Abstract
Fruit constitutes a major component of human diets, providing fiber, vitamins, and phytonutrients. Carotenoids are a major class of compounds found in many fruits, providing nutritional benefits as precursors to essential vitamins and as antioxidants. Although recent gene isolation efforts and metabolic engineering have primarily targeted genes involved in carotenoid biosynthesis, factors that regulate flux through the carotenoid pathway remain largely unknown. Characterization of the tomato high-pigment mutations (hp1 and hp2) suggests the manipulation of light signal transduction machinery may be an effective approach toward practical manipulation of plant carotenoids. We demonstrate here that hp1 alleles represent mutations in a tomato UV-DAMAGED DNA-BINDING PROTEIN 1 (DDB1) homolog. We further demonstrate that two tomato light signal transduction genes, LeHY5 and LeCOP1LIKE, are positive and negative regulators of fruit pigmentation, respectively. Down-regulated LeHY5 plants exhibit defects in light responses, including inhibited seedling photomorphogenesis, loss of thylakoid organization, and reduced carotenoid accumulation. In contrast, repression of LeCOP1LIKE expression results in plants with exaggerated photomorphogenesis, dark green leaves, and elevated fruit carotenoid levels. These results suggest genes encoding components of light signal transduction machinery also influence fruit pigmentation and represent genetic tools for manipulation of fruit quality and nutritional value.
Fleshy fruit ripening is a developmental process that has evolved to allow plants to use animal vectors for seed distribution. Ripening-associated color development assists in this process by rendering fruit attractive to participating organisms. One of the best-characterized systems of fruit color change and ripening is tomato (1).
The major pigments of ripe tomato fruit are carotenoids, including β-carotene and lycopene. β-Carotene is a pro-vitamin A compound, and lycopene has been associated through epidemiological studies with reduced incidence of degenerative diseases, including heart disease and cancer (2, 3). Although recent gene isolation efforts and metabolic engineering have primarily targeted genes involved in carotenoid synthesis (4-7), the molecular basis of factors that regulate flux through the carotenoid pathway remain largely unknown. Indeed, genetic analysis suggests that numerous loci in addition to carotenoid biosynthesis genes contribute to the variation in fruit pigmentation observed in tomato (8). Accumulating physiological evidence also suggests that light is involved in the fruit-ripening process, primarily impacting pigmentation (9, 10).
Although most tomato pigmentation mutants represent defects in carotenoid synthesis, several light-hyperresponsive mutants displaying elevated fruit carotenoid levels have been described in tomato. Specifically, mutants carrying the monogenic recessive high pigment (nonallelic hp1 and hp2) mutations are characterized by their exaggerated photoresponsiveness and increased fruit pigment (11-14). The high fruit pigmentation of these mutants is interesting in that the entire repertoire of fruit carotenoids is elevated (15), as opposed to the shifting of carotenoid profiles typical of biosynthetic mutants. This phenotypic feature has resulted in commercial utilization of these genes in breeding programs targeting fruit color and nutrient quality and suggests that targeted manipulation of light signal transduction may be a strategy for enhancement of fruit color and nutritional quality.
Although numerous genes encoding light-signaling functions have been identified in Arabidopsis (16, 17), their potential impact on fleshy fruit pigmentation and nutritional quality remains largely untested. Genetic analysis of the hp2 alleles revealed mutations in a negative regulator of light signal transduction, DE-ETIOLATED1 (DET1), originally defined in Arabidopsis (19, 20). Here we demonstrate that hp1 is a mutation in a tomato UV-DAMAGED DNA-BINDING PROTEIN 1 (DDB1) homolog whose Arabidopsis counterpart interacts with DET1 (21). We have additionally used RNA interference (RNAi)-mediated gene repression to repress two putative light signal transduction genes, LeHY5 and LeCOP1LIKE, which antagonistically regulate fruit pigmentation in tomato. These results demonstrate that genes encoding components of light signaling represent genetic tools for manipulation of fruit color and nutritional value.
Materials and Methods
Plant Growth Conditions. Tomato plants were grown in a naturally illuminated greenhouse under standard conditions (26°C day, 18°C night; 12 h watering cycle). Transgenic generation 1 (T1) segregating populations for LeHY5 and LeCOP1LIKE constructs were planted in the greenhouse and transplanted into the field 35 days later. For hypocotyl measurements, T2 populations and controls were germinated in water-agar in sterile jars under continuous white light or darkness at 25°C.
