Abstract
Diversity Arrays Technology (DArT) can detect and type DNA variation at several hundred genomic loci in parallel without relying on sequence information. Here we show that it can be effectively applied to genetic mapping and diversity analyses of barley, a species with a 5,000-Mbp genome. We tested several complexity reduction methods and selected two that generated the most polymorphic genomic representations. Arrays containing individual fragments from these representations generated DArT fingerprints with a genotype call rate of 98.0% and a scoring reproducibility of at least 99.8%. The fingerprints grouped barley lines according to known genetic relationships. To validate the Mendelian behavior of DArT markers, we constructed a genetic map for a cross between cultivars Steptoe and Morex. Nearly all polymorphic array features could be incorporated into one of seven linkage groups (98.8%). The resulting map comprised ≈385 unique DArT markers and spanned 1,137 centimorgans. A comparison with the restriction fragment length polymorphism-based framework map indicated that the quality of the DArT map was equivalent, if not superior, to that of the framework map. These results highlight the potential of DArT as a generic technique for genome profiling in the context of molecular breeding and genomics.
Although 50 years have passed since the structure of DNA was deciphered (1), the study of DNA variation emerged as a field of scientific endeavor only in the last 25 years. Two groups of technologies were developing in parallel from the very beginning: DNA sequencing and molecular markers. DNA sequencing technology developed quickly from proof of concept (2, 3) to an automated process (4), enabling the field of genomics. Molecular marker technologies progressed rapidly as well. Based on the Southern blot technique (5), Botstein et al. (6) developed the restriction fragment length polymorphism (RFLP) technique as a method for creating genetic linkage maps.
Development of the PCR technique spawned two important molecular marker techniques: amplified fragment length polymorphism (AFLP) (7) and simple sequence repeats (8). Thousands of studies using molecular markers in plants, including hundreds in barley, have been published but are not referenced because of space limitations.
DNA sequencing and molecular marker technologies started to merge when the accumulated sequence data began to yield information on sequence variation among different accessions of the same species. It was soon noted that single-nucleotide polymorphism (SNP) is the most abundant marker type, promising nearly unlimited supply of markers (9). Many alternatives were developed for the SNP assay (primer extension, selective ligation) and the platform to type assays in high throughput (DNA chip, printed and self-assembling arrays, matrix-assisted laser desorption ionization/time-of-flight mass spectroscopy) (10-14).
For humans and a limited number of model organisms, the throughput of SNP assays has increased impressively, and assay costs have decreased correspondingly. Yet discovering sequence polymorphism in nonmodel species is difficult, which is particularly true for many crops with limited resources and often complex, polyploid genomes. We have developed Diversity Arrays Technology (DArT) to enable whole-genome profiling of such crops without the need of sequence information. DArT is based on microarray hybridizations that detect the presence versus absence of individual fragments in genomic representations as described by Jaccoud et al. (15).
For our initial proof-of-concept study, we selected a species with a simple genome (rice) and used AFLP-like complexity reduction methods to generate genomic representations (15). Here we apply a non-AFLP version of DArT to barley, a species with a complex genome nearly twice as large as the human genome and 13 times larger than that of rice (16). We show that DArT can be used to effectively create a medium-density genetic map, a result that points to its potential as a generic technique for high-throughput genome profiling of plants.
Materials and Methods
DArT Protocol. Preparation of genomic representations. Genomic representations were generated by cutting 100 ng of a mixture of DNA samples from a group of barley cultivars (cvs.) with 2 units of both PstI and one of the frequent cutters listed in Table 1 (NEB, Beverly, MA). A PstI adapter (5′-CAC GAT GGA TCC AGT GCA-3′ annealed with 5′-CTG GAT CCA TCG TGC A-3′) was ligated with T4 DNA ligase (NEB). A 1-μl aliquot of the ligation product was used as a template in 50-μl amplification reactions with DArT-PstI primer (5′-GAT GGA TCC AGT GCA G-3′) and a program applicable to all plant species tested so far: 94°C for 1 min, followed by 30 cycles of 94°C for 20 sec, 58°C for 40 sec, 72°C for 1 min, and 72°C for 7 min.
Table 1. Number of unique clones and polymorphism levels in PstI-based DArT libraries differing in the enzyme used for codigestion.
| Estimated no. of unique clones
|
||||
|---|---|---|---|---|
| Codigesting enzyme | Empirical* | Rice† | Hordeum plus Triticum† | Polymorphism level, %‡ |
| AluI | 3,100 | 12,000 | 18,000 | 7.0 |
| ApoI | 4,900 | 78,000 | 88,000 | 6.9 |
| BanII | 16,500 | 130,000 | 122,000 | 3.4 |
| Bsp 12861 | 6,600 | 77,000 | 61,000 | 2.9 |
| BstNI | 5,800 | 80,000 | 61,000 | 10.0 |
| HaeIII | 2,000 | 46,000 | 27,000 | 3.5 |
| MseI | 4,100 | 42,000 | 36,000 | 4.0 |
| RsaI | 3,600 | 42,000 | 43,000 | 8.6 |
| TaqI | 3,000 | 50,000 | 52,000 | 10.4 |
See Materials and Methods for a description of procedures.
