Abstract
We have generated transgenic rice plants expressing the Datura stramonium adc gene and investigated their response to drought stress. We monitored the steady-state mRNA levels of genes involved in polyamine biosynthesis (Datura adc, rice adc, and rice samdc) and polyamine levels. Wild-type plants responded to the onset of drought stress by increasing endogenous putrescine levels, but this was insufficient to trigger the conversion of putrescine into spermidine and spermine (the agents that are believed to protect plants under stress). In contrast, transgenic plants expressing Datura adc produced much higher levels of putrescine under stress, promoting spermidine and spermine synthesis and ultimately protecting the plants from drought. We demonstrate clearly that the manipulation of polyamine biosynthesis in plants can produce drought-tolerant germplasm, and we propose a model consistent with the role of polyamines in the protection of plants against abiotic stress.
Abiotic stresses such as drought represent some of the most significant constraints to agricultural productivity. Transgenic approaches can be used in combination with conventional breeding strategies to create crops with enhanced drought tolerance, and one way in which this can be achieved is through the manipulation of polyamine metabolism. Polyamines are small, ubiquitous, nitrogenous compounds that have been implicated in a variety of stress responses in plants (1). The link between polyamines and abiotic stress was first documented through putrescine accumulation in response to suboptimal potassium levels in barley (2). Since then, a connection has been suggested between increased putrescine levels and abiotic stress (3). Similar phenomena have been described in animals, e.g., during ischemic and postischemic responses in neurons (4). The physiological role of putrescine in abiotic stress responses is a matter of controversy. It has been very difficult to establish a direct cause-and-effect relationship between increased putrescine levels in plants and abiotic stress. Elevated putrescine might be the cause of stress-induced injury or, alternatively, a protective response resulting from stress (5).
The genetic manipulation of polyamine metabolism has become a valuable tool for studying their physiological roles in plants (6). Plant polyamine content has been modulated by the overexpression/down-regulation of arginine decarboxylase (adc), ornithine decarboxylase (odc), and S-adenosylmethionine decarboxylase (samdc) (6-10). Overexpression of heterologous adc or odc cDNAs in plants generally results in the production of high levels of putrescine (11-13). In most cases, this is accompanied by a relatively small increase in spermidine and spermine concentrations (7, 14). Such findings suggest that the levels of spermidine and spermine are under strict homeostatic regulation (15). Therefore, the study of plants transformed with genes involved in polyamine biosynthesis may shed light on the importance of polyamines, their role in the acquisition of stress tolerance, and relevant stress tolerance mechanisms. In this report, we discus the response of transgenic rice plants with increased putrescine levels to drought stress. We exposed plants expressing the Datura stramonium adc gene to drought stress induced by 20% polyethylene glycol (PEG). We monitored transcript and polyamine levels as well as physiological responses in these plants compared with nontransgenic controls. We propose a model consistent with a mechanism linking polyamine metabolism to drought tolerance and possibly tolerance of other forms of abiotic stress.
Materials and Methods
Gene Transfer and Plant Regeneration. The 2,916-bp D. stramonium adc cDNA, containing the 5′ untranslated sequence and coding region, was excised as an XhoI fragment from pBluescriptII SK+/- (GenBank accession no. AJ251819), trimmed with exonuclease, and digested with SmaI. The SmaI/blunt-end fragment was sub-cloned into the SmaI site of pAL76 (16), which contains the maize ubiquitin 1 (Ubi-1) promoter and first intron and an Agrobacterium tumefaciens nos transcriptional terminator. The plasmid was named Ubi:Dadc. Rice transformation, selection, and plant regeneration were as described in refs. 17 and 18.
Drought Stress Treatment. We carried out preliminary studies using 2-month-old, nontransgenic plants to determine the optimum age of plants for subsequent stress experiments, a suitable PEG concentration, and the most appropriate stress duration. Treatment with 20% PEG (Mr 8,000) (19) for 6 days was sufficient to give a clear phenotypic response; therefore, these conditions were chosen for subsequent experiments with wild-type and transgenic plants.
Plants were grown in the greenhouse at 26 ± 2°C for a 12-h photoperiod (900 μmohn m-2 s-1 photosynthetically active radiation) at 80% relative humidity. After 60 days, the water was replaced with the PEG solution. Leaf samples were collected 0, 3, and 6 days after PEG treatment. The PEG solution was then replaced with water. The plants were allowed to recover for 48 h, and final samples were collected.
PCR Analysis. Genomic DNA was extracted from leaves as described (20). PCR amplification to detect the Datura adc transgene was carried out in a total volume of 25 μl [50 ng of genomic DNA/1× Roche PCR buffer (50 mM KCl/10 mM Tris·HCl, pH 8.3)/1.5 mM MgCl2/200 μM of each dNTP/50 nM of each primer/1.25 units of Roche TaqDNA polymerase]. The forward primer (pDadc-1, 5′-CGCCGCTGTTTCCCCTCCTC-3′) started at position 459 in the Datura adc cDNA. The sequence of the reverse primer, pDadc-2, was 5′-CATACCAGACTCATCCAGCT-3′. We carried out 35 amplification cycles: denaturation (94°C, 40 s), annealing (58°C, 30 s), and extension (72°C, 60 s). The 0.8-kb product was visualized after agarose gel electrophoresis [1% Tris-borate/EDTA (TBE)].