Plasmid Construction and Tomato Transformation. DNA manipulations were carried out by using standard procedures (22). Sequences from LeCOP1LIKE cDNA (GenBank accession no. AW625993) and LeHY5 cDNA (GenBank accession no. AI897283) for construction of RNAi vectors were amplified from plasmid DNA by PCR with primers designed for the LeCOP1LIKE gene (CLXbaIF, 5′-GTACAAAAATCTAGATTACATTAGATTTGTTGATGAACG-3′; CLSfiIR1, 5′-GTACGGCCATCTTGGCCAATGTGTTACCTTGTTGTATTGGCC-3′; CLSacIF, 5′-GTACAAAAAGAGCTCTTACATTAGATTTGTTGATGA ACG-3′; and CLSfiIR2, 5′-GTACGGCCTAGATGGCCAATGTGTTACCTTGTTGTATTGGCC-3′) or the LeHY5 gene (HY5XbaIF, 5′-GTACAAAAATCTAGAGGAGATGGGCGGAGAAGCGACGGG-3′; HY5SfiIR1, 5′-GTACGGCCATCTTGGCCCGGTTAAGAGTAGCAAGTATGC-3′; HY5SacIF, 5′-GTACAAAAAGAGCTCGGAGATGGGCGGAGAAGCGACGGG-3′; and HY5SfiIR2, 5′-GTACGGCCTAGATGGCCCGGTTAAGAGTAGCAAGTATGC-3′), introducing unique restriction sites at the product ends. To generate sequences with dyad symmetry, PCR products were made as follows: One product incorporated sequence in primer design to include an XbaI site at the 5′ end and SfiI site (top strand sequence GGCCATCTTGGCC) at the 3′ end. The second product incorporated sequences in primer design to include a SacI site at the 5′ end and a different SfiI site (top strand sequence GGCCTAGATGGCC) at the 3′ end. After digestion with SfiI, the two products were ligated. Dimers were digested with XbaI and SacI and ligated into vector pBI121. Constructs were introduced into tomato (cv. Ailsa Craig) by means of Agrobacterium transformation (23). Primers designed to the NPTII (Kanr) marker of pBI121 for confirmation of integration are 5′-GGCAATTACCTTATCCGCAA-3′ and 5′-AGAACTCGTCAAGAAGGCGA-3′.
Isolation and Hybridization of Nucleic Acids. Genomic DNA extraction, digestion, and hybridization were as described (24). Total RNA was purified as described (25). Total RNA (15 μg) was separated on denaturing agarose gels and blotted to Hybond N membrane and hybridized to radio-labeled probes by using protocols from Amersham Pharmacia. PCR fragments were labeled by random priming (26).
Isolation and Sequencing of cDNAs and Genomic Clones. As part of an ongoing effort to map light signal transduction HP1 candidates, a tomato DDB1 homolog (cLHT22D18, GenBank accession no. AW617366) was identified in the EST collection after publication of the Arabidopsis gene (21). Then 5′-RACE (Clontech) was performed with total tomato leaf RNA as template and with the oligonucleotide 5′-GATGTAGCCCGATCAATCTGCAATCTGG-3′ (5′ end of cLHT22D18). A 0.9-kb PCR product containing the 5′ end of the LeDDB1/HP1 cDNA was obtained by RT-PCR using the cLHT22D18-derived nested primer (5′-CGCTCTGTTGCGATGAAGAGAAGATC-3′) and the provided anchor primer (5′-TTCTAGAATTCAGCGGCCGC(dT)30-3′). The resulting product was cloned into pGEM-T (Promega) and sequenced. Full-length first-strand cDNA from the normal near-isolines (cvs. Ailsa Craig and GT) and the nearly isogenic hp1 and hp1w (also known as WB3) mutants, respectively, was synthesized by RT-PCR (Marathon kit, Clontech). LeDDB1/HP1 full-length cDNA was PCR amplified with primers 5′-ATTCAACTGAAACAAGTATTAGGGTT-3′ and 5′-GGGTTTGCAAATGATTCTTTTTCAACG-3′ by using PfuUltra DNA Polymerase (Stratagene) and cloned in pGEM-T. Six independent clones from each genotype were sequenced. Tomato genomic bacterial artificial chromosome clones from a 10× Lycopersicon cheesmannii genomic library (J.V. and J.G., unpublished data) were obtained by using 32P-labeled cLEG23L24 cDNA as probe (22). The LeDDB1/HP1 region of bacterial artificial chromosome clone LA483-94G3 was sequenced in steps by using cDNA-derived primers and resulting sequences to design subsequent primers. Sequencing was performed so that both strands were sequenced across the 23.5 kb of LeDDB1/HP1. The GenBank accession no. AY531660 is for Hp1 cDNA (including hp1 and hp1w alleles) and no. AY531661 is for Hp1 genomic.
Chlorophyll Assays. Tissue was ground in liquid N2. Total chlorophyll was extracted into 80% acetone, and chlorophyll a and b content was calculated by using MacKinney's coefficients, in which chlorophyll a equals 12.7(A663) - 2.69(A645) and chlorophyll b equals 22.9(A645) - 4.48(A663).
Extraction and Purification of Carotenoids. Fruit pericarp was homogenized in tetrahydrofuran in the presence of magnesium carbonate (1% of the sample weight). The resulting homogenate was extracted first with methanol/5% (wt/vol) butylated hydroxytoluene and subsequently with tetrahydrofuran until the tissue became colorless. Pigments were transferred to petroleum ether by adding 1/5 vol of ether and 1/10 vol of 25% NaCl to the combined tetrahydrofuran/methanol/butylated hydroxytoluene extract. The resulting ether fraction was dried and resuspended in methyl tert-butyl ether/methanol/butylated hydroxytoluene for HPLC analysis on a Dionex HPLC with photodiode array detector.