The numbers shown were obtained by in silico analysis (see Materials and Methods) based on bacterial artificial chromosome (BAC) sequences extrapolated from a random set of 327 BAC clones of rice (39 Mbp in total) to the whole genome or from a mixed set of Hordeum and Triticum BAC clones (1.6 Mbp in total) to the whole genome.
Percentage of clones polymorphic between cvs. Clipper and Sahara.
Preparation of arrays. Libraries of genomic representations were prepared essentially as by Jaccoud et al. (15). Individual clones were grown in 384-well plates containing LB medium supplemented with 100 mg·liter-1 ampicillin and a “freezing mix” (unpublished observation). Small aliquots of the cultures were used as templates to amplify inserts according to Jaccoud et al. (15).
We used two types of arrays for DArT fingerprinting: “discovery arrays” and “polymorphism-enriched arrays.”
Discovery arrays contained inserts amplified from random clones of DArT libraries. The amplification reactions were dried, dissolved in diluted print buffer A (Vanderbilt University, South Nashville, TN), and spotted in triplicate on Polysine (Menzel Gläser, Braunschweig, Germany) or SuperChip poly-L-lysine slides (Erie Microarray, Portsmouth, NH) by using a MicroGrid II arrayer (Biorobotics, Cambridge, U.K.). A single-replicate format was chosen for large arrays. After being printed, slides were heated to 80°Cfor2h, incubated in hot water (95°C) for 2 min, and dried by centrifugation.
A polymorphism-enriched PstI/BstNI array was produced from 1,920 candidate polymorphic clones. They were printed together with 1,152 control features derived from 64 nonpolymorphic (control) clones so that each group of spots printed by a particular pin contained the same number of each of the 64 control clones (MicroGrid II arrayer). The two groups of clones had been identified during a preliminary diversity analysis of Australian barley varieties by using an array of 7,680 PstI/BstNI clones from a library prepared from cvs. Alexis, Amaji Nijo, Chebec, Clipper, Galleon, Harrington, Haruna Nijo, Sahara, Sloop, and WI2585.
Naming of clones. Each clone (marker) was given a preliminary name, which will be revised in the future with a more generally applicable naming system. The name contains information on array type (Bd and Br, PstI/BstNI discovery and rearrayed libraries, respectively; Td, PstI/TaqI discovery library), plate location (plate number and well position), source of the “1” allele (S, cv. Steptoe; M, cv. Morex), and the between-allele variance in relative hybridization intensity as a percentage of the total variance (see Image analysis and polymorphism scoring).
Fingerprinting of DNA samples. Genomic representations of individual barley lines were generated by using the same complexity reduction method as the one used to generate the respective array. Genomic representations were concentrated 10-fold by precipitation with 1 vol of isopropanol, denatured and labeled with 1 μl of 500 μM cy3-labeled random decamers and the exo- Klenow fragment of Escherichia coli DNA polymerase I (NEB). Labeled representations, called targets, were added to 50 μl of a 50:5:1 mixture of ExpressHyb buffer (Clontech), 10 g·liter-1 herring sperm DNA, and the cy5-labeled polylinker fragment of the plasmid used for library preparation (as a reference) (15). The samples were denatured and hybridized to microarrays overnight at 65°C. Slides were washed according to Jaccoud et al. (15) and scanned on an Affymetrix 428 (Santa Clara, CA) or Tecan LS300 (Grödig, Austria) confocal laser scanner.
Image analysis and polymorphism scoring. A typical experiment consisted of 96 slides simultaneously hybridized with 96 genomic representations from up to 96 barley lines. dartsoft, a software package developed in-house, was used to both identify and score the markers that were polymorphic within such an experiment (C. Cayla, G. Uszynski, D.J., P.W., and A. Kilian, unpublished data).
dartsoft automatically localized the spots in all scanner image pairs (cy3, cy5) generated in an experiment, rejected those with weak reference signals, and computed and normalized background-subtracted relative hybridization intensities (calculated as log[cy3target/cy5reference]). The software then compared the relative intensity values for each individual clone across slides by using a combination of fuzzy C-means clustering at a “fuzziness” level of 1.5 (17) and ANOVA: If two clusters (alleles) could be distinguished and the between-cluster variance in relative intensity was at least 80% of the total variance, the clone was called polymorphic and scored as 0 or 1. A clone was incorporated into the 0/1 scoring table of a particular experiment if it was scored with a probability of P > 0.95 in at least 90% of the slides (scoring probabilities were estimated by the clustering algorithm). Individual calls with P < 0.95 were scored as missing. Slides with <90% of the identified polymorphic markers scored at P > 0.95 were rejected (typically 5%).
Experiments Performed. Optimization of complexity reduction methods. Nine 768-clone libraries of PstI fragments, each differing in the frequent cutter used for codigestion, were produced from a mixture of genomic DNA of cvs. Clipper and Sahara (18). Corresponding targets, prepared from the two cvs. and 20 Clipper × Sahara doubled haploid (DH) lines were hybridized to arrays containing triplicate spots of clones from these libraries.