DNA and RNA Gel Blot Analyses. After KpnI digestion and electrophoresis on a 0.8% TBE agarose gel (21), 15 μg of DNA was transferred to a positively charged nylon membrane (Roche). Nucleic acids were fixed by UV crosslinking. Filters were washed in 2× SSC for 30 min and prehybridized at 42°C for 2 h by using the DIG-easy hybridization solution (Roche). The Datura adc PCR product described above was used as the probe. Labeling of the probe, hybridization, washes, and chemiluminescent detection were carried out as described in ref. 10. Membranes were then incubated with CSPD Chemiluminescent Substrate (Roche) and exposed to x-ray film (Fuji) for 30 min at 37°C.
Total RNA was extracted by using the RNeasy Plant Mini Kit (Qiagen, London). Denatured RNA (30 μg) from leaves was fractionated by 1.2% agarose-formaldehyde gel electrophoresis by using 1× Mops buffer (21). Hybridization with the Datura adc probe was carried out as described above. Probes for the rice adc and samdc genes were generated by PCR as described (10), and reprobing was performed as described in ref. 22. Membranes were exposed to x-ray film for 30 min at 37°C in the case of the rice adc and samdc genes and for 20 min in the case of the Datura adc gene.
RNA experiments were carried out twice by using independently isolated RNA samples. Steady-state mRNA hybridization signals were quantified by using quantityone quantification software (Bio-Rad), and the resulting values were normalized by using values obtained from RNA loading levels. Averages (mean of two samples ± SE) of duplicate hybridization signals were used to generate graphs. Blots represent typical results of two independent experiments.
Polyamine Analysis. Polyamine analysis and quantification were as described in refs. 14 and 23. Results were expressed as nmol g-1 fresh weight (fw).
Statistical Analysis. Hpt-transformed plants and wild type (average of three samples each from six independent lines; n = 36) were used as controls for polyamine content. Hpt-resistant transformants and wild-type values were not significantly different (P > 0.05) in terms of polyamine levels. As control values for polyamine content under drought stress we used hpt-transformed plants and wild-type controls (average of three samples each from two independent lines; n = 12). The hpt transformants and wild-type values were not significantly different (P > 0.05) under drought stress. To determine polyamine content in plants transformed with the Ubi:Dadc gene we used the average value of three samples from each plant (n = 3). To determine polyamine contents in transgenic plants under drought stress we used the average value of three samples for each time point (n = 3). The stress experiment was performed twice by using progeny plants (R1) derived from the same clone(s). We analyzed the data by two-way ANOVA followed by a t test using the residual mean square in the ANOVA as the estimate of variability.
Results
The Polyamine Profile of Transgenic Rice Plants Expressing Ubi:Dadc Is Altered Compared with Wild-Type Plants. We generated 50 independent transgenic rice lines containing Ubi:Dadc, and we confirmed the presence of the transgene by genomic DNA gel blot analysis. Each line showed a unique integration pattern, confirming that the plants originated from independent transformation events (see Fig. 1A). Transgene expression was monitored at the mRNA level. Although 84% (42 of 50) of the plants accumulated the Datura adc transcript, this was expressed at different levels in different lines (see Fig. 1B).
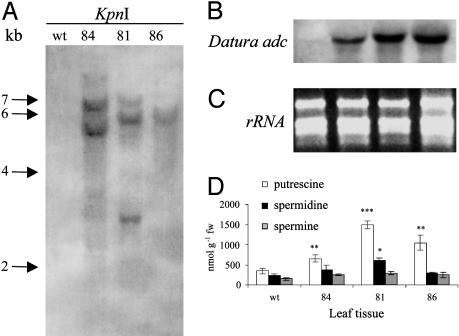
Fig. 1.
Molecular and biochemical characterization of transgenic plants carrying the Dadc transgene. (A) Gel blot analysis of KpnI-digested genomic rice DNA. 84, 81, and 86 represent independent transgenic lines; wt, wild type. (B) Gel blot of total leaf RNA from wild-type and independent lines containing Ubi:Dadc. Blots in A and B were probed with a 0.8-kb DIG-labeled PCR product from Ubi:Dadc. (C) Ethidium bromide gel demonstrating equal loading of total RNA. (D) Polyamine content. Values are means ± SE for control lines (n = 36) and means ± SE in transgenic lines (n = 3). Significance of data for putrescine and spermidine is as follows: ***, P < 0.001; **, 0.01 > P > 0.001; *, 0.05 > P > 0.01; remaining values were not significantly different from wild type at P > 0.05.
We measured the free polyamine content in transgenic leaves to determine whether expression of the Ubi:Dadc construct had any effect on the titers of putrescine, spermidine, and spermine. Our results indicated that putrescine levels were increased significantly in most of the Dadc-expressing lines (65%; 27 of 42 plants). Increases varied from 2-fold in plant 84 (648.02 ± 88 nmol g-1 fw, P < 0.01) to 4-fold in plant 81 (1,495.72 ± 100 nmol g-1 fw, P < 0.001) compared with wild-type plants and hpt-transformants (344.72 ± 66 nmol g-1 fw; Fig. 1D). Plant 81 also exhibited a significant increase in spermidine levels (605.38 ± 68 nmol g-1 fw, P < 0.05) compared with wild type (235.78 ± 38 nmol g-1 fw). Three more plants with putrescine levels similar to plant 81 had a significant increase in spermidine content. No significant variation (P > 0.05) in spermidine was observed in any of the remaining lines (see Fig. 1D). No significant variation in spermine was observed in any of the lines (Fig. 1D).
