Abstract
As individual success often comes at the expense of others, interactions between the members of a species are frequently antagonistic, especially in the context of reproduction. In theory, this conflict may be reduced in magnitude when kin interact, as cooperative behaviour between relatives can result in increased inclusive fitness. Recent tests of the potential role of cooperative behaviour between brothers in Drosophila melanogaster have proved to be both exciting and controversial. We set out to replicate these experiments, which have profound implications for the study of kin selection and sexual conflict, and to expand upon them by also examining the potential role of kinship between males and females in reproductive interactions. While we did observe reduced fighting and courtship effort between competing brothers, contrary to previous studies we did not detect any fitness benefit to females as a result of the modification of male antagonistic behaviours. Furthermore, we did not observe any differential treatment of females by their brothers, as would be expected if the intensity of sexual conflict was mediated by kin selection. In the light of these results, we propose an alternative explanation for observed differences in male–male conflict and provide preliminary empirical support for this hypothesis.
Keywords: sexual conflict, kin selection, inclusive fitness, sexual selection, aggression, social behaviour
1. Introduction
An individual's fitness is defined by the size of their contribution of alleles to the next generation [1]. As such, there is often strong selection on individuals for traits and/or behaviours that maximize fitness via outcompeting rivals [2–4]. Such ‘selfish’ strategies are favoured by selection when an individual is surrounded by unrelated conspecifics, because fitness is strongly associated with the ‘direct’ transmission of genetic material from parent to offspring [5,6]. However, when the competitors also include relatives, it may be beneficial (and thus adaptive) to act less antagonistically towards those individuals. This is because the reproductive success of one's relatives also results in ‘indirect’ fitness benefits owing to the increased transmission of alleles inherited from a common ancestor [7]. Thus, according to kin selection theory, it is hypothesized that individuals should act more altruistically towards their relatives to maximize their inclusive fitness, as long as the net benefit(s) outweigh costs [7,8].
Several theoretical studies have attempted to identify how the predictions of kin selection theory may interact with those of sexual conflict theory [8–10]. According to these models, there are circumstances in which altruistic behaviour towards relatives should occur, both in intra-sexual interactions [9,10] and in inter-sexual interactions [8]. When competing against same-sex relatives, one adaptive strategy is to reduce the amount of harm inflicted on these ‘rivals', thereby maximizing potential inclusive fitness benefits via the enhanced success of relatives and the shared reproductive resource [10]. When the potential for mating exists between relatives, it is further hypothesized that there exists an optimal level for inbreeding, where it can be beneficial to mate with a relative [8]. In these situations, where an individual benefits from the increased transmission of alleles owing to shared ancestry with a related mate, a reduction in the level of inter-sexual harm on this relative and thus a direct gain (owing to lifetime reproductive success (LRS) of a related mate) is also predicted to occur [8].
Inspired by these predictions, Carazo et al. [11] attempted to empirically determine whether the presence of familial bonds between potential male rivals modulated the expression of antagonistic behaviour and subsequently influenced fitness outcomes using the model species Drosophila melanogaster. In D. melanogaster, males often fight with each other for pre-copulatory access to females [12–14], and competition continues in the post-copulatory realm via the effects of the sperm and accessory gland products (ACPs) of rival males, which are transferred in their ejaculates [15]. Females are often harmed by males, as a direct result of harassment by courting males [16–20], the physical harm associated with mating [16,21,22] and/or the toxic side effects of the ACPs [23–27]. With a wide variety of well-documented intra- and inter-sexual interactions, this species is well suited to the study of sexual conflict. In a series of assays, Carazo et al. [11] experimentally housed a single adult female fly with three males (all unrelated to her) where the relatedness between these rivals was experimentally manipulated. Groups comprise: (i) full-sibling males, (ii) unrelated males, or (iii) two brothers and a third unrelated male. Male behavioural traits, such as the frequency of male–male fighting, the intensity of courtship and mating rate were measured, as were a number of male fitness variables. The longevity, reproductive lifespan and LRS of the females were measured. They observed that when some (or all) males in a group were related, fighting frequencies and courtship intensities were significantly lower than in groups of unrelated males. Furthermore, females housed with groups of brothers lived longer, and produced more offspring than females housed with groups of unrelated males, a finding that Carazo et al. [11] attributed to differences in harm associated with copulatory behaviours. Overall, Carazo et al. [11] concluded that male flies modulated their intra-sexual behaviour to act less selfishly towards relatives in order to benefit via indirect fitness gains in a manner theoretically consistent with the predictions of kin selection and inclusive fitness models [8,10].
The results of Carazo et al. [11]—which have potentially broad implications for the understanding of sexual selection and the evolution of fitness-maximizing strategies [28]—have been received with great interest [28] and some scepticism. Hollis et al. [29] argued that by failing to properly control for developmental and social familiarity between relatives, the results of Carazo et al. [11] cannot be clearly interpreted as arising due to kin selection alone. Specifically, Hollis et al. [29] argued that increased male–male aggression in the unrelated males treatment could be an artefact of the pre-trial developmental conditions. Whereas the relatives all developed in the same vial, the unrelated individuals were all obtained from different vials. Thus, the difference ascribed to perception of kinship might be owing to social/developmental familiarity. In order to test this theory, Hollis et al. [29] conducted their own set of experiments where relatedness and developmental familiarity were independently manipulated. They found that females housed with sets of brothers had greater LRS, but that this effect was only manifested if those brothers had also shared a pre-trial developmental environment. Brothers reared in separate vials had the same effect on female LRS as sets of unrelated (and unfamiliar) males. As Hollis et al. [29] did not measure male–male aggression or courtship intensity, the functional changes responsible for the differences in female LRS could only be inferred. A follow-up experiment by Carazo et al. [30] has also reported higher rates of male–male fighting (but not courtship intensity) when males are unrelated and unfamiliar to each other (reared in separate vials) compared with groups of brothers raised in the same environment. Most recently, a study by Chippindale et al. [31] that used a similar protocol to that of Carazo et al. [11] failed to find any significant differences in the longevity, reproductive lifespan or LRS of females housed with three brothers or three unrelated males.