Microscopy. For conventional microscopy, samples were fixed for 0.5 h at 22°C plus 1.5 h at 4°C in 2.5% glutaraldehyde and 0.25 M sucrose in 0.1 M sodium cacodylate buffer (pH 6.8). Tissue was postfixed with 1% osmium tetroxide at 4°C overnight in the same buffer. Samples were dehydrated in a graded alcohol series and embedded in Spurr's resin. Blocks were sectioned on a Reichert OmU2 ultramicrotome. For plastid counts, ≈50 cells from each block were observed and at least 100 cells for each genotype were counted. Ultrathin sections were picked up on copper grids and contrasted with lead and uranyl acetate. Sections were viewed with a Tecnai 12 Biotwin transmission electron microscope (FEI, Hilsboro, OR). Digital images were taken with a Gatan multiscan camera (model 791, Pleasanton, CA).
Results
The Tomato HIGH-PIGMENT1 Gene Encodes a DDB1 Homolog. We have shown that hp1 maps to chromosome 2 near the 45S ribosomal repeat (12) and have since generated F2 mapping populations from crosses of the L. esculentum hp1/hp1 mutant to L. cheesmannii and Lycopersicon pennellii introgression lines (27) harboring introgressions spanning the hp1 locus. Putative light-signaling genes were mapped in the L. pennellii introgression lines to define approximate map positions (Y.L. and J.G., unpublished data). A subset of these mapping results is shown for chromosome 2 in Fig. 1. Putative light signal transduction loci that localized to chromosome 2 were mapped to high resolution in an F2 population segregating for hp1 (Fig. 1).
Fig. 1.
Genetic mapping of HP1 candidates. (Left) Mapping on the L. pennellii introgression lines of chromosome 2. The chromosome is drawn as an open bar and introgressed segments as solid bars to the right. The bins defined by the introgressions are designated by capital letters to the left. The hp1 locus and bin locations of candidate gene sequences are indicated to the right. (Right) Fine mapping of the hp1 locus and candidate genes in an F2 population of 7,850 individuals derived from the cross L. esculentum hp1/hp1 × L. pennellii introgression line IL2-1 (Hp1/Hp1); the number of recombinants in the F2 population between indicated loci is shown on the left.
Two tomato cDNAs with homology to DDB1 (cLEG23L24) and DIMINUTO/DWARF1 (DIM1, involved in brassinosteroid synthesis, which negatively impacts light signaling, ref. 28) showed linkage to hp1 (Fig. 1). Expression analysis and sequencing of tomato DIM homolog alleles from normal and hp1 lines revealed no expression or allelic differences, respectively (data not shown). Tomato cDNA cLEG23L24 cosegregated with hp1 (<0.1 centimorgan), and a full-length cDNA sequence was recovered by RACE-PCR. RTPCR products were also recovered from two independent and allelic hp1 mutants (11, 13), and subsequent sequencing revealed unique point mutations in each allele resulting in amino acid substitutions (Fig. 2).
Fig. 2.
Point mutations at the hp1 locus.
Tomato DDB1/HP1 genomic sequences were initially isolated from L. cheesmannii. The L. cheesmannii sequence was used to design PCR primers for overlapping regions of the corresponding L. esculentum (cv. Ailsa Craig) sequence. Alignment of the tomato DDB1 cDNA and genomic sequences revealed 18 introns, identical to the number in Arabidopsis DDB1A (21). The coding sequence of LeDDB1/HP1 in L. esculentum and L. cheesmannii is identical. Comparative protein sequence analysis between Arabidopsis DDB1A and the tomato homolog shows 86% identity and 92% similarity.
Tight genetic linkage of cLEG23L24 and hp1 combined with the presence of independent point mutations in two separate hp1 mutants strongly suggests that HP1 encodes a protein homologous to human DBB1 (29) and an Arabidopsis counterpart, DDB1A, whose product has been shown to interact with DET1 (21). Analysis of hp1 hp2 double mutants also suggests interaction of the corresponding proteins in tomato. Fig. 3A shows the seedling phenotype of the hp1 hp2 double mutant displays a synergistic effect. A similar effect has been described for the Arabidopsis det1 ddb1a double mutant, and it has been shown that the Arabidopsis DET1 and DDB1A proteins are capable of interaction (21). Viewed in the context of the Arabidopsis results, it seems most plausible that the HP1 and HP2 proteins are involved in the same pathway(s) and interact comparably to DET1 and DDB1A.
Fig. 3.
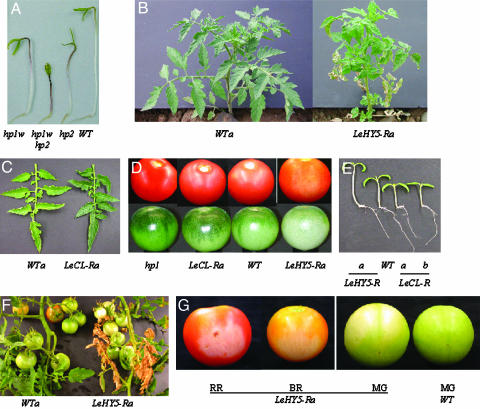
Normal, mutant, and transgenic plant phenotypes. R represents the presence of the relevant RNAi construct. (A) Representative 6-day (5-dark-grown days followed by 1 day in light) seedlings showing the additive effect of the hp1 hp2 double mutant. (B) Representative field-grown plants of LeHY5-deficient (LeHY5-Ra) and a nontransgenic segregant (WTa) 15 days after field transplant. (C) Mature leaves from representative field-grown plants from a LeCOP1LIKE-deficient line (LeCL-Ra and a nontransgenic segregant (WTa). (D) Mature green and red ripe fruits from field-grown plants of hp1, LeCOP1LIKE-deficient (LeCL-Ra), WT Ailsa Craig (WT), and LeHY5- deficient (LeHY5-Ra) lines. (E) Seedlings of LeHY5-deficient (Ra), WT Ailsa Craig (WT), and two LeCOP1LIKE-deficient (Ra and Rb) lines grown in white light. (F) Field-grown mature plants of LeHY5-deficient (Ra) and a nontransgenic segregant (WTa). (G) Representative mature green (MG), breaker (BR), and red ripe (RR) fruits, sun-exposed side forward, from field-grown plants of WT Ailsa Craig (WT) and a LeHY5-deficient (Ra) line.
