Abstract
Early pig farmers in Europe imported Asian pigs to cross with their local breeds in order to improve traits of commercial interest. Current genomics techniques enabled genome-wide identification of these Asian introgressed haplotypes in modern European pig breeds. We propose that the Asian variants are still present because they affect phenotypes that were important for ancient traditional, as well as recent, commercial pig breeding. Genome-wide introgression levels were only weakly correlated with gene content and recombination frequency. However, regions with an excess or absence of Asian haplotypes (AS) contained genes that were previously identified as phenotypically important such as FASN, ME1, and KIT. Therefore, the Asian alleles are thought to have an effect on phenotypes that were historically under selection. We aimed to estimate the effect of AS in introgressed regions in Large White pigs on the traits of backfat (BF) and litter size. The majority of regions we tested that retained Asian deoxyribonucleic acid (DNA) showed significantly increased BF from the Asian alleles. Our results suggest that the introgression in Large White pigs has been strongly determined by the selective pressure acting upon the introgressed AS. We therefore conclude that human-driven hybridization and selection contributed to the genomic architecture of these commercial pigs.
Keywords: hybridization, adaptive introgression, selection, domestication, Sus scrofa, commercial breeding
1. Background
Introgression and the subsequent selection on introgressed variants is thought to be a widespread phenomenon among many species [1]. Introgression can occur naturally, due to mixture of populations in hybrid zones or occasional invasions. Then, selection for introgressed haplotypes can occur, a process known as adaptive introgression [2]. However, introgression can also be human-driven through hybridization, either accidental or on purpose [3,4]. Domestic animals are a clear example of species that have experienced population admixture due to human interference [5]. The introduction of and selection on novel alleles into a population has been observed in, for example, chickens [6], cagebirds [7], and cattle [8]. Also in pigs, human-mediated hybridization has introduced haplotypes that cause desired effects on phenotypes [9].
Pig farming has undergone a true metamorphosis from first domestication to the intensified industry we know today. Pigs were domesticated independently leading to separate European and Asian domestic pigs some 10 000 years ago [10,11]. Subsequent selection and breeding resulted in highly distinct breeds on these continents [12,13]. Especially in Europe, farmers were herding their swine in surrounding forests, and it was not before the onset of the Industrial Revolution, during the eighteenth century, that pigs were kept in sties and became an important farm animal [14]. Pigs were selected based on their phenotypes regarding traits of commercial interest.
Over the last 100 years, with the improvements in performance recording and making use of genetic evaluation methods based on pedigree information, breeding programmes have achieved remarkable genetic progress in reducing backfat (BF) for carcass quality and improving growth rate for production efficiency. Since the 1990s, using the same traditional breeding strategies (pedigree based), genetic progress has also been observed in reproduction traits, especially litter size (LS) [15]. Part of the success of these breeding programmes can also be attributed to the introduction of genes from Chinese breeds into commercial European breeds. About three centuries ago, with the intensification of global trade, farmers in Europe realized that Chinese pigs possessed particular characteristics that would be beneficial to introduce into their breeding stock. Therefore, pigs from Chinese breeds were imported into Europe and multiple crosses between European and Asian breeds were made with the purpose of combining beneficial traits, such as BF and LS from Asian pigs, and body length from European pigs [14,16].
With the advent of genomic selection [17], genetic progress is expected to speed up even more. The design of a 60 K SNPchip for pigs in 2009 [18] and the publication of the pig reference genome in 2012 [19] greatly contributed to the applicability of these techniques in pig breeding. This genomic information can also be used to pinpoint regions in the genome that have been under selective pressure. The resulting changes at the DNA level have been detected as selective sweeps in a multitude of breeds [20,21]. Interestingly, some of these loci have been identified as not only being under selection, but also introgressed. Asian alleles at the EDNRB [20], IGF2 [22], and KITLG [23] locus have proved effects on phenotypes (e.g. meat content and coat colour) of European commercial breeds.
With current genomics techniques, it has become possible to trace back the haplotypes that were introduced during the Industrial Revolution. In Bosse et al. [9], we examined the occurrence of Asian haplotypes (AS) in a population of European pigs that belong to the commercial Large White breed. Our results showed that AS are widely present in the genomes of these commercial pigs and highlighted the effect of the introgressed Asian variant of the AHR gene on LS. Since the AS were introduced for a specific purpose, the effect on the phenotype should co-occur with the presence of Asian variants in regions of the genome that are associated with the traits known to have been under selection. However, how much influence the Asian introgression had on the selective history of commercial traits remains unknown.
We hypothesize that the introgression landscape in commercial breeds is shaped mostly by artificial selection and therefore most introgressed regions should have an effect on commercial traits (BF and LS). Also, an absence of AS in some parts of the Large White genomes could be the result of purifying selection. In this study, we examine in more detail the introgression signatures that we identified previously in Bosse et al. [9] and show that the majority of the regions we tested had a significant effect on BF in a commercial Large White population. These findings have important implications for the knowledge of natural and human-driven evolutionary forces shaping genomes after hybridization.
