEM. Low sample sizes are noted on the plot above strain name. The lower left panel shows summary data (for females only) in tabular format (mean, SD, SEM, number of mice tested (N), coefficient of variation (CV), min–max range, and Z-score). The lower right panel shows the individual animal data when clicking on the SM/J region of the plot (arrow). To generate this figure: From the homepage, search on “IGF,” from results page click on “List these traits/measurements,” click on “insulin-like growth factor 1 (serum IGF-1),” age 6 mo for the Yuan1 project. The plot shown in this figure will be near the top of the page; scroll down for summary data (click on “Magnitude order/details” to get the detailed view shown in this figure); click on the original plot directly or click on the strain mean links in the original summary table for your strain of interest to generate individual animal data (shown). “eps” was selected as the “Result type” after selecting “Detail options” just above the plot; image was downloaded for publication-quality figure.
Analyzing Sets of Measurements
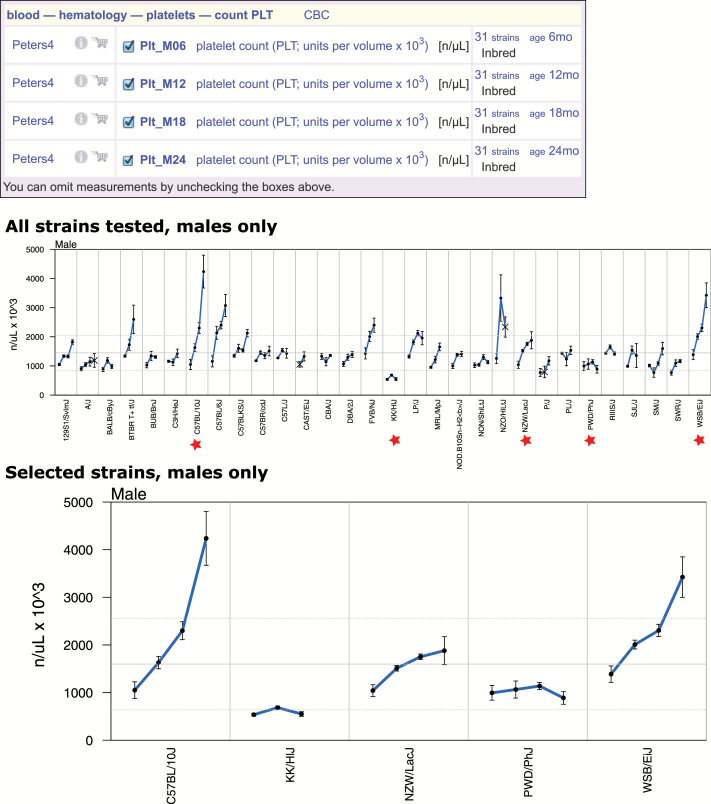
Measurements collected at multiple ages can be viewed in a single plot as shown in Figure 3. In this example, we show platelet counts over the following ages: 6, 12, 18, 24 months, for males of all strains tested (Peters4). At a glance, users can readily see that there are a wide range of age-related phenotypic profiles across these strains, for example, compare KK/HlJ to C57BL/10J. Users may highlight strains, omit strains, or show only selected strains. In this example, we have selected a subset of strains for more detailed viewing (lower panel).
Figure 3.
Strain comparison for platelet count across multiple ages. The top panel identifies the measurements that are plotted (see Fig. 2 legend for more information about measurements and their attributes). The middle panel shows the profiles for males of all strains tested for ages 6, 12, 18, and 24 mo. Error bars are standard error of the mean. Various detail options are available, including adjusting the color of the data points, changing the size of the data points, choosing the color and thickness of error bars, highlighting strains, omitting strains, or showing selected strains only. The lower panel shows a more detailed plot of five strains we selected (red stars). Data are from the Peters4 study. To generate these figures: From the homepage, “Phenotype,” then select “Blood—hematology,” then “platelets,” scroll down to Peters4, click on “age comparison” to arrive at the middle panel, scroll down further for plotting options and the ability to select specific strains. “eps” was selected as the “Result type”; image was downloaded for publication-quality figure.
