Abstract
The development of saturated linkage maps using transferable markers, restriction fragment length polymorphisms, and micro-satellites has provided a foundation for fruit tree genetics and breeding. A Prunus reference map with 562 such markers is available, and a further set of 13 maps constructed with a subset of these markers has allowed genome comparison among seven Prunus diploid (x = 8) species (almond, peach, apricot, cherry, Prunus ferganensis, Prunus davidiana, and Prunus cerasifera); marker colinearity was the rule with all of them. Preliminary results of the comparison between apple and Prunus maps suggest a high level of synteny between these two genera. Conserved genomic regions have also been detected between Prunus and Arabidopsis. By using the data from different linkage maps anchored with the reference Prunus map, it has been possible to establish, in a general map, the position of 28 major genes affecting agronomic characters found in different species. Markers tightly linked to the major genes responsible for the expression of important traits (disease/pest resistances, fruit/nut quality, self-incompatibility, etc.) have been developed in apple and Prunus and are currently in use for marker-assisted selection in breeding programs. Quantitative character dissection using linkage maps and candidate gene approaches has already started. Genomic tools such as the Prunus physical map, large EST collections in both Prunus and Malus, and the establishment of the map position of high numbers of ESTs are required for a better understanding of the Rosaceae genome and to foster additional research and applications on fruit tree genetics.
The major temperate fruit tree crops, apple (Malus × domestica), peach (Prunus persica), cherry (Prunus avium and Prunus cerasus), plum (Prunus domestica and Prunus salicina), apricot (Prunus armeniaca), almond (Prunus dulcis), pear (Pyrus communis), quince (Cydonia oblonga), and loquat (Eriobotrya japonica), belong to the Rosaceae family. This also includes some other important crops such as strawberry (Fragaria × ananassa) and rose (Rosa spp.). Most of these species are woody perennials with a long intergeneration period due to their juvenile phase and large plant sizes, which make them poorly suited organisms for classical genetic analysis. On the other hand, fruit trees have some advantageous features such as a long life, the existence of efficient methods of vegetative reproduction, the possibility of making interspecific crosses (frequent at the congeneric level), and a small basic genome; e.g., wild strawberry (Fragaria vesca) has a haploid genome size of 164 Mbp (1), and peach has a haploid genome size of 290 Mbp (2). Until recently, only very limited information existed on the genetics of phenotypic characters of simple inheritance; only 31 major genes had been described in peach (3), the best characterized of the Prunus species, or three genes in almond (4).
The breeding methods used in these species have undergone very few changes over the last 50 years, and the incorporation of alleles of interest from wild or exotic materials into elite breeding lines has rarely produced new commercial cultivars. Methods based on knowledge provided by advances in molecular genetics, notably molecular markers, promise faster and more efficient approaches to cultivar improvement. Early selection with molecular markers allows an accurate screening of seedlings several years before the characters can be evaluated in the field, makes possible the accumulation of different resistance factors in a genotype of interest, or shortens the number of generations needed to recover the genotype of the cultivated species after a cross with an exotic genotype or wild species. Overall, these methods result in savings of space, and even more importantly, of time, factors already crucial in herbaceous species, and still more important in woody perennials. Given the poor level of genetic knowledge of these species, the research effort needed to bring them to the level of other important crops has been considerable, particularly in the last decade. Results are now emerging, some important tools (markers, maps, DNA sequences, and quantitative trait loci, QTLs) have been developed and made available to researchers, and applications at the breeding program level have been started. In this paper, we summarize the main results obtained for this group of crops with special emphasis on the members of the Prunus genus, which have experienced a particularly fast growth.
Methods: Linkage Map Construction
The Prunus Reference Map. As a result of a European project, a saturated linkage map of 246 markers (235 RFLPs and 11 isozymes) in an almond (cv. Texas) × peach (cv. Earlygold) F2 progeny was constructed (5). This map (the TxE map) detected the expected eight linkage groups (G1 to G8), with a total distance of 491 centimorgans (cM). A set of 96 simple sequence repeats (SSRs) has been recently added to this map (6). In this paper, we provide the map position of 220 additional markers [89 SSRs, five sequence-tagged sites, and 126 restriction fragment length polymorphisms (RFLPs) obtained mainly with Arabidopsis thaliana probes with high level of sequence conservation with rice; ref. 7]. The current map has 562 markers, covering 519 cM (average density, 0.92 cM per marker; largest gap, 7 cM). This map is shown in Fig. 4, which is published as supporting information on the PNAS web site. Most of the probes used for the RFLPs of this map are sequenced and publicly available. The majority (87%) of the TxE loci correspond to a known DNA sequence, and 37% of these sequences correspond to a putative protein.
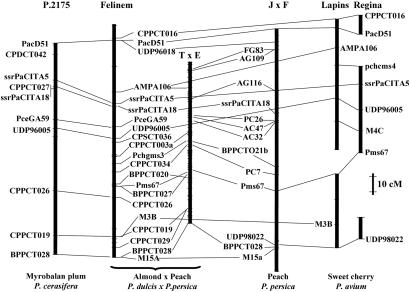
Fig. 1.