Together, these studies offer an intriguing (albeit controversial) preliminary perspective on the role for relatedness/kin selection in behaviour modulation, particularly because their findings are in some cases at odds with one another. Moreover, many aspects of the role of kin selection and its relation to the study of sexual conflict remain unknown and untested. Of considerable importance is the role of inter-sexual relatedness: what happens when males encounter their sisters as potential mates? The logic invoked by Carazo et al. [11] and Pizzari et al. [10] for behavioural modification driven by kin selection in intra-sexual interactions should also apply for inter-sexual interactions. In a species such as D. melanogaster where there is considerable inter-locus sexual conflict [15–21], small changes in the intensity of inter-sexual interactions have the potential to dramatically affect male and female LRS. Thus, determining how kinship and social dynamics might influence sexual conflict has important implications for understanding sexual coevolution in this species, as well as for social evolution in general [28].
In order to more fully understand the role of inter- and intra-sexual relatedness on reproductive behaviour and fitness, we conducted a series of experiments designed to replicate and elaborate upon the first Carazo et al. [11] study. Using D. melanogaster, we examined variation in behaviour and fitness under a number of possible mating and social settings that differed in the potential for male–male competition, the degree of relatedness between males and the relatedness of male(s) to a target female. We measured the effects of these combinations in a number of ways: by quantifying male fighting frequencies, courtship intensities, female longevities, female reproductive lifespans and LRS. We designed this study to provide our own assessment for the role (if any) for relatedness and kin selection in inter- and intra-sexual selection.
2. Material and methods
(a). Experimental population and culturing protocol
All flies used in this study originated from the large, outbred, wild-type Ives (hereafter ‘IV’) population of D. melanogaster, which was created using a sample of wild-caught flies from South Amherst, MA, USA in 1975 and has been maintained under standardized culture conditions since 1980 [32]. Our population was obtained from the laboratory of Adam Chippindale (Queen's University, Kingston, Canada) in 2011. These flies are housed in vials containing a standard banana/agar/killed-yeast medium, and develop at a controlled density of approximately 100 eggs per vial. Flies are raised at 25°C and 60% humidity, on a 12 L : 12 D diurnal light cycle, and this population is maintained on a discrete (non-overlapping) 14-day generation culture cycle; flies are cultured en masse using light CO2 anaesthesia [33].
From the IV population, we created 45 familial lineages for use in our assays. Each lineage was created by first mating a virgin female to a single, randomly selected, unrelated male. In each of the following 12 generations, a single virgin brother and sister were mated to propagate the lineage. Subsequently, the size of each lineage was increased to 100 adults per generation and cultured every 14 days on non-overlapping generations. Before the start of the assay, a replicate culture of each lineage, temporally offset by 7 days, was established, which permitted continuous access to young adult males (less than 4 days post-eclosion) from each lineage throughout the course of the experiment. The protocol for the creation of familial lineages resulted in individuals in each familial lineage having a high degree of relatedness (at least r = 0.9255, as per Falconer [1]). We used these highly related familial lineages in our experiment in order to maximize the probability that any kin-related changes to sexual interactions would be detected. Furthermore, if there exists preference for mating with relatives, as a number of past studies have suggested [33,34], and if this preference is driven by genetic factors, inbreeding should enhance these preferences. The use of highly inbred lines, however, does come with the caveat that fitness-associated traits often exhibit directional dominance [35–37], with inbreeding resulting in reduced fitness in offspring. If kin recognition is such a trait, our inbreeding procedure may have (ironically) resulted in flies with reduced ability to detect their kin.
(b). Mating treatments and fly handling
We began by collecting five adult females as virgins (less than 2 h of eclosion) from each of the 45 familial lineages, which were held individually in vials containing standard medium. At the same time, adult males were also collected as virgins (less than 2 h of eclosion) from all familial lineages. Males were assigned to one of five different experimental treatments (electronic supplementary material, figure S1) compiled in replicate for each family (n = 45) and were held in these groups for a maximum of 72 h post-eclosion until the start of the experiment. While we did not control for larval social familiarity, all triple-male groups received comparable adult socialization prior to introduction to the female, regardless of their treatment. Additionally, if larval familiarity is required for kin recognition, allowing brothers to develop in the same environment should increase their ability to modulate their behaviour when encountering each other.
Our treatments were designed to test the following questions: (i) does the relatedness of a group of males alter intra-sexual behavioural interactions and/or their effects on a target female? and (ii) does relatedness between the sexes alter the inter-sexual interactions between a single male or group of males and a target female, or influence a female's fitness or longevity? The experimental treatments were: (i) ‘related-pair’, a single male from the same familial lineage as the target female; (ii) ‘unrelated-pair’, a single male from a different familial lineage than the target female; (iii) ‘all-related’, three males related to each other (same familial lineage) and to the target female; (iv) ‘males-related’, three males related to each other but from a different familial lineage than the target female; and (v) ‘all-unrelated’, three males unrelated (different familial lineages) to either each other or to the target female. In treatments involving unrelated males (both single- and triple-male treatments) combinations of lineages were assigned randomly and were equally represented. To better study the consequences arising from male–male competition, we ensured that for each set of treatments for individual females of a given familial lineage, the same lineage of males was used in the unrelated-pair treatment as in the males-related treatment (electronic supplementary material, figure S1).
(c). Longevity and male behaviour experiment
The experiment began by combining unanaesthetized males and females into a single vial by lightly tapping the male(s) into each female's vial. Following the combination of flies, all vials (n = 45 × 5 = 225) were placed horizontally in a quiet, temperature-and-humidity-controlled room. Vials were observed and behaviours scored daily in five sessions beginning at 9.00. In each session, vials were scanned for a period of 5 s each, during which counts were made of the number of instances of copulation, courtship or fighting between males. In the case of fighting, fights involving two males were scored as a single event, while cases of all three males fighting with each other were recorded as double events. The numbers of each type of event scored during daily sessions were summed prior to analysis. All vials were observed on a daily basis until the death of the target female, at which time the date of each death (female longevity) was recorded.