Correlations Across Multiple Measurements
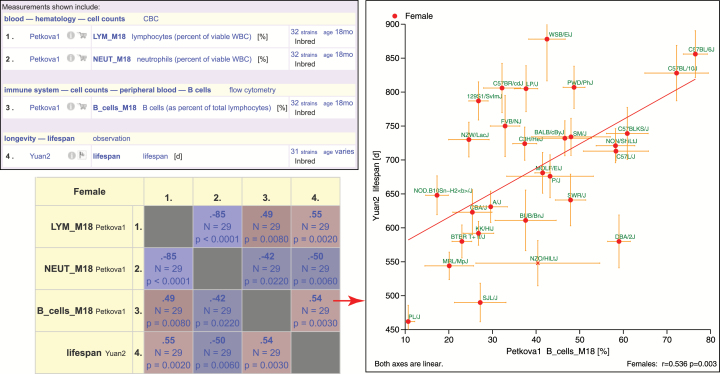
Multiple measurements can be correlated where a color-coded correlations matrix is generated as shown in Figure 4. In this example, we chose three peripheral blood leukocyte (PBL) measurements at 18 months of age from Petkova1 to correlate against life span from Yuan2. Because these measurements are from different projects, users must select (flag) measurements across projects. Users can collect MPD measurements from various labs and subject areas into a shopping cart where they can be analyzed together. Users’ flag measurements of interest by simply clicking on the shopping cart icon (Figure 2). Statistics are provided in each cell of the correlations matrix. These results can also be viewed as a scatterplot matrix (thumbnail images as cells) or in tabular format which can be copy/pasted into other desktop applications. Detailed scatterplots can be accessed by clicking on individual cells of the matrix, as shown. In this example, we found that the percentage of B cells at 18 months of age correlates to life span in females (r = .536, p = .003). In addition to correlation matrices and scatterplots, users can query the database to find correlations to selected measurements across the entire MPD (not shown). For example, life span data can be used to find compelling correlations across JAX NSC measurements as well as across thousands of other measurements in MPD. This capability may help identify biomarkers of aging. In addition to phenotype versus phenotype correlations, users can correlate phenotype data and gene expression data by starting with a phenotype of interest or a gene of interest (not shown).
Figure 4.
Correlations matrix and scatterplot of selected peripheral blood leukocytes and life span. The top right panel identifies measurements of interest (see Fig. 2 legend for more information about measurements and their attributes). This matrix was generated from various flagged measurements (shopping cart, see Fig. 2, circled in red). The lower left panel shows the correlation matrix for females with Pearson correlation coefficient, sample size (N), and p value for each pair-wise result (individual cells of the matrix). Red color values are for positive correlations, blue for negative. The more intense colors indicate higher coefficients. The right panel shows a detailed scatterplot resulting from clicking on the associated cell of the matrix (arrow). Life span is on the y-axis, B cell percentage (18 mo) is on the x-axis. PBL data are from Petkova1 and life span data are from Yuan2. To generate this figure: The shopping cart feature must be used to flag measurements of interest across multiple projects. In this example, measurements are selected from Yuan2 and Petkova1. Search on “Yuan2,” click on the project, then click on the shopping cart (example circled in Fig. 2) for life span data (a pop-up window indicates that a measurement has been added to the collection). Then go to the Petkova1 project by searching on “Petkova1.” Click on “Apply tools” to the left of the listing of measurements. Select (check boxes) Petkova1 measurements shown in the screenshot at the top of this figure. Then scroll all the way down to see measurements in the collection (shopping cart). Select “life span” from the Yuan2 project. Then click on “Next” near the top of the page to go to the Mouse Phenome Database toolbox. Click on “Pheno correlations matrix” near the middle of the page. To generate the scatterplot, click on the indicated cell then scroll down and select “females.” “eps” was selected as the “Result type”; image was downloaded for publication-quality figure.