Comparison among the SSR maps of linkage group 1 in the almond × peach Prunus reference map (TxE), peach (cv. Ferjalou Jalusia × cv. Fantasia = FxJ), almond × peach (cv. Felinem), P. cerasifera (P.2175), and cherry (cv. Lapins and cv. Regina). Positions of anchor loci between maps are indicated by connecting lines. Only marker positions as in ref. 6 are indicated for the TxE map.
The existence of the TxE map has been very useful for the Prunus research community, providing a highly polymorphic population for linkage studies, establishing a common terminology for linkage groups, and providing a set of transferable markers (anchor markers) of known map position that have facilitated the development of framework maps in other crosses. It has also allowed the location of different major genes and QTLs in a unique map, the search for markers to saturate specific genome regions, or the establishment of map comparisons with other Prunus species. Thirteen additional maps obtained with progenies including species such as almond, peach, apricot, cherry, Prunus davidiana, Prunus cerasifera, and Prunus ferganensis have been constructed with a set of anchor markers selected from the TxE map (8-18). Details on these maps and their relationship to the TxE map are provided in Table 2, which is published as supporting information on the PNAS web site.
Apple Maps. The first detailed apple map was constructed by a European consortium using the F1 progeny of the cross cv. Prima × cv. Fiesta (the PxF map) (19). A total of 290 markers [including 124 RFLPs, 10 SSRs, 17 isozymes, 133 random amplified polymorphic DNA, and six markers of other origins) were distributed among 17 linkage groups in cv. Prima and cv. Fiesta, as expected from the apple chromosome number (x = 17). A more saturated map with 840 molecular markers, including 129 SSRs as the only codominant markers, was recently constructed with the F1 progeny between cv. Fiesta and cv. Discovery (the FxD map) (20). The high number of SSRs on this map promises a rapid transfer of this information to other apple-segregating populations, allowing its widespread use by Malus geneticists and breeders.
Results and Discussion: Comparative Mapping
Prunus Synteny. Comparing the positions of anchor markers (RFLPs, SSRs, and isozymes) of the TxE map with those of 13 maps constructed with other Prunus populations, it can been shown that the genomes of the diploid (2n = 16) species, peach, almond, apricot, cherry, P. davidiana, P. cerasifera, and P. ferganensis, are essentially collinear (Fig. 1, Figs. 5-7, which are published as supporting information on the PNAS web site, and Table 2). The comparison between TxE and P. cerasifera, and TxE and cherry, is shown in Figs. 5 and 7, respectively. The occasional divergences between maps of different species can generally be attributable to the mapping of different duplicates of RFLPs or SSRs that have more than one copy in different regions of the Prunus genome. Moreover, order inversions affect almost always pairs of loci that are close together in the TxE map (≤10 cM), suggesting that they are more probably caused by errors in the assignment of marker order than to inversion of chromosome fragments. These results are in agreement with the pattern of crossability between species of this genus, where interspecific crosses are usually possible, directly or through bridge species, and hybrids are often fertile. Thus, at the genome level, the Prunus genus can be treated as a single genetic entity.
Only one major chromosomal rearrangement has been documented, consisting of a reciprocal translocation between G6 and G8 that was demonstrated in the F2 progeny of almond (cv. Garfi) × peach (cv. Nemared) (8). The same translocation was later found in the peach F2 cv. Akame × cv. Juseitou (18). Although it has not been determined which of the parents had the standard or translocated configurations in either case, the most reasonable hypothesis is that it occurs within the peach germplasm (8). Given that one of the parents of each cross is a red-leaved peach (cv. Nemared and cv. Juseitou) and that the gene that determines red vs. green leaf color (Gr/gr) is located close to the translocation breakpoint, a possible relationship between the cytogenetic and the morphological phenotypes cannot be discarded.
Prunus and Malus Map Comparisons. Whereas most Prunus species are diploids, apple and pear are allotetraploid species with 2n = 34 (21). The PxF apple map (19) clearly showed the allopolyploid nature of the apple, where the RFLP or SSR markers studied had disomic segregations. Duplicate RFLPs mapping to two map positions were also frequent, and it was possible to establish some of the homoeologous pairs of chromosomes and to also detect some possible rearrangements between the original ancestral genomes. However, this was not so evident when SSR markers were used (20), partly because only a small proportion of them had more than one copy, indicating that the two component genomes were divergent enough to impede the identification of both homoeologous SSRs when the same primers were used.
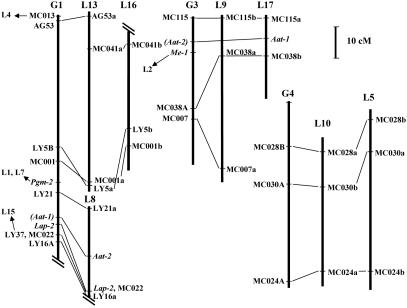
A total of 30 loci (24 RFLPs detected by the same probes and 6 isozyme genes) of the TxE Prunus map have homologous counterparts in the PxF apple map (19), enabling us to make a first comparison between the genomes (Fig. 2). In three of the Prunus linkage groups (G1, G3, and G4) there were three or more anchors with apple linkage groups (L5, L8, L9, L10, L13, and L17). Marker order was generally identical, and one linkage group of Prunus usually corresponded to two of the apple linkage groups considered as homoeologous. These results provide a preliminary indication for a high level of synteny between the Prunus genome and the two component genomes of apple. An interesting observation is that approximately half of linkage group 1 of Prunus aligns with one pair of homoeologous linkage groups in apple (L13 and L16) and the rest with a third group (L8). G1 is longer and much more populated with markers than the other Prunus linkage groups, indicating its likely correspondence with the long chromosome 1 (22), but apple does not have such a large chromosome in its karyotype (23). These results suggest that, if one of the ancestral genomes of Malus is close to the Prunus genome, the long chromosome has split into two in apple or that two ancestral Prunus chromosomes fused after the separation between these two genera.