In each of the first 3 days of the experiment, all living flies were transferred (after the completion of observations) using light anaesthesia to new vials containing fresh media. The numbers of eggs laid in these three sets of vials were immediately counted. After 14 days, the numbers of eclosed adults in the vials were also counted. Data from egg-to-adult viability yielded from the first 3 days of the experiment were later used to correct for differences between vials in offspring numbers resulting from inbreeding depression. For the remainder of the experiment, flies were transferred (with light anaesthesia) to new vials every second day. Eggs laid during these time periods were not counted, but the numbers of offspring eclosing from these vials 14 days later were recorded. These numbers were adjusted using the vial-specific egg-to-adult survivorship rates to generate estimated total egg values. Based on the final date of viable offspring production, we were also able to quantify a female's reproductive lifespan. Following the procedure of Carazo et al. [11], all males were replaced, on average, every 7 days to ensure male co-ageing did not impact female mortality or fecundity. This was accomplished by replacing all old males every 6th or 8th day of the experiment (corresponding to the closest date of transfer of female flies to new vials) with virgin adult males collected from one of the two temporally offset sets of familial lineage populations. This protocol of vial-transfer and male-replacement was continued for each vial until the death of the target female.
(d). Statistical analysis
All statistical analyses were performed using R (v. 3.2.0, the R Foundation for Statistical Computing [38]). When analysing cumulative totals for courtship and fighting events, we calculated the daily rate for each of these events in order to control for any potential confounds associated with differences in female longevity; this was done for estimated egg production rates (eggs per day) as well, although estimated lifetime totals were also analysed. Estimates for total egg production and egg production rate were compared across treatments. Total egg estimates were compared for the entire lifespan of the female, and for the first 3 days of the experiment—a time period that is evolutionarily relevant to the female owing to the nature of the IV population culture protocol. Fighting and courtship variables were compared as daily rates, while copulation events were compared as lifetime totals. All data were assessed for normality and homogeneity of variance, using the Shapiro–Wilk test and Levene's Test, respectively, in order to determine whether data met parametric assumptions.
To compare behaviour and fitness metrics for the three triple-male treatments (n = 45 each), data were analysed using general linear models (GLMs) constructed with quasi-poisson error distributions. A model was created for each class of behavioural or fitness response, with treatment as an independent factor. The significance of treatment was determined using the Anova function (in the car package), with type II sums of squares. Following the identification of significant differences between treatments, contrast analysis was used to test a priori hypotheses about differences between treatments. Specifically, we contrasted data for: (i) groups of brother (all-related and males-related) were contrasted against the all unrelated treatment; (ii) the all-related treatment against the males-related treatment; and (iii) and the males-related and the all-unrelated treatments against the all-related treatment. Respectively, these contrasts allowed us to assess: (i) the effects of intra-sexual relatedness, regardless of male relation to a focal female; (ii) the effects of inter-sexual relatedness when males are all related to one another; and (iii) the effects of inter-sexual relatedness, regardless of male intra-sexual relatedness.
We created GLM models using data from the related-pair, unrelated-pair, all-related and males-related treatments (n = 45 each) to assess the effects of both relatedness and exposure to multiple males. In these models, we used estimated egg numbers or behaviour traits as the response variable, with inter-sexual relatedness, the number of males as factors and their interactions as independent variables, and specified a quasi-poisson error distribution. Significance of these terms was determined as noted previously using likelihood ratio (LR) tests. To examine whether the presence of male rivals caused equivalent changes to behaviour and fitness outcomes across treatments differing in inter-sexual relatedness, data from single-male treatments (related-pair and unrelated-pair) were also compared with their corresponding triple-male treatments (all-related and males-related). This was accomplished using paired t-tests or paired Wilcox signed-rank tests when data did not meet parametric assumptions.
For both comparisons of intra-sexual and inter-sexual relatedness, female longevity and reproductive lifespan were assessed via survivorship analysis modelling (using the survreg function in the survival package), with number of males, inter-sexual relatedness and their interactions as independent variables.
3. Results
(a). Effects of male intra-sexual relatedness
The daily courtship rate of males in the triple-male groups differed significantly between treatments, both over the entire lifespan (henceforth ‘full-term’) of the female (LR χ22 = 10.202, p = 0.006), as well as within the first 3 days of the assay (LR χ22 = 8.141, p = 0.017). Post hoc contrast analyses revealed that overall, brothers courted females less frequently than did males unrelated to each other (estimate ± s.e. = 0.340 ± 0.106, z = 3.200, p = 0.004). This phenomenon was of medium effect size (Cliff's delta = 0.343, 95% confidence interval (CI) = [0.141, 0.518]). No differences arising from variation in inter-sexual relatedness were found for courtship frequency in either the contrast of all-related against males-related (est. ± s.e. = −0.023 ± 0.065, z = −0.356, p = 0.928), or the contrast of all-related against males-related and all-unrelated (est. ± s.e. = 0.135 ± 0.110, z = 1.226, p = 0.419). Daily fighting rate also differed (figure 1) between treatments (LR χ22 = 6.941, p = 0.031). Contrast analyses of all-unrelated against all-related and males-related revealed that unrelated males fought among themselves more frequently than did related males (est. ± s.e. = 0.448 ± 0.172, z = 2.612, p = 0.022). Effect size for this difference in fighting rate was small (Cliff's delta = 0.245, 95% CI = [0.051, 0.422]). Again treatments did not differ in the contrast of all-related against males-related (est. ± s.e. = 0.055 ± 0.107, z = 0.520, p = 0.853) or the contrast of all-related against males-related and all-unrelated combined (est. ± s.e. = 0.3078 ± 0.183, z = 1.682, p = 0.197), indicating that inter-sexual relatedness did not modulate male aggressiveness in the presence or the absence of control for male–male relatedness. Table 1 summarizes the values of these behaviours and fitness variables across treatments.
