Abstract
The ubiquitin proteasome system (UPS) is a major regulator of protein processing, trafficking, and degradation. While protein ubiquitination is utilized for many cellular processes, one major function of this system is to target proteins to the proteasome for degradation. In schizophrenia, studies have found UPS transcript abnormalities in both blood and brain, and we have previously reported decreased protein expression of ubiquitin-associated proteins in brain. To test whether the proteasome is similarly dysregulated, we measured the protein expression of proteasome catalytic subunits as well as essential subunits from proteasome regulatory complexes in 14 pair-matched schizophrenia and comparison subjects in superior temporal cortex. We found decreased expression of Rpt1, Rpt3, and Rpt6, subunits of the 19S regulatory particle essential for ubiquitin-dependent degradation by the proteasome. Additionally, the α subunit of the 11S αβ regulatory particle, which enhances proteasomal degradation of small peptides and unfolded proteins, was also decreased. Haloperidol-treated rats did not have altered expression of these subunits, suggesting the changes we observed in schizophrenia are likely not due to chronic antipsychotic treatment. Interestingly, expression of the catalytic subunits of both the standard and immunoproteasome were unchanged, suggesting the abnormalities we observed may be specific to the complexed state of the proteasome. Aging has significant effects on the proteasome, and several subunits (20S β2, Rpn10, Rpn13, 11Sβ, and 11Sγ) were significantly correlated with subject age. These data provide further evidence of dysfunction of the ubiquitin-proteasome system in schizophrenia, and suggest that altered proteasome activity may be associated with the pathophysiology of this illness.
Introduction
The ubiquitin proteasome system (UPS) is a critical cellular process that consists of initial ubiquitination of proteins followed by trafficking of those proteins to a large multimeric complex referred to as the proteasome (Voges et al, 1999). The proteasome is the primary intracellular machinery responsible for degradation of cellular proteins (Rock et al, 1994), a process essential for maintaining cellular homeostasis and responding to cellular stress. Interestingly, transcript expression of proteins associated with the UPS have been reported to be abnormal in schizophrenia (Vawter et al, 2001; Middleton et al, 2002; Altar et al, 2005; Chu et al, 2009; Bousman et al, 2010a, 2010b; Arion et al, 2015), suggesting that this system may be associated with the pathophysiology of this illness.
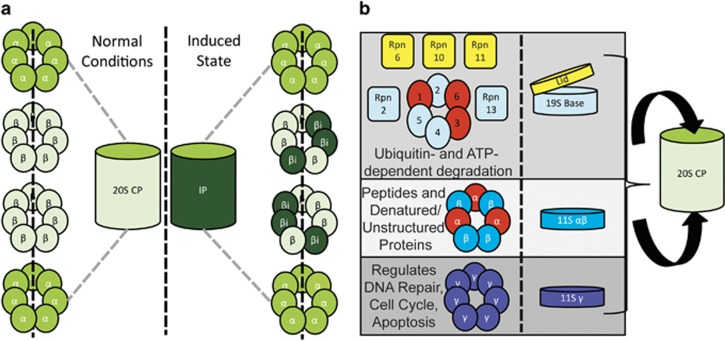
The proteasome consists of a core complex, which is regulated by a variety of modifications. The core of the mammalian proteasome is the 20S common proteasome (CP), which consists of four heptameric rings, stacked in an α-β-β-α conformation (Zwickl et al, 1992; Groll et al, 1997; Unno et al, 2002) (Figure 1). The α rings contain seven unique but similar subunits that are necessary for the assembly of the 20S CP, regulate access to the β subunits, and have binding capabilities for a plethora of regulatory particles (RP) (Zwickl et al, 1994; Voges et al, 1999; Groll et al, 2000; Unno et al, 2002). The β rings consist of seven different β subunits, three of which, β1, β2, and β5, have catalytic activities and perform the basic proteolytic function of the proteasome (Groll et al 1997, 1999; Heinemeyer et al, 1997; Unno et al, 2002). However, fully folded proteins are unable to access the catalytic core of the 20S CP (Groll et al, 2000). To facilitate the entry of proteins marked for degradation, multimeric complexes known as RPs are required (Bajorek and Glickman, 2004). These complexes regulate both the ability of proteins to enter the catalytic core of the proteasome, as well as the rate and efficiency of proteolytic activity (Voges et al, 1999). The primary RP of the proteasome is the 19S RP, which is important for ubiquitin-dependent degradation of proteins (Voges et al, 1999; Bar-Nun and Glickman, 2012). The 19S RP has two major types of subunits, the AAA-ATPases (Rpt), which unfold proteins in an ATP-dependent manner to allow them access to the 20S CP, and non-ATPases (Rpn), which perform a myriad of activities such as maintaining structural integrity, recognizing polyubiquitination, and deubiquitinating proteins, all of which are essential for the degradation of ubiquitinated proteins (Glickman et al, 1998; Voges et al, 1999; Bar-Nun and Glickman, 2012; Tanaka, 2013).
Figure 1.
Structure, organization, and modifications of the proteasome. (a) The 20S CP consists of four heptameric rings containing seven α subunits and seven β subunits. Three of the β subunits are responsible for the proteolytic activity of the proteasome. These subunits are replaced under conditions of increased immune response or oxidative stress, resulting in the immunoproteasome (IP). (b) Proteolytic activity of the 20S CP can be regulated by three main complexes, the 19S, 11S αβ, and 11S γ. These regulatory particles bind to the α subunits of the 20S CP and can cap one or both ends. The 20S CP can also be bound by both the 19S and 11S αβ, resulting in a chimeric proteasome. The 19S RP consists of a lid and base. The lid components measured in this study are Rpn6, 10, and 11, and the base components are Rpn2 and 13 as well as the hexameric ring of AAA-ATPases (Rpt1-6) that unfold proteins for access to the 20S CP. The 11S αβ RP is a hetero-heptameric ring formed by three α subunits and four β subunits, while the 11Sγ is a mono-heptameric ring. The major functions of these regulatory particles when they are bound to the proteasome are listed. Subunits marked in red were significantly decreased in schizophrenia in this study.