Fig. 2.
Comparison between Prunus (5) and apple (21) linkage maps. Only the position of anchor loci is shown. Linkage groups in Prunus are noted as G and apple groups are noted as L followed by a number. The positions of markers in parenthesis in Prunus were inferred from other maps. Marker positions in apple were obtained by using the maps of both parents of the F1 cross cv. Prima × cv. Fiesta. Two parallel oblique lines indicate that only a fragment of the linkage group is included. Arrows pointing to the left in the Prunus map are anchors to markers located in the indicated linkage groups of the apple map.
Prunus and Arabidopsis Comparisons. By using the sequence of 177 DNA probes, corresponding to 227 mapped loci in the reference TxE Prunus map and highly homologous (tblastx value <10-15) to Arabidopsis genome sequences, it was possible to compare the Prunus map with the Arabidopsis genome sequence (6). The 227 Prunus loci (corresponding to a density of 2.6 cM per marker in this map) detected 703 loci in the Arabidopsis genome. Syntenic blocks between these two species were accepted when a minimum of three homologous marker loci was located within 1.2 Mb or less (≤1%) of the Arabidopsis genome and within a distance of 6 cM (1%) of the Prunus map. Blocks meeting these criteria but containing gaps >1% of either genome were also rejected. With these stringent criteria there were a total of 37 syntenic regions spanning 23% of the Prunus map and 17% of the Arabidopsis genome. The longest syntenic region covered 25 cM, including 13 homologous markers, in G2 of Prunus and corresponded to a region of 5.4 Mbp (16 markers) in chromosome 5 of Arabidopsis. The comparison allowed the detection of duplicated regions in the Prunus genome (those that corresponded to the same Arabidopsis block) between and within chromosomes, suggesting a pattern of duplication similar to that found in Arabidopsis (24). Although the comparison of these two genomes with a larger number of markers would probably have shown a more complete pattern of similarity and a higher number of conserved regions, these results provide evidence for a considerable level of conservation between these two distant genomes. The observed homology is clearly lower than that found in confamilial or congeneric comparisons, but still sufficient to facilitate strategies for marker saturation of specific regions (25) or candidate gene search in Prunus based on the Arabidopsis sequence.
Rosaceae Macrosynteny Compared to That of Other Plant Families.Comparative mapping in the three plant families where it has been more extensively studied, Solanaceae (26), Poaceae (27), and Brassicaeae (28), consistently indicates that genome evolution within a family consists of a limited amount of chromosome restructuring due to inversion and translocation events that lead to the conservation of large chromosomal fragments in the genomes of its constituent species. Results available so far in the Rosaceae indicate the same trend. The six members of the Prunus genus studied have a genome without any major chromosomal rearrangements, indicating their close relationship. Similar results have been found in congeneric species of other families having the same chromosome number, such as Pinus (29), Eucalyptus (30), and some of the wild relatives of wheat or barley, such as Aegilops tauschii and Aegilops speltoides (27). This trend can also be inferred from the colinearity of the various interspecific maps of tomato and some of its wild relatives (http://soldb.cit.cornell.edu). More distant comparisons, such as Prunus and Malus, and possibly among the two constituent genomes of Malus, easily allows detection of large colinear blocks, even with a small number of markers as used here, and also obvious major genome rearrangements, such as that detected between linkage group 1 of Prunus and groups L8, L13, and L16 of apple. Comparisons beyond the family level (Prunus-Arabidopsis) result in a more fragmentary pattern of similarity, where generally only small DNA conserved regions can be recognized.
Marker-Assisted Selection. Mapping major genes. Some important agronomic characters in fruit trees behave as major genes, including disease resistances, as well as flower, vegetative, or fruit or nut quality traits. Their simple inheritance makes of them obvious targets to the search of tightly linked markers for early selection, particularly for characters that require complex analysis, such as some pest or disease inoculation, or that cannot be evaluated until the plant has reached the adult stage, such as fruit characters or the self-incompatibility genotype.