2. Material and methods
(a). Introgression data
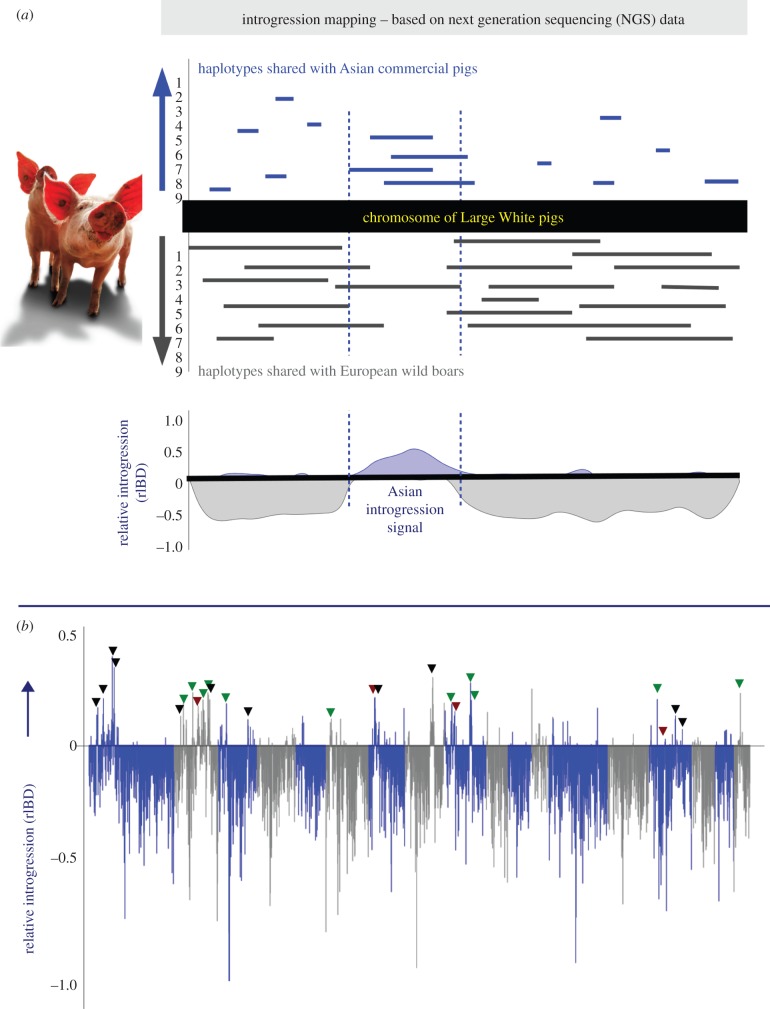
The analyses in this paper build upon the dataset and results that were obtained in Bosse et al. [9,24]. Briefly, the proportion of Asian introgressed haplotypes in a population of Large White pigs was assessed over all autosomes. Introgression mapping was performed on a group of nine Large White pigs and the background of their haplotypes was assigned to be European or Asian [9]. In bins of 10 kb over the genome, the relative Asian introgression signal in the Large White population was obtained (figure 1a). We assessed whether the Large White haplotypes were identical by descent (IBD) with Asian and/or European haplotypes and calculated the relative frequency of IBD with AS in the population for each bin (rIBD). We used these genome-wide rIBD signals to understand the details of the Asian introgression (see electronic supplementary material, S1, for more details about the introgression mapping).
Figure 1.
The principle of introgression mapping. The purpose of introgression mapping is to determine what the background is of haplotypes in a particular part of the genome. (a) Haplotypes that were shared with either Asian or European pigs were mapped to the genome for all nine Large White pigs. The total numbers of overlapping haplotypes were counted and the relative introgression signal (rIBD) was obtained by taking the difference of haplotype frequencies as described in Bosse et al. [9,24]. (b) The y-axis displays the rIBD signal averaged in bins of 1 Mb, and the x-axis contains the physical position for all 18 autosomes. Regions that were selected for the commercial trait analysis are indicated, with green triangles indicating included regions, black triangles indicating selected regions with strong 'Hardy–Weinberg disequilibrium, and red triangles indicating regions discarded because of allele frequencies.
(b). Genome characteristics
To assess the correlation between gene density, recombination frequency, and introgression signal, we averaged the rIBD in 1 Mb bins and counted the number of genes within each bin. The recombination map from Tortereau et al. [25] was used to obtain the recombination frequency per Mb. To test whether the probability of introgression decreases with an increase in the number of genes in a region, we used the Pearson's product-moment correlation in R.
(c). Selection of regions
We used approximately 400 regions of introgression previously identified by Bosse et al. [9] with a Z-transformed rIBD (ZrIBD) of more than two (electronic supplementary material, table S3). The regions were extended by one 10 kb bin at a time to the left and right flank of each identified region of introgression, until the threshold of more than two ZrIBD was no longer met and/or the rIBD value for one particular 10 kb bin was less than 0. We found 33 regions of introgression that were longer than 150 kb and checked whether they physically overlapped with markers on the Illumina Porcine 60 K iSelect Beadchip (60 K chip). Three regions were discarded because they contained less than three segregating markers on the chip. Electronic supplementary material, table S1, contains the list of 30 regions that were included in the further analyses.
(d). Genotyping and phasing
We genotyped a total of 9 970 pigs and wild boar for 488 markers on the 60 K chip that fell within the 30 identified regions of introgression. Phasing of haplotypes was performed independently for each region with Beagle v. 3.3.2 [26], using the genotype information for all individuals. After the phasing step, we used haplotype data from three groups: Asian (N = 448), European wild (N = 920), and European commercial (N = 18 572). Because the introgression analysis was performed on Large White pigs, we extracted only those individuals from the European commercial group that were known to be purebred Large Whites, leaving us with a total of 4 764 Large White haplotypes.
(e). Determination of haplotype origin
The total number of observed reference haplotypes in the group of European wild boar was 920, and the total number of observed Asian reference haplotypes was 448. For each of the 4 764 haplotypes observed in Large White animals, we determined the Asian (AS) or European (EU) origin based on the frequencies of this haplotype in the European and/or Asian group of animals. For each region, we counted the number of unique haplotypes among the 448 haplotypes in the Asian group. Because we have unequal sampling, the number of unique haplotypes in a random sample of 448 haplotypes from the European group was also counted. Then, for each unique haplotype that was observed within the group of Large White haplotypes, we counted the number of times the haplotype was observed in the European and in the Asian group. To avoid a bias due to the (generally) higher diversity in Asia, we adjusted the counts for the amount of diversity in the Asian and European groups. This adjustment was achieved by calculating the ratio of unique haplotypes in both AS and EU groups. The number of times that particular haplotype was observed in the European group was multiplied by the proportion of unique EU compared to AS haplotypes, and the number of times the haplotype was observed in the Asian reference group was multiplied by the proportion of total observed AS. We then checked whether the corrected number of observed haplotypes in the European and Asian reference groups differed by at least a factor of four. If so, the haplotype was assigned to the group in which it was observed most. If not, it was assigned to the group for which both backgrounds were considered (‘Both’).