Find Mouse Models: Criteria-Fit tool
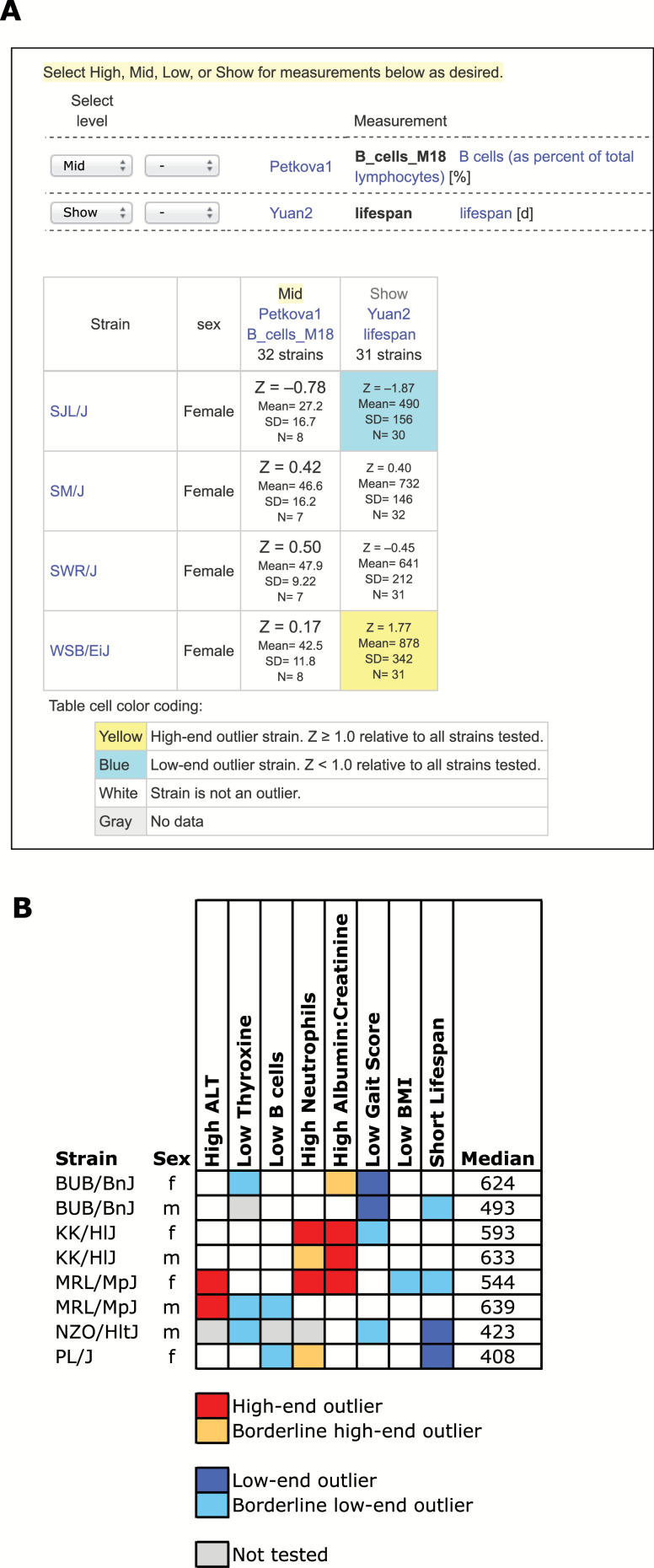
A criteria-fit tool is available to identify mouse strains that best fit a user’s criteria, based on Z-scores of available data (Figure 5A). In this example, we wanted to find mouse strains (females) with long or short life spans but with average percentages of B cells. Using the MPD criteria-fit tool, we have easily identified relevant mouse models: SJL/J with average B cell percentage and short life span, and WSB/EiJ with average B cells and long life span. This tool helps choose optimal strains across multiple phenotypes for designing well informed experiments as further illustrated in Figure 5B where we identified a small number of strains that would be useful for testing compounds that might increase longevity and improve health span. For example, MRL/MpJ females show declining liver function with age (high alanine aminotransferase), high neutrophils (inflammation), declining kidney function (albumin:creatine ratio), low BMI, and a relatively short life span (554 days). For reference, median life span for WSB/EiJ females is 964 days (Yuan2).