In humans, DDB1 has been shown to participate in the initial damage response resulting from UV exposure by damage recognition, factor recruitment, and/or chromatin remodeling (30). Mutations altering this repair complex are the basis of xeroderma pigmentosum disease, resulting in increased incidence of skin cancer (29, 31, 32). Isolation of tomato and Arabidopsis DDB1 homologs participating in light response supports recent data suggesting light signal transduction in plants may involve chromatin remodeling (33).
Generation of Transgenic Plants Deficient in LeHY5 and LeCOP1LIKE Expression. To further test our hypothesis that manipulation of light signaling in fruit could be an effective means of modifying nutrient quality, we identified candidate positive and negative regulators of light response from the tomato EST collection. A candidate positive regulator of light signaling is a tomato HY5 homolog (GenBank accession no. AI897283), and a COP1-like gene was selected as a candidate negative regulator (GenBank accession no. AW625993). The later was chosen both to test our broader hypothesis on the potential for manipulating light signaling for nutrient modification and to test the function of an uncharacterized tomato COP1 family member.
LeHY5 encodes a predicted protein of 146 aa including a basic leucine zipper domain with characteristic leucine repeats. The LeHY5-predicted peptide shares 78% amino acid identity with Arabidopsis HY5. A blast search of predicted peptide sequences revealed that LeHY5 has a higher level of identity with HY5 (E = 1e-93) than any other predicted sequence in the Arabidopsis genome.
A blast search of the Arabidopsis peptide database revealed that LeCOP1LIKE has highest similarity (E = 8e-96) to a putative WD-repeat protein (GenBank accession no. NM_124604) also similar to COP1. It should be noted that tomato LeCOP1 (GenBank accession no. AF029984) is more homologous to Arabidopsis COP1 than LeCOP1LIKE and presumably represents the tomato COP1 ortholog. Functional characterization of LeCOP1 is ongoing (R. E. Kendrick and C.B., unpublished data). Functional characterization of LeCOP1LIKE presented an opportunity to characterize a participant in light signaling not yet reported in Arabidopsis.
To dissect the roles of LeHY5 and LeCOP1LIKE in fruit pigmentation and nutrient quality, we generated transgenic tomato plants expressing LeHY5 or LeCOP1LIKE RNAi. Constructs were introduced into normal tomatoes by Agrobacterium-mediated TDNA transfer. LeHY5 (36 of 41) and LeCOP1LIKE (25 of 28) primary transgenics (T0) resulted in PCR amplification with primers designed to the NPTII (kanamycin resistance) marker. DNA gel-blot analysis with the NPTII gene as probe confirmed PCR results (data not shown). In T1-segregating populations, we observed a correlation of transgene integration with LeHY5 progeny having pale green immature fruits and leaves (Fig. 3 B and D). In contrast, LeCOP1LIKE transformants had darker green leaves and fruits compared with both progeny segregating out the transgene and normal plants (Fig. 3 C and D).
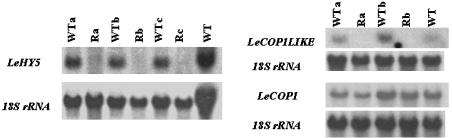
To ensure that observed phenotypes correlated with reduced endogenous transcript, total RNA was harvested from immature fruits. RNA gel-blot analysis using the LeHY5 cDNA as probe indicated a reduction in endogenous LeHY5 transcript levels in transformants (Fig. 4). Using the 3′ UTR of LeCOP1LIKE as probe, we also performed RNA gel-blot analysis demonstrating that LeCOP1LIKE mRNA was substantially reduced in transformed plants vs. nontransformed controls (Fig. 4).
Fig. 4.
Effects of LeHY5 (Left) and LeCOP1LIKE (Right) RNAi on gene expression. RNA gel-blots for LeHY5 and LeCOP1LIKE RNAi T1 progeny are shown. R represents the presence of the indicated transgene, whereas a, b, and c indicate progeny derived from independent transformation events. 32P-labeled probes to LeHY5 and LeCOP1LIKE were derived from sequences not used in the RNAi constructs to prevent cross-hybridization with transgene RNA. LeCOP1 is the most similar known homolog of LeCOP1LIKE and was used to demonstrate specificity of LeCOP1LIKE RNAi repression. All filters were stripped and reprobed with an 18S rRNA gene to control for RNA loading.
Because the WD-repeat domain is conserved within at least 10 members of the LeCOP1LIKE gene family in the tomato genome (www.tigr.org) and was included in the LeCOP1LIKE RNAi construct, it was possible that homologous transcripts also could be targets of RNAi. To test this hypothesis, we conducted RNA gel-blot analysis using LeCOP1 cDNA as probe. This gene is the closest available homolog of LeCOP1LIKE and is unaffected in the down-regulated LeCOP1LIKE lines (Fig. 4).