Although ATP- and ubiquitin-dependent degradation have been considered the main functions of the proteasome, there are other regulatory particles that promote ubiquitin- and ATP- independent degradation. The 11S RPs, heptameric complexes that cap the proteasome similar to the 19S RP (Figure 1), are associated with ubiquitin-independent degradation (Gray et al, 1994; Realini et al, 1997; Voges et al, 1999). One of these, the 11Sγ RP, is localized to the nucleus and regulates the degradation of a variety of proteins, including key regulators of apoptosis and the cell cycle (Mao et al, 2008). Another 11S RP is a heptameric ring of 11S α and β subunits which enhances the catalytic activity of the 20S proteasome, but only permits processing of peptides and unfolded proteins (Realini et al, 1997; Voges et al, 1999). Oxidative conditions recruit both of the 11S RPs as well as another modification, the immunoproteasome (IP) (Pickering et al, 2010; Pickering and Davies, 2012). The IP is characterized by the replacement of the 20S CP catalytic β subunits with inducible β subunits, β1i, β2i, and β5i, resulting in diminished caspase-like activity and higher chymotrypsin- and trypsin-like activity (Aki et al, 1994; Hisamatsu et al, 1996). Together, these factors result in a complex and heterogeneous proteasome population within the cell, and alterations in these complexes can have significant effects on cell health and protein processing.
Previous studies of the UPS have primarily focused on abnormalities of transcript expression (Vawter et al, 2001; Middleton et al, 2002; Altar et al, 2005; Chu et al, 2009; Bousman et al, 2010a, 2010b; Arion et al, 2015), but studies of protein expression of elements of the proteasome have not been reported. We recently reported dysfunction of ubiquitin and ubiquitin-like systems in the superior temporal gyrus in schizophrenia (Rubio et al, 2013). We focused on the superior temporal gyrus because it is a region essential for auditory language processing and has been shown to have gray matter volume reductions and disturbances of activity functionally correlated to the severity of positive symptoms in patients with schizophrenia (McCarley et al, 1993; Menon et al, 1995; Pearlson 1997; Rajarethinam et al, 2000). In this previous report, we found decreased levels of total and free ubiquitin protein, and several ubiquitin-activating enzymes and ligases (Rubio et al, 2013). In proteins of molecular weights of 40 and 70 kDA, K48-linked polyubiquitination, the major posttranslational modification, which targets proteins to the proteasome, was also decreased in the superior temporal gyrus in schizophrenia (Rubio et al, 2013). Decreased ubiquitination and K48-linked polyubiquitination suggests that proteins are either (i) being degraded more quickly because of an increase in proteasome activity and/or abundance or (ii) not being targeted to the proteasome for degradation through ubiquitination. To explore these possibilities, we undertook this study in which we measured the protein expression of proteasome subunits of the 20S CP, as well as subunits that modify the proteasome to facilitate ubiquitin-dependent (19S RP) and -independent (11S RPs; IP) degradation in the same subjects in which we previously found decreased ubiquitination (Rubio et al, 2013).
Materials and methods
Subjects
Postmortem brain samples were obtained from the Mount Sinai/Bronx Veterans Administration (VA) Medical Center Department of Psychiatry Brain Collection. Assessment, consent, and postmortem procedures were performed for all subjects as has been previously described (Powchik et al, 1998; Purohit et al, 1998). Fourteen subjects with schizophrenia based on DSM-III-R criteria were pairwise matched with 14 comparison subjects by sex, age, tissue pH, and postmortem interval (Table 1). All subjects died of natural causes; neurodegenerative disorders were ruled out by neuropathologic examination, and none of the subjects had a history of alcoholism and/or substance abuse.
Table 1. Paired Subject Characteristics.
| Pair | Subject | Sex/Age | Rx | pH | PMI (h) |
|---|---|---|---|---|---|
| 1 | Comparison | M/59 | 6.7 | 21 | |
| Schizophrenia | M/57 | 1 | 6.4 | 21 | |
| 2 | Comparison | M/70 | 6.1 | 6.7 | |
| Schizophrenia | M/70 | 0 | 6.4 | 7.2 | |
| 3 | Comparison | M/73 | 6.2 | 15 | |
| Schizophrenia | M/73 | 0 | 6.5 | 7.9 | |
| 4 | Comparison | M/75 | 6.4 | 5.0 | |
| Schizophrenia | M/78 | 1 | 6.8 | 26 | |
| 5 | Comparison | M/76 | 6.3 | 2.9 | |
| Schizophrenia | M/80 | 1 | 6.4 | 15 | |
| 6 | Comparison | M/93 | 6.3 | 4.2 | |
| Schizophrenia | M/92 | 0 | 6.7 | 18 | |
| 7 | Comparison | M/95 | 6.5 | 4.1 | |
| Schizophrenia | M/97 | 1 | 6.5 | 9.3 | |
| 8 | Comparison | F/66 | 6.9 | 23 | |
| Schizophrenia | F/62 | 1 | 6.7 | 24 | |
| 9 | Comparison | F/73 | 7.0 | 3.0 | |
| Schizophrenia | F/70 | 1 | 6.5 | 13 | |
| 10 | Comparison | F/74 | 6.3 | 4.8 | |
| Schizophrenia | F/75 | 1 | 6.5 | 22 | |
| 11 | Comparison | F/79 | 6.4 | 10 | |
| Schizophrenia | F/77 | 1 | 6.0 | 10 | |
| 12 | Comparison | F/80 | 6.6 | 3.8 | |
| Schizophrenia | F/81 | 0 | 6.7 | 15 | |
| 13 | Comparison | F/85 | 7.3 | 8.0 | |
| Schizophrenia | F/84 | 1 | 6.8 | 22 | |
| 14 | Comparison | F/89 | 6.7 | 2.3 | |
| Schizophrenia | F/89 | 1 | 6.2 | 10 |
Abbreviations: PMI, postmortem interval; Rx, 0=off antipsychotic medications for 6 weeks or more prior to death, 1=treated with antipsychotic medications at time of death.