(f). Cleaning of the data
Introgression regions in which the genotypes showed strong deviation from the Hardy–Weinberg equilibrium (p < 0.00001) and the frequency of AS was less than 0.60 or greater than 0.99 were excluded from further analysis. As we evaluated regions with strong Asian introgression signals, we expected that the AS should be higher in frequency than European haplotypes. We assumed that regions where the frequency of AS was less than 0.60 were observed due to possible errors during the phasing procedures and therefore they were excluded. We excluded regions where the frequency of AS were greater than 0.99 because this means that these regions were fixed, which does not allow to test the association of these regions with the evaluated phenotypes. In order to test the independence of the evaluated regions, we also estimated the pairwise Pearson's correlations (r) for all regions using the recoded haplotypes. When two regions showed r > 0.80, the shortest regions was excluded. After cleaning procedures, a total 1 384 animals with haplotype information on 11 regions were available for further steps (see electronic supplementary material S2, for more details).
(g). Breeding values and association analyses
In this study, we evaluated the traits of BF and LS. Deregressed estimated breeding values (DEBV) were used as a response variable for each trait under study. The estimated breeding value (EBV) was separately deregressed for each trait using the methodology described by Garrick et al. [27]. The EBV of each animal was obtained from the routine genetic evaluation by Topigs Norsvin using an animal model (pedigree based). The model for BF included genetic line, sex, herd-year-month, and weight as fixed effects, and an additive genetic effect (animal) and a common litter effect as random effects. For LS, the model included genetic line, parity number, interval weaning-pregnancy (days), whether more than one insemination procedure was performed (yes or no), and herd-year-season, while the random effects consisted of service sire, a permanent effect to account for the repeated observations of a single sow, and an additive genetic effect (animal). The reliabilities per animal for the purpose of deregression were extracted from the genetic evaluation based on the methodology of Tier & Meyer [28]. The heritabilities used for the deregression were also extracted from the routine genetic evaluation.
Association analyses were performed using the software ASReml [29] applying the following model:
where DEBVij is the observed DEBV for the animal j, μ is the overall DEBV mean of the population, Ri is the count of AS of the region i, aj is the additive genetic effect estimated using a pedigree-based relationship matrix, and eij the residual error. The weighting factor w was used in the association analyses to account for the differences in the amount of offspring information available for the estimation of the DEBV [27]. To ensure the quality of the DEBV, only animals with a w higher than zero and a reliability of the DEBV greater than 0.20 were used. The reliability of the DEBV was obtained according to Garrick et al. [27]. Association analyses were performed per region. In addition, a combined analysis was performed where R represented the count of AS summed over all regions.
3. Results and discussion
(a). Effect of introgression on commercial traits
We hypothesized that the pattern of introgression in the Large White population is mainly determined by artificial selection acting upon the AS. Following this rationale, the Asian introgression should persist mainly in those regions of the genome where the Asian variant has a favourable effect on a phenotype of interest. To test this, we extracted haplotypes in the introgressed regions and estimated the effect of their origin (European or Asian) on two commercially important traits: BF and LS. A total of 2 382 individuals from the commercial Large White line were genotyped for markers on the 60 K SNPchip [18] that covers those regions. More specifically, we extracted 11 regions that had an introgression signal that persisted over more than 150 Kbp from the data presented by Bosse et al. [9] and that passed our thresholds for data cleaning (see Material and methods and electronic supplementary material for more details). The further analyses of these regions were based on this selection of 60 K markers.
(b). Effects per region
We evaluated whether these 11 regions were significantly associated with the traits of BF and LS. None of these regions were found to have a significant effect on LS (table 1). However, six of these 11 regions showed a significant association with BF (table 1). For all these significant regions, we observed an increase in BF when a European haplotype was replaced by an Asian haplotype.
Table 1.
Effect of AS on backfat (BF) and litter size (LS) in introgression regions. An animal model was used to estimate the effect of AS on the DEBV for the two traits. p-values of significant regions (p < 0.10) are given in italic. Effect for BF is in mm BF per Asian haplotype, and effect for LS is in number of piglets per Asian haplotype. ‘All combined’ indicates the regression analysis over all 11 regions.
| BF |
LS |
|||
|---|---|---|---|---|
| region | p-value | effect | p-value | effect |
| 15_1 | 0.560 | −0.07 | 0.110 | 0.21 |
| 18_1 | 0.024 | 0.15 | 0.617 | −0.03 |
| 2_2 | 0.002 | 0.22 | 1.000 | 0.00 |
| 2_4 | 0.420 | 0.07 | 0.471 | 0.07 |
| 2_6 | 0.027 | 0.15 | 0.203 | 0.09 |
| 2_7 | 0.011 | 0.17 | 0.234 | −0.09 |
| 3_1 | 0.527 | −0.05 | 0.729 | 0.03 |
| 6_1 | 0.299 | −0.42 | 0.348 | −0.41 |
| 9_1 | 0.823 | −0.05 | 1.000 | 0.00 |
| 9_3 | 0.081 | 0.14 | 0.590 | 0.05 |
| 9_5 | 0.054 | 0.13 | 1.000 | 0.00 |
| all combined | <0.001 | 0.09 | 0.610 | 0.01 |
Most introgressed regions were identified on chromosome 2 (figure 1b; electronic supplementary material, figure S1a) with the strongest effect in the gene-dense region 2_2 (0.22 mm of increase in BF). This region contains multiple genes coding for intercellular adhesion molecules (ICAMs) that have been shown to have an effect on obesity [30]. Whether the effect of BF is caused by these genes is however unclear, since the region contains a total of 39 annotated genes in the current Ensembl release 76.