LeHY5 and LeCOP1LIKE Antagonistically Modify Hypocotyl Growth. To determine the in vivo roles of LeHY5 and LeCOP1LIKE in tomato photomorphogenic responses, we examined the effects of down-regulated gene expression on hypocotyl elongation. Seed was germinated from three LeHY5 (a, b, and c) and two LeCOP1LIKE (a and b)T2 lines in continuous white light or darkness, respectively, for 7 days with normal seed as control. When germinated in light, LeHY5-repressed seedlings showed a significant increase in hypocotyl growth, whereas LeCOP1LIKE demonstrated inhibited hypocotyl growth (Figs. 3E and 5A). Dark-grown seedlings showed no significant difference between LeHY5-repressed lines and the control, whereas LeCOP1LIKE had a small repressive effect. The LeHY5 results are consistent with HY5 loss-of-function mutants in Arabidopsis (34), suggesting that LeHY5 is a functional homolog of HY5.
Fig. 5.
Quantitation of LeHY5 and LeCOP1LIKE repression effects. Bars indicate SE. R indicates the presence of the indicated transgene. (A) Seedling hypocotyl lengths of WT Ailsa Craig (WT), three LeHY5-deficient (HY5-Ra, -b, and -c), and two LeCOP1LIKE-deficient (CL-Ra and -b) lines grown in white light or darkness. (B) Chlorophyll content in mature leaves of three LeHY5 (HY5-Ra,-b, and -c) and two LeCOP1LIKE (CL-Ra and b) RNAi lines. FW, fresh weight. (C) Chlorophyll content in mature green fruits of the same lines indicated in B. L and S designate pericarp tissues taken from the side of the fruit exposed to natural light and shade, respectively. (D) Total carotenoids in red ripe fruits of the same lines indicated in B. AU, arbitrary units.
The phenotype in LeCOP1LIKE-repressed seedlings is similar to that observed for the hp1 and hp2 mutations (11, 19, 35) in that little, if any, phenotypic variation is observed in the dark. The data for LeHY5- and LeCOP1LIKE-repressed lines suggest that these genes represent antagonistic components of tomato light signal transduction useful in assessing the potential for manipulating this regulatory pathway for modified fruit color and nutrient quality.
Fruit Chlorophyll Accumulation. To assess the impact of modified light signaling on maturing plant tissues, we examined the effects of reduced LeHY5 and LeCOP1LIKE expression on chlorophyll levels in T1 leaves and immature fruits. When grown in the greenhouse, LeHY5 RNAi leaves were paler than controls, whereas LeCOP1LIKE RNAi leaves were darker (data not shown). We observed more pronounced differences in greening of field-grown plants. LeHY5 RNAi plants had considerably lighter leaves and immature fruits than nontransformed progeny (Fig. 3 B and D). In contrast, LeCOP1LIKE RNAi plants had considerably darker leaves and fruit as compared with progeny that had segregated for transgene absence (Fig. 3 C and D). Fig. 5B shows that all three LeHY5 transgenic populations resulted in 24-31% reduction in leaf chlorophyll in transformed plants as compared with nontransgenic segregants. LeHY5 RNAi immature fruit showed greater reduction in chlorophyll accumulation, with the most severe phenotype associated with the side of the fruit exposed to direct sunlight (Fig. 5C). In addition, RNAi lines from the two LeCOP1LIKE transgenic populations demonstrated significantly higher leaf and fruit chlorophyll than corresponding nontransformed controls.
LeHY5 and LeCOP1LIKE Repression Significantly Modifies Fruit Carotenoid Content. We conducted HPLC analysis of carotenoids for three LeHY5- and two LeCOP1LIKE T1-segregating populations. Samples from three different ripe fruits from each plant were subjected to HPLC analysis. Total carotenoid levels in LeHY5-deficient fruit were consistently decreased (12-32%) compared with normal controls, whereas LeCOP1LIKE repression increased total fruit carotenoids 25-43% (Fig. 5D). This result is consistent with our hypothesis that modification of light signal transduction machinery may be an effective means of modifying fruit carotenoid content and associated nutrient quality.
LeHY5 Deficiency Causes Cell Death in Response to High-Intensity Irradiation. We observed cell death phenotypes in most of the field-grown LeHY5-deficient transformants at various developmental stages. This phenotype likely represents the impact of field intensity light exposure. Approximately 15 days after seedlings were transplanted into the field, plants with LeHY5 repression became pale and lower leaves displayed premature cell death (Fig. 3B). Although aging leaves in LeHY5-deficient plants succumbed before maturity, new leaf and shoot growth continued and led to production of flowers and mature fruit (Fig. 3F). The cell death phenotype occurred only in fruit tissues exposed to direct sunlight (Fig. 3G). This phenotype was greatly reduced in LeHY5 repression plants grown in the greenhouse, further supporting a role of light intensity on the severity of this response (data not shown). A recent report by Ulm et al. (36) demonstrates a similar response in Arabidopsis hy5 mutants.