Tissue Preparation
Samples were obtained at autopsy and sliced into 0.8–1-cm slabs in the coronal plane, which were then dissected into 1-cm3 cubes. The tissue used in this study was from the full thickness of the left superior temporal gyrus and stored at −80 °C until use (Powchik et al, 1998; Purohit et al, 1998; Katsel et al, 2005). Samples were homogenized using a Power Gen 125 homogenizer (Thermo Fisher Scientific, Rockford, Illinois) in RIPA buffer (50 mM Tris HCl (pH 7.4), 150 mM NaCl, 0.5 mM EGTA, 1% Triton X-100, 1% sodium deoxycholate, 0.5% SDS) containing deubiquitinase inhibitors (5 mM N-ethylmaleimide, 50 mM iodoacetamide), and protease and phosphatase inhibitor tablets (Complete Mini and Phostop, respectively, Roche Diagnostics, Manheim, Germany). Protein concentration was determined using a BCA assay kit (Thermo Fisher Scientific) and homogenates were aliquoted and stored at −80 °C until use.
Rodent Antipsychotic Drug Treatment
Male Sprague-Dawley rats (250 g) were housed in pairs for the duration of the study (9 months), during which they were treated with either haloperidol decanoate (28.5 mg/kg, n=10) or vehicle (sesame oil, n=10) every 3 weeks, for a total of 12 intramuscular injections. The animals were killed by decapitation and the brains immediately removed. The left anterior cortex was dissected on wet ice, snap frozen on dry ice, and stored at −80 °C. This study was approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham.
Western Blot Analysis
Western blot analyses were performed as previously described (Rubio et al, 2012; Rubio et al, 2013). Ultrapure water and a reducing buffer were used to dilute samples to a concentration of 2 μg/μl and denatured at 70 °C. Twenty micrograms of protein were loaded in each lane of a 4–12% gradient polyacrylamide bis-tris gel (Invitrogen, Carlsbad, California). Samples were run in duplicate for each subject, with matched comparison and schizophrenia subjects in alternating lanes. After electrophoresis, samples were transferred to PVDF membrane and then incubated with Li-Cor blocking buffer (Lincoln, Nebraska) or 5% bovine serum albumin in phosphate-buffered solution for 1 h at room temperature followed by an overnight incubation in primary antisera diluted in Li-Cor blocking buffer containing 0.1% Tween-20 or 5% bovine serum albumin buffer containing 0.1% Tween-20 at 4 °C (Table 2). All antibodies were optimized such that detection was within the linear range of the assay and each primary antibody was present in excess. β-Tubulin expression was used as a loading control, and blots were incubated with β-tubulin primary antisera for an hour at room temperature instead of overnight. No significant difference in β-tubulin expression was observed between diagnostic groups.
Table 2. Antibodies Used for Western Blot Analysis.
| Antibody | Host | Dilution | Buffer | Company | Catalogue # | Location |
|---|---|---|---|---|---|---|
| 20S β1 | Mouse | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW8140 | Farmingdale, NY |
| 20S β2 | Mouse | 1 : 1000 | Li-Cor | Enzo Life Sciences | BML-PW9300 | Farmingdale, NY |
| 20S β5 | Rabbit | 1 : 5000 | Li-Cor | Enzo Life Sciences | BML-PW8895 | Farmingdale, NY |
| 19S Rpt1 | Mouse | 1 : 2000 | BSA | Enzo Life Sciences | BML-PW8825 | Farmingdale, NY |
| 19S Rpt2 | Rabbit | 1 : 2000 | Li-Cor | Enzo Life Sciences | BML-PW8305 | Farmingdale, NY |
| 19S Rpt3 | Mouse | 1 : 2000 | BSA | Enzo Life Sciences | BML-PW8765 | Farmingdale, NY |
| 19S Rpt4 | Mouse | 1 : 2000 | BSA | Enzo Life Sciences | BML-PW8830 | Farmingdale, NY |
| 19S Rpt5 | Mouse | 1 : 2000 | BSA | Enzo Life Sciences | BML-PW8770 | Farmingdale, NY |
| 19S Rpt6 | Rabbit | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW8320 | Farmingdale, NY |
| 19S Rpn2 | Mouse | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW9270 | Farmingdale, NY |
| 19S Rpn6 | Rabbit | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW8370 | Farmingdale, NY |
| 19S Rpn10 | Rabbit | 1 : 1000 | BSA | Abcam | Ab137109 | Cambridge, MA |
| 19S Rpn11 | Rabbit | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW9625 | Farmingdale, NY |
| 19S Rpn13 | Rabbit | 1 : 1000 | BSA | Novus Biologicals | NBP1-30447 | Littleton,CO |
| 11S α | Rabbit | 1 : 2000 | Li-Cor | Enzo Life Sciences | BML-PW8185 | Farmingdale, NY |
| 11S β | Rabbit | 1 : 1000 | BSA | Thermo Scientific | PA5-17332 | Rockford, IL |
| 11S γ | Rabbit | 1 : 2000 | Li-Cor | Enzo Life Sciences | BML-PW8190 | Farmingdale, NY |
| IP β1i | Rabbit | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW8205 | Farmingdale, NY |
| IP β2i | Rabbit | 1 : 1000 | BSA | Upstate | 09-276 | Temecula, CA |
| IP β5i | Mouse | 1 : 1000 | BSA | Enzo Life Sciences | BML-PW8845 | Farmingdale, NY |
| β-Tubulin | Mouse | 1 : 20 000 | Li-Cor | Upstate | 05-661 | Temecula, CA |
| β-Tubulin | Rabbit | 1 : 5000 | Li-Cor | Novus Biologicals | NB600-936 | Littleton, CO |
Data Analysis
Odyssey 3.0 analytical software (Li-Cor) was used to determine integrated intensity values. Boxes were manually placed around each band at the expected molecular weight of each protein, and intra-lane background was subtracted. The value of each band was normalized to the in-lane value of β-tubulin. Data from two lanes were averaged for analysis. To determine whether data were normally distributed, a D'Agostino & Pearson omnibus normality test (GraphPad Prism 6, La Jolla, CA, USA) was performed before all comparisons. Normally distributed data from the human subjects were analyzed using paired Student's t-tests, and data that were not normally distributed were analyzed using Wilcoxon matched-pairs signed rank tests (GraphPad Prism 6). Correlations between protein expression and subject age were performed for all data, with Pearson correlation coefficients (r) calculated for normally distributed data, and Spearman's rank correlation coefficients (ρ) calculated for data not normally distributed. Proteins measured in haloperidol-treated rats were analyzed with either unpaired t-tests or Mann Whitney U tests. For all analyses, α=0.05.