The number of unique clones in each of these libraries was estimated based on an evaluation of redundancy levels within the group of polymorphic clones. Potential replicates were identified by comparing their segregation patterns in the group of DHs. A truncated Poisson distribution was fitted to the redundancy classes (19) to estimate the number of unique polymorphic clones. The total number of unique clones was estimated by extrapolating to all clones (nonpolymorphic + polymorphic) based on the percentage of polymorphic clones within a library. The resulting estimates were corrected by a factor that accounted for tightly linked unique clones that cosegregated because of the limited resolution provided by the 20 DHs. This factor (1.65) had been measured for a subset of polymorphic clones by comparing the estimated redundancy level with the actual redundancy level (determined by fingerprinting the cloned inserts with a mixture of MspI and Sau3AI).
The numbers of fragments in the nine genomic representations were estimated in silico by counting the number of PstI fragments produced from a genomic input sequence, which fell into the empirically observed amplifiable range of 0.4-1 kb and lacked recognition sites for the codigesting enzyme (vector nti and mathcad).
Analysis of genetic relationships among barley lines. PstI/BstNI representations were prepared from a range of cvs., two landraces, and two accessions of Hordeum spontaneum and were hybridized in duplicate to the polymorphism-enriched PstI/BstNI array (see Preparation of arrays above). Consistent 0/1 scores were used as input for the restdist and neighbor programs of the phylip 3.6 software package to construct an Unweighted Pair Group Method with Algorithmic Mean dendrogram based on Felsenstein's modification of the Nei/Li restriction fragment distance (20). Clade strength was tested by 1,000 bootstrap analyses performed with the seqboot program (21).
Creation of a DArT linkage map. PstI/BstNI and PstI/TaqI targets were prepared from 94 DH lines derived from a cross between cvs. Steptoe and Morex. A first set of PstI/BstNI targets were hybridized to a corresponding discovery array containing triplicate spots of clones from a 3,840-clone library prepared from a mixture of Steptoe and Morex DNA. A second set of PstI/BstNI targets was hybridized to the polymorphism-enriched PstI/BstNI array (see Preparation of arrays above). The PstI/TaqI targets were hybridized in duplicate to arrays containing single replicates of clones from an 8,448-clone PstI/TaqI library prepared from cvs. Alexis, Amaji Nijo, Chebec, Clipper, Galleon, Harrington, Haruna Nijo, Morex, Sahara, Sloop, Steptoe, and WI2585. Scoring data from the three sets of hybridizations were combined to construct a linkage map with map manager qtxb19 and a linkage criterion of P < 10-5 (22).
Results and Discussion
Optimization of Complexity Reduction Methods. DArT detects DNA polymorphism by comparing the composition of genomic representations of different genotypes through hybridizations to microarrays (15). Fig. 3, which is published as supporting information on the PNAS web site, gives a graphical representation of the procedure. We addressed several key issues to develop a robust technology.
The first was the exact type of complexity reduction method to use to maximize the number of polymorphic clones in DArT libraries. We produced nine 768-clone PstI libraries from two genetically distant cvs. (Clipper and Sahara) (18), each of them prepared by amplifying PstI fragments digested with a different frequent cutter (AluI, ApoI, BanII, Bsp1286I, BstNI, HaeIII, MseI, RsaI, or TaqI). The libraries were evaluated for the frequency of polymorphisms between the two cvs. as well as the number of unique clones (Table 1).
The polymorphism rates varied more than 3-fold among the libraries tested (2.9-10.4%, average 6.3%; P < 0.01 as determined by a χ2 test) and were weakly negatively correlated with the estimated numbers of unique clones in the libraries (r2 = -0.37; Table 1).
In silico estimates of the number of unique clone, although closely correlated with the empirical estimates (r2 = 0.86), were approximately 10× larger (Table 1). This was expected because each of the recognition sites at the two ends of PstI fragments contains two CWG motifs (W = A or T). If symmetrically methylated at the cytosine residue, each of these motifs prevents PstI cutting. Assuming that methylated CWG motifs are randomly distributed in the genome, a 10-fold lower-than-predicted number of PstI fragments would suggest that 44% of these motifs were methylated [(1 - 0.44)4 = 0.1]. This value is not far from the 53% estimate obtained from the probably hypermethylated 5S rRNA clusters of diploid rye (23), but is significantly lower than the >80% measured for hexaploid wheat (24). Both ploidy level and nonrandom distribution of methylated CWG motifs could account for these differences. We assumed the empirical estimates were sufficiently accurate for the purpose of this study and expanded the two libraries with the highest polymorphism levels (PstI/TaqI and PstI/BstNI). Together, the libraries were expected to contain ≈900 clones polymorphic between the two genetically distant cvs. (Table 1).
In parallel, we measured the scoring reproducibility for clones from one of the selected libraries. From a single cv., we generated duplicate PstI/TaqI fingerprints of 27 DNA extracts sampled at three growth stages and three environmental conditions. The genotype call rate was 99% (similar to the average rate for this report, which was 98.0% ± 1.3%). The scoring reproducibility, computed from the 27 duplicate analyses, was 99.9% (Table 2, which is published as supporting information on the PNAS web site). The vast majority of markers (97%) scored identical for all DNA preparations; the remaining 3% gave consistently different results for different DNA samples, perhaps reflecting developmentally regulated changes in DNA methylation (Table 2; see also Stability of Methylation Patterns). Such markers would typically not be included in a properly formatted genotyping array.