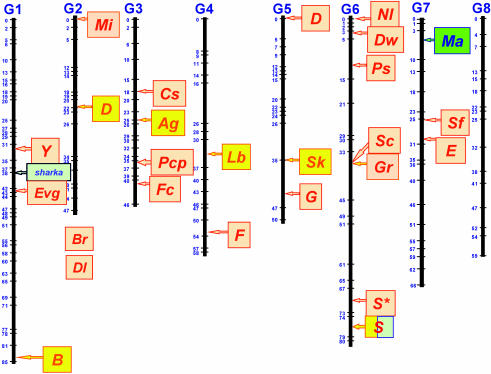
With the high level of synteny between the genome of Prunus crops, and the existence of a reference map, a considerable number of genes studied in different populations of various species have been integrated in a single map. The approximate position of 28 of these genes on TxE is provided in Fig. 3, and their description is in Table 1. Although in most cases the location of these genes has been established in low-density maps, their position can be further defined by using the information provided by the network of maps available for Prunus, and additional markers can be found in the regions of interest and used for marker-assisted selection without further development. One of the characters currently selected by molecular methods in breeding programs is the gametophytic self-incompatibility of almond, apricot, and cherry. This character is encoded by a highly polymorphic locus (S/s), located in the distal part of G6 (41), which was first studied as a stylar ribonuclease isozyme (43). When sequences of the polymorphic S-RNase gene at the S locus were determined (44), a wave of species-specific or even allele-specific DNA markers was developed (45), allowing earlier and more accurate selection of the most common self-incompatibility or self-compatibility alleles.
Fig. 3.
Approximate position of 28 major genes mapped in different populations of apricot (blue background), peach (orange background), almond or almond × peach (yellow background), and Myrobalan plum (green background) on the framework of the Prunus reference map (5). Gene abbreviations correspond to: Y, peach flesh color; B, almond/peach petal color; sharka, plum pox virus resistance; B, flower color in almond x peach; Mi, nematode resistance from peach; D, almond shell hardness; Br, broomy plant habit; Dl, double flower; Cs, flesh color around the stone; Ag, anther color; Pcp, polycarpel; Fc, flower color; Lb, blooming date; F, flesh adherence to stone; D, non-acid fruit in peach, Sk, bitter kernel; G, fruit skin pubescence; Nl, leaf shape; Dw, dwarf plant; Ps, male sterility; Sc, fruit skin color; Gr, leaf color; S*, fruit shape; S, self-incompatibility (almond and apricot); Ma, nematode resistance from Myrobalan plum; E, leaf gland shape; Sf, resistance to powdey mildew. Genes Dl and Br are located on an unknown position of G2.
Table 1. Description of 28 major genes affecting morphological or agronomic characters in different Prunus crops that can be located on the reference map.
| LG* | Characters | Species | Symbol | Populations† | Ref. |
|---|---|---|---|---|---|
| G1 | Fruit flesh color (white/yellow) | Peach | Y | Padre × 54P455 | 9 |
| Sharka resistance | Apricot | sharka | Lito × Lito | 15 | |
| Evergrowing | Peach | Evg | Empress op op dwarf × P1442380 | 31 | |
| Flower color | Almond × peach | B | Garfi × Nemared | 32 | |
| G2 | Root-knot nematode resistance | Peach | Mi‡ | P.2175 × Felinem, Akame × Juseitou, Lowell × Nemared, Garfi × Nemared; Padre × 54P455 | 9, 18, 32, 33, 34§ |
| Shell hardness | Almond | D | Ferragnès × Tuono | 35 | |
| Broomy (or pillar) growth habit | Peach | Br | Various progenies | 36 | |
| Double flower | Peach | Dl | NC 174RL × P1 | 37 | |
| G3 | Flesh color around the stone | Peach | Cs | Akame × Jusetou | 18§ |
| Anther color (yellow/anthocyanic) | Almond × peach | Ag | Texas × Earlygold | 38 | |
| Polycarpel | Peach | Pcp | Padre × 54P455 | 9 | |
| Flower color | Peach | Fc | Akame × Jusetou | 18§ | |
| G4 | Blooming time | Almond | Lb | D.3.5 × Bertina | 14 |
| Flesh adhesion (clingstone/freestone) | Peach | F | (P. ferganensis × IF310828)BCI; Akame × Juseitou | 11, 18§ | |
| G5 | Non-acid fruit | Peach | D | Ferjalou Jalousia × Fantasia | 17, 39, 40 |
| Kernel taste (bitter/sweet) | Almond | Sk | Padre × 54P455 | 9 | |
| Skin hairiness (nectarine/peach) | Peach | G | Ferjalou Jalousia × Fantasia; Padre × 54P455 | 9, 39, 40 | |
| G6 | Leaf shape (narrow/wide) | Peach | Nl | Akame × Juseitou | 18§ |
| Plant height (normal/dwarf) | Peach | Dw | Akame × Juseitou | 18§ | |
| Male sterility | Peach | Ps | Ferjalou Jalousia × Fantasia | 39 | |
| Fruit shape (flat/round) | Peach | S* | Ferjalou Jalousia × Fantasia | 39, 40 | |
| Self-incompatibility | Almond | S | Ferragnès × Tuono; D.3.5 × Bertina; Padre × 54P455 | 9, 41 | |
| Apricot | S | Lito × Lito | 15 | ||
| G6-G8 | Fruit skin color | Peach | Sc | Akame × Juseitou | 18§ |
| Leaf color (red/green) | Peach | Gr | Garfi × Nemared; P.2175 × Felinem, Akame × Juseitou | 18, 32, 33§ | |
| G7 | Root-knot nematode resistance | Myrobalan plum | Ma | P.2175 × Felinem | 33, 42 |
| Resistance to powdery mildew | Peach | Sf | (P. ferganensis × IF310828)BCI | 11 | |
| Leaf gland (reniform/globose) | Peach | E | (P. ferganensis × IF310828)BCI | 11 |
LG, linkage group; G6-G8 genes located close to the translocation breakpoint between these two linkage groups.