Figure 1.
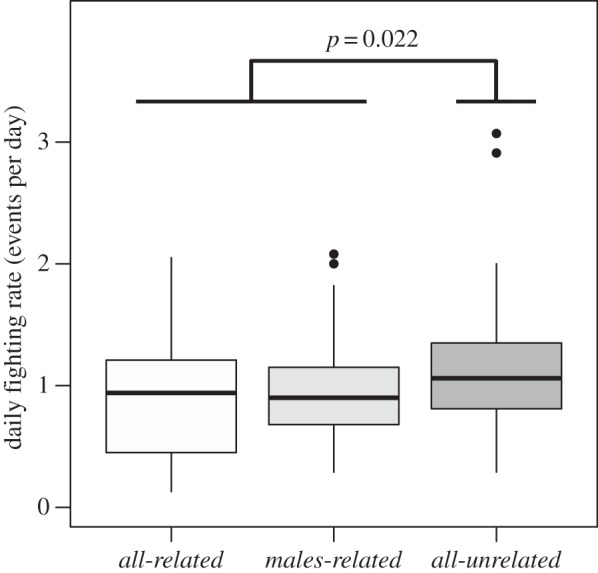
Boxplots showing distribution of daily fighting rates observed in vials of D. melanogaster containing one female and a trio of males, for treatments differing in the type of inter- and intra-sexual relatedness. In ‘all-related’ vials the males are related to each other and to the female, in ‘males-related’ vials the males are related to each other but not to the female, and in ‘all-unrelated’ vials all flies are from different familial lineages. While statistically significant, male–male relatedness is only a marginal predictor of overall fighting rates between treatments (Akaike information criterion (AIC) = 194.841, adjusted R2 = 0.042). The box encloses values between the first and third quartiles of the data (the inter-quartile range, IQR), while the horizontal bar within the box indicates the median. Whiskers extend from the box to largest/smallest values that are within 1.5 × the IQR of the box. Values outside that range are outliers and are indicated by circles.
Table 1.
Summary statistics (mean ± s.e.) from behavioural and life-history data collected from our five treatments (n = 45 for each): related-pair, a female with a single sibling male; unrelated-pair, a female with a single unrelated male; all-related, a female with three males related to her and each other; males-related, a female with a three males related to each other but not to her; and all-unrelated, a female with a group of entirely unrelated males, either to her or each other.
| variable | related-pair | unrelated-pair | all-related | males-related | all-unrelated |
|---|---|---|---|---|---|
| behaviour | |||||
| courtships (events day−1) | 1.08 ± 0.07 | 0.95 ± 0.06 | 2.15 ± 0.11 | 2.10 ± 0.09 | 2.51 ± 0.10 |
| fighting (events day−1) | n.a. | n.a. | 0.91 ± 0.07 | 0.96 ± 0.06 | 1.17 ± 0.08 |
| copulations (lifetime) | 0.42 ± 0.12 | 0.51 ± 0.13 | 0.78 ± 0.16 | 0.84 ± 0.17 | 0.62 ± 0.13 |
| fitness | |||||
| longevity (days) | 29.38 ± 1.03 | 28.44 ± 1.41 | 24.20 ± 1.14 | 25.00 ± 1.00 | 24.82 ± 1.18 |
| reproductive lifespan (days) | 23.51 ± 1.44 | 23.47 ± 1.58 | 18.64 ± 1.54 | 18.00 ± 1.48 | 19.47 ± 1.32 |
| egg production | |||||
| lifetime (total) | 154.70 ± 18.12 | 163.10 ± 20.81 | 117.90 ± 14.38 | 109.60 ± 12.69 | 140.10 ± 16.42 |
| lifetime (eggs day−1) | 4.92 ± 0.49 | 5.25 ± 0.54 | 4.48 ± 0.48 | 4.23 ± 0.42 | 5.36 ± 0.55 |
| short-term (total) | 19.51 ± 1.93 | 19.11 ± 1.51 | 18.84 ± 1.85 | 18.60 ± 1.70 | 24.00 ± 1.27 |
Despite the observed differences in courtship and fighting frequencies between treatments, no differences were found between these treatments for mean number of copulations observed (LR χ22 = 1.142, p = 0.565). Furthermore, no significant differences were observed between treatments for estimated total egg production either in the full-term (LR χ22 = 2.336, p = 0.311) or in the first 3 days of the assay (LR χ22 = 0.475, p = 0.789); egg production rate was also found not to differ between treatments (LR χ22 = 2.937, p = 0.230). Finally, using survivorship modelling, we detected no differences between treatments in female longevity (χ22 = 1.13, p = 0.57, n = 135) or reproductive lifespan (χ22 = 0.49, p = 0.78, n = 135) for triple-male treatments.
(b). Effects of inter-sexual relatedness
The effects of inter-sexual relatedness were assessed via the paired comparison of the two paired single-male treatments—related-pair versus unrelated-pair, and the correspondingly paired three-male treatments—all-related versus males-related. In these analyses, treatments were paired according to female lineage. The results of these analyses are presented in the electronic supplementary material, table S1, and revealed no significant differences between treatments differing in male–female relatedness for any of the variables considered. We also analysed whether the number of males in a vial and inter-sexual relatedness (and the interaction between these two factors) were associated with differences in behaviour or fitness variables. The rate of courtship was significantly lower in single-male vials than in triple-male vials (F1,176 = 171.603, p < 2 × 10−16). However, there was no significant effect of the inter-sexual relatedness of flies (F1,176 = 1.150, p = 0.285), or the interaction between these factors (F1,176 = 0.243, p = 0.623). The number of males a female was exposed to significantly affected lifespan (χ21 = 19.409, p = 1.055 × 10−5), with female exposure to trios of male experiencing earlier mortality than those housed with single males. Female longevity did not differ based on inter-sexual relatedness (χ21 = 0.001, p = 0.976) or the interaction of relatedness and number of males (χ21 = 0.144, p = 0.704). Reproductive lifespan was significantly lower in triple-male groups (χ21 = 7.020, p = 0.008) than in single-male groups (figure 2), but there was no significant effect of inter-sexual relatedness (χ21 = 0.024, p = 0.876) or the interaction (χ21 = 0.102, p = 0.749).