Region 2_7 is the region with the strongest introgression signal on chromosome 2, and therefore it was used as an example of the applied method in electronic supplementary material, figure S1. The substitution of a European haplotype for an AS in this region on average increased BF by 0.17 mm. As can be seen in the Ensembl annotation for this region (electronic supplementary material, figure S1b), one candidate gene, COMMD10, lies within the peak of region 2_7. COMMD proteins contain a conserved and unique 'COMM' domain involved in cellular homeostasis including copper and the NFkβ pathway, and at least 10 COMM family members exist that are conserved in all vertebrates [31]. Murgiano et al. [32] found another COMMD gene differentially expressed in longissimus lumborum muscle samples between Large White and Casertana pigs and suggested that COMMD negatively regulates NFkβ signalling which in turn can result in triggering the adipogenetic cascades.
The other significant region (2_6) on this chromosome contains seven candidate genes for increased BF (CAST, ERAP1, ERAP2, LNPEP, LIX1 RIOK2, and RGMB). LNPEP is an insulin-regulated amino peptidase which acts as a membrane protein associated with glucose transporter vesicles in cultured mouse adipocytes according to Larance et al. [33]. CAST, calpastatin, has no direct known function in fat synthesis, but interestingly it is a well-known locus involved in meat tenderness in multiple species and studies [34], and meat quality traits of pigs in general [35].
Regions 9_3 and 9_5 are both located on chromosome 9, in the vicinity of the AHR gene that was identified in Bosse et al. [9], with an effect of approximately 0.15 mm of BF per Asian haplotype (table 1). Region 9_3 overlaps with the AHR gene, suggesting that this gene is involved in multiple biological processes, as discussed by Denison et al. [36] and Hernandez-Ochoa et al. [37]. Region 9_5, in addition to its proximity to AHR, contains two TWIST neighbour genes (TWISTNB) that are involved in transcription and the TMEM196 gene coding for transmembrane protein 196. The last significant region 18_1 on chromosome 18 contains only one gene, protection of telomeres1 (POT1), that has previously been shown to have a higher expression level in multiple tissues from the fat-type Wujin pigs, compared with Large White pigs, including the longissimus dorsi muscle [38].
In summary, for the 11 regions with a strong introgression signal, the AS displayed a significant positive effect on BF in the majority of regions. Zooming into the genes in the BF-associated regions, a multitude of candidate genes could be identified that possibly caused the effect on BF. We suggest further experiments that focus on these specific genes to confirm their role in the accumulation of BF in pigs. Selecting those regions with the most important signal of introgression might have introduced some bias making the results less representative. In electronic supplementary material, S4, we discuss why we believe the potential bias is minor in our analysis.
(c). Additive effect over regions
The effect of the AS is an increase in BF in all significant regions, suggesting that the AS could have an additive effect on BF over all regions. We examined the association of AS with BF and LS combining all regions (summing the count of AS of all regions). We performed regression of the counts of AS (ranging from 9 to 22, electronic supplementary material, figure S2) on both BF and LS. For LS, the association test was not significant, but for BF the association was even stronger than when individual regions were analysed (table 1). For BF, an additive effect of 0.09 mm of BF was observed per Asian haplotype that replaced a European haplotype (table 1). The overall phenotypic standard deviation of BF in the Large White population was 1.61 mm. Analysing all 11 regions together, an individual that contains only AS (n = 22) will show 1.98 mm of BF more than an animal that contains only European haplotypes, which means 1.23 phenotypic standard deviations of BF. To demonstrate that the associations between the introgression regions and BF found in this study are significantly different from those expected by chance, we performed a theoretical exercise which is described in detail in electronic supplementary material, S5.
Chinese breeds were thought to be superior for the traits of fatness and LS according to the early European pig farmers, and these traits were artificially selected after introgression [14]. Müller et al. [39] showed that the amount of BF observed in Meishan pigs was 2.38-fold the amount of BF observed in Pietrain pigs (32.83 mm and 13.78 mm, respectively). Haley et al. [40] comparing LS between Meishan and Large White sows at first parity, showed that the Meishan sows had an average of 28% more piglets than Large White sows (13.03 piglets and 10.20 piglets, respectively). Our initial hypothesis was, therefore, that the regions with a strong introgression signal would have a significant effect on both BF and LS. However, our results showed that regions of introgression only have a significant effect on BF. Selection signatures for complex traits do not necessarily leave a sweeplike signature in the genome [41,42]. This could explain why none of the introgressed regions display a significant association with LS. Indeed, if we look at the previous finding for the AHR gene [9], it is one particular Asian allele rather than all AS at that locus that have the effect on LS. Another explanation why the introgressed AS have no effect on LS could be that the specific loci that are involved in a complex trait like LS contain genes that are also involved in other (life-history) traits. The pleiotropic nature of these genes may restrict the selection on AS, resulting in less obvious signatures in the genome than for BF-related genes, although this is speculative.
Our results demonstrate that by screening a population for signals of introgression, regions can be pinpointed where introduced haplotypes have an effect on selected traits. Popular methods that are developed to detect selective sweeps in a population, like Fst [43] or homozygosity tests, use an increase or decrease of genetic variation as a signal. Ongoing selection for introduced haplotypes that are genetically more diverse or distant than haplotypes from the source population, will not be picked up by these methods. We therefore suggest consideration of alternative methods (such as a test for the background of haplotypes as we have described in the manuscript) when the studied population has a known history of admixture, or when the goal is to screen specifically for adaptive introgression.