All genotypes used as parents of the crosses are named cultivars with the exceptions of 54P455, Empress op op dwarf, P1442380, P.2175, NC174RL, P1, D.3.5, and IF310821, which are breeding lines or plant introductions.
One or two genes of nematode resistance and one QTL with have been described in this linkage group by the authors cited.
Information relevant to the map position of genes segregating in cv. Akame × cv. Juseitou comes also from T. Yamamoto (personal communication).
Markers near the two genes of resistance to root-knot nematodes are also used for the selection of resistant Prunus rootstocks. Markers tightly linked to a resistance gene (Ma/ma) from Myrobalan plum have been identified (42). This gene, located on G7, and another one from peach cv. Nemared (Mi/mi) mapping to G2, have been screened with markers in a search for rootstocks that pyramid both resistance genes in a three-way (peach, almond, and Myrobalan plum) progeny (33). Possible targets for marker-assisted selection are a major gene involved in Sharka (Plum Pox Virus) resistance (15) located in G1 and a major gene for shell hardness in almond on G2 (35).
Markers linked to disease and pest resistance genes have also been described in apple. Among the most important are several resistance genes to scab (Venturia inaequalis) (46-48), powdery mildew (Podosphaera leucotricha) (49, 50), fire blight (Erwinia amylovora) (51), or the rosy leaf curling aphid (Disaphis devecta) (52) and woolly aphid (Eriosoma lanigerum) (51). Use of marker-assisted selection for disease resistance is becoming increasingly widespread in apple as a means of early selection (48, 50), to pyramid resistance genes (53), and as a strategy to limit the important linkage drag associated with the use of exotic materials for the introgression of resistance genes (54).
Mapping QTLs. Quantitatively inherited characters constitute the bulk of the variability selected during the breeding process in fruit trees as in most cultivated species. Characters related with plant growth and architecture, yield, blooming and harvesting times, and fruit quality, are usually of quantitative nature, and they have been analyzed with the help of molecular markers in several fruit species. The long period from seed to fruiting in trees is often a problem because there is a long time from the moment the experiments are conceived until the results can be obtained. The use of populations already existing can be a solution to this problem, although they rarely conform the requirements of this kind of experiment, e.g., uniformity (age, rootstock, plantation stand, etc.), population size, and lack of selection. On the other hand, vegetative reproduction allows every population to be immortalized, making it possible to study the characters of interest for as many years and in as many different environments as needed.
Polygenic resistance loci may contribute to the development of cultivars with effective and durable resistance to biotic stress in fruit trees. P. davidiana, P. kansuensis and Prunus mira, all closely related with peach, are possible sources of resistance genes for some of its most important pests and diseases such as the peach aphid (Aphis gossipii), leaf curl (Taphrina deformans), powdery mildew (Sphaeroteca pannosa var. persicae), or sharka virus (55, 56). Various populations of peach × P. davidiana crosses with different levels of introgression of the P. davidiana genome into the cultivated peach (F1, F2, or BC2) have been analyzed, giving the position of QTLs for all of them (55-57). For example, 13 QTLs explaining up to a 65% of the total phenotypic variation for powdery mildew resistance in plants exposed to the disease in different times and environments have been identified (56)
Several major genes (Vf, Vm, Vr, Vg, Vb, Vbj, and Va) coming from Asian Malus species confer race-specific resistance of apple to scab (58). These resistances are ephemeral, and all have been overcome by the disease, some of them even before being used in commercial cultivars. Then, there is a need for finding durable sources of resistance and introgressing them into new cultivars. One possible way is to incorporate several sources of resistance, either by pyramiding major genes or by combining the effects of major genes with QTLs that confer partial resistance. Both strategies require molecular markers linked to these genes to be achieved. Markers exist for three of the major resistances, and QTLs have already been identified; eight and two QTLs involved in leaf and fruit scab resistance, respectively, one of them in common, were found in the FxD population, the majority of them coming from the cv. Fiesta parent (59).
QTLs for blooming, ripening, and fruit quality characters have also been detected in both peach (17, 40) and apple (60, 61). Some the QTLs involved in the inheritance of fruit quality components or blooming time were located in regions of the genome where major genes had been described previously, such as the D/d gene, responsible for low fruit acidity in peach (40), the Ma/ma gene, coding for the malic acid content in apple fruit (60), or the Lb/lb gene, which determines blooming time of almond (14).