Figure 2.
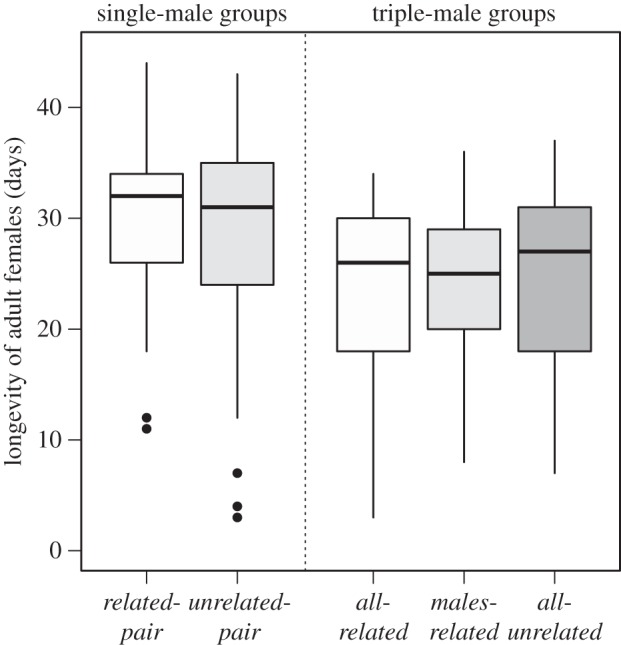
Boxplots showing distribution of female longevities (in days) observed in vials of D. melanogaster containing one female and male(s), in treatments differing in the number of males and/or the type of inter- and intra-sexual relatedness. Vials in the ‘related-pair’ consist of a single male and female from the same familial lineage; while in the ‘unrelated-pair’ treatment, the flies are from different lineages. In ‘all-related’ vials the males are related to each other and to the female, in ‘males-related’ vials the males are related to each other but not to the female and in ‘all-unrelated’ vials the all flies are from different familial lineages. Shading of boxes indicates whether the male(s) in a vial are related to the female (white) or are unrelated (grey). Boxplot components are as described in figure 1.
We observed a significant effect of the number of males in a vial on the lifetime production of eggs (F1,176 = 7.250, p = 0.008). Females housed with three males produced fewer eggs when compared with single-male groups. However, total egg production was not affected by either inter-sexual relatedness (F1,176 < 0.001, p = 0.998) or its interaction with male quantity (F1,176 = 0.247, p = 0.620). When egg production rates were compared, we found no significant effect of the number of males in a vial (F1,176 = 2.265, p = 0.134), the relation of these males to the female (F1,176 = 0.006, p = 0.940) or the interaction of these factors (F1,176 = 0.363, p = 0.548). Similarly, no significant differences were detected for short-term total egg production (number of males: F1,176 = 0.113, p = 0.738; inter-sexual relatedness: F1,176 = 0.034, p = 0.855; interaction: F1,176 = 0.002, p = 0.965).
4. Discussion
In the quest to maximize fitness, selection has often favoured the evolution of traits by which an individual benefits by acting antagonistically towards others in the population (be they males or females) [12–14,16,19,21,25]. It has been hypothesized that the magnitude of this conflict may be relaxed in the presence of kin, as an individual can benefit indirectly via the reproductive success of relatives [9,10]. A recent high-profile paper by Carazo et al. [11] attempted to test this prediction using D. melanogaster (an important model species for the study of sexual selection and sexual conflict), and found evidence of harm modulation. Their results have been controversial [29,31], and motivated our attempt to replicate and extend the experiments of Carazo et al. [11,30]. In our study, we set out to examine the potential role of kinship in intra-sexual interactions and fitness consequences, and further investigate inter-sexual interactions as this factor that has not previously been considered. Our assays revealed higher fighting and courtship rates in groups of unrelated males compared to groups of brothers, consistent with the observations of Carazo et al. [11] and (partially) Carazo et al. [30]. Similar results have been interpreted [11,30] as an indicator of altruistic behaviour between kin for the purpose of increasing inclusive fitness. However, we did not observe any decrease in the magnitude of harm inflicted on females when males were related. Females in our triple-male treatments did not differ in their longevity, or in the length of their reproductive lifespan. Furthermore, intra-sexual kinship had no effect on female reproductive output, measured in the currency of egg production, in either the short-term or over a female's entire lifespan.
Additionally, we found no effect of kinship on any of the inter-sexual interactions or fitness outcomes in our paired comparisons of single or triple-male treatments (electronic supplementary material, table S1). While the number of males had a profound impact on the rate of intra-sexual behaviour and female longevity (figure 2), most fitness-related indices were unaffected by either kinship between male(s) and the female in the vial, or the interaction between kinship and the number of males (table 1). In other words, males acted equally as antagonistically towards their sisters as they did towards unrelated females. In their study, Carazo et al. [11] had suggested that males modulate aggression when housed with kin to reduce the magnitude of harm done to brothers and to shared mates, thereby increasing indirect fitness gains. However, our study, like that of Chippindale et al. [31], reveals no such fitness benefit. Additionally, the logic invoked to predict reduced intra-sexual aggression between brothers should also theoretically apply for inter-sexual interactions, as a male mating with a relative stands to gain twofold by acting less aggressively towards his sister—both directly via increased LRS from a mate with greater fecundity, and indirectly via the transmission of shared alleles inherited from the female. Therefore, the benefits of preferential treatment towards opposite-sex kin are potentially greater than cooperative behaviour between same-sex kin. However, the differences we see in the fitness of females exposed to a single male versus three males (figure 2) indicates that males are not reducing the magnitude of harm they cause to sisters in any meaningful way.