4. Introgression and genome characteristics
(a). General patterns
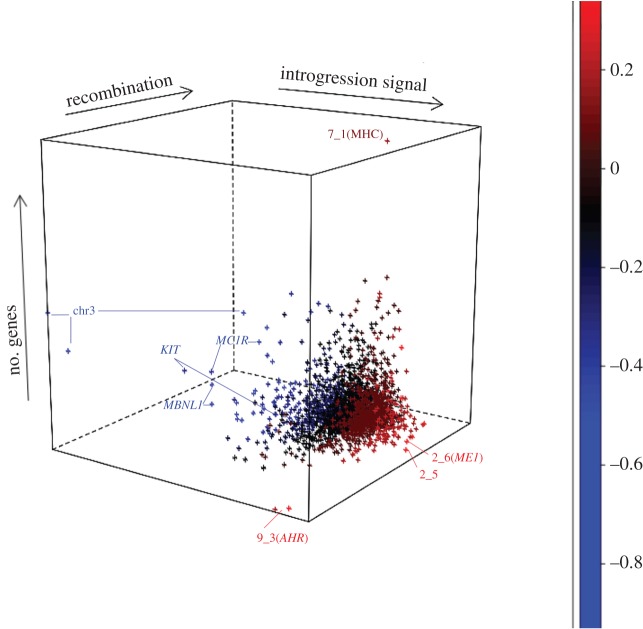
Our hypothesis was that artificial selection is the main factor in shaping the introgression pattern in Large White pigs. According to Hedrick et al. [2], the probability of an introgressed haplotype to be maintained in a population is strongly increased when it has some selective advantage. The results of the association analysis support the hypothesis that introgression signals are enriched for associations with commercially interesting traits. In line with this hypothesis, general genome characteristics like gene density and recombination frequency should contribute little to the introgression pattern. To assess whether the introgression signal is correlated with gene density or recombination frequency, the rIBD was averaged over 1 Mb bins over the genome. We found a very modest significant negative correlation of −0.05, as well as a significant correlation between rIBD and the (log-transformed) recombination frequency of 0.10 (figure 2). In a recent study on Neanderthal introgression in modern humans, gene deserts were enriched for Neanderthal ancestry [44]. This finding suggests that purifying selection removed the majority of Neanderthal haplotypes from the population and that introgressed haplotypes mainly occur in regions with relaxed selection pressure. In pigs, these general patterns in the genome explain only a fraction of the variation in introgression signatures. The circumstances of introgression in these two species are however very different, since the admixture in commercial pigs has been deliberate, and selection for some of the introgressed haplotypes is expected to be positive because of the known differences between Asian and European pigs.
Figure 2.
rIBD and genome characteristics. For each 1 Mb bin in the genome, the rIBD signal was plotted against the number of genes and the recombination frequency in the bin. Colouration is based on the rIBD signal so that bright red indicates bins with the strongest Asian introgression signal, and blue bins indicate the strongest under-representation of AS. Number of genes per bin ranged from 0 to 128 and recombination frequency ranged from −16.6 to 3.3 (log2-transformed) cM/Mb. Bins containing interesting genes that are discussed in the main text are indicated.
Although the purpose of the introgression of AS was to maintain beneficial variants in the population, probably not all introduced AS had the desired effect on the European stock. Selection pressure on AS could have been either positive or purifying, and should have resulted in either an excess or an absence of the Asian variants, depending on the location in the genome and the associated genes in those regions. Commercial pig breeds are known to be under strong artificial selection, and our results have shown that indeed a majority of the strongly introgressed regions we tested had a significant effect on BF. Therefore, we expect that regions with an excess or absence of AS contain genes of commercial interest. The 1% extreme tails of the introgression distribution (electronic supplementary material, table S2) were scanned for genes that are known to be related to commercially important traits. Introgressed regions should contain genes that have an effect on traits that are present in the Asian breeds and that had a selective advantage in the European breeds. By contrast, within those regions that do not contain AS, we expect to identify genes that have an effect on traits that are typical for European breeds. For the regions with lack of introgression signal, it would be interesting to create experimental crosses to introduce AS and test whether we can validate that indeed phenotypes of commercial interest are affected by locus-specific Asian introgression.
(b). Fat-related genes
The two 1 Mb bins containing the highest rIBD scores are both on chromosome 1. When we look for candidate genes in the first bin, the malic enzyme 1 (ME1) gene has previously been described as an important quantitative trait locus (QTL) region for BF and meat quality [45–47]. This bin overlaps with region 1_6 from the association analysis, but that region was discarded because of an excess of heterozygotes. The second bin contains AMD1 (cell proliferation, polyamine synthesis pathway) and zeta catalytic subunit DNA polymerase (rev3 L in human) as candidate genes. This bin overlaps with region 1_5 from the association analysis, but it was also discarded due to an excess of heterozygotes.
On the beginning of chromosome 12, we identified another bin with a high rIBD signal, hinting at a locus that contains Asian introgressed haplotypes. This region overlaps the FASN gene that was previously described as fatty acid synthase and an important gene involved in fat deposition [42]. In addition to the genes described in this paragraph, our 11 regions that were used in the association analysis can be found in the 25 bins that span the top 1% of introgressed regions in the genome. These findings hint at a prominent role for the selection of fatness in shaping the introgression landscape in Large White pigs.
The reason for the observation that more heterozygous individuals are observed than one would expect based on the Hardy–Weinberg equilibrium remains unclear. Apart from technical issues, these regions are potentially very interesting if the signal is genuine. Balancing selection or a heterozygote advantage can result in more heterozygous individuals than expected. Also, if selection for a previously low-frequency haplotype is ongoing, an excess of heterozygous individuals could be observed. As shown by Merks et al. [15], BF was included in the selection index of breeding companies over 100 years ago, and nowadays leanness is preferred in commercial pigs. This switch in preference and direction of selection could result in more heterozygosity in some regions. Further experiments for the regions with an excess of heterozygotes should indicate what causes this peculiar pattern.