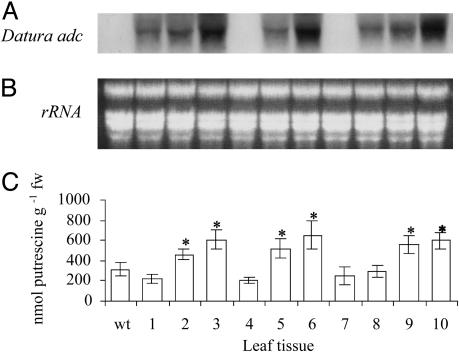
We determined the steady-state mRNA and putrescine levels in 20 phenotypically normal progeny plants (R1) from a number of clones with elevated putrescine levels. The progeny from plant 84 were typical and were selected for more detailed studies. We observed that the Ubi:Dadc transgene segregated with an ≈3:1 ratio, confirming that the adc transgene had integrated at a single genetic locus. R1 progeny had rather simple integration patterns that were identical to those of the primary transformants. Progeny from the same line accumulated the Datura adc transcript at different levels (Fig. 2A). The putrescine content was measured in the R1 progeny. Six of the 10 transgenic plants had a significant increase in putrescine content, ranging from 1.5- to 2-fold (450.36 ± 54 nmol g-1 fw to 650.16 ± 140 nmol g-1 fw) compared with wild type (315.67 ± 66 nmol g-1 fw; Fig. 2C). All 10 progeny plants were used in the stress experiments described below. Transgenic plants with 1.5- to 2-fold increases in putrescine compared with the wild type showed very similar stress responses. We selected progeny plant 84-2 for illustration purposes. This plant was homozygous for the transgene (as determined by screening 25 individual R2 progeny for the presence of the Dadc transgene).
Fig. 2.
Molecular and biochemical characterization of R1 progeny of clone 84. (A) Gel blot analysis of total RNA from leaf tissue. wt, wild type. Numbers represent different R1 progeny from plant 84. A 0.8-kb DIG-labeled PCR probe from Datura adc cDNA was used. (B)UV fluorescence of an ethidium bromide-stained gel showing equal loading of total RNA from plants used in the hybridizations shown in A.(C) Cellular putrescine content. Significance of data for putrescine is as follows: *, 0.05 > P > 0.01; remaining values are not significantly different from wild type at P > 0.05.
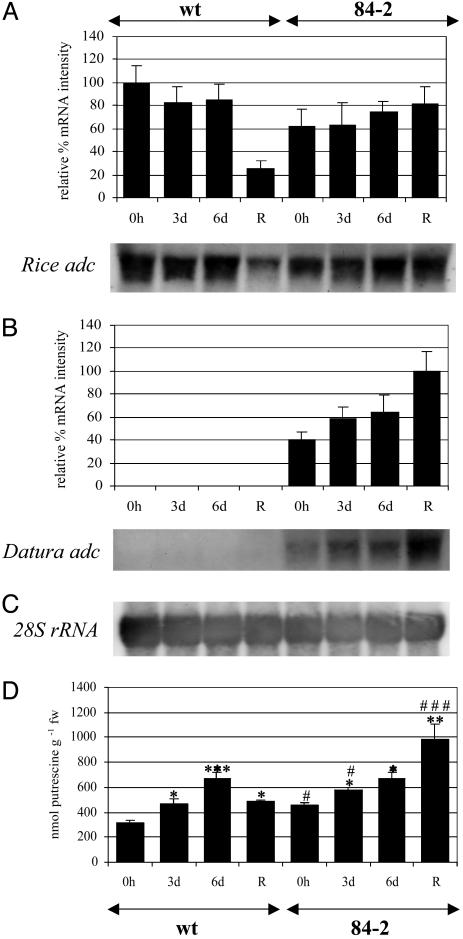
Differential Expression of the Endogenous and Heterologous adc Genes in Rice. The steady-state level of the rice adc mRNA in wild-type plants did not change after 3 or 6 days of drought stress (Fig. 3A). Two days after stress withdrawal, the rice adc mRNA declined sharply to ≈25% of the level at t = 0 (Fig. 3A). The steady-state level of the rice adc mRNA in plant 84-2 was comparable to the levels observed in wild-type plants after 3 or 6 days of stress (Fig. 3A). Two days after stress withdrawal, the rice adc steady-state mRNA level in transgenic plants was maintained at levels similar to those measured at t = 0 and t = 6 days after stress induction (Fig. 3A). Steady-state Dadc mRNA levels were increased progressively under drought and remained high for 2 days after stress withdrawal (Fig. 3B).
Fig. 3.
Molecular and biochemical characterization of wild-type and transgenic plants under drought stress. (A) Normalization of rice adc mRNA hybridization signals and quantitation as described in Materials and Methods. (B) Normalization of Datura adc mRNA hybridization signals and quantitation as described in Materials and Methods. wt, wild type (progeny from plant 84-2). The 400-bp and 800-bp DIG-labeled PCR probes from rice and Datura adc cDNAs were used. Each blot represents a typical result of two independent experiments. (C) Membrane was reprobed with the 28S rRNA probe to demonstrate equal loadings. (D) Putrescine content. Values are means ± SE for control lines (n = 12) and means ± SE for transgenic lines (n = 3). Significance of data for putrescine among plants of the same line is designated by *, and significance between transgenic lines and wild-type plants at the same time period is designated by #. Significance of data for putrescine is as follows: ***, P < 0.001; **, 0.01 > P > 0.001; *, 0.05 > P > 0.01; remaining values were not significantly different from wild type at P > 0.05.