Candidate gene approaches have proven useful for finding associations between genes involved in relevant metabolic pathways and major genes or QTLs in fruit trees. Several resistance gene analogs (RGAs) have been mapped in Prunus (9) and are placed in similar genomic positions as genes or QTLs that determine sharka resistance (15, 55) or root-knot nematode resistance (18, 33), suggesting that they may belong to resistance gene clusters that include the gene or genes involved in resistance. RGAs selected from an apple EST database (62) have provided markers closer to genes of resistance to scab (Vf), powdery mildew (Pl2 and PlMIS) and woolly aphid (Er3) than the best markers available. One of the 12 candidates involved in fruit sugar and organic acid metabolic pathways studied in peach (18) mapped to the same region that a QTL located on G6. This gene, PRUpe;Vp2, is a vacuolar pyrophosphatase involved in the establishment of an electrochemical gradient across the tonoplast, which may be implicated in the sucrose accumulation to the vacuole.
Prospects of Marker Research in Fruit Trees
An international consortium led by Albert Abbott at Clemson University (Clemson, SC) has developed additional tools for the characterization of the Prunus genome. Using the RFLPs on the TxE map and a BAC library of peach cv. Nemared, a physical map has been partially assembled. A growing collection of ESTs from peach and almond based on cDNA libraries has been released to public databases, and >3,800 peach putative unigenes have been detected. Approximately 2,000 of these unigenes have been assigned to specific bacterial artificial chromosomes that contain them, and twice as many unigenes are expected to be placed by the end of the project. A Rosaceae database (www.genome.clemson.edu/gdr) has recently been created with the objective of assembling all this information and make it available worldwide to researchers working in this group of species.
The progress made during the last decade on the genetic knowledge of the cultivated species of the Rosaceae, and particularly of peach as its more logical model, has been enormous. However, to allow the application of the newly generated information to the production of improved cultivars, some important topics need to be addressed in the near future. The comparative mapping between the most important genera of this family, like Prunus, Malus, Pyrus, Fragaria, and Rosa, should be undertaken in detail. Another important aspect that needs additional development concerns the use of exotic germplasm. Some of these genera, like Prunus or Malus, include a large number of intercompatible species, meaning that an enormous gene pool is available for fruit breeding. Little use has been made of this variability because of the slowness of the classical breeding methods. Genomic methodologies may make it possible to discover genes of interest from exotic materials of the well characterized existing germplasm collections (www.ecpgr.cgiar.org/workgroups/prunus/prunus.htm) and to introgress them into cultivated ones with methods already proposed and in use in annual species (63, 64), but adapted to the characteristics of woody perennials.
Supplementary Material
Acknowledgments
We gratefully acknowledge A. G. Abbott, J.-M. Audergon, C.-E. Durel, D. Esmenjaud, S. Gardiner, C. Gessler, L. Gianfranceschi, M. Kellerhals, J. Kervella, G. J. King, F. Laigret, C. Maliepaard, R. Quarta, S. Sansavini, R. Testolin, K. Tobutt, and T. Yamamoto for providing useful information. This work was supported in part by European Union Projects AIR3 CT93-1585, BIO4 CT97-2170, and FAIR6-CT-98-4139.
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: QTL, quantitative trait locus; cM, centimorgan; SSR, simple sequence repeat; RFLP, restriction fragment length polymorphism.
References
- 1.Akiyama, Y., Yamamoto, Y., Ohmido, N., Ohshima, M.& Fukui, K. (2001) Cytologia 66, 431-436. [Google Scholar]
- 2.Baird, W. V., Estager, A. S. & Wells, J. K. (1994) J. Am. Soc. Hort. Sci. 199, 1312-1316. [Google Scholar]
- 3.Monet, R., Guye, A., Roy, M. & Dachary, N. (1996) Agronomie 16, 321-329. [Google Scholar]
- 4.Socias i Company, R. (1998) Theor. Appl. Genet. 96, 588-601. [Google Scholar]
- 5.Joobeur, T., Viruel, M. A., de Vicente, M. C., Jauregui, B., Ballester, J., Dettori, M. T., Verde, I., Truco, M. J., Messeguer, R., Batlle, I., et al. (1998) Theor. Appl. Genet. 97, 1034-1041. [Google Scholar]