(c). Pigmentation genes
Skin pigmentation is an important trait for modern pig breeders, and therefore we expect a distinct introgression signal at pigmentation loci (electronic supplementary material, figure S3). In the top 1% of bins with low Asian introgression are two regions that have previously been identified as important candidate regions for coat colouration in pigs, containing the KIT gene and the MC1R gene. The KIT gene is a very well-known gene that is involved in coat colour [23]. European pigs contain a copy number variable dominant white allele, mostly in homozygous form [21,48]. We clearly see European haplotypes surrounding this gene rather than Asian, suggesting selection against introgression at this locus. Also present in this peak region is MAP9, a gene involved in mitotic spindle formation (electronic supplementary material, figure S3). MC1R is known to be involved in pigmentation in multiple species including pigs. No introgression is expected in Large Whites for this region based on previous results [49], and indeed we see a clear lack of introgression at this locus (electronic supplementary material, figure S3). Interestingly, among those bins that contain the highest introgression signal in the Large Whites, were two other genes found to be involved in pigmentation (TYR and RAB38). RAB38 and TYR are both involved in the pigmentation patterns of skin, eyes, and hair, according to a multitude of different studies [50,51]. Tyrosinase seems to have some temperature-dependent colouration patterns, but different forms exist. RAB38 lies within region 9_1 and has been identified by Wilkinson et al. [20] as well as a region in Large White pigs that is introgressed and selected.
(d). Morphology
The 1 Mb bin covering MBNL1 (muscleblind-like splicing regulator 1 gene) has the lowest introgression signal. In humans and mice, this gene has been shown to be associated with muscle dystrophy [52]. Since European commercial pigs and Asian pigs are known to be very different in terms of muscle content, selection for muscle-related traits in European pigs might select against Asian variants in this region. In the introgression peak at the very end of chromosome 8, we identified another interesting gene, BMP3 (bone morphogenesis protein 3), that has previously been identified as being involved in growth restriction in humans [53] and a mutation in this gene has an effect on the skull shape of zebrafish and dogs [54]. This region was also found to be introgressed and selected in Large White pigs by Wilkinson et al. [20].
5. Concluding remarks
With this work, we demonstrate that the introgression landscape in Large White pigs seems to be strongly determined by the selective pressure acting upon the introgressed AS. The majority of the regions we tested with a high frequency in AS turn out to have an effect on BF, and many of these regions overlap with previously identified fat-related genes, and therefore we conclude that artificial selection on fatness influenced the introgression landscape in Large White pigs. To investigate whether this is expected behaviour of introgressed haplotypes under selection, we propose future simulation studies on the introgression landscape of populations under a neutral scenario and with selection pressure. We then hypothesize that introgressed haplotypes will be elevated to high frequency due to positive selection, and other introgressed haplotypes will quickly be removed because of purifying selection. The fact that the proportion of Asian material is relatively similar for most European commercial breeds [19,24] suggests that the introgression occurred before the establishment of modern breeds. Many generations after the introgression, AS could have been purged if they had a selective disadvantage. On a genome-wide scale, however, we observed a general pattern of low frequency of AS, suggesting a more neutral scenario for the introgressed genetic material in this Large White pig population. Regions with an excess or absence of AS indeed contain genes where the Asian variants are thought to have an effect on phenotypes of interest, and therefore we illustrate that human-driven introgression and selection may have broadly shaped the genomic architecture of this commercial pig breed. Extending this study to more commercial traits and different breeds will provide more insight into the process of selective introgression. Our findings provide a unique insight about how the selection history of pig breeding influenced the genomic haplotype patterns of the commercial pigs that we know today. How general this introgression pattern is, should be pointed out by future studies on other organisms that likely experienced introgression and consecutive selection.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The DNA samples that we used for genotyping were provided by Dr Ning Li, China Agricultural University, China; Dr Alain Ducos, UMR INRA-ENVT, France; Sem Genini, Parco technologico Padano, Italy; Dr Gono Semiadi, Puslit Biologi, Indonesia; Dr Naohiko Okumura, Staff Institute 446–1 Ippaizuka, Japan; Dr Alan Archibald, Roslin Institute and the Royal (Dick) School of Veterinary Studies, University of Edinburgh, Scotland; Topigs Norsvin Research Center, The Netherlands; Dr Oliver Ryder, San Diego Zoo, USA; Cheryl L. Morris, PhD, Omaha's Henry Doorly Zoo, USA. We thank Bert Dibbits for laboratory work, B. van de Water for graphics production, and Yogesh Paudel and Egbert Knol for valuable discussion. We also thank Oscar Gaggiotti, Pablo Orozco-ter Wengel, and an anonymous reviewer for all their valuable comments on our manuscript.
Data accessibility
All BAM files of the individuals used in the previous studies have been deposited in the European Nucleotide Archive (ENA) under the accession no. ERP001813. The relevant phenotype and haplotype data are available from the Dryad accession doi:10.5061/dryad.g40f0 linked to this article.
Authors' contributions
M.A.M.G., O.M., H.-J.M., J.B., M.B., and M.S.L. conceived and designed the experiments. M.S.L., M.B., and R.P.M.A.C. performed the experiments and data analysis was done by M.B., M.S.L., H.-J.M., and L.A.F.F. M.A.M.G., R.P.M.A.C., B.H., and M.S.L. provided samples, genotypes, and phenotypes. M.S.L. and M.B. wrote the paper, with input from J.B., M.A.M.G., H.-J.M., O.M., B.H., and L.A.F.F.
Competing interests
We have no competing interest.