This data suggested that the robustness of scoring would be sufficient to accurately evaluate genetic relationships among lines and to build a high-quality genetic map.
DArT Fingerprints Reflect Genetic Relationships. We analyzed DNA from 33 barley cvs. and two accessions of wild barley (H. spontaneum) on DArT arrays containing a selection of 1,920 candidate PstI/BstNI polymorphisms and 1,158 control features. A total of 383 polymorphic clones were identified. The scoring table is available as Table 3, which is published as supporting information on the PNAS web site. None of these polymorphisms came from the group of the 1,158 control features, a result that underscored the reproducibility of DArT assays and validated our procedure of selecting polymorphisms.
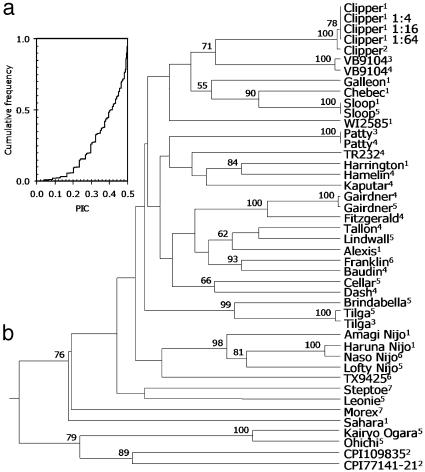
Polymorphism information content (PIC) values of the 383 identified polymorphic markers ranged from 0.04 to 0.50, with a median value of 0.42 (average 0.38), fairly high for randomly selected biallelic loci (Fig. 1) (25).
Fig. 1.
Genetic relationships among a group of barley cvs. and two accessions of wild barley (H. spontaneum). (a) Cumulative distribution function of the PIC values of the 383 PstI/BstNI markers identified (25). (b) Unweighted Pair Group Method with Algorithmic Mean dendrogram constructed from 383 PstI/BstNI markers based on the modified Nei/Li restriction fragment distance matrix (20, 21). A single DNA sample of cv. Clipper was assayed several times at various dilutions. Bootstrap support values (1,000 replicates) are shown if >50%. Superscript numbers correspond to suppliers of DNA samples. DNA samples were provided by (1) Peter Langridge, University of Adelaide, Adelaide, Australia; (2) Tony Brown (Commonwealth Scientific and Industrial Research Organisation, Canberra, Australia), (3) David Poulsen (Queensland Department of Primary Industries, Brisbane, Australia), (4) Mehmet Cakir (Murdoch University, Perth, Australia), (5) Harsh Raman (NSW Agriculture, Orange, Australia), (6) Haobing Li (University of Tasmania, Hobart, Australia), and (7) Evans Lagudah (Commonwealth Scientific and Industrial Research Organisation).
The dendrogram in Fig. 1 displays the genetic relationships among the genotypes analyzed. Although the diversity analysis presented here serves primarily as an example of DArT performance, a few observations can be made from the dendrogram. As expected, the two H. spontaneum accessions and the two landraces from South East Asia (Ohichi and Kairo Ogara) were fairly distant from most of the cvs. The scores of these four genotypes were biased toward “0” (P ≤ 0.05, computed based on the distribution of the percentage of 0 scores across genotypes), indicating that their alleles were underrepresented on the array. Not surprisingly, the genotypes clustered together. Incorporation of clones from these genotypes into the array would increase its resolution power for this kind of germplasm.
All other lines had statistically indistinguishable percentages of 0 scores, suggesting the genetic diversity sampled during DArT library preparation was sufficient to resolve genetic relationships among the cultivated varieties. The four Japanese cvs. clustered together, with cvs. Haruna Nijo and Naso Nijo being the most similar among all genotypes analyzed. Another group in the dendrogram contained cvs. that have cv. Triumph in their pedigree (cvs. Alexis, Fitzgerald, Franklin, Lindwall, Gairdner, Baudin, and Tallon). We conclude that DArT markers tend to group together the expected lines.
In the same experiment we reevaluated more thoroughly the two aspects of DArT data consistency investigated in the previous subchapter. Consistency of the platform itself was tested through duplicate analysis of all DNA samples. We obtained 35 pairs of conflicting scores among 16,739 individual comparisons, indicating a scoring reproducibility of 99.8%: a very good result, particularly because we typed DNA samples from seven different sources of various levels of quality and concentration.
To evaluate more precisely how the amount of DNA per assay affects data quality, we fingerprinted a series of 4-fold dilutions of a single DNA sample of cv. Clipper (100 to 1.5 ng per assay). The fact that we obtained identical scores for all markers suggested that the DArT platform tolerates well differences in DNA quantity. Even very small amounts, equivalent to <1,000 barley cells, were sufficient for the highly multiplexed DArT assay.