Putrescine levels in wild-type plants under drought stress increased significantly by day 3, reaching a maximum by day 6 (1.5-fold increase; 667.97 ± 47 nmol g-1 fw, P < 0.05; Fig. 3D). Two days after stress withdrawal wild-type plants showed a small but significant increase in putrescine (490.87 ± 11 nmol g-1 fw, P < 0.05; Fig. 3D) compared with the same plants at t = 0 (309.65 ± 28 nmol g-1 fw). In contrast, transgenic plants expressing the Ubi:Dadc transgene showed a significant and progressive increase in putrescine levels upon stress induction (456.77 ± 25 nmol g-1 fw at t = 0; 573.92 ± 28 nmol g-1 fw after 3 days, P < 0.05; 673.75 ± 49 nmol g-1 fw after 6 days, P < 0.05; and 985.87 ± 115 nmol g-1 fw after stress withdrawal, P < 0.01; all compared with t = 0 transgenic plant; Fig. 3D).
When statistical comparisons were made between wild-type and transgenic lines over the same time period, a small but significant increase in putrescine content was observed by 3 days after stress induction. This value was higher in the transgenic plants (573.92 ± 28 nmol g-1 fw compared with 471.15 ± 36 nmol g-1 fw in the wild type, P < 0.05; Fig. 3D). Interestingly, both transgenic and wild-type populations contained very similar putrescine levels 6 days after the onset of drought stress (no significant difference at P > 0.05; Fig. 3D). After recovery, a 2-fold increase in putrescine content was observed in the transgenic plants compared with the wild type (985.87 ± 115 nmol g-1 fw compared with 490.87 ± 11 nmol g-1 fw, P < 0.001).
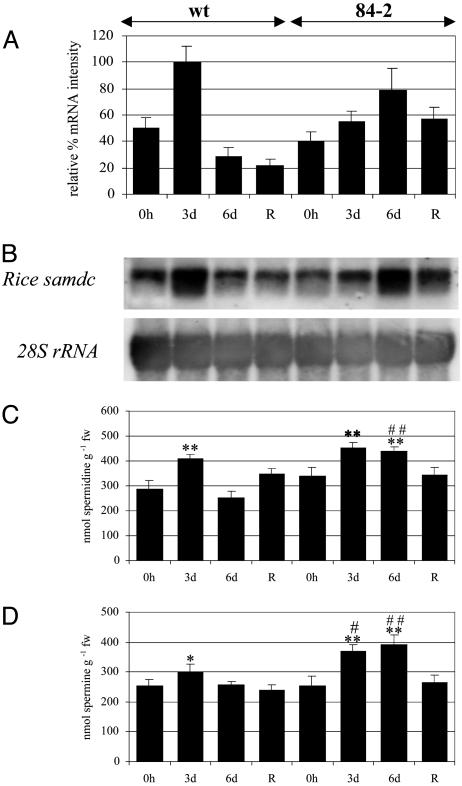
Differential Expression of the Rice samdc Gene in Wild-Type and Transgenic Plants Under Drought Stress. The steady-state levels of rice samdc mRNA increased 3 days after the induction of drought stress in wild-type plants but declined by day 6, stabilizing 2 days after stress withdrawal (Fig. 4A). In the Dadc-transgenic plants, the steady-state levels of rice samdc mRNA were similar to those of the wild type at t = 0 but were slightly up-regulated after 3 days of drought stress, reaching a maximum after 6 days. The levels of rice samdc mRNA had declined in these plants 2 days after stress withdrawal compared with levels measured after 6 days of drought stress (Fig. 4A).
Fig. 4.
Molecular and biochemical characterization of wild-type and transgenic plants under drought stress. (A) Normalization of rice samdc mRNA hybridization signals and quantitation as described in Materials and Methods. Each blot represents a typical result of two independent experiments. (B) Membrane was reprobed with the 28S rRNA probe to determine equal loadings. Spermidine (C) and spermine (D) levels in wild-type and transgenic plants after different periods of drought stress are shown. Values are means ± SE for wild type (n = 12) and means ± SE in transgenic lines (n = 3). Significance of data among the same line is designated by *, and significance between transgenic lines and wild-type controls at the same time period is designated by #. Significance of data for spermidine and spermine is as follows: **, 0.01 > P > 0.001; *, 0.05 > P > 0.01; remaining values were not significantly different from wild type at P > 0.05.
The spermidine content in wild-type plants increased significantly after 3 days of drought stress (409.75 ± 14 nmol g-1 fw, compared with 287.37 ± 33 nmol g-1 fw at t = 0, P < 0.01; Fig. 4C). The spermidine content also increased in Dadc-transgenic plants after 3 days (450.17 ± 24 nmol g-1 fw, compared with 340.90 ± 43 nmol g-1 fw at t = 0, P < 0.01; Fig. 4C). A small but significant increase in spermine levels was observed in wild-type (302.5 ± 22 nmol g-1 fw, P < 0.05) and transgenic (370.15 ± 21 nmol g-1 fw, P < 0.05) plants after 3 days of drought stress, compared with 254.1 ± 12 nmol g-1 fw and 253.2 ± 32 nmol g-1 fw at t = 0, respectively (Fig. 4D). The spermidine and spermine contents in Dadc-transgenic plants had increased significantly by t = 6 days compared with the transgenic control plants (440.91 ± 14 nmol g-1 fw and 392.15 ± 30 nmol g-1 fw, respectively; both P < 0.01; compared with 340.9 ± 43 nmol g-1 fw for spermidine and 253.2 ± 33 nmol g-1 fw for spermine at t = 0; Fig. 4D).