It is possible that fruit flies are unable to recognize female kin. This would account for the equal treatment of all potential mates regardless of shared relation. However, a study by Tan et al. [39] suggests that male D. melanogaster are capable of differentiating between females of different genetic or environmental backgrounds, which could theoretically include an ability to recognize individuals with whom they share a genetic or environmental background as well. A number of other studies do indeed report that D. melanogaster are capable of kin recognition, as individuals were observed to preferentially mate with relatives [33,34], which necessitates the ability to recognize kin. In our assay, we used flies with a high degree of relatedness, to increase the likelihood that differences between individuals would be easier to perceive, and so that the gains from indirect fitness would be maximized. Yet we found no evidence of inter-sexual kin-related modulation of antagonistic behaviour, and little evidence of intra-sexual modulation—save for differences in fighting and courtship rates. But then how can we account for these observations (as well as those of Carazo et al. [11,30])? Our results have led us to question the plausibility of the kinship-modulated conflict explanation of Carazo et al. [11] and instead consider other reasons why levels of fighting and courtship may vary between treatments, independent of intra-sexual relatedness. Here we propose one potential alternative explanation, and present preliminary data in support of this hypothesis.
When considering groups of unrelated flies, it is almost inevitable that they will be more variable than groups of related flies. If variation in behaviour, such as aggression level or courtship intensity, has a genetic basis—something that has been extensively documented in D. melanogaster [13,40–43]—it is worth considering the possibility that the increased rates of fighting and/or courtship by genetically heterogeneous groups of males are the product of combining different genotypes together, and thus novel social environments [44,45]. Indeed, Saltz [13] reported that the overall aggressiveness of a group of male D. melanogaster is influenced by the genetic composition of that group, or rather, by indirect genetic effects (IGEs). Further evidence for IGEs is provided by Kent et al. [46], who report that chemical signalling employed by this species also varies based on the genotypes of neighbours; similar effects of genetic social environment have been documented in mice [47]. Therefore, if genetically determined aggression levels vary within our population, it is possible that social heterogeneity, rather than kinship, may be responsible for the between-treatment variation seen in overall group aggression.
To begin to test this hypothesis, we made use of the fact that for each of our 45 all-unrelated vials, we knew the familial identities of the three males that made up each trio, and that we also had made independent measurements of how members of each family acted when housed with others of the same sex and lineage (i.e. the phenotypic values measured from the males-related vials). Thus, we were able to create a synthetic dataset comprised a response variable (the fighting rate values observed in the 45 all-unrelated vials), and three independent variables (corresponding to the fighting rate values of the three families that made up each specific trio, ranked from most to least aggressive within each trio). This ranking was essential to understand the phenotypic composition of fighting rates in each combination of males, but it resulted in multicollinearity of the variables. Thus, we first performed a principal component analysis (PCA) on these three variables (using the princomp function in R) to convert them into a set of orthogonal, linearly uncorrelated variables (see table 2 for PCA loadings). Using the re-scored data, we created a model to determine whether we could predict the observed phenotypic variation in the all-unrelated vials (response variable). Our original model included all three principle components (and their interactions) as independent factors and was subsequently simplified by the sequential removal of all non-significant parameters, until we reached the minimum adequate model. Despite the limitations in our dataset, this model (table 3) was quite effective at describing a sizable fraction of the variation in fighting values observed in groups of unrelated males. In our model, we saw that in the synthetic group trios, when the range of ‘fighting phenotypes' was reduced (i.e. male aggression levels were more similar to each other; electronic supplementary material, figure S2), and when the male of intermediate aggressiveness displayed a relatively high aggression level within that range (electronic supplementary material, figure S3), there was a correspondingly higher frequency of overall conflict in the all-unrelated vial (table 3). In essence, when trios comprise males with relatively similar levels of aggression, and the aggression phenotypes of the two most aggressive lineages were even more similar, this was associated with a greater frequency of fighting within the all-unrelated trio. This model (table 3) was able to predict group phenotypic variation in the all-unrelated fighting values. This preliminary analysis provides some tentative support for social composition as a driver of the behavioural differences observed in groups of unrelated individuals, but more research on this subject is warranted. Though we did not set out to deliberately create combinations that differed in their aggressive behaviours, our work, similar to the work of Billeter et al. [48], Carazo et al. [11,30] and Saltz [13], does highlight the complexities associated with IGEs and is suggestive of a fruitful avenue for future research.
Table 2.
Loadings of three related-males fighting variables on principal components for the 45 all-unrelated combinations trios of males in our ‘synthetic’ dataset. (In PC1, all the loadings are positively correlated and are of similar magnitude, an inevitable result of the ranking of males by their lineage's aggressiveness in the creation of the dataset. Variation in PC2 is primarily defined by the range of aggressiveness phenotypes in the trio—large positive values when they are very different, and negative values when they are closer in magnitude. Variation in PC3 is primarily defined by the similarity in the level of aggressiveness of the intermediate and low aggressive lineages. The large positive PC values occur when their phenotypes are closer in magnitude.)
| variable | PC1 | PC2 | PC3 |
|---|---|---|---|
| most aggressive family | 0.555 | 0.797 | 0.239 |
| intermediate aggressive family | 0.599 | −0.184 | −0.780 |
| least aggressive family | 0.577 | −0.576 | 0.579 |
| eigenvalue | 2.174 | 0.489 | 0.337 |
| % of variance explained | 0.725 | 0.163 | 0.112 |
Table 3.