The second aspect of data consistency reevaluated was the reproducibility of DArT fingerprints obtained from different DNA preparations of the same cv. We obtained six pairs of DNA samples, each from two different individuals of the same cv. Two of these pairs were identical for all 383 markers (cvs. Patty and Sloop). Very few differences were observed among the other pairs of DNA samples: from 2/383 (0.5%), in the case of cv. Gairdner, to 5/383 (1.3%) for cv. VBg104. This is a high level of “biological” reproducibility, bearing in mind that the average difference between pairs of different cvs. was 41% of all polymorphisms, with a range of 15-63%.
We suspect that the differences observed between DNA samples from the same cv. were mainly due to genetic heterogeneity within those cvs., although instability of allelic states of DArT markers in plants grown in different environments could not be excluded as an additional source of variation (see previous subchapter). For example, cv. Tilga, for which four markers scored differently in the above comparison, is known for its phenotypic heterogeneity (H. Raman, personal communication). Heterogeneity has been observed at the molecular level in many barley cvs. by using marker technologies, such as RFLP, which evaluated fewer loci than DArT (26).
Assembly of a DArT Linkage Map. We selected a DH population from a cross between two six-row barleys, cvs. Steptoe and Morex, to map DArT markers and validate their Mendelian behavior. This cross had previously been used to create a comprehensive molecular linkage map of barley (27) and currently has 953 markers.
By using the quality thresholds specified in Materials and Methods, we identified 969 segregating polymorphisms of a total of ≈20,000 PstI/BstNI and PstI/TaqI clones. A comparison with estimates of the number of unique clones (Table 1) indicated that we assayed DArT markers with roughly 2.5-fold redundancy. This redundancy level not only enabled us to identify and type most of the clones polymorphic between Steptoe and Morex but also created a stringent test for the platform's performance: map expansion as a result of occasional miss-scores would be easier to detect if each marker was assayed repeatedly.
A linkage analysis of the 969 DArT markers plus three RFLP markers from the framework (FW) map to bridge gaps >28 centimorgans (cM), created seven linkage groups containing 90-170 markers each. The groups were 138-198 cM long and spanned a total of 1,137 cM. Twelve markers (1.2%) failed to incorporate and were removed from the data set. The remaining 957 markers fell into 279 segregation patterns (Table 4, which is published as supporting information on the PNAS web site). Fifty-three of these segregation patterns comprised both maternal and paternal markers (Table 5, which is published as supporting information on the PNAS web site). There should have been a similar number of cases in which different markers from the same parent cosegregated. We therefore estimated the total number of unique markers to be in the vicinity of 279 + (2 × 53) = 385.
Map Quality. To benchmark the performance of DArT markers we compared the DArT map with the existing Steptoe × Morex RFLP FW map, from which we removed 18 RFLP markers that were uninformative for our set of DHs (http://wheat.pw.usda.gov/ggpages/SxM/smbasev2.map). For the remaining 204 markers we only retained the scores for the 94 DHs used in our mapping experiment, which resulted in 199 unique segregation patterns (Table 6, which is published as supporting information on the PNAS web site). We assembled the FW map under identical conditions as those used for DArT markers. The resulting map had seven linkage groups spanning 1,195 cM, 5% longer than the DArT map. The length of the DArT map, therefore, indicated a low level of scoring errors.
We compared the frequency of double crossovers (DCO) in each of the two data sets. For this analysis we removed markers with redundant segregation patterns (not knowing whether they were identical or just cosegregating) because, otherwise, DCO adjacent to blocks of identically scored markers would have been undetectable. We then calculated for both maps the percentage of unique markers that introduced DCO. In the DArT map, 13.2% of the estimated 385 unique markers introduced DCO (2.9% created two or three DCO). In the FW set, 20.6% (42/204) of the markers introduced DCO (5.4% created two to four DCO). We conclude that automatically scored DArT markers appear to introduce less DCO than manually scored RFLP markers.
Having independently evaluated the two sets of markers, we merged the two datasets to assemble a joint map. We obtained 416 unique segregation patterns in seven linkage groups, each containing both DArT and FW markers. There were 65 loci with a DArT marker(s) cosegregating with FW markers. The size of the linkage groups obtained varied slightly depending on the parameters used for map assembly, but the shortest combined map was just <1,400 cM. There was virtually no difference between the two marker sets in logarithm of odds score statistics (Table 7, which is published as supporting information on the PNAS web site).