Figure 5.
Find mouse models that meet your criteria. (A) For this example, we wanted to identify strains that have an average percentage of B cells at 18 mo, but that have relatively long or short life spans. This result was generated from measurements previously flagged (shopping cart, see Figure 2). The chosen measurements for this example are shown near the top. A user must select the desired “level”’ for each measurement based on Z-score: high, mid, low, or simply show. In this case, we want to see a “mid” range for B cells and “show” life span so that we can identify strains with long versus short life spans. Blue highlighting indicates low-end outlier strains, yellow indicates high-end outliers. Results are truncated and shown as a screenshot. B cell data are from Petkova1, life span data are from Yuan2. (B) For this more complicated example, we wanted to identify mouse strains with multiple phenotypes relevant to health span so that we can make an informed decision for choosing optimal strains to test compounds that might extend life span. We used the same tool illustrated in Fig 5A, but present the results of the analysis more compactly in Excel for ease of viewing. All mice were 18–20 mo of age. Phenotypes accessed and criteria imposed: high alanine aminotransferase (ALT) (Yuan3), low thyroxine (T4) (Yuan3), low B cell percentage (Petkova1), high neutrophil percentage (Petkova1), high albumin:creatinine ratio (Korstanje1), low gait score (Xing2), low BMI (Ackert1), and short life span (Yuan2). High and low-end outliers are greater than ±2.0 SD from the mean while borderline outliers are greater than ±1.25 SD. To generate these figures: (A) Select measurements across projects as detailed in Fig. 4 legend. Select measurements shown in this figure and click on “Next.” In the toolbox, select “Find strains that best fit your criteria.” From the pulldown menus select indicated levels for each measurement. For output, adjust the pulldown menus to: “all, alphabetical,” “females,” “outliers colored.” Click on “Go.” Scroll all the way down next to the color key at the bottom of the page to see the strains in this example. (B) Add indicated measurements across projects (listed above) to the shopping cart as detailed in Fig. 4 legend. Follow directions above for (A). Summarize results of selected strains in Excel by color-coding.
Conclusions
JAX NSC projects available through MPD (Table 1) are an important resource for the aging community, allowing selection of the best laboratory strains for future hypothesis-driven research as well as the generation of new hypotheses based on MPD’s extensive datasets across multiple phenotypes. Characterization of age-related phenotypes by JAX NSC is ongoing, including life span in other reference populations. These data will be added to MPD in the future. MPD accepts data submissions including protocols and individual animal experimental data from studies of inbred strains, hybrids, mutants, and mapping populations. Information about how to contribute data is located on the MPD website (http://phenome.jax.org/db/q?rtn=docs/dataguide). By placing primary data in a single repository, users may conveniently access and compare results across studies. Large data submissions from major aging studies including the NIA Intervention Testing Program (40) and a collaborative project on the QTL analysis of age-related phenotypes centered at Penn State (PI: G. McClearn) along with CC and DO data from ongoing JAX NSC studies will enable investigators to place their data in the context of known variation and to test hypotheses about relationships among phenotypic variants across life span (41).
Funding
This work was supported by the National Institutes of Health to The Jackson Laboratory Nathan Shock Center of Excellence in the Basic Biology of Aging (AG038070) and to the Mouse Phenome Database (DA028420).