When we compared the spermidine content of wild-type and transgenic plants after 3 days of stress, no significant variation was observed (P > 0.05). A 1.7-fold increase in spermidine content was observed after 6 days between transgenic (440.91 ± 14 nmol g-1 fw) and wild-type (253.27 ± 24 nmol g-1 fw, P < 0.01; Fig. 4C) plants at the same time point. For spermine, a small but significant increase was observed between transgenic (370.15 ± 21 nmol g-1 fw) and wild-type (302.5 ± 22 nmol g-1 fw, P < 0.05; Fig. 4D) plants after 3 days of stress. After 6 days of stress, a 1.5-fold increase in spermine was observed in transgenic (392.15 ± 30 nmol g-1 fw) and wild-type (255.75 ± 14 nmol g-1 fw, P < 0.01; Fig. 4D) plants at the same time point.
Plants Expressing Ubi:Dadc Are Phenotypically Normal Under Drought Stress, Whereas Wild-Type Plants Under the Same Conditions Are Severely Affected. All transgenic plants, irrespective of the putrescine levels, were phenotypically normal. Subtle differences were observed between transgenic plants with higher levels of putrescine and wild-type plants. The transgenic plants had a slightly reduced growth rate and flowered 4-5 days later than wild-type plants did. However, these observations were not statistically significant (P > 0.05).
After 3 and 6 days of drought stress, all wild-type and hpt-transformed plants had wilted and showed drought-induced rolling of leaves. Such symptoms were absent from Dadc-transgenic plants, which exhibited significant putrescine accumulation during the same period. After the 6-day drought stress period, the phenotype of transgenic plants (84-2, 84-9, and 84-10) was indistinguishable from nonstressed control plants (Fig. 5). Transgenic plants, with 2- to 4-fold higher levels of putrescine, set seed in a manner that was indistinguishable from wild-type plants under normal growing conditions.
Fig. 5.
Response of rice plants to drought stress. (A) Phenotype of 2-month-old wild-type (WT) and transgenic plants (84-2 and 84-9 lineages) growing in soil after drought stress (6 days). (B) Close-up of rice leaves (wild type on the left and 84-2 on the right).
Discussion
Many reports link polyamines and abiotic stress in plants, but they do not provide unequivocal evidence for the involvement of polyamines in abiotic stress responses. They do provide strong circumstantial evidence that polyamines protect plants from abiotic stress, but they do not establish a cause-and-effect relationship. For the past several years we have been investigating molecular, biochemical, and physiological aspects of the polyamine biosynthetic pathway in plants, using rice as a model. We created a diverse collection of transgenic germplasm expressing various genes in the polyamine biosynthetic pathway, including adc, odc, and samdc (7, 9, 13, 24). Here we report transgenic plants overexpressing the Datura adc gene under the control of the strong monocot maize Ubi-1 promoter (16). These plants were generated to investigate the role of polyamines in the response to abiotic stress, in particular drought stress, which is a major constraint in rice productivity, mostly in rain-fed agro-ecosystems.
Expression of Ubi:Dadc Under Drought Stress Provides a Steady-State Putrescine Pool Available for Subsequent Steps in the Polyamine Pathway. Many plants accumulate specific amino acids or their derivatives in response to environmental stresses (25, 26). The accumulation of putrescine has been widely reported in monocotyledonous and dicotyledonous plants but is most pronounced in cereals where the putrescine pool represents a major sink for carbon and nitrogen (27). We found that wild-type rice plants subjected to PEG-induced drought stress responded by increasing cellular putrescine levels significantly (Fig. 3D), without any changes in the steady-state rice adc mRNA levels (Fig. 3A). In agreement with ref. 28, putrescine accumulation as a result of increased ADC enzyme activity did not appear to involve a substantial net change in the steady-state levels of adc mRNA. Flores and Galston (29) suggested that the primary event in this stress-induced phenomenon occurs very rapidly and requires de novo protein synthesis. This response was attributed to translational or posttranslational regulation of ADC, a mechanism that would not involve a net change in steady-state adc mRNA levels. The authors suggested a role for putrescine in plants under stress, which extends beyond its involvement as a simple precursor for the higher polyamines along the pathway (28). Putrescine accumulation in tissues under stress is also a consequence of the reduction in the rate of spermidine and spermine synthesis (29). Such accumulation can be toxic to certain cells. Whether toxicity is a direct result of putrescine accumulation or an indirect response to changes in the kinetics and/or products of its catabolism remains to be investigated (30). DiTomasso et al. (31) suggested that the basis of putrescine toxicity is the presence of an apoplastic diamine oxidase that catalyzes the formation of oxidation products, which most probably damage plasma membranes. In our experiments, the physiological stress responses of wild type, negative segregants, and transformants that did not exhibit significant accumulation of putrescine in their leaves manifested as progressive wilting and rolling of leaves (Fig. 5). Detached leaves from plants subjected to high osmoticum showed a massive accumulation of putrescine, but conversion to spermidine and spermine was very slow and mesophyll protoplasts isolated from such leaves were incapable of cell division. In contrast, dicotyledonous plants that readily regenerate from mesophyll protoplasts show a very different response. Putrescine levels are reduced, whereas the levels of spermidine and spermine increase significantly (29). Putrescine accumulation in the Dadc-transgenic plants is also a consequence of transgene expression. The Ubi-1 promoter, driving Dadc expression in our transgenic plants, is known to possesses a number of stress-responsive elements (16) that boost transgene expression under drought stress (32, 33). Transgene expression under such conditions would provide a constant supply of putrescine, thus maintaining a near constant steady-state pool of this compound in the transgenic plants.