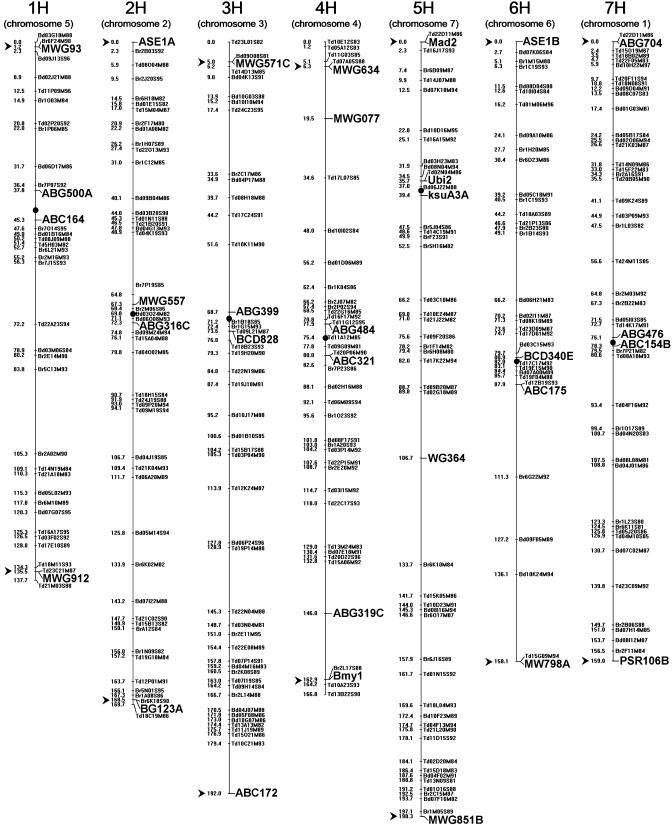
For this report, we incorporated only the most distal (telomeric) and two centromeric FW markers of each chromosome into the DArT map. The resulting map was 1,182 cM long (Fig. 2). It had fewer 10- to 20 cM-long gaps than the FW map (2.7 ± 1.4 per chromosome versus 3.7 ± 1.6 for the FW map), and a similar number of gaps >20 cM (<1 per chromosome; these numbers were derived after removing the three gap-bridging FW markers from the DArT map).
Fig. 2.
DArT linkage map for a Steptoe × Morex DH population displaying markers with unique segregation patterns. Approximate centromere locations are shown as black dots. Telomeric and centromeric FW markers that were added to the DArT map are highlighted in larger font; telomeric markers are also designated by arrows. Also highlighted in larger fonts are three additional FW markers on chromosomes 4H and 5H that were retained to facilitate map construction. Chromosome numbers according to the old nomenclature are given in brackets.
To evaluate genome coverage provided by the DArT map we analyzed the locations of the most distal DArT markers in relationship to the most distal FW markers and a set of telomeric markers, mapped either for Steptoe × Morex or Harrington × TR306 (28). For nine of 10 chromosome arms with telomeric markers identified, the most distal DArT marker was either cosegregating or within 2 cM. The telomeric marker for the long arm of chromosome 4H was 4 cM distal to the most distal group of DArT markers. The remaining four chromosome arms had DArT markers within 2 cM from the most distal RFLP marker, except for 3HL, in which ABC172 was 12 cM distal to a DArT (or any other RFLP) marker.
Based on the above comparative analyses of map length, DCO events, logarithm of odds scores, and genome coverage, we conclude that the quality of the DArT map was equivalent, if not superior, to that of the RFLP-based FW map.
Stability of Methylation Patterns. The vast majority of DArT markers (98.8%) could be solidly incorporated into a genetic linkage map (Fig. 2 and Table 7). However, 12 DArT markers (1.2%), could not be incorporated and ≈1.1% of the DArT markers introduced multiple (two or three) apparent DCO events. Given the high level of scoring reproducibility of DArT (see Optimization of complexity reduction methods) it is unlikely that the occurrence of these DCO events was due to scoring errors. We suspect that unstable cytosine methylation caused non-Mendelian behavior of a small percentage of the markers, although we could not rule out a contribution of gene conversion events (29). Although a few reports indicate some level of instability of methylation patterns (30, 31), most indicate that they are stable, both in dicots and monocots (32-35). In barley, Mendelian behavior of de novo created methylation polymorphism was observed over several generations (P. Devaux, personal communication). We expect that some of the Mendelian-type DArT markers may be due to stable methylation polymorphisms.
Extensive use of the methylation-sensitive PstI enzyme in AFLP technology, especially for species with large genomes, underscores its value for genetic mapping and diversity studies (31, 34, 36-38). PstI-based AFLP markers tend to cluster less and have higher PIC values than those generated with methylation-insensitive enzymes (34, 38, 39). Consistent with these findings, PstI-based DArT markers did not cluster significantly, and their average PIC value was identical to that of PstI-based AFLP markers (40).
DArT Versus Other Microarray-Based Genotyping Techniques. Apart from DArT, solid phase-based genotyping appears to be restricted to a few model species with sequenced genomes. The high-density gene chip designed to type SNPs in genomic representations of human DNA, for example, is based on comprehensive sequence information (12). Oligonucleotide arrays revealing single feature polymorphisms in whole-genome hybridizations of yeast and Arabidopsis (41, 42) are also based on comprehensive sequence information. Large and polyploid genomes may not be amenable to the whole-genome hybridization approach. It also remains to be seen whether the development of sequence-based arrays could become affordable for a broad range of agricultural species.
By contrast, DArT is independent of investment in genome sequencing and can be fine-tuned to detect polymorphism in genomes of virtually any size, including the 16,000 Mbp genome of hexaploid wheat as ongoing work in our laboratory has shown. DArT is flexible enough to design genotyping arrays for a variety of applications; it is by no means restricted to PstI-based or even restriction enzyme-based complexity reduction methods. We have successfully tested methods to enrich for different classes of genomic sequences or distinct types of DNA variation (SNP/insertion-deletion or methylation variation). In addition, markers from different complexity reduction methods (for example, the two used in this report) can be typed simultaneously, either by mixing genomic representations before labeling or by multicolor detection.