Results
AAA-ATPase Subunits of the 19S Regulatory Particle are Decreased in Subjects with Schizophrenia
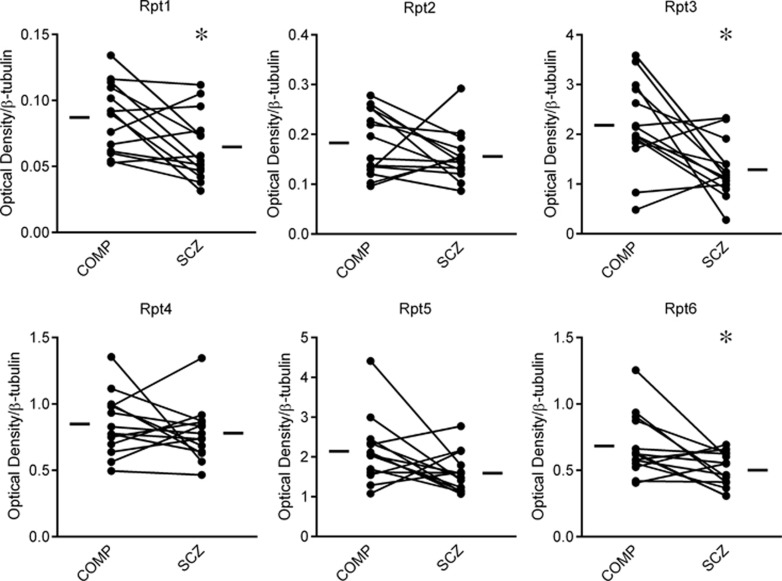
The six AAA-ATPase subunits of the 19S RP were measured in the superior temporal cortex in paired schizophrenia and comparison subjects. Rpt1, Rpt3, and Rpt6 were decreased in schizophrenia (t(13)=2.77, p<0.05; t(13)=3.13, p<0.05; t(13)=2.69, p<0.05, respectively), whereas the other subunits Rpt2, Rpt4, and Rpt5 were unchanged (Figure 2). In addition to the Rpt subunits, five Rpn subunits of the 19S RP, Rpn2, Rpn6, Rpn10, Rpn11, and Rpn13, were also measured. None of these subunits were abnormally expressed in schizophrenia (Table 3).
Figure 2.
19S regulatory particle (RP) subunit protein expression in schizophrenia. Protein expression of 19S Rpt subunits was measured in paired comparison (COMP) and schizophrenia (SCZ) subjects. Decreased expression was observed for Rpt1, Rpt3, and Rpt6, whereas no change was observed in Rpt2, Rpt4, and Rpt5. These data reflect decreased expression of 19S RP subunits specific to the AAA-ATPases responsible for unfolding proteins to allow them entrance to the proteasome. *p<0.05.
Table 3. Protein Levels of Non-ATPase Subunits of the 19S RP.
| Protein | Comparison | Schizophrenia | Test statistic | p |
|---|---|---|---|---|
| 19S Rpn2 | 0.16±0.03 | 0.21±0.05 | W=39 | 0.24 |
| 19S Rpn6 | 0.31±0.05 | 0.24±0.05 | t(13)=1.08 | 0.30 |
| 19S Rpn10 | 0.67±0.23 | 0.49±0.23 | W=−25 | 0.46 |
| 19S Rpn11 | 0.12±0.01 | 0.14±0.01 | t(13)=1.10 | 0.29 |
| 19S Rpn13 | 0.45±0.04 | 0.38±0.05 | t(13)=1.59 | 0.14 |
t: paired t-test; W: Wilcoxon matched pairs signed rank test; data are presented as means±SEM.
The Catalytic β Subunits of the Standard 20S CP and the IP are Unchanged in Schizophrenia
Protein levels of the 20S catalytic β subunits, β1, β2, and β5, were measured in the superior temporal cortex in paired schizophrenia and comparison subjects. No differences were observed for these subunits (Table 4). Additionally, the IP subunits, β1i, β2i, and β5i, were also measured and no differences were found (Table 4).
Table 4. Protein Levels of 20S CP and IP Catalytic β Subunits.
| Protein | Comparison | Schizophrenia | Test statistic | p |
|---|---|---|---|---|
| 20S β1 | 0.02±0.003 | 0.02±0.003 | t(13)=0.30 | 0.77 |
| 20S β2 | 0.81±0.13 | 0.58±0.08 | t(13)=1.87 | 0.08 |
| 20S β5 | 0.27±0.03 | 0.25±0.02 | t(13)=0.53 | 0.61 |
| 20S β1i | 0.31±0.05 | 0.24±0.04 | t(13)=1.08 | 0.30 |
| 20S β2i | 0.63±0.23 | 0.43±0.15 | W=−43 | 0.19 |
| 20S β5i | 0.08±0.01 | 0.09±0.01 | t(13)=1.28 | 0.22 |
t: paired t-test; W: Wilcoxon matched pairs signed rank test; data are presented as means±SEM.
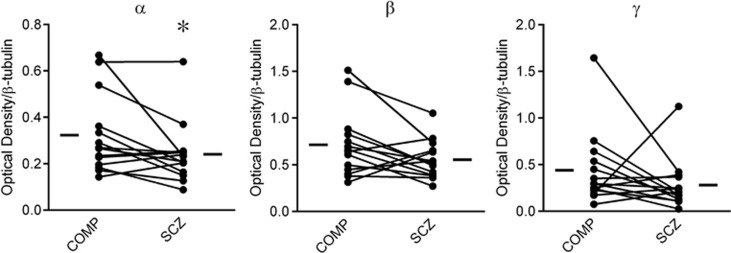
The 11S α Subunit is Decreased in Schizophrenia
Protein levels of the three 11S subunits were assessed in schizophrenia and comparison subjects. The α subunit was decreased in schizophrenia (W(14)=−65, p<0.05), whereas the β and γ subunits were unchanged (Figure 3).
Figure 3.
11S regulatory particle (RP) subunit protein expression. Protein expression of 11S RP subunits was measured in paired comparison (COMP) and schizophrenia (SCZ) subjects. Expression of the 11S α subunit was decreased, whereas expression of the β and γ subunits were unchanged, suggesting a potential abnormality in the 11S αβ RP, but not the 11S γ RP. *p<0.05.