- 6.Aranzana, M. J., Pineda, A., Cosson, P., Dirlewanger, E., Ascasibar, J., Cipriani, G., Ryder, C. D., Testolin, R., Abbott, A., King, G. J., et al. (2003) Theor. Appl. Genet. 106, 819-825. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez, I., Graziano, E., Gebhardt, C., Barakat, A., Berry, S., Arús, P., Delseny, M. & Barnes, S. (2003) Plant Biotechnol. J. 1, 91-99. [DOI] [PubMed] [Google Scholar]
- 8.Jáuregui, B., de Vicente, M. C., Messeguer, R., Felipe, A., Bonnet, A., Salesses, G. & Arús, P. (2001) Theor. Appl. Genet. 102, 1169-1176. [Google Scholar]
- 9.Bliss, F. A., Arulsekar, S., Foolad, M. R., Becerra, V., Gillen, A. M., Warburton, M. L., Dandekar, A. M., Kocsisne, G. M. & Mydin, K. K. (2002) Genome 45, 520-529. [DOI] [PubMed] [Google Scholar]
- 10.Foulongne, M., Pascal, T., Arús, P. & Kervella, J. (2003) Theor. Appl. Genet. 107, 227-238. [DOI] [PubMed] [Google Scholar]
- 11.Dettori, M. T., Quarta, R. & Verde, I. (2001) Genome 44, 783-790. [PubMed] [Google Scholar]
- 12.Viruel, M. A., Messeguer, R., de Vicente, M. C., Garcia-Mas, J., Puigdomènech, P., Vargas, F. & Arús, P. (1995) Theor. Appl. Genet. 91, 964-971. [DOI] [PubMed] [Google Scholar]
- 13.Joobeur, T., Periam, N., de Vicente, M. C., King, G. & Arús, P. (2000) Genome 43, 649-655. [PubMed] [Google Scholar]
- 14.Ballester, J., Socias i Company, R., Arús, P. & de Vicente, M. C. (2001) Plant Breed. 120, 268-270. [Google Scholar]
- 15.Vilanova, S., Romero, C., Abbott, A. G., Llacer, G. & Badenes, M. L. (2003) Theor. Appl. Genet. 107, 239-247. [DOI] [PubMed] [Google Scholar]
- 16.Lambert, P., Hagen L. S., Arús, P. & Audergon, J. M. (2003) Theor. Appl. Genet. 108, 1120-1130. [DOI] [PubMed] [Google Scholar]
- 17.Etienne, C., Rothan, C., Moing, A., Plomion, C., Bodenes, C., Svanella-Dumas, L., Cosson, P., Pronier, V., Monet, R. & Dirlewanger, E. (2002) Theor. Appl. Genet. 105, 145-159. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto, T., Shimada, T., Imai, T., Yaegaki, H., Haji, T., Matsuta, N., Yamaguchi, M. & Hayashi, T. (2001) Breed. Sci. 51, 271-278. [Google Scholar]
- 19.Maliepaard, C., Alston, F. H., van Arkel, G., Brown, L. M., Chevreau, E., Dunemann, F., Evans, K. M., Gardiner, S., Guilford, P., Heusden, A. W. V., et al. (1998) Theor. Appl. Genet. 97, 60-73. [Google Scholar]
- 20.Liebhard, R., Koller, B., Gianfranceschi, L. & Gessler, C. (2003) Theor. Appl. Genet. 106, 1497-1508. [DOI] [PubMed] [Google Scholar]
- 21.Chevreau, E. & Laurens, F. (1987) Theor. Appl. Genet. 75, 90-95. [Google Scholar]
- 22.Salesses, G. & Mouras, A. (1977) Ann. Amelior. Plantes 27, 363-368. [Google Scholar]
- 23.Bouvier, L., Lespinasse, Y. & Schuster, M. (2000) Acta Hort. 538, 321-324. [Google Scholar]
- 24.Blanc, G., Barakat, A., Guyot, R., Cooke, R. & Delseny, M. (2000) Plant Cell 12, 1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsin-Mei, K., Liu, J., Doganlar, S. & Tanksley, S. D. (2001) Genome 44, 470-475. [DOI] [PubMed] [Google Scholar]
- 26.Doganlar, S., Frary, A., Daunay, M-C., Lester, R. N. & Tanksley S. D. (2003) Genetics 161, 1697-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devos, K. M. & Gale, M. D. (2000) Plant Cell 12, 637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukens, L., Zou, F., Lydiate, D., Parkin, I. & Osborn, T. (2003) Genetics 164, 359-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komulainen, P., Brown, G. R., Mikkonen, M., Karhu, A., García-Gil, M. R., O'Malley, D., Lee, B., Neale, D. B. & Sovalainen, O. (2003) Theor. Appl. Genet. 107, 667-678. [DOI] [PubMed] [Google Scholar]
- 30.Marques, C. M., Brondani, R. P. V., Grattapaglia, D. & Sederoff, R. (2002) Theor. Appl. Genet. 105, 474-478. [DOI] [PubMed] [Google Scholar]
- 31.Wang, Y., Georgi, L. L., Reighard, G. L., Scorza, R. & Abbott, A. G. (2002) J. Hered. 93, 352-358. [DOI] [PubMed] [Google Scholar]
- 32.Jáuregui, B. (1998) Ph.D. thesis (Universitat de Barcelona, Barcelona)
- 33.Claverie, M., Bosselut, N. Lecouls, A. C., Voisin, R., Poizat, C., Dirlewanger, E., Kleinhentz, M., Lafargue, B., Laigret, F. & Esmenjaud, D. (2004) Theor. Appl. Genet. 108, 765-773. [DOI] [PubMed] [Google Scholar]
- 34.Lu, Z. X., Sosinski, B., Reighard, G. L., Baird, W. V. & Abbott, A. G. (1998) Genome 41, 199-207. [Google Scholar]
- 35.Arús, P., Ballester, J., Jáuregui, B., Joobeur, T., Truco M. J. & de Vicente, M. C. (1998) Acta Hort. 484, 325-330. [Google Scholar]
- 36.Scorza, R., Melnicenco, L., Dang, P. & Abbott, A. G. (2002) Acta Hort. 592, 285-289. [Google Scholar]
- 37.Chaparro, J. X., Werner, D. J., O'Malley, D. & Sederoff, R. R. (1994) Theor. Appl. Genet. 87, 805-815. [DOI] [PubMed] [Google Scholar]
- 38.Joobeur, T. (1998) Ph.D. thesis (Universitat de Lleida, Lleida, Spain).