Stress-Induced Spermidine and Spermine Accumulation in Transgenic Plants Confers Tolerance to Drought Stress. In previous experiments, endogenous levels of spermidine and spermine in detached oat leaf segments under osmotic stress declined sharply 6 h after stress induction (34). This response was attributed, at least in part, to activation of the polyamine catabolic pathway. The decline in spermidine and spermine levels resulted in chlorophyll loss and leaf senescence. When guazatine, an inhibitor of polyamine oxidase activity, was added to the incubation buffer, endogenous spermidine and spermine levels increased significantly and prevented chlorophyll loss in osmotically treated oat leaf segments, thus delaying senescence (34). We observed a remarkably similar reduction in the content of cellular spermidine and spermine in wild-type plants after 6 days of drought stress compared with the increases we observed after 3 days (Fig. 4 C and D). This finding correlated extremely well with the wilting and rolling of leaves (Fig. 5). Transgenic plants that exhibited drought tolerance behaved similarly to wild-type plants up to 3 days after the onset of stress, with spermidine and spermine levels increasing in a similar fashion under the same conditions. However, in contrast to the dramatic reduction in the levels of these two polyamines seen in wild type after day 3, the transgenic plants maintained high spermidine and spermine levels for the duration of the experiment (Fig. 4 C and D). This finding correlated perfectly with the drought tolerant phenotype of transgenic plants compared with wild type (Fig. 5).
In both transgenic and wild-type plants under drought stress, the spermidine and spermine content correlated with rice samdc steady-state mRNA profile (Fig. 4). When Li and Chen (35) exposed rice seedlings to drought stress using 15% PEG, this resulted in up-regulation of rice samdc mRNA after 3 days of stress. However, the investigators were not able to correlate samdc mRNA levels with the polyamine content of these plants. The accumulation of spermidine and spermine in leaf segments of mustard subjected to osmotic stress was correlated with an increase in the steady-state mustard samdc mRNA. However, the phenotype of these mustard plants was not described (36). Activation of the rice samdc gene pushes the pathway forward by using the steady-state putrescine pool that is generated by transgene expression. The net result is an increase in the levels of spermidine and spermine in the transgenic plants.
Differential Response of Wild-Type and Dadc-Expressing Transgenic Plants After Drought Stress Withdrawal. The two Arabidopsis thaliana adc genes (adc1 and adc2) (28, 37) were among the genes down-regulated by rehydration after dehydration in experiments using full-length cDNA microarrays to monitor profiles of Arabidopsis gene expression under drought stress (38). In our experiments, steady-state mRNA levels of the endogenous adc gene declined to very low levels in wild-type plants 2 days after the stress was removed (Fig. 3A) and correlated well with a significant reduction in putrescine. However, putrescine levels in wild-type plants did not return to the normal physiological levels measured at t = 0, i.e., before these plants were subjected to drought stress (Fig. 3D). In contrast, putrescine levels peaked at this time point in the Dadc-expressing plants, whereas endogenous levels of the rice adc mRNA in the transgenic plants also remained at high levels, as did the Dadc mRNA (Fig. 3 A, B, and D). This striking differential behavior in the transcript profiles of the rice and Datura adc mRNAs in wild-type and transgenic plants during their transition from drought stress conditions to recovery, as well as concomitant changes in putrescine levels, perhaps reflect the differential rates at which sensitive and tolerant plants return to their ground state after the stress is removed. Because putrescine levels are at their maximum in wild-type plants 6 days after stress induction, adc transcript levels need to be reduced immediately after the stress is removed, most likely through a feedback inhibition mechanism, to allow the plants to reduce their putrescine levels and attain a physiologically normal state. Because the Dadc gene is constantly active, transgenic plants are not able to respond in a manner similar to their wild-type counterparts, and this results in high adc transcript and putrescine levels in these plants. The fact that the rice adc transcript levels in transgenic plants is not reduced is most likely a consequence of the complete saturation of the system as a result of Dadc expression.
Steady-state mRNA levels for the rice samdc gene remained relatively unchanged in the wild-type plants 2 days after the stress was removed. Spermidine and spermine levels in these plants were not significantly different from the levels at t = 0 (Fig. 4 A, C, and D). In contrast, the steady-state rice samdc mRNA levels in transgenic plants remained substantially higher compared with those at t = 0 (in either transgenic plants or wild type; Fig. 4A). This differential behavior of the rice samdc transcript can be explained through a mechanism similar to that discussed earlier for adc.