Minimum adequate model (AIC = 68.439, F3,41 = 4.982, p = 0.005, adjusted R2 = 0.214) created to explain phenotypic variation in observed daily fighting rates of trios of males in all-unrelated vials.
| variable | d.f. | SS | F | p-value |
|---|---|---|---|---|
| PC2 | 1 | 1.431 | 6.079 | 0.018 |
| PC3 | 1 | 0.041 | 0.172 | 0.681 |
| PC2 × PC3 | 1 | 2.048 | 8.697 | 0.005 |
| residuals | 41 | 9.655 | — | — |
In summary, our experiment tested and extended the predictions of the kin selection model for conflict modulation. In our population, evidence for any modulation of conflict owing to shared relatedness is tenuous. While we did detect differences in intra-sexual interactions between related males, we did not observe any such effect of kinship on inter-sexual interactions, which are theoretically a more likely target for modulation by kin selection. Furthermore, in spite of altered male–male interactions, we did not observe any mediating effect on fitness outcomes for these groups. Because of the success with which our alternative model is able to predict male–male aggression between non-kin, we propose that the dynamics and structures of social groups (IGEs) may be more important factors both in our experiment and those of Carazo et al. [11,30] and should be considered in future studies. Our results join the ranks of many other studies documenting the importance of social structure and their effects on conflict and other social behaviours [13,10,47,48].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We would like to thank all the members of the Long laboratory at WLU for their contributions to this project, with particular thanks to Oluwapamile Olayemi, D. Felice, T. Balasubramaniam, A. Legere, K. Schang, H. Malek, N. Jivraj and S. MacDonald. We would also like to thank Dr S. Ramsay and Dr M. Costea for their support and input.
Data accessibility
Data available online at dryad: http://dx.doi.org/10.5061/dryad.q28b2.
Competing interests
The authors declare no competing interests.
Funding
This research was funded through a Natural Sciences and Engineering Research Council of Canada grant held by T.A.F.L., and by funding from Wilfrid Laurier University.
References
- 1.Falkner DS. 1981. Introduction to quantitative genetics, 2nd edn Essex, UK: Longman Scientific & Technical. [Google Scholar]
- 2.Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hoang A, Hill CE, Beerli P, Kingsolver JG. 2001. Strength and tempo of directional selection in the wild. Proc. Natl Acad. Sci. USA 98, 9157–9160. ( 10.1073/pnas.161281098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kingsolver JG, Hoekstra HE, Hoekstra JM, Berrigan D, Vignieri SN, Hill CE, Hoang A, Gilbert P, Beerli P. 2001. The strength of phenotypic selection in natural populations. Am. Nat. 157, 245–261. ( 10.1086/319193) [DOI] [PubMed] [Google Scholar]
- 4.West SA, Pen I, Griffin AS. 2002. Cooperation and competition between relatives. Science 5, 72–75. ( 10.1126/science.1065507) [DOI] [PubMed] [Google Scholar]
- 5.Brockelman WY. 1975. Competition, the fitness of offspring, and optimal clutch size. Am. Nat. 109, 677–699. ( 10.1086/283037) [DOI] [Google Scholar]
- 6.Kingsolver JG, Pfenning DW. 2007. Patterns and power of phenotypic selection in nature. BioScience 57, 561–572. ( 10.1641/B570706) [DOI] [Google Scholar]
- 7.Hamilton WD. 1964. The genetical coevolution social behaviour I. J. Theor. Biol. 7, 1–16. ( 10.1016/0022-5193(64)90038-4) [DOI] [PubMed] [Google Scholar]
- 8.Kokko H, Ots I. 2006. When not to avoid inbreeding. Evolution 60, 467–475. ( 10.1111/j.1365-294X.2011.05325.x.) [DOI] [PubMed] [Google Scholar]
- 9.Díaz-Muñoz SI, DuVal EH, Krakauer AH, Lacey EA. 2014. Cooperating to compete: altruism, sexual selection and causes of male reproductive cooperation. Anim. Behav. 88, 67–78. ( 10.1016/j.anbehav.2013.11.008) [DOI] [Google Scholar]
- 10.Pizzari T, Biernaskie JM, Carazo P. 2015. Inclusive fitness and sexual conflict: how population structure can modulate the battle of the sexes. BioEssays 37, 155–166. ( 10.1002/bies.201400130) [DOI] [PubMed] [Google Scholar]
- 11.Carazo P, Tan CKW, Allen F, Wigby S, Pizzari T. 2014. Within-group male relatedness reduced harm to females in Drosophila. Nature 505, 672–675. ( 10.1038/nature12949) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yurkovic A, Wang O, Basu AC, Kravits EA. 2006. Learning and memory associated with aggression in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 103, 17 519–17 524. ( 10.1073/pnas.0608211103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saltz JB. 2013. Genetic composition of social groups influences male aggressive behaviour and fitness in natural genotypes of Drosophila melanogaster. Proc. R. Soc. B 280, 20131926 ( 10.1098/rspb.2013.1926) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saleem S, Ruggles PH, Abbott WKA, Carney GE. 2014. Sexual experience enhances Drosophila melanogaster males mating behavior and success. PLoS ONE 9, e96639 ( 10.1371/journal.pone.0096639) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birkhead TR, Pizzari T. 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3, 262–273. ( 10.1038/nrg774) [DOI] [PubMed] [Google Scholar]
- 16.Partridge L, Fowler K. 1990. Non-mating costs of exposure to males in female Drosophila melanogaster. J. Insect Physiol. 36, 419–425. ( 10.1016/0022-1910(90)90059-O) [DOI] [Google Scholar]
- 17.McGraw LA, Gibson G, Clark AG. 2004. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr. Biol. 14, 1506–1514. ( 10.1016/j.cub.2004.08.028) [DOI] [PubMed] [Google Scholar]