- 39.Dirlewanger, E., Pronier, V., Parvery, C., Rothan, C., Guye, A. & Monet, R. (1998) Theor. Appl. Genet. 97, 888-895. [Google Scholar]
- 40.Dirlewanger, E., Moing, A., Rothan, C., Svanella, L., Pronier, V., Guye A., Plomion, C. & Monet, R. (1999) Theor. Appl. Genet. 98, 18-31. [DOI] [PubMed] [Google Scholar]
- 41.Ballester, J., Boskovic, R., Batlle, I., Arús, P., Vargas, F. & de Vicente, M. C. (1998) Plant Breed. 117, 69-72. [Google Scholar]
- 42.Lecouls, A. C., Rubio-Cabetas, M. J., Minot, J. C., Voisin, R., Bonnet, A., Salesses, G., Dirlewanger, E. & Esmenjaud, D. (1999) Theor. Appl. Genet. 99, 328-335. [Google Scholar]
- 43.Boskovic, R. & Tobutt, K. R. (1996) Euphytica 90, 245-250. [Google Scholar]
- 44.Tao, R., Yamane, H., Sassa, H., Mori, H., Gradziel, T. M., Dandekar, A. M. & Sugiura, A. (1997) Plant Cell Physiol. 38, 304-311. [DOI] [PubMed] [Google Scholar]
- 45.Tamura, M., Ushijima, K., Hirano, H. S., Tao, R., Gradziel, T. M. & Dandekar, A. M. (2000) Theor. Appl. Genet. 101, 344-349. [Google Scholar]
- 46.Xu, M. L. & Korban, S. S. (2002) Plant Mol. Biol. 50, 803-818. [DOI] [PubMed] [Google Scholar]
- 47.Cheng, F. S., Weeden, N. F., Brown, S. K., Aldwinckle, H. S., Gardiner, S. E. & Bus, V. G. (1998) Genome 41, 208-214. [Google Scholar]
- 48.Hemmat, M., Brown, S. K. & Weeden, N. F. (2002) J. Am. Soc. Hort. Sci. 127, 365-370. [Google Scholar]
- 49.Markussen, T., Kruger, J., Schmidt, H. & Dunemann, F. (1995) Plant Breed. 114, 530-534. [Google Scholar]
- 50.Evans, K. M. & James, C. M. (2003) Theor. Appl. Genet. 106, 1178-1183. [DOI] [PubMed] [Google Scholar]
- 51.Bus, V., Ranatunga, C., Gardiner, S., Basset, H. & Rikkerink, E. (2000) Acta Hort. 538, 541-547. [Google Scholar]
- 52.Cevik, V. & King, G. J. (2002) Theor. Appl. Genet. 105, 346-354. [DOI] [PubMed] [Google Scholar]
- 53.Tartarini, S. & Sansavini, S. (2003) Acta Hort. 622, 129-140. [Google Scholar]
- 54.King, G. J., Tartarini, Brown, L. M., Gennari, F. & Sansavini, S. (1999) Theor. Appl. Genet. 99, 1039-1046. [Google Scholar]
- 55.Foulongne, M. (2002) Ph.D. thesis (Université de la Mediterranée, Marseille, France).
- 56.Foulongne, M., Pascal, T., Pfeiffer, F.& Kervella, J. (2003) Mol. Breed. 12, 33-50. [Google Scholar]
- 57.Dirlewanger, E., Pascal, T., Zuger, C. & Kervella, J. (1996) Theor. Appl. Genet. 93, 909-919. [DOI] [PubMed] [Google Scholar]
- 58.Williams, E. B. & Kuç, J. (1969) Annu. Rev. Phytopathol. 7, 223-246. [Google Scholar]
- 59.Liebhard, R., Koller, B., Patocci, A., Kellerhals, M., Pfammatter, W., Jermini, M. & Gessler, C. (2003) Phytopathology 93, 493-501. [DOI] [PubMed] [Google Scholar]
- 60.King, G. J., Lynn, J. R., Dover, C. J., Evans, K. M., Seymour, G. B. (2001) Theor. Appl. Genet. 102, 1227-1235. [Google Scholar]
- 61.Liebhard, R., Kellerhals, M., Pfammatter, W., Jermini, M. & Gessler, C. (2003) Plant Mol. Biol. 52, 511-526. [DOI] [PubMed] [Google Scholar]
- 62.Gardiner, S., Murdoch, J., Meech, S., Rusholme, R., Bassett, H., Cook, M., Bus, V., Rikkerink, E., Gleave, A., Crowhurst, R., et al. (2003) Acta Hort. 622, 141-151. [Google Scholar]
- 63.Tanksley, S. D., Young, N. D., Paterson, A. H. & Bonierbale, M. W. (1989) BioTechnology 7, 257-263. [Google Scholar]
- 64.Tanksley, S. D., Grandillo, S., Fulton, T. M., Zamir, D., Eshed, Y., Petiard, V., Lopez, J. & Beck-Bunn, T. (1996) Theor. Appl. Genet. 92, 213-224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.