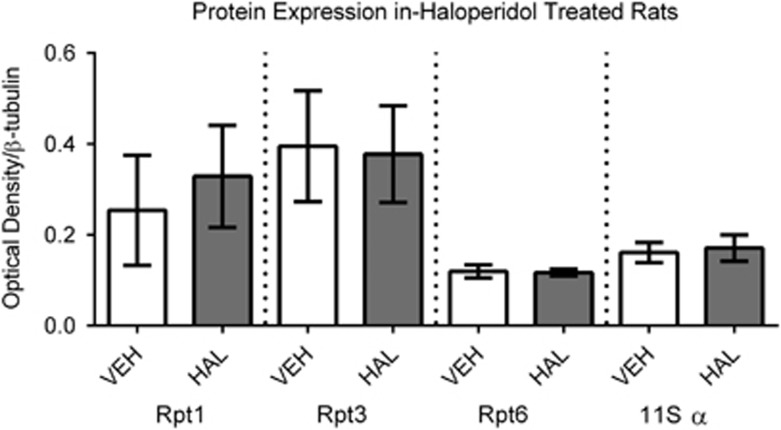
Haloperidol-Treatment did not Alter Protein Levels of 11S α, Rpt1, Rpt3, or Rpt6
Protein levels of the proteins found to be abnormal in schizophrenia were measured in rats chronically treated with haloperidol. No changes due to antipsychotic treatment were observed for the 11S α subunit, or the 19S Rpt1, Rpt3, and Rpt6 subunits (Figure 4).
Figure 4.
Proteasome regulatory particle subunits in haloperidol-treated rats. Protein expression of subunits found decreased in schizophrenia were measured in rats chronically treated with haloperidol. No change was observed due to treatment for 11S α, Rpt1, Rpt3, and Rpt6, suggesting that changes observed in these subunits in schizophrenia were not due to antipsychotic treatment effects.
Multiple Proteasome Subunits are Significantly Correlated with Age
Relationships between subject age and protein levels of proteasome subunits were determined. 20S CP β2, Rpn10, 11S β, and 11S γ were negatively correlated with age, whereas Rpn13 was positively correlated with age. None of the other subunits measured in this study were significantly associated with age (Table 5).
Table 5. Proteasome Subunit Expression Correlated with Subject Age.
| Proteasome subunits | Correlation coefficient | p-value |
|---|---|---|
| 20S β1 | r=0.10 | |
| 20S β2 | ρ=−0.50 | <0.01 |
| 20S β5 | r=0.02 | |
| 19S Rpt1 | r=0.18 | |
| 19S Rpt2 | r=0.17 | |
| 19S Rpt3 | r=−0.26 | |
| 19S Rpt4 | r=0.20 | |
| 19S Rpt5 | ρ=0.01 | |
| 19S Rpt6 | ρ=−0.05 | |
| 19S Rpn2 | ρ<0.01 | |
| 19S Rpn6 | r=0.16 | |
| 19S Rpn10 | ρ=−0.41 | 0.03 |
| 19S Rpn11 | ρ<0.01 | |
| 19S Rpn13 | r=0.63 | <0.01 |
| 11S α | ρ=0.04 | |
| 11S β | ρ=−0.48 | <0.01 |
| 11S γ | ρ=−0.41 | 0.03 |
| IP β1i | ρ=−0.21 | |
| IP β2i | ρ=−0.03 | |
| IP β5i | r=0.26 |
r: Pearson correlation coefficient; ρ: Spearman rank order correlation coefficent; only significant values of p (<0.05) are shown.
Discussion
In this study, abnormalities in protein expression of the proteasome regulatory complex subunits were found in superior temporal cortex of subjects with schizophrenia. Of the six Rpt subunits of the 19S RP, half (Rpt1, Rpt3, and Rpt6) were decreased in schizophrenia. Additionally, the α subunit of the 11S αβ RP was decreased. Catalytic subunits of the 20S CP and the IP were also assayed and were unchanged in schizophrenia. These changes suggest an abnormal pattern of proteasome subtypes consistent with fewer 19S RP- and 11S αβ RP- capped proteasomes. Altered expression of these complexes may influence protein degradation, protein synthesis, cellular energetics, and synaptic plasticity.
Protein degradation is an essential part of normal cellular activity. Targeting proteins to the proteasome through polyubiquitination provides specificity for determining which proteins are degraded. The most efficient type of proteasome performing ubiquitin-dependent degradation are proteasomes doubly capped by 19S RPs, also known as 26S proteasomes (Glickman et al, 1998; Tanaka, 2013) (Figure 1). The Rpt subunits are essential for the function of 26S proteasomes (Bar-Nun and Glickman, 2012; Tanaka, 2013). Given the integral role of ATPases in 26S function, the decrease in Rpt subunit protein expression found in this study is consistent with fewer 19S-capped proteasomes in schizophrenia, despite normal levels of Rpn subunits. Fewer 19S-capped proteasomes could diminish the cellular capacity for degrading ubiquitinated proteins, which parallels our previous finding of decreased markers of ubiquitination within the same subjects in schizophrenia (Rubio et al, 2013). This decrease in ubiquitination may reflect a compensatory mechanism for the diminished ability of the cell to degrade proteins expressing this posttranslational modification. Alternatively, the relative decrease in 19S proteasome complexes we found may be the result of the cell funneling its resources into less energetically demanding degradation mechanisms, such as lysosomal degradation, which is targeted by K63-linked polyubiquitination (MacGurn et al, 2012), or degradation by other proteasome types, which require neither ubiquitination nor ATP (Voges et al, 1999).