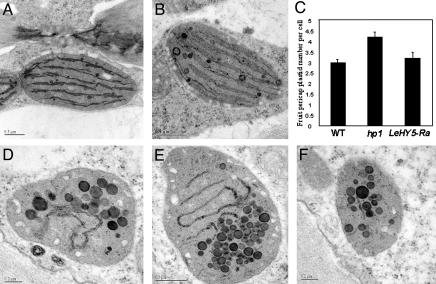
Alteration of Light Signaling in Fruit Impacts Plastid Number and Integrity. To more fully characterize the impact of manipulating light signaling in fruit, we examined fruit pericarp plastids (immature green stage, 20 days post anthesis) from normal, hp1, and LeHY5 RNAi lines by transmission electron microscopy (Fig. 6). Comparison of normal to hp1 plastids indicates typical thylakoid structure and grana stacking in the mutant and nearly isogenic normal lines. However, plastid counts indicates a 30% increase in plastid number in hp1. Increased plastid numbers in hp1 is consistent with previous molecular and microscopy studies (12, 15). LeHY5 RNAi lines had normal plastid numbers but clear deficiencies in both organization and abundance of thylakoids. In addition, LeHY5 repressed lines had abundant osmium staining plastiglobuli (lipid droplets), presumably representing continued lipid synthesis in the absence of thylakoid formation. These abnormalities in thylakoid organization are consistent with the fact that Arabidopsis HY5 is required for transcription of the CAB gene, whose product is necessary for thylakoid organization (37, 38). In summary, these results suggest that hyper light activation as observed from mutation in a negative regulator of light signaling (LeDDB1/HP1) increases carotenoid accumulation in part by means of increased plastid numbers. In contrast, repression of a positive regulator of light signaling (LeHY5) has little impact on basic plastid number but results in considerable reduction of thylakoid organization, which may result in reduced capacity for carotenoid accumulation.
Fig. 6.
Transmission electron microscopy of tomato fruit pericarp chloroplasts. Typical thylakoids including grana stacks can be observed in WT (cv. Ailsa Craig) (A) and hp1 (B). (D-F) LeHY5 RNAi lines showing deficiencies in both organization and abundance of thylakoids and accumulation of plastoglobuli. Samples shown are from LeHY5-Ra.(C) Relative plastid numbers in these same lines.
Discussion
Carotenoids are components of photosynthetic membranes in all plants, algae, and cyanobacteria and serve critical functions in plant biology, including light harvesting, quenching of photooxidation, and coloring of flowers and fruits. Dietary carotenoids derived from plants serve roles as necessary nutrients (e.g., pro-vitamin A) and antioxidants. Epidemiological studies indicate that high dietary intake of carotenoid-rich foods can decrease the incidence of degenerative diseases, including coronary heart disease, certain cancers, and macular degeneration.
Although the genes responsible for carotenoid biosynthesis are largely defined, little is known about the molecular mechanisms that regulate carotenoid accumulation. Indeed, previous efforts to manipulate carotenoid accumulation, particularly early in the pathway, have resulted in plant phenotypes having a negative impact on crop performance as a result of consequences on phytohormones (e.g., abscisic acid and gibberellic acid) synthesized from the same precursors (39). Although “golden rice” was a clear technical success with the potential for real-world benefit (40), the realization of lower than anticipated carotenoid levels may reflect uncharacterized regulatory constraints on synthesis and/or accumulation. Phenotypic analysis of the tomato hp mutations (12) suggests that light signal transduction regulates the carotenoid pathway in ways that can have an impact on total fruit carotenoid content. Here we describe the cloning of the tomato gene altered in the hp1 mutant and demonstrate that it is homologous to sequences involved in light response. We also report the isolation of two tomato light signal transduction genes (LeHY5 and LeCOP1LIKE) and confirm the potential for manipulation of light signaling for improved nutrient quality.
HP1 Is a DDB1 Homolog. Genetic mapping of tomato putative light signal transduction loci indicated that a tomato DDB1 gene was indeed linked to the hp1 locus (Fig. 1). Sequencing of RT-PCR products from two separate allelic mutations in hp1 indicated that each had a unique point mutation resulting in amino acid substitution (Fig. 2). This result has been recently confirmed in an independent study (41). Analysis of hp1 hp2 double mutant phenotypes suggests synergistic effects on light signaling (Fig. 3A), consistent with documented interaction of Arabidopsis DET1 and DDB1A (21). Efforts to complement the hp1 mutant with the normal tomato LeDDB1/HP1 cDNA under direction of the cauliflower mosaic virus-35S promoter failed to yield viable plants, whereas numerous simultaneous and unrelated transformation experiments resulted in many transformants, suggesting that ec-topic LeDDB1/HP1 expression may be lethal (data not shown).
LeHY5 and LeCOP1LIKE Encode Positive and Negative Regulators of Light Signaling, Respectively. Manipulation of LeHY5 and LeCOP1LIKE results in altered photomorphogenic response and fruit carotenoid content. Arabidopsis HY5 has been genetically defined as a positive regulator of light signaling that acts downstream of photoreceptors (42, 43). HY5 has been shown to encode a basic leucine zipper transcription factor required for light regulation of cell elongation, proliferation, and chloroplast development (34). Repression of LeHY5 by RNAi results in elongated hypocotyls and reduced chlorophyll similar to Arabidopsis hy5 mutations, suggesting that LeHY5 encodes a similar positive regulator of light signaling in tomato (Fig. 3).