Concluding Remarks. We conclude that DArT can be used to create medium-density genetic maps for plants with complex genomes and no sequence information available. By using a properly formatted genotyping array, the generation of a linkage map would typically take only 3 days. This throughput enables routine use of DArT in plant breeding programs; e.g., for exhaustive fingerprinting of germplasm, quantitative trait locus identification, genome background screening, simultaneous marker-assisted selection of several loci, or accelerated introgression of selected genomic regions. Integration of DArT maps would be straightforward provided they are developed with the same array. High-density maps for map-based cloning and chromosome-landing approaches (43) could be rapidly built by pyramiding data from a limited number of independent arrays. We suggest that DArT opens significant opportunities for plant breeding to benefit from whole-genome profiling, particularly in the context of improving traits with complex inheritance.
Supplementary Material
Acknowledgments
We thank the Australian Grains Research and Development Corporation and our colleagues Richard Jefferson, Peter Sharp, Neil Howes, and Bill Rathmell for ongoing support and interest; our colleagues listed in the legend to Fig. 1 for donating DNA samples of barley; Arnis Druka for providing us with an unpublished barley sequence; Harsh Raman and Evans Lagudah for information on some barley cvs.; and Nicola Flanagan for critical comments on the manuscript.
This report was presented at the international Congress, “In the Wake of the Double Helix: From the Green Revolution to the Gene Revolution,” held May 27-31, 2003, at the University of Bologna, Bologna, Italy. The scientific organizers were Roberto Tuberosa, University of Bologna, Bologna, Italy; Ronald L. Phillips, University of Minnesota, St. Paul, MN; and Mike Gale, John Innes Center, Norwich, United Kingdom. The Congress web site (www.doublehelix.too.it) reports the list of sponsors and the abstracts.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DArT, Diversity Arrays Technology; RFLP, restriction fragment length polymorphism; AFLP, amplified fragment length polymorphism; SNP, single-nucleotide polymorphism; cv., cultivar; cM, centimorgan; PIC, polymorphism information content; DCO, double crossovers; FW, framework.
References
- 1.Watson, J. D. & Crick, F. H. (1953) Nature 170, 737. [DOI] [PubMed] [Google Scholar]
- 2.Maxam, A. M. & Gilbert, W. (1977) Proc. Natl. Acad. Sci. USA 74, 560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanger, F., Nicklen, S. & Coulson, A. R. (1977) Proc. Nat. Acad. Sci. USA, 74, 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith, L. M., Sanders, J. Z., Kaiser, R. J., Hughes, P., Dodd, C., Connell, C. R., Heiner, C., Kent, S. B. H. & Hood, L. E. (1986) Nature 321, 674-679. [DOI] [PubMed] [Google Scholar]
- 5.Southern, E. M. (1975) J. Mol. Biol. 98, 503-517. [DOI] [PubMed] [Google Scholar]
- 6.Botstein, D., White, R. L., Skolnick, M. & Davis, R. W. (1980) Am. J. Hum. Genet. 32, 314-331. [PMC free article] [PubMed] [Google Scholar]
- 7.Vos, P., Hogers, R., Bleeker, M., Reijans, M., van der Lee, T., Hornes, M., Frijters, A., Pot, J., Peleman, J., Kuiper, M., et al. (1995) Nucleic Acids Res. 23, 4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber, J. & May, P. E. (1989) Am. J. Hum. Genet. 44, 388-396. [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X. C., Stern, D., Winkler, J., Lockhart, D. J., Morris, M. S., Fodor, S. P., et al. (1996) Science 274, 610-614. [DOI] [PubMed] [Google Scholar]
- 10.Ross, P., Hall, L., Smirnov, I. & Haff, L. (1998) Nat. Biotechnol. 16, 1314-1315. [DOI] [PubMed] [Google Scholar]
- 11.Landegren, U., Kaiser, R., Sanders, J. & Hood, L. (1988) Science 241, 1077-1080. [DOI] [PubMed] [Google Scholar]
- 12.Dong, S., Wang, E., Hsie, L., Cao, Y., Chen, X. & Gingeras, T. R. (2001) Genome Res. 11, 1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilles, P. N., Wu, D. J., Foster, C. B., Dillon, P. J. & Chanock, S. J. (1999) Nat. Biotechnol. 17, 365-370. [DOI] [PubMed] [Google Scholar]
- 14.Bray, M. S., Boerwinkle, E. & Doris, P. A. (2001) Hum. Mutat. 17, 296-304. [DOI] [PubMed] [Google Scholar]
- 15.Jaccoud, D., Peng, K., Feinstein, D. & Kilian, A. (2001) Nucleic Acids Res. 29, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arumunganathan, K. & Earle, E. D. (1991) Plant Mol. Biol. Rep. 9, 208-219. [Google Scholar]
- 17.Bezdek, J. C. (1981). Pattern Recognition with Fuzzy Objective Function Algorithms (Plenum, New York).