BDNF and myelin basic protein, two proteins that have been consistently shown to be decreased in schizophrenia (Martins-de-Souza et al, 2009; Jindal et al, 2010; Green et al, 2011; Zhang et al, 2012) are examples of proteins not degraded through ubiquitin-dependent proteasome activity. BDNF is degraded in the lysosome (Evans et al, 2011) and myelin basic protein can be degraded by metalloproteinases and the proteasome in an ubiquitin-independent manner (Chandler et al, 1995; Belogurov et al, 2015). Accordingly, changes in these and other proteins in schizophrenia are consistent with a model of increased non-26S proteasome degradation. Additionally, abnormalities in glutamate and GABA receptor modifications and trafficking may be, in part, the result of altered degradation activity. Neurotransmitter receptors are primarily degraded by the lysosome (MacGurn et al, 2012). An exception to this is receptor subunit processing in the endoplasmic reticulum, where misfolded proteins and uncomplexed subunits are recognized and exported for proteasomal degradation through the endoplasmic reticulum-associated degradation pathway (MacGurn et al, 2012). This process is dependent on ubiquitination, and facilitated by interactions between endoplasmic reticulum-associated degradation machinery and 19S AAA-ATPases (Lipson et al, 2008; Zemoura and Benke, 2014). If this process is disrupted, decreased 26S proteasome activity may result in the retention of misfolded proteins and/or abnormal incorporation of neurotransmitter receptor subunits into complexes that are then trafficked to the synapse. This may be a contributing factor to the abnormal N-glycosylation and subcellular localization of both glutamate and GABA receptor subunits that we have previously reported in schizophrenia (Hammond et al, 2010; Tucholski et al, 2013a, b; Mueller et al, 2014, 2015).
Degradation of proteins can be accomplished by several proteasome types other than the 26S, including uncapped 20S CP, the IP, and 11S γ RP-capped proteasomes. Proteasomes capped with the 11S αβ, however, can only degrade smaller peptides and not fully folded proteins (Realini et al, 1997; Voges et al, 1999). These proteasome types are recruited under times of cell stress, including oxidative stress (Pickering et al, 2010; Pickering and Davies, 2012). While the 26S proteasome is usually considered more efficient than these other complexes, both uncapped 20S CP and the IP have been shown to be more capable of degrading oxidized proteins (Pickering et al, 2010). Markers of increased oxidative stress such as increased lipid peroxidation, reactive oxygen species, and decreased antioxidant levels have been observed in postmortem brain in schizophrenia (Bošković et al, 2011; Gonzalez-Liencres et al, 2014; Reyazuddin et al, 2014; Rajasekaran et al, 2015). This may suggest that, in schizophrenia, there is an environment of oxidative stress, which would lead to the recruitment of the IP and 11S αβ RP in response to the increased oxidative load. However, we found normal levels of IP subunits and decreased 11S α subunit expression, which may instead suggest a diminished capacity to respond to oxidative stress in schizophrenia.
Synaptic plasticity may also be impacted by alterations in proteasome complex availability. Proteasome activity is essential for the developmental effects of BDNF (Leal et al, 2014; Santos et al, 2015), a protein downregulated in schizophrenia (Jindal et al, 2010; Green et al, 2011; Zhang et al, 2012). Both activation and localization of proteasomes to synapses is essential for regulating synaptic strength and dendritic pruning, and phosphorylation of Rpt6 is necessary for activity-dependent localization of the proteasome to the synapse (Djakovic et al, 2012; Ertürk et al, 2014). The decreased expression of Rpt6 that we observed could reflect dysregulation of proteasome localization. This may be associated with dendritic pruning abnormalities, which are a distinct characteristic of schizophrenia (Garey et al, 1998; Glantz and Lewis, 2000; Sweet et al, 2009).
In schizophrenia, several studies have found abnormalities of UPS-associated transcripts in both blood as well as in multiple brain regions (Vawter et al, 2001; Middleton et al, 2002; Altar et al, 2005; Chu et al, 2009; Bousman et al, 2010a, 2010b; Arion et al, 2015). The protein data we observed are consistent with these findings in that there appears to be decreased expression of molecular components of the UPS in schizophrenia. Interestingly, although abnormal expression of genes encoding UPS proteins have been identified in these studies, the individual genes found to be abnormal varies from study to study. These experiments were all performed in different tissue, from blood to various brain regions including the prefrontal cortex, thalamus, middle temporal cortex, and hippocampus. Regional differences in both transcript and protein abnormalities has been commonly observed in schizophrenia (Glantz and Lewis, 1997; Oni-Orisan et al, 2008), which may account for some of this across-study variability. Additionally, Arion et al (2015) observed changes in proteasome transcripts that were specific to dorsolateral prefrontal cortex layer V pyramidal neurons and not to layer III pyramidal neurons, suggesting that cell-specific expression patterns may also contribute to this variability.
Proteasome activity is known to decrease with age, and this decrease appears to be due in part to posttranslational modifications and proteasome complex composition (Hayashi and Goto, 1998; Bulteau et al, 2002; Husom et al, 2004). Although our subjects were well-matched for age, we examined the relationship between subject age and proteasome subunit expression. Unsurprisingly, we found that multiple proteasome subunits were significantly associated with subject age (Table 5).
This study investigated the proteasome by measuring proteins in postmortem tissue from geriatric subjects, resulting in several limitations in interpreting these data. The pathophysiology of subjects who are at later stages of the illness may not reflect abnormalities associated with earlier stages of the disorder, thus our findings may not generalize to younger subjects. As many of the schizophrenia subjects were receiving antipsychotic medication at the time of death, we tested whether long-term treatment of rats with haloperidol could produce similar changes in proteasome subunits and found no alterations, suggesting that our findings in schizophrenia are not likely due to chronic antipsychotic treatment. Finally, we examined protein levels of these subunits, but not how they are physically complexed within the cell or how proteasome activity may be altered. Although decreased abundance of regulatory particle subunits suggests decreased number of complexes containing these subunits, there are alternative possibilities. It is conceivable that the proteasome is more efficiently complexed in schizophrenia, resulting in relatively normal levels of complexes and proteasome activity. This might suggest a reduction in the ability of the cell to respond to stress by recruiting new proteasomes, rather than a more generalized abnormality of proteasome activity. Additionally, the three proteolytic activities of the proteasome, trypsin-like, chymotrypsin-like, and peptidylglutamyl-peptide hydrolyzing (or caspase-like), work in concert, but can be independently regulated by different modifications, including the ones investigated in this study and others. For example, posttranslational modifications including phosphorylation, O-GlcNAcylation, and S-glutathionylation have been shown to alter proteasome function without necessarily affecting subunit abundance (Ishii et al, 2005; Aiken et al, 2011; Xu et al, 2012). Further analysis in patients with schizophrenia on the state of different proteasome complexes, posttranslational modifications, and assays of trypsin-, chymotrypsin-, and caspase-like activity will be necessary to fully understand how these protein abnormalities affect cellular function and potentially contribute to the pathophysiology of schizophrenia.