A combination of genetic and biochemical approaches has implicated that Arabidopsis HY5 functionally interacts with COP/DET/FUS gene products, which are pleiotropic negative regulators of photomorphogenesis (38, 42-45). Arabidopsis COP1 is a well-characterized photomorphogenesis repressor capable of directly interacting with HY5 to facilitate targeted degradation of HY5 in the dark. The region of COP1 physically interacting with HY5 encompasses the WD repeat (46). The function of WD-repeat domains in photomorphogenic repressors is reiterated by the discovery of another WD-repeat protein SPA1 that also acts as a negative regulator of photomorphogenic development (47-49).
Sequence alignment suggests a tomato homolog of the Arabidopsis COP1 gene more similar to COP1 than LeCOP1LIKE (GenBank accession no. AW6259993) and which is currently the focus of functional characterization (C.B., unpublished data). Here we demonstrate that LeCOP1LIKE functions as a repressor of photomorphogenesis in tomato. LeCOP1LIKE repression additionally elevates the level of carotenoids during fruit ripening (Fig. 5D). Targeting genes operating in signaling pathways that have evolved to modulate activity of pathways of interest in biologically viable ways (as with LeCOP1LIKE and light signaling) may ultimately prove equally or more effective in yielding viable crops with targeted metabolite alterations than direct pathway manipulation.
Acknowledgments
We are grateful to S. Caldwell and M. V. Parthasarathy of the Cornell Integrated Microscopy Center for assistance with electron microscopy and plastid counts. We especially thank Y. Liu, R. McQuinn, R. Alba, Z. Fei, X. Tang, B. Anible, and R. White for technical assistance. This work was supported by grants from the U.S. Department of Agriculture National Research Initiative (00-35300-9356) and the National Science Foundation (DBI-0116076 and DBI-0211875) to J.G. and colleagues and from the European Union (QLK5-CT-2000-00357) to C.B.
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: RNAi, RNA interference.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY531660 and AY531661).
References
- 1.Hirschberg, J. (2001) Curr. Opin. Plant Biol. 4, 201-218. [DOI] [PubMed] [Google Scholar]
- 2.Hwang, E. S. & Bowen, P. E. (2002) Integr. Cancer Ther. 1, 121-132. [DOI] [PubMed] [Google Scholar]
- 3.Murtaugh, M. A., Ma, K. N., Benson, J., Curtin, K., Caan, B. & Slattery, M. L. (2004) Am. Epidemiol. 159, 32-41. [DOI] [PubMed] [Google Scholar]
- 4.Ronen, G., Carmel-Goren, L., Zamir, D. & Hirschberg, J. (2000) Proc. Natl. Acad. Sci. USA 97, 11102-11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacson, T., Ronen, G., Zamir, D. & Hirschberg, J. (2002) Plant Cell 14, 333-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romer, S., Fraser, P. D., Kiano, J. W., Shipton, C. A., Misawa, N., Schuch, W. & Bramley, P. M. (2000) Nat. Biotechnol. 18, 666-669. [DOI] [PubMed] [Google Scholar]
- 7.Rosati, C., Aquilani, R., Dharmapuri, S., Pallara, P., Marusic, C., Tavazza, R., Bouvier, F., Camara, B. & Giuliano, G. (2000) Plant J. 24, 413-419. [DOI] [PubMed] [Google Scholar]
- 8.Liu, Y. S., Gur, A., Ronen, G., Causse, M., Damidaux, R., Buret, M., Hirschberg, J. & Zamir, D. (2003) Plant Biotechnol. J. 1, 195-207. [DOI] [PubMed] [Google Scholar]
- 9.Alba, R., Cordonnier-Pratt, M. M. & Pratt, L. H. (2000) Plant Physiol. 123, 363-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannoni, J. (2004) Plant Cell, in press. [DOI] [PMC free article] [PubMed]
- 11.Peters, J. L., van Tuinen, A., Adamse, P., Kendrick, R. E. & Koornneef, M. (1989) J. Plant Physiol. 134, 661-666. [Google Scholar]
- 12.Yen, H. C., Shelton, B. A., Howard, L. R., Lee, S., Vrebalov, J. & Giovannoni, J. (1997) Theor. Appl. Genet. 95, 1069-1079. [Google Scholar]
- 13.Kerckhoffs, L. H. J., De Groot, N. A. M. A., Van Tuinen, A., Schreuder, M. E. L., Nagatani, A., Koornneef, M. & Kendrick, R. E. (1997) J. Plant Physiol. 150, 578-587. [Google Scholar]
- 14.van Tuinen, A., Cordonnier-Pratt, M. M., Pratt, L. H., Verkerk, R., Zabel, P. & Koorneef, M. (1997) Theor. Appl. Genet. 94, 115-122. [DOI] [PubMed] [Google Scholar]
- 15.Cookson, P. J., Kiano, J. W., Shipton, C. A., Fraser, P. D., Romer, S., Schuch, W., Bramley, P. M. & Pyke, K. A. (2003) Planta 217, 896-903. [DOI] [PubMed] [Google Scholar]
- 16.Neff, M. M., Fankhauser, C. & Chory, J. (2000) Genes Dev. 14, 257-271. [PubMed] [Google Scholar]