- 18.Rice WR, Stewart AD, Morrow EH, Linder JE, Orteiza N, Byrne PG. 2006. Assessing sexual conflict in the Drosophila melanogaster laboratory model system. Phil. Trans. R. Soc. B 361, 287–299. ( 10.1098/rstb.2005.1787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long TAF, Pischedda A, Stewart AD, Rice WR. 2009. A cost of sexual attractiveness to high-fitness females. PLoS Biol. 7, e10000254 ( 10.1371/journal.pbio.1000254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maklakov AA, Immler S, Løvlie H, Flis I, Friberg U. 2013. The effect of sexual harassment on lethal mutation rate in female Drosophila melanogaster. Proc. R. Soc. B 280, 20121874 ( 10.1098/rspb.2012.1874) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fowler K, Partridge L. 1989. A cost of mating in female fruitflies. Nature 338, 760–761. ( 10.1038/338760a0) [DOI] [Google Scholar]
- 22.Dukas R, Jongsma K. 2012. Costs to females and benefits to males from forced copulations in fruit flies. Anim. Behav. 84, 1177–1182. ( 10.1016/j.anbehav.2012.08.021) [DOI] [Google Scholar]
- 23.Chapman T, Liddle LF, Kalb JM, Wolfner MF, Partridge L. 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373, 241–244. ( 10.1038/373241a0) [DOI] [PubMed] [Google Scholar]
- 24.Wolfner MF. 2002. The gifts that keep on giving: physiological functions and evolutionary dynamics of male seminal proteins in Drosophila. Heredity 88, 85–93. ( 10.1038/sj.hdy.6800017) [DOI] [PubMed] [Google Scholar]
- 25.Wigby S, Chapman T. 2005. Sex peptide causes mating costs in female Drosophila melanogaster. Curr. Biol. 15, 316–321. ( 10.1016/j.cub.2005.01.051) [DOI] [PubMed] [Google Scholar]
- 26.Swanson WJ. 2003. Sex peptide and the sperm effect in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 100, 9643–9644. ( 10.1073/pnas.1834127100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ram KR, Wolfner MF. 2007. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr. Comp. Biol. 47, 427–445. ( 10.1093/icb/icm046) [DOI] [PubMed] [Google Scholar]
- 28.Pitnick S, Pfenning DW. 2014. Brotherly love benefits females. Nature 505, 626–627. ( 10.1038/nature12853) [DOI] [PubMed] [Google Scholar]
- 29.Hollis B, Kawecki TJ, Keller L. 2015. No evidence that within-group male relatedness reduces harm to females in Drosophila. Ecol. Evol. 5, 979–983. ( 10.1002/ece3.1417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carazo P, Perry JC, Johnson F, Pizzari T, Wigby S. 2015. Relative male Drosophila melanogaster reared together as larvae fight less and sire longer lived daughters. Ecol. Evol. 5, 2787–2797. ( 10.1002/ece3.1549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chippindale AK, Berggren M, Alpern J, Montgomerie R. 2015. Does kin selection moderate sexual conflict in Drosophila? Proc. R. Soc. B 282, 20151417 ( 10.1098/rspb.2015.1417) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose MR, Charlesworth B. 1981. Genetics of life history in Drosophila melanogaster. I. Sib analysis of adult females. Genetics 97, 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loyau A, Cornuau JH, Clobert J, Danchin É. 2012. Incestuous sisters: mate preference for brother over unrelated males in Drosophila melanogaster. PLoS ONE 7, e51293 ( 10.1371/journal.pone.0051293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robinson SP, Kennington WJ, Simmons LW. 2012. Assortative mating for relatedness in a large naturally occurring population of Drosophila melanogaster. J. Evol. Biol. 25, 716–725. ( 10.1111/j.1420-9101.2012.02466.x) [DOI] [PubMed] [Google Scholar]
- 35.Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics, 4th edn Essex, UK: Pearson Education Limited. [Google Scholar]
- 36.Lynch M, Walsh B. 1998. Genetics and analysis of quantitative traits. Sunderland, MA: Sinauer Associates, Inc. [Google Scholar]
- 37.DeRose MA, Roff DA. 1999. A comparison of inbreeding depression in life-history and morphological traits in animals. Evolution 53, 1288–1292. [DOI] [PubMed] [Google Scholar]
- 38.R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R-project.org/) [Google Scholar]
- 39.Tan CKW, Løvlie H, Greenway E, Goodwin SF, Pizzari T, Wigby S. 2013. Sex-specific responses to sexual familiarity, and the role of olfaction in Drosophila. Proc. R. Soc. B 280, 20131691 ( 10.1098/rspb.2013.1691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majerus MEN, Weir PO'D. 1982. Female mating preference is genetic. Nature 300, 521–523. ( 10.1038/300521a0) [DOI] [PubMed] [Google Scholar]
- 41.Cobb M, Ferveur J-F. 1995. Evolution and genetic control of mate recognition and stimulation in Drosophila. Behav. Process. 35, 35–54. ( 10.1016/0376-6357(95)00052-6) [DOI] [PubMed] [Google Scholar]
- 42.Sokolowski MB. 2001. Drosophila: genetics meets behaviour. Nat. Rev. Genet. 2, 879–890. ( 10.1038/35098592) [DOI] [PubMed] [Google Scholar]
- 43.Anholt RRH. 2010. Making scents of behavioural genetics: lessons from Drosophila. Genet. Res. (Camb). 92, 349–359. ( 10.1017/S0016672310000492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf JB, Brodie ED III, Cheverud JM, Moore AJ, Wade MJ. 1998. Evolutionary consequences of indirect genetic effects. Trends Ecol. Evol. 13, 64–69. ( 10.1016/S0169-5347(97)01233-0) [DOI] [PubMed] [Google Scholar]
- 45.Bijma P, Wade MJ. 2008. The joint effects of kin, multilevel selection and indirect genetic effects on response to genetic selection. J. Evol. Biol. 21, 1175–1188. ( 10.1111/j.1420-9101.2008.01550.x) [DOI] [PubMed] [Google Scholar]
- 46.Kent C, Azanchi R, Smith B, Formosa A, Levine JD. 2008. Social context influences chemical communication in D. melanogaster. Curr. Biol. 18, 1384–1389. ( 10.1016/j.cub.2008.07.088) [DOI] [PubMed] [Google Scholar]
- 47.Wilson AJ, Gelin U, Perron MC, Réale D. 2009. Indirect genetic effect and the evolution of aggression in a vertebrate system. Proc. R. Soc. B 276, 533–541. ( 10.1098/rspb.2008.1193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Billeter J-C, Jagadeesh S, Stepek N, Azanchi R, Levine JD. 2012. Drosophila melanogaster females change mating behaviour and offspring production based on social context. Proc. R. Soc. B 279, 2417–2425. ( 10.1098/rspb.2011.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available online at dryad: http://dx.doi.org/10.5061/dryad.q28b2.