References
- 1. Brown SD, Moore MW. The International Mouse Phenotyping Consortium: past and future perspectives on mouse phenotyping. Mamm Genome. 2012;23:632–640. doi:10.1007/s00335-012-9427-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ringwald M, Iyer V, Mason JC, et al. The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res. 2011;39(Database issue):D849–D855. doi:10.1093/nar/gkq879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Churchill GA, Airey DC, Allayee H, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi:10.1038/ng1104-1133 [DOI] [PubMed] [Google Scholar]
- 4. Chesler EJ, Miller DR, Branstetter LR, et al. The Collaborative Cross at Oak Ridge National Laboratory: developing a powerful resource for systems genetics. Mamm Genome. 2008;19:382–389. doi:10.1007/s00335-008-9135-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Churchill GA, Gatti DM, Munger SC, Svenson KL. The Diversity Outbred mouse population. Mamm Genome. 2012;23:713–718. doi:10.1007/s00335-012-9414-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yalcin B, Adams DJ, Flint J, Keane TM. Next-generation sequencing of experimental mouse strains. Mamm Genome. 2012;23:490–498. doi:10.1007/s00335-012-9402-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Keane TM, Goodstadt L, Danecek P, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477:289–294. doi:10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wong K, Bumpstead S, Van Der Weyden L, et al. Sequencing and characterization of the FVB/NJ mouse genome. Genome Biol. 2012;13:R72. doi:10.1186/gb-2012-13-8-r72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ananda G, Takemon Y, Hinerfeld D, Korstanje R. Whole-genome sequence of the C57L/J mouse inbred strain. G3 (Bethesda). 2014;4:1689–1692. doi:10.1534/g3.114.012997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boguski MS. Comparative genomics: the mouse that roared. Nature. 2002;420:515–516. doi:10.1038/420515a [DOI] [PubMed] [Google Scholar]
- 11. Yang H, Ding Y, Hutchins LN, et al. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6:663–666. doi:10.1038/nmeth.1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan R, Peters LL, Paigen B. Mice as a mammalian model for research on the genetics of aging. ILAR J. 2011;52:4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindeman RD, Tobin JD, Shock NW. Association between blood pressure and the rate of decline in renal function with age. Kidney Int. 1984;26:861–868. [DOI] [PubMed] [Google Scholar]
- 14. Lindeman RD, Tobin J, Shock NW. Longitudinal studies on the rate of decline in renal function with age. J Am Geriatr Soc. 1985;33:278–285. [DOI] [PubMed] [Google Scholar]
- 15. Boggs DR, Patrene KD. Hematopoiesis and aging III: anemia and a blunted erythropoietic response to hemorrhage in aged mice. Am J Hematol. 1985;19:327–338. [DOI] [PubMed] [Google Scholar]
- 16. Van Zant G, Holland BP, Eldridge PW, Chen JJ. Genotype-restricted growth and aging patterns in hematopoietic stem cell populations of allophenic mice. J Exp Med. 1990;171:1547–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen J, Astle CM, Harrison DE. Development and aging of primitive hematopoietic stem cells in BALB/cBy mice. Exp Hematol. 1999;27:928–935. [DOI] [PubMed] [Google Scholar]
- 18. Chen J, Astle CM, Harrison DE. Hematopoietic senescence is postponed and hematopoietic stem cell function is enhanced by dietary restriction. Exp Hematol. 2003;31:1097–1103. [DOI] [PubMed] [Google Scholar]
- 19. Blizard DA, Klein LC, Cohen R, McClearn GE. A novel mouse-friendly cognitive task suitable for use in aging studies. Behav Genet. 2003;33:181–189. [DOI] [PubMed] [Google Scholar]
- 20. McClearn GE. Genetics of behavioral aging: animal models and the human condition. Exp Aging Res. 2002;28:453–476. doi:10.1080/ 03610730290080425 [DOI] [PubMed] [Google Scholar]
- 21. Stavnes K, Sprott RL. Effects of age and genotype on acquisition of an active avoidance response in mice. Dev Psychobiol. 1975;8:437–445. doi:10.1002/dev.420080508 [DOI] [PubMed] [Google Scholar]
- 22. Charlesworth B. Evolutionary mechanisms of senescence. Genetica. 1993;91:11–19. [DOI] [PubMed] [Google Scholar]
- 23. Miller RA, Harper JM, Dysko RC, Durkee SJ, Austad SN. Longer life spans and delayed maturation in wild-derived mice. Exp Biol Med (Maywood). 2002;227:500–508. [DOI] [PubMed] [Google Scholar]