- 18.Karakousis, A., Barr, A. R., Kretschmer, J. M., Manning, S., Jefferies, S. P., Chalmers, K. J., Islam, A. K. M. & Langridge, P. (2003) Aust. J. Agric. Res. 54, 1137-1140. [Google Scholar]
- 19.Cohen, A. C., Jr. (1960) Biometrics 16, 203-211. [Google Scholar]
- 20.Nei, M. & Li., W. H. (1979) Proc. Natl. Acad. Sci. USA 76, 5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein, J. (1989) Cladistics 5, 164-166. [Google Scholar]
- 22.Manly, K. F, Cudmore, R. H., Jr. & Meer, J. M. (2001) Mamm. Genome 12, 930-932. [DOI] [PubMed] [Google Scholar]
- 23.Fulnecek, J., Matyasek, R. & Kovarík, A. (2002) Mol. Genet. Genomics 268, 510-517. [DOI] [PubMed] [Google Scholar]
- 24.Gruenbaum, Y., Naveh-Many, T., Cedar, H. & Razin, A. (1981) Nature 292, 860-862. [DOI] [PubMed] [Google Scholar]
- 25.Anderson, J. A., Churchill, G. A., Autrique, J. E., Tanksley, S. D. & Sorrells, M. E. (1993) Genome 36, 181-186. [DOI] [PubMed] [Google Scholar]
- 26.Devaux, P., Kilian, A. & Kleinhofs, A. (1993) Mol. Gen. Genet. 241, 647-649. [DOI] [PubMed] [Google Scholar]
- 27.Kleinhofs, A., Kilian, A., Saghai Maroof, M. A., Biyashev, R. M., Hayes, P., Chen, F. Q., Lapitan, N., Fenwick, A., Blake, T. K., Kanazin, V., et al. (1993) Theor. Appl. Genet. 86, 705-712. [DOI] [PubMed] [Google Scholar]
- 28.Kilian, A., Kudrna, D. & Kleinhofs, A. (1999) Genome 42, 412-419. [DOI] [PubMed] [Google Scholar]
- 29.Haubold, B., Kroymann, J., Ratzka, A., Mitchell-Olds, T. & Wiehe, T. (2002) Genetics 161, 1269-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox, M. R. & Ellis, T. H. (2001) Mol. Genet. Genomics 265, 497-507. [DOI] [PubMed] [Google Scholar]
- 31.Isidore, E., van Os, H., Andrzejewski, S., Bakker, J., Barrena, I., Bryan, G. J., Caromel, B., van Eck, H., Ghareeb, B., de Jong, W., et al. (2003) Genetics 165, 2107-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messeguer, R., Ganal, M. W., Steffens, J. C. & Tanksley, S. D. (1991) Plant Mol. Biol. 16, 753-770. [DOI] [PubMed] [Google Scholar]
- 33.Cervera, M. T., Ruiz-Garcia, L. & Martinez-Zapater, J. M. (2002) Mol. Genet. Genomics 268, 543-552. [DOI] [PubMed] [Google Scholar]
- 34.Vuylsteke, M., Mank, R., Antonise, R., Bastiaans, E., Senior, M. L., Stuber, C. W., Melchinger, A. E., Luebberstedt, T., Xia, X. C., Stam, P., et al. (1999) Theor. Appl. Genet. 99, 921-935. [Google Scholar]
- 35.Ashikawa, I. (2001) Plant Mol. Biol. 45, 31-39. [DOI] [PubMed] [Google Scholar]
- 36.Manifesto, M. M., Schlatter, A. R., Hopp, H. E., Suárez, E. Y. & Dubcovsky, J. (2001) Crop Sci. 41, 682-690. [Google Scholar]
- 37.Wu, X. L., Larson, S. R., Hu, Z. M., Palazzo, A. J., Jones, T. A., Wang, R. R., Jensen, K. B. & Chatterton, N. J. (2003) Genome 46, 627-646. [DOI] [PubMed] [Google Scholar]
- 38.Pradhan, A. K., Gupta, V., Mukhopadhyay, A., Arumugam, N., Sodhi, Y. S. & Pental, D. (2003) Theor. Appl. Genet. 106, 607-614. [DOI] [PubMed] [Google Scholar]
- 39.Young, W. P., Schupp, J. M. & Keim, P. (1999) Theor. Appl. Genet. 99, 785-792. [Google Scholar]
- 40.Vuylsteke, M., Mank, R., Brugmans, B., Stam, P. & Kuiper, M. (2000) Mol. Breeding 6, 265-276. [Google Scholar]
- 41.Winzeler, E. A., Richards, D. R., Conway, A. R., Goldstein, A. L., Kalman, S., McCullough, M. J., McCusker, J. H., Stevens, D. A., Wodicka, L., Lockhart, D. J., Davis, R. W. (1998) Science 281, 1194-1197. [DOI] [PubMed] [Google Scholar]
- 42.Borevitz, J. O., Liang, D., Plouffe, D., Chang, H. S., Zhu, T., Weigel, D., Berry, C. C., Winzeler, E. & Chory, J. (2003) Genome Res. 13, 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanksley, S. D., Ganal, M. W. & Martin, G. B. (1995) Trends Genet. 11, 63-68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.