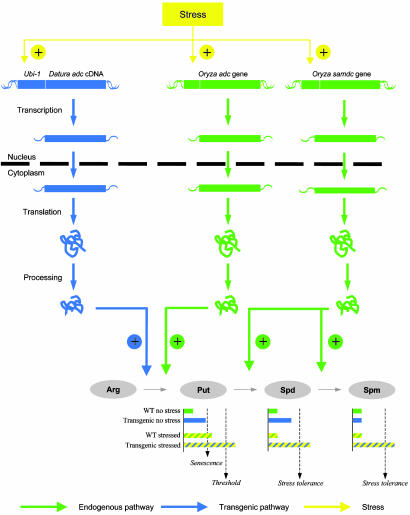
A Unified Model That Explains the Involvement of Polyamines in Abiotic Stress Response in Plants. Galston et al. (39) proposed a model that attempts to explain how ADC activity is regulated by spermine under osmotic stress. Using a detached oat leaf system, the authors postulated that, upon the onset of osmotic stress, a signal activates transcription of the oat adc gene. The translation product of the adc mRNA is an inactive precursor protein with a molecular mass of ≈60 kDa. This precursor is cleaved to produce an N-terminal fragment and a 24-kDa C-terminal fragment containing the ADC active site (40, 41). This active ADC form catalyzes the decarboxylation of arginine, leading to the accumulation of putrescine. The physiological response to increased putrescine levels includes chlorophyll loss and accelerated senescence (34). In the model proposed by Galston et al. (39), exogenously applied spermine can inhibit the posttranslational processing of the inactive ADC precursor molecule. The consequence of this inhibition is a decrease in ADC activity and a concomitant prevention of excess putrescine accumulation. Oat leaf segments exposed to spermine were able to retain chlorophyll after 72 h under osmotic stress (34). In the rice whole-plant system, we showed that endogenous spermidine and spermine accumulation resulting from adc transgene expression has a similar effect. Expression of the heterologous adc transgene driven by the maize Ubi-1 promoter, which is known to be activated by stress (32, 33), would augment the putrescine pool to levels that extend beyond the critical threshold required to initiate the conversion of excess putrescine to spermidine and spermine (14). Spermidine and spermine de novo synthesis in transgenic plants under drought stress is corroborated by the activation of the rice samdc gene. Transcript levels for rice samdc reach a maximum 6 days after stress induction. Such increases in the endogenous spermidine and spermine pools of transgenic plants not only regulate the putrescine response but also exert an anti-senescence effect at the whole-plant level, resulting in phenotypically normal plants. Wild-type plants, however, are not able to raise their spermidine and spermine levels after 6 days of drought stress and consequently exhibit the classical drought-stress response (Figs. 5 and 6).
Fig. 6.
Unified model explaining the role of polyamines in the response of wild-type and transgenic plants expressing Dadc under abiotic stress conditions. The model is considered in more detail in Discussion. Histograms show relative polyamine levels measured in each type of plant. Once putrescine levels exceed the threshold shown, the synthesis of spermidine and spermine is triggered, resulting in protection against drought and other forms of stress.
Our results are thus consistent with an emerging picture in which the temporal profile of transcripts and corresponding polyamines are implicated in the response of wild-type and transgenic plants to drought stress.
Acknowledgments
We thank Dr. T. Michael (Institute of Food Research, Norwich, U.K.) for the kind gift of the Datura adc and the rice samdc cDNAs, Dr. T. Sasaki (Rice Genome Research Program, Tsukuba, Japan) for the kind gift of the rice adc cDNA, and Marta Rafel for technical assistance. We also acknowledge long-term support from the Rockefeller Foundation (New York).
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: adc, arginine decarboxylase; PEG, polyethylene glycol; samdc, S-adenosylmethionine decarboxylase; Ubi-1, ubiquitin promoter; fw, fresh weight.
References
- 1.Bajaj, S., Targolli, J., Liu, L. F., Ho, T. H. D. & Wu, R. (1999) Mol. Breeding 5, 493-503. [Google Scholar]
- 2.Richards, F. J. & Coleman, R. G. (1952) Nature 170, 460. [DOI] [PubMed] [Google Scholar]
- 3.Bouchereau, A., Aziz, A., Larher, F. & Martin-Tanguy, J. (1999) Plant Sci. 140, 103-125. [Google Scholar]
- 4.Koening, H., Goldstone, A. D., Lu, C. Y. & Trout, J. J. (1990) Stroke 21, Suppl. III, 98-103. [PubMed] [Google Scholar]
- 5.Reggiani, R., Aurisano, N., Mattana, M. & Bertani, A. (1993) J. Plant Physiol. 142, 94-98. [Google Scholar]
- 6.Kakkar, R. K. & Sawhney, V. K. (2002) Physiol. Plant. 116, 281-292. [Google Scholar]
- 7.Lepri, O., Bassie, L., Safwat, G., Thu-Hang, P., Trung-Nghia, P., Hölttä, E., Christou, P. & Capell, T. (2001) Mol. Gen. Genet. 266, 303-312. [DOI] [PubMed] [Google Scholar]