- 24. Liao CY, Rikke BA, Johnson TE, Diaz V, Nelson JF. Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell. 2010;9:92–95. doi:10.1111/j.1474-9726.2009.00533.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yuan R, Tsaih SW, Petkova SB, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. 2009;8:277–287. doi:10.1111/j.1474-9726.2009.00478.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grubb SC, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2014;42(Database issue):D825–D834. doi:10.1093/nar/gkt1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leduc MS, Hageman RS, Meng Q, et al. Identification of genetic determinants of IGF-1 levels and longevity among mouse inbred strains. Aging Cell. 2010;9:823–836. doi:10.1111/j.1474-9726.2010.00612.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yuan R, Meng Q, Nautiyal J, et al. Genetic coregulation of age of female sexual maturation and lifespan through circulating IGF1 among inbred mouse strains. Proc Natl Acad Sci U S A. 2012;109:8224–8229. doi:10.1073/pnas.1121113109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sinke AP, Caputo C, Tsaih SW, et al. Genetic analysis of mouse strains with variable serum sodium concentrations identifies the Nalcn sodium channel as a novel player in osmoregulation. Physiol Genomics. 2011;43:265–270. doi:10.1152/physiolgenomics.00188.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petkova SB, Yuan R, Tsaih SW, Schott W, Roopenian DC, Paigen B. Genetic influence on immune phenotype revealed strain-specific variations in peripheral blood lineages. Physiol Genomics. 2008;34:304–314. doi:10.1152/physiolgenomics.00185.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xing S, Tsaih SW, Yuan R, et al. Genetic influence on electrocardiogram time intervals and heart rate in aging mice. Am J Physiol Heart Circ Physiol. 2009;296:H1907–H1913. doi:10.1152/ajpheart.00681.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tsaih SW, Pezzolesi MG, Yuan R, Warram JH, Krolewski AS, Korstanje R. Genetic analysis of albuminuria in aging mice and concordance with loci for human diabetic nephropathy found in a genome-wide association scan. Kidney Int. 2010;77:201–210. doi:10.1038/ki.2009.434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noordmans GA, Caputo CR, Huang Y, et al. Genetic analysis of mesangial matrix expansion in aging mice and identification of Far2 as a candidate gene. J Am Soc Nephrol. 2013;24:1995–2001. doi:10.1681/ASN.2012080838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang Y, Caputo CR, Noordmans GA, et al. Identification of novel genes associated with renal tertiary lymphoid organ formation in aging mice. PLoS One. 2014;9:e91850. doi:10.1371/journal.pone.0091850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wooley CM, Xing S, Burgess RW, Cox GA, Seburn KL. Age, experience and genetic background influence treadmill walking in mice. Physiol Behav. 2009;96:350–361. doi:10.1016/j.physbeh.2008.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan R, Gatti DM, Krier R, Malay E, Schultz D, Peters LL, et al. Genetic regulation of female sexual maturation and longevity through circulating IGF1. J Gerontol A Biol Sci Med Sci. 2014. doi:10.1093/gerona/glu114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berndt A, Cario CL, Silva KA, Kennedy VE, Harrison DE, Paigen B, Sundberg JP. Identification of fat4 and tsc22d1 as novel candidate genes for spontaneous pulmonary adenomas. Cancer Res. 2011;71:5779–5791.doi:10.1158/0008-5472.CAN-11-1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sundberg JP, Berndt A, Sundberg BA, et al. The mouse as a model for understanding chronic diseases of aging: the histopathologic basis of aging in inbred mice. Pathobiol Aging Age Relat Dis. 2011;1:7179–7187. doi:10.3402/pba.v1i0.7179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paigen K, Eppig JT. A mouse phenome project. Mamm Genome. 2000;11:715–717. [DOI] [PubMed] [Google Scholar]
- 40. Miller RA, Harrison DE, Astle CM, et al. An Aging Interventions Testing Program: study design and interim report. Aging Cell. 2007;6:565–575. doi:10.1111/j.1474-9726.2007.00311.x [DOI] [PubMed] [Google Scholar]
- 41. Moeller M, Hirose M, Mueller S, et al. Inbred mouse strains reveal biomarkers that are pro-longevity, antilongevity or role switching. Aging Cell. 2014;13:729–738. doi:10.1111/acel.12226 [DOI] [PMC free article] [PubMed] [Google Scholar]