Funding
This project was financially supported by European Research Council under the European Community's Seventh Framework Programme (FP7/2007–2013)/ERC grant agreement no. 249894 and the Breed4Food consortium. The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Currat M, Ruedi M, Petit RJ, Excoffier L. 2008. The hidden side of invasions: massive introgression by local genes. Evolution 62, 1908–1920. ( 10.1111/j.1558-5646.2008.00413.x) [DOI] [PubMed] [Google Scholar]
- 2.Hedrick PW. 2013. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 22, 4606–4618. ( 10.1111/mec.12415) [DOI] [PubMed] [Google Scholar]
- 3.Crispo E, Moore JS, Lee-Yaw JA, Gray SM, Haller BC. 2011. Broken barriers: human-induced changes to gene flow and introgression in animals. BioEssays 33, 508–518. ( 10.1002/bies.201000154) [DOI] [PubMed] [Google Scholar]
- 4.Harrison RG, Larson EL. 2014. Hybridization, introgression, and the nature of species boundaries. J. Hered. 105, 795–809. ( 10.1093/jhered/esu033) [DOI] [PubMed] [Google Scholar]
- 5.Larson G, Burger J. 2013. A population genetics view of animal domestication. Trends Genet. 29, 197–205. ( 10.1016/j.tig.2013.01.003) [DOI] [PubMed] [Google Scholar]
- 6.Eriksson J, et al. 2008. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet. 4, e1000010 ( 10.1371/journal.pgen.1000010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rheindt FE, Edwards SV. 2011. Genetic introgression: an integral but neglected component of speciation in birds. Auk 128, 620–632. ( 10.1525/auk.2011.128.4.620) [DOI] [Google Scholar]
- 8.Flori L, Thevenon S, Dayo GK, Marcel S, Sylla S, Berthier D, Moazami-Goudarzi K, Gautier M. 2014. Adaptive admixture in the West African bovine hybrid zone: insight from the Borgou population. Mol. Ecol. 23, 3241–3257. ( 10.1111/mec.12816) [DOI] [PubMed] [Google Scholar]
- 9.Bosse M, et al. 2014. Genomic analysis reveals selection for Asian genes in European pigs following human-mediated introgression. Nat. Commun. 5, 4392 ( 10.1038/ncomms5392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larson G, et al. 2005. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science 307, 1618–1621. ( 10.1126/science.1106927) [DOI] [PubMed] [Google Scholar]
- 11.Larson G, et al. 2007. Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc. Natl Acad. Sci. USA 104, 15 276–15 281. ( 10.1073/pnas.0703411104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kijas J, Andersson L. 2001. A phylogenetic study of the origin of the domestic pig estimated from the near-complete mtDNA genome. J. Mol. Evol. 52, 302–308. [DOI] [PubMed] [Google Scholar]
- 13.Megens H-J, Crooijmans R, San Cristobal M, Hui X, Li N, Groenen M. 2008. Biodiversity of pig breeds from China and Europe estimated from pooled DNA samples: differences in microsatellite variation between two areas of domestication. Genet. Sel. Evol. 40, 103–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White S. 2011. From globalized pig breeds to capitalist pigs: a study in animal cultures and evolutionary history. Environ. History 16, 94–120. ( 10.1093/envhis/emq143) [DOI] [Google Scholar]
- 15.Merks J, Mathur P, Knol E. 2012. New phenotypes for new breeding goals in pigs. Animal 6, 535–543. ( 10.1017/S1751731111002266) [DOI] [PubMed] [Google Scholar]
- 16.Jones G, Rothschild M, Ruvinsky A. 1998. Genetic aspects of domestication, common breeds and their origin. In The genetics of the pig (eds M Rothschild, A Ruvinsky), pp. 17–50. New York, NY: CAB International.
- 17.Meuwissen T, Hayes B, Goddard M. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos AM, et al. 2009. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE 4, e6524 ( 10.1371/journal.pone.0006524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groenen MA, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491, 393–398. ( 10.1038/nature11622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson S, et al. 2013. Signatures of diversifying selection in European pig breeds. PLoS Genet. 9, e1003453 ( 10.1371/journal.pgen.1003453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rubin C-J, et al. 2012. Strong signatures of selection in the domestic pig genome. Proc. Natl Acad. Sci. USA 109, 19 529–19 536. ( 10.1073/pnas.1217149109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ojeda A, et al. 2008. Selection in the making: a worldwide survey of haplotypic diversity around a causative mutation in porcine IGF2. Genetics 178, 1639–1652. ( 10.1534/genetics.107.084269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okumura N, Matsumoto T, Hamasima N, Awata T. 2008. Single nucleotide polymorphisms of the KIT and KITLG genes in pigs. Anim. Sci. J. 79, 303–313. ( 10.1111/j.1740-0929.2008.00531.x) [DOI] [Google Scholar]
- 24.Bosse M, Megens HJ, Madsen O, Frantz LA, Paudel Y, Crooijmans RP, Groenen MA. 2014. Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol. Ecol. 23, 4089–4102. ( 10.1111/mec.12807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tortereau F, et al. 2012. A high density recombination map of the pig reveals a correlation between sex-specific recombination and GC content. BMC Genomics 13, 586 ( 10.1186/1471-2164-13-586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097. ( 10.1086/521987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrick DJ, Taylor JF, Fernando RL. 2009. Deregressing estimated breeding values and weighting information for genomic regression analyses. Genet. Sel. Evol. 41, 44 ( 10.1186/1297-9686-41-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tier B, Meyer K. 2004. Approximating prediction error covariances among additive genetic effects within animals in multiple-trait and random regression models. J. Anim. Breed. Genet. 121, 77–89. ( 10.1111/j.1439-0388.2003.00444.x) [DOI] [Google Scholar]
- 29.Gilmour AR, Gogel B, Cullis B, Thompson R. 2009. ASReml user guide release 3.0. Hemel Hempstead, UK: VSN International Ltd. [Google Scholar]