- 17.Wang, H. & Deng, X. W. (2003) Trends Plant Sci. 8, 172-178. [DOI] [PubMed] [Google Scholar]
- 18.Pepper, A., Delaney, T., Washburn, T., Poole, D. & Chory, J. (1994) Cell 78, 109-116. [DOI] [PubMed] [Google Scholar]
- 19.Mustilli, A. C., Fenzi, F., Ciliento, R., Alfano, F. & Bowler, C. (1999) Plant Cell 11, 145-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin, I., Frankel, P., Gilboa, N., Tanny, S. & Lalazar, A. (2003) Theor. Appl. Genet. 106, 454-460. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder, D. F., Gahrtz, M., Maxwel, B. B., Cook, R. K., Kan, J. M., Alonso, J. M., Ecker, J. R. & Chory, J. (2002) Curr. Biol. 12, 1462-1472. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 23.Fillatti, J. J., Kiser, J., Rose, R. & Comai, L. (1987) Bio/Technology 5, 726-730. [Google Scholar]
- 24.Zamir, D. & Tanksley, S. D. (1988) Mol. Gen. Genet. 213, 254-261. [Google Scholar]
- 25.Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., Schuch, W. & Giovannoni, J. (2002) Science 296, 343-346. [DOI] [PubMed] [Google Scholar]
- 26.Tanksley, S. D., Ganal, M. W., Prince, J. P., de Vicente, M. C., Bonierbale, M. W., Broun, P., Fulton, T. M., Giovannoni, J. J., Grandillo, S., Martin, G. B., et al. (1992) Genetics 132, 1141-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan, Q., Liu, Y. S., Budai-Hadrian, O., Sela, M., Carmel-Goren, L., Zamir, D. & Fluhr, R. (2000) Genetics 155, 309-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klahre, U., Noguchi, T., Fujioka, S., Takatsuto, S., Yokota, T., Nomura, T., Yoshida, S. & Chua, N. H. (1998) Plant Cell 10, 1677-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu, G. & Chang, E. (1988) Science 242, 564-567. [DOI] [PubMed] [Google Scholar]
- 30.Wakasugi, M., Kawashima, A., Morioka, H., Linn, S., Sancar, A., Mori, T., Nikaido, O. & Matsunaga, T. (2002) J. Biol. Chem. 277, 1637-1640. [DOI] [PubMed] [Google Scholar]
- 31.Kazantsev, A., Mu, D., Nichols, A. F., Zhao, X., Linn, S. & Sancar, A. (1996) Proc. Natl. Acad. Sci. USA 93, 5014-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keeney, S., Chang, G. J. & Linn, S. (1993) J. Biol. Chem. 268, 21293-21300. [PubMed] [Google Scholar]
- 33.Benvenuto, G., Formiggini, F., Laflamme, P., Malakhov, M. & Bowler, C. (2002) Curr. Biol. 12, 1529-1534. [DOI] [PubMed] [Google Scholar]
- 34.Oyama, T., Shimura, Y. & Okada, K. (1997) Genes Dev. 11, 2983-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters, J. L., Szell, M. & Kendrick, R. E. (1998) Plant Physiol. 117, 797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulm, R., Baumann, A., Oravecz, A., Mate, Z., Adam, E., Oakeley, E. J., Schafer, E. & Nagy, F. (2004) Proc. Natl. Acad. Sci. USA 101, 1397-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chattopadhyay, S., Ang, L. H., Puente, P., Deng, X. W. & Wei, N. (1998) Plant Cell 10, 673-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maxwell, B. B., Andersson, C. R., Poole, D. S., Kay, S. A. & Chory, J. (2003) Plant Physiol. 133, 1565-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fray, R., Wallace, A., Fraser, P., Valero, D., Hedden, P., Bramley, P. & Grierson, D. (1995) Plant J. 8, 693-701. [Google Scholar]
- 40.Beyer, P., Al-Babili, S., Ye, X., Lucca, P., Schaub, P., Welsch, R. & Potrykus, I. (2002) J. Nutr. 132, 506S-510S. [DOI] [PubMed] [Google Scholar]
- 41.Lieberman, M., Seveg, O., Gilboa, N., Lalazar, A. & Levin, I. (2004) Theor. Appl. Genet. 108, 1574-1581. [DOI] [PubMed] [Google Scholar]
- 42.Koornneef, M., Rolff, E. & Spruit, C. J. P. (1980) Z. Pflanzenphysiol. 100, 147-160. [Google Scholar]
- 43.Ang, L. H. & Deng, X. W. (1994) Plant Cell 6, 613-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chory, J., Peto, C., Feinbaum, R., Pratt, L. & Ausubel, F. (1998) Cell 58, 991-999. [DOI] [PubMed] [Google Scholar]
- 45.Pepper, A. E. & Chory, J. (1997) Genetics 145, 1125-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holm, M., Hardtke, C. S., Gaudet, R. & Deng, X. W. (2001) EMBO J. 20, 118-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoecker, U., Tepperman, J. M. & Quail, P. H. (1999) Science 284, 496-4999. [DOI] [PubMed] [Google Scholar]
- 48.Seo, H. S., Yang, J. Y., Ishikawa, M., Bolle, C., Ballesteros, M. L. & Chua, N. H. (2003) Nature 424, 995-999. [DOI] [PubMed] [Google Scholar]
- 49.Saijo, Y., Sullivan, J. A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U. & Deng, X. W. (2003) Genes Dev. 17, 2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]