- 8.Mehta, R. A., Cassol, T., Li, N., Ali, N., Handa, A. K. & Mattoo, A. K. (2002) Nat. Biotechnol. 20, 613-618. [DOI] [PubMed] [Google Scholar]
- 9.Thu-Hang, P., Bassie, L., Sawfat, G., Trung-Nghia, P. & Capell, T. (2002) Plant Physiol. 129, 1744-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trung-Nghia, P., Bassie, L., Safwat, G., Thu-Hang, P., Lepri, O., Rocha, P., Christou, P. & Capell, T. (2003) Planta 218, 125-134. [DOI] [PubMed] [Google Scholar]
- 11.DeScenzo, R. A. & Minocha, S. C. (1993) Plant Mol. Biol. 22, 113-127. [DOI] [PubMed] [Google Scholar]
- 12.Bastola, D. R. & Minocha, S. C. (1995) Plant Physiol. 109, 63-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Capell, T., Escobar, C., Lui, H., Burtin, D., Lepri, O. & Christou, P. (1998) Theor. Appl. Genet. 97, 246-254. [Google Scholar]
- 14.Bassie, L., Noury, M., Lepri, O., Lahaye, T., Christou, P. & Capell, T. (2000) Transgenic Res. 9, 33-42. [DOI] [PubMed] [Google Scholar]
- 15.Bhatnagar, P., Minocha, R. & Minocha, S. C. (2002) Plant Physiol. 128, 1455-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen, A. H. & Quail, P. H. (1996) Transgenic Res. 5, 213-218. [DOI] [PubMed] [Google Scholar]
- 17.Sudhakar, D., Duc, L. T., Bong, B. B., Tinjuangjun, P., Maqbool, S. B., Valdez, M., Jefferson, R. & Christou, P. (1998) Transgenic Res. 7, 289-294. [Google Scholar]
- 18.Valdez, M., Cabrera-Ponce, J. L., Sudhakhar, D., Herrera-Estrella, L. & Christou, P. (1998) Ann. Bot. 82, 795-801. [Google Scholar]
- 19.Perez-Molphe-Bach, E., Gidekel, M., Segura-Nieto, M., Herrera-Estrella, L. & Ochoa-Alejo, N. (1996) Physiol. Plant. 96, 284-290. [Google Scholar]
- 20.Edwards, K., Johnstone, C. & Thompson, C. (1991) Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) in Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 2nd Ed.
- 22.Lahaye, T., Rueger, S., Toepsch, S., Thalhammer, J. & Schulze-Lefert, P. (1996) Biotechniques 21, 1067-1072. [DOI] [PubMed] [Google Scholar]
- 23.Lepri, O., Bassie, L., Thu-Hang, P., Christou, P. & Capell, T. (2002) Theor. Appl. Genet. 105, 594-603. [DOI] [PubMed] [Google Scholar]
- 24.Noury, M., Bassie, L., Lepri, O., Kurek, I., Christou, P. & Capell, T. (2000) Plant Mol. Biol. 43, 357-544. [DOI] [PubMed] [Google Scholar]
- 25.Bohnert, H. J. & Jensen, R. G. (1996) Trends Biotechnol. 14, 89-97. [Google Scholar]
- 26.Holmberg, N. & Bülow, L. (1998) Trends Plant Sci. 3, 61-66. [Google Scholar]
- 27.Slocum, R. D. & Weinstein, L. H. (1990) in Polyamines and Ethylene: Biochemistry, Physiology, and Interactions, eds. Flores, H. E., Arteca, R. N. & Shannon, J. C. (Am. Soc. Plant), pp. 157-165.
- 28.Watson, M. B. & Malmberg, R. L. (1996) Plant Physiol. 111, 1077-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flores, H. E. & Galston, A. W. (1984) Plant Physiol. 75, 102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masgrau, C., Altabella, T., Farras, R., Flores, D., Thompson, A. J., Besford, R. T. & Tiburcio, A. F. (1997) Plant J. 11, 465-473. [DOI] [PubMed] [Google Scholar]
- 31.DiTomasso, J. M., Shaff, J. E. & Kochian, L. V. (1989) Plant Physiol. 90, 988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cornejo, M. J., Luth, D., Blankenship, K. M., Anderson, O. D. & Blechl, A. E. (1993) Plant Mol. Biol. 23, 567-581. [DOI] [PubMed] [Google Scholar]
- 33.Takimoto, I., Christensen, A. H., Quail, P. H., Uchimiya, H. & Toki, S. (1994) Plant Mol. Biol. 26, 1007-1012. [DOI] [PubMed] [Google Scholar]
- 34.Capell, T., Campos, J. L. & Tiburcio, A. F. (1993) Phytochemistry 32, 785-788. [Google Scholar]
- 35.Li, Z. Y. & Chen, S. Y. (2000) J. Plant Physiol. 156, 386-393. [Google Scholar]
- 36.Mo, H. & Pua, E. C. (2002) Physiol. Plant. 114, 439-449. [DOI] [PubMed] [Google Scholar]
- 37.Watson, M. W., Yu, W., Galloway, G. L. & Malmberg, R. L. (1997) Plant Physiol. 114, 1569. [Google Scholar]
- 38.Oono, Y., Seki, M., Nanjo, T., Narusaka, M., Fujita, M., Satoh, R., Satou, M., Sakurai, T., Ishida, J., Akiyama, K., et al. (2003) Plant J. 34, 868-887. [DOI] [PubMed] [Google Scholar]
- 39.Galston, A. W., Kaur-Sawhney, R., Altabella, T. & Tiburcio, A. F. (1997) Bot. Acta 110, 197-207. [Google Scholar]
- 40.Malmberg, R. L., Smith, K. E., Bell, E. & Cellino, M. L. (1992) Plant Physiol. 100, 146-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malmberg, R. L. & Cellino, M. L. (1994) J. Biol. Chem. 269, 2703-2706. [PubMed] [Google Scholar]