- 30.Dong ZM, Gutierrez-Ramos J-C, Coxon A, Mayadas TN, Wagner DD. 1997. A new class of obesity genes encodes leukocyte adhesion receptors. Proc. Natl Acad. Sci. USA 94, 7526–7530. ( 10.1073/pnas.94.14.7526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maine GN, Burstein E. 2007. COMMD proteins: COMMing to the scene. Cell. Mol. Life Sci. 64, 1997–2005. ( 10.1007/s00018-007-7078-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murgiano L, D'Alessandro A, Egidi MG, Crisa A, Prosperini G, Timperio AM, Valentini A, Zolla L. 2010. Proteomics and transcriptomics investigation on longissimus muscles in Large White and Casertana pig breeds. J. Proteome Res. 9, 6450–6466. ( 10.1021/pr100693h) [DOI] [PubMed] [Google Scholar]
- 33.Larance M, et al. 2005. Characterization of the role of the Rab GTPase-activating protein AS160 in insulin-regulated GLUT4 trafficking. J. Biol. Chem. 280, 37 803–37 813. ( 10.1074/jbc.M503897200) [DOI] [PubMed] [Google Scholar]
- 34.Ropka-Molik K, Bereta A, Tyra M, Różycki M, Piórkowska K, Szyndler-Nędza M, Szmatoła T. 2014. Association of calpastatin gene polymorphisms and meat quality traits in pig. Meat. Sci. 97, 143–150. ( 10.1016/j.meatsci.2014.01.021) [DOI] [PubMed] [Google Scholar]
- 35.Ciobanu D, et al. 2004. New alleles in calpastatin gene are associated with meat quality traits in pigs. J. Anim. Sci. 82, 2829–2839. [DOI] [PubMed] [Google Scholar]
- 36.Denison MS, Soshilov AA, He GC, DeGroot DE, Zhao B. 2011. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicol. Sci. 124, 1–22. ( 10.1093/toxsci/kfr218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez-Ochoa I, Karman BN, Flaws JA. 2009. The role of the aryl hydrocarbon receptor in the female reproductive system. Biochem. Pharmacol. 77, 547–559. ( 10.1016/j.bcp.2008.09.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yong WJ, Jing L, Jiugang Z, Lei C, Yonggang L. 2012. A novel porcine gene, POT1, differentially expressed in the longissimus muscle tissues from Wujin and Large White pigs. Cytokine 59, 22–26. ( 10.1016/j.cyto.2012.03.028) [DOI] [PubMed] [Google Scholar]
- 39.Müller E, Moser G, Bartenschilager H, Geldermann H. 2000. Trait values of growth, carcass and meat quality in Wild Boar, Meishan and Pietrain pigs as well as their crossbred generations. J. Anim. Breed. Genet. 117, 189–202. ( 10.1046/j.1439-0388.2000.00239.x) [DOI] [Google Scholar]
- 40.Haley C, Lee G, Ritchie M. 1995. Comparative reproductive performance in Meishan and Large White pigs and their crosses. Anim. Sci. 60, 259–267. ( 10.1017/S1357729800008420) [DOI] [Google Scholar]
- 41.Kemper KE, Saxton SJ, Bolormaa S, Hayes BJ, Goddard ME. 2014. Selection for complex traits leaves little or no classic signatures of selection. BMC Genomics 15, 246 ( 10.1186/1471-2164-15-246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heidaritabar M, Vereijken A, Muir WM, Meuwissen T, Cheng H, Megens HJ, Groenen MA, Bastiaansen JW. 2014. Systematic differences in the response of genetic variation to pedigree and genome-based selection methods. Heredity 113, 503–513. ( 10.1038/hdy.2014.55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir BS, Cockerham CC. 1984. Estimating F-statistics for the analysis of population-structure. Evolution 38, 1358–1370. ( 10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- 44.Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507, 354–357. ( 10.1038/nature12961) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vidal O, Varona L, Oliver M, Noguera J, Sanchez A, Amills M. 2006. Malic enzyme 1 genotype is associated with backfat thickness and meat quality traits in pigs. Anim. Genet. 37, 28–32. ( 10.1111/j.1365-2052.2005.01366.x) [DOI] [PubMed] [Google Scholar]
- 46.Bartz M, Kociucka B, Mankowska M, Switonski M, Szydlowski M. 2013. Transcript level of the porcine ME1 gene is affected by SNP in its 3′ UTR, which is also associated with subcutaneous fat thickness. J. Anim. Breed. Genet. 131, 271–278. [DOI] [PubMed] [Google Scholar]
- 47.Ramírez O, Quintanilla R, Varona L, Gallardo D, Díaz I, Pena R, Amills M. 2014. DECR1 and ME1 genotypes are associated with lipid composition traits in Duroc pigs. J. Anim. Breed. Genet. 131, 46–52. ( 10.1111/jbg.12035) [DOI] [PubMed] [Google Scholar]
- 48.Paudel Y, Madsen O, Megens H-J, Frantz LA, Bosse M, Bastiaansen JW, Crooijmans RPMA, Groenen MAM. 2013. Evolutionary dynamics of copy number variation in pig genomes in the context of adaptation and domestication. BMC Genomics 14, 449 ( 10.1186/1471-2164-14-449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang M, Larson G, Ribeiro HS, Li N, Andersson L. 2009. Contrasting mode of evolution at a coat color locus in wild and domestic pigs. PLoS Genet. 5, e1000341 ( 10.1371/journal.pgen.1000341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loftus SK, et al. 2002. Mutation of melanosome protein RAB38 in chocolate mice. Proc. Natl Acad. Sci. USA 99, 4471–4476. ( 10.1073/pnas.072087599) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.del Marmol V, Beermann F. 1996. Tyrosinase and related proteins in mammalian pigmentation. FEBS Lett. 381, 165–168. ( 10.1016/0014-5793(96)00109-3) [DOI] [PubMed] [Google Scholar]
- 52.Kanadia RN, et al. 2003. A muscleblind knockout model for myotonic dystrophy. Science 302, 1978–1980. ( 10.1126/science.1088583) [DOI] [PubMed] [Google Scholar]
- 53.Bonnet C, et al. 2010. Microdeletion at chromosome 4q21 defines a new emerging syndrome with marked growth restriction, mental retardation and absent or severely delayed speech. J. Med. Genet. 47, 377–384. ( 10.1136/jmg.2009.071902) [DOI] [PubMed] [Google Scholar]
- 54.Schoenebeck JJ, et al. 2012. Variation of BMP3 contributes to dog breed skull diversity. PLoS Genet. 8, e1002849 ( 10.1371/journal.pgen.1002849) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All BAM files of the individuals used in the previous studies have been deposited in the European Nucleotide Archive (ENA) under the accession no. ERP001813. The relevant phenotype and haplotype data are available from the Dryad accession doi:10.5061/dryad.g40f0 linked to this article.