In summary, we examined the protein expression of proteasome subunits in the superior temporal cortex of patients with schizophrenia and comparison subjects. We found decreases in essential regulatory subunits, suggesting reduced expression of the 19S RP and 11S αβ RP. These findings point to abnormal proteasome complexes in schizophrenia, which may have broad effects on protein degradation, protein synthesis, and synapse dynamics.
Funding and disclosure
This work was funded by the National Institutes of Health Grants MH53327 (JHMW), and MH064673 and MH066392 (VH). The authors declare no conflict of interest.
Acknowledgments
We would like to acknowledge Dr Rosalinda Roberts and the Alabama Brain Bank for providing materials used to optimize the antibody conditions used in this study.
References
- Aiken CT, Kaake RM, Wang X, Huang L (2011). Oxidative stress-mediated regulation of proteasome complexes. Mol Cell Proteomics 10: R110.006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aki M, Shimbara N, Takashina M, Akiyama K, Kagawa S, Tamura T et al (1994). Interferon-γ induces different subunit organizations and functional diversity of proteasomes. J Biochem 115: 257–269. [DOI] [PubMed] [Google Scholar]
- Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y et al (2005). Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry 58: 85–96. [DOI] [PubMed] [Google Scholar]
- Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A (2015). Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry e-pub ahead of print 6 January 2015. doi:10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed]
- Bajorek M, Glickman MH (2004). Keepers at the final gates: regulatory complexes and gating of the proteasome channel. Cell Mol Life Sci 61: 1579–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Nun S, Glickman MH (2012). Proteasomal AAA-ATPases: structure and function. Biochim Biophys Acta 1823: 67–82. [DOI] [PubMed] [Google Scholar]
- Belogurov A, Kuzina E, Kudriaeva A, Kononikhin A, Kovalchuk S, Surina Y et al (2015). Ubiquitin-independnet proteasomal degradation of myelin basic protein contributes to development of neurodegenerative autoimmunity. FASEB J 29: 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bošković M, Vovk T, Plsničar BK, Grabnar I (2011). Oxidative stress in schizophrenia. Curr Neuropharmacol 9: 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousman CA, Chana G, Glatt SJ, Chandler SD, May T, Lohr J et al (2010. a). Positive symptoms of psychosis correlate with expression of ubiquitin proteasome genes in peripheral blood. Am J Med Genet B Neuropsychiatr Genet 153B: 1336–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E et al (2010. b). Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: Convergent pathway analysis findings from two independent samples. Am J Med Genet B Neuropsychiatr Genet 0: 494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulteau AL, Szweda LI, Friguet B (2002). Age-dependent declines in proteasome activity in the heart. Arch Biochem Biophys 397: 298–304. [DOI] [PubMed] [Google Scholar]
- Chandler S, Coates R, Gearing A, Lury J, Wells G, Bone E (1995). Matrix metalloproteinases degrade myelin basic protein. Neurosci Lett 201: 223–226. [DOI] [PubMed] [Google Scholar]
- Chu TT, Liu Y, Kemether E (2009). Thalamic transcriptome screening in three psychiatric states. J Hum Genet 54: 665–675. [DOI] [PubMed] [Google Scholar]
- Djakovic SN, Marquez-Lona EM, Jakawich SK, Wright R, Chu C, Sutton MA et al (2012). Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J Neurosci 32: 5126–5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk A, Wang Y, Sheng M (2014). Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J Neurosci 34: 1672–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans SF, Irmady K, Ostrow K, Kim T, Nykajaer A, Saftig P et al (2011). Neuronal brain-derived neurotrophic factor is synthesized in excess, with levels regulated by sortilin-mediated trafficking and lysosomal degradation. J Biol Chem 286: 29556–29567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garey LJ, Ong WY, Patel TS, Kanani M, Davis A, Mortimer AM et al (1998). Reduced dendritic spine density on cerebral cortical pyramidal neurons in schizophrenia. J Neurol Neurosurg Psychiatry 65: 446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA (1997). Reduction in synaptophysin immunoreactivity in the prefrontal cortex of subjects with schizophrenia. Regional and diagnostic specificity. Arch Gen Psychiatry 54: 943–952. [DOI] [PubMed] [Google Scholar]
- Glantz LA, Lewis DA (2000). Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry 57: 65–73. [DOI] [PubMed] [Google Scholar]
- Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z et al (1998). A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9 singnalosome and eIF3. Cell 94: 615–623. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Liencres C, Tas C, Brown EC, Erdin S, Onur E, Cubukcoglu Z et al (2014). Oxidative stress in schizophrenia: A case-control study on the effects on social cognition and neurocognition. BMC Psychiatry 14: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray CW, Slaughter CA, DeMartino GN (1994). PA28 activator protein forms regulatory caps on proteasome stacked rings. J Mol Biol 236: 7–15. [DOI] [PubMed] [Google Scholar]
- Green MJ, Matheson SL, Shepherd A, Weickert CS, Carr VJ (2011). Brain-derived neurotrophic factor levels in schizophrenia: a systematic review with meta-analysis. Mol Psychiatry 16: 960–972. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Köhler A, Moroder L, Rubin DM, Huber R et al (2000). A gated channel into the proteasome core particle. Nat Struct Biol 7: 1062–1067. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Löwe J, Stock D, Bochtler M, Bartunik HD et al (1997). Structure of 20S proteasome from yeast at 2.4Å resolution. Nature 386: 463–471. [DOI] [PubMed] [Google Scholar]
- Groll M, Heinemeyer W, Jäger S, Ullrich T, Bochtler M, Wolf DH et al (1999). The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc Natl Acad Sci USA 96: 10976–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JC, McCullumsmith RE, Funk AJ, Haroutunian V, Meador-Woodruff JH (2010). Evidence for abnormal forward trafficking of AMPA receptors in frontal cortex of elderly patients with schizophrenia. Neuropsychopharmacology 35:2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Goto S (1998). Age-related changes in the 20S and 26S proteasome activities in the liver of male F344 rats. Mech Ageing Dev 102: 55–66. [DOI] [PubMed] [Google Scholar]
- Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH (1997). The active sites of the eukaryotic 20S proteasome and their involvement in subunit precursor processing. J Biol Chem 272: 25200–25209. [DOI] [PubMed] [Google Scholar]
- Hisamatsu H, Shimbara N, Saito Y, Kristensen P, Hendil KB, Fujiwara T et al (1996). Newly identified pair of proteasomal subunits regulated reciprocally by interferon γ. J Exp Med 183: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husom AD, Peters EA, Kolling EA, Fugere NA, Thompson LV, Ferrington DA (2004). Altered proteasome function and subunit composition in aged muscle. Arch Biochem Biophys 421: 67–76. [DOI] [PubMed] [Google Scholar]
- Ishii T, Sakurai T, Usami H, Uchida K (2005). Modification of proteasome: Identification of an oxidation-sensitive subunit in 26S proteasome. Biochemistry 44: 13893–13901. [DOI] [PubMed] [Google Scholar]
- Jindal RD, Pillai AK, Mahadik SP, Eklund K, Montrose DM, Keshavan MS (2010). Decreased BDNF in patients with antipsychotic naïve first episode schizophrenia. Schizophr Res 119: 47–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsel P, Davis KL, Gorman JM, Haroutunian V (2005). Variations in differential gene expression patterns across multiple brain regions in schizophrenia. Schizophr Res 77: 241–252. [DOI] [PubMed] [Google Scholar]
- Leal G, Afonso PM, Salazar IL, Duarte CB (2014). Regulation of hippocampal synaptic plasticity by BDNF. Brain Research 76: 696–708. [DOI] [PubMed] [Google Scholar]
- Lipson C, Alalouf G, Bajorek M, Rabinovich E, Atir-Lande A, Glickman M et al (2008). A proteasomal ATPase contributes to dislocation of endoplasmic reticulum-associated degradation (ERAD) substrates. J Biol Chem 283: 7166–7175. [DOI] [PubMed] [Google Scholar]
- MacGurn JA, Hsu PC, Emr SD (2012). Ubiquitin and membrane protein turnover: From cradle to grave. Annu Rev Biochem 81: 231–259. [DOI] [PubMed] [Google Scholar]
- Martins-de-Souza D, Wagner FG, Schmitt A, Maccarrone G, Hunyadi-Gulyås E, Eberlin MN et al (2009). Proteomic analysis of dorsolateral prefrontal cortex indicates the involvement of cytoskeleton, oligodendrocyte, energy metabolism and new potential markers in schizophriena. J Psychiatr Res 43: 978–986. [DOI] [PubMed] [Google Scholar]
- Mao I, Liu J, Li X, Luo H (2008). REGγ, a proteasome activator and beyond? Cell Mol Life Sci 65: 3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarley RW, Shenton ME, O'Donnell BF, Faux SF, Kikinis R, Nestor PG et al (1993). Auditory P300 abnormalities and left posterior superior temporal gyrus volume reduction in schizophrenia. Arch Gen Psychiatry 50: 190–197. [DOI] [PubMed] [Google Scholar]
- Menon RR, Barta PE, Aylward EH, Richards SS, Vaughn DD, Tien AY et al (1995). Posterior superior temporal gyrus in schizophrenia: grey matter changes and clinical correlates. Schizophr Res 16: 127–135. [DOI] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P (2002). Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 22: 2718–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TM, Haroutunian V, Meador-Woodruff JH (2014). N-glycosylation of GABAA receptor subunits is altered in schizophrenia. Neuropsychopharmacology 39: 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller TM, Remedies CE, Haroutunian V, Meador-Woodruff JH (2015). Abnormal subcellular localization of GABA(A) receptor subunits in schizophrenia brain. Translational Psychiatry 5 (e-pub ahead of print; doi:10.1038/tp.2015.102). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oni-Orisan A, Kristiansen LV, Haroutunian V, Meador-Woodruff JH, McCullumsmith RE (2008). Altered vesicular glutamate transporter expression in the anterior cingulate cortex in schizophrenia. Biol Psychiatry 63: 766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson GD (1997). Superior temporal gyrus and planum temporale in schizophrenia: A selective review. Prog Neuropsychopharmacol Biol Psychiatry 21: 1203–1229. [DOI] [PubMed] [Google Scholar]
- Pickering AM, Davies KJ (2012). Differential roles of proteasome and immunoproteasome regulators Pa28αβ, Pa28γ and Pa200 in the degradation of oxidized proteins. Arch Biochem Biophys 523: 181–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ (2010). The immunoproteasome, the 20S proteasome, and the PA28αβ proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J 432: 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powchik P, Davidson M, Haroutunian V, Gabriel SM, Purohit DP, Perl DP et al (1998). Postmortem studies in schizophrenia. Schizophr Bull 24: 325–341. [DOI] [PubMed] [Google Scholar]
- Purohit DP, Perl DP, Haroutunian V, Powchik P, Davidson M, Davis KL (1998). Alzheimer disease and related neurodegenerative diseases in elderly patients with schizophrenia: a postmortem neuropathologic study of 100 cases. Arch Gen Psychiatry 55: 205–211. [DOI] [PubMed] [Google Scholar]
- Rajarethinam RP, DeQuardo JR, Nalepa R, Tandon R (2000). Superior temporal gyrus in schizophrenia: a volumetric magnetic resonance imaging study. Schizophr Res 41: 303–321. [DOI] [PubMed] [Google Scholar]
- Rajasekaran A, Venkatasubramanian G, Berk M, Debnath M (2015). Mitochondrial dysfunction in schizophrenia: Pathways, mechanisms and implications. Neurosci Biobehav Rev 48: 10–21. [DOI] [PubMed] [Google Scholar]
- Realini C, Jensen CC, Zhang Z, Johnston SC, Knowlton JR, Hill CP et al (1997). Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J Biol Chem 272: 25483–25492. [DOI] [PubMed] [Google Scholar]
- Reyazuddin M, Azmi SA, Islam N, Rizvi A (2014). Oxidative stress and level of antioxidant enzymes in drug-naïve schizophrenics. Indian J Psychiatry 56: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L et al (1994). Inhibitors of the proteasome block degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78: 761–771. [DOI] [PubMed] [Google Scholar]
- Rubio MD, Haroutunian V, Meador-Woodruff JH (2012). Abnormalities of the Duo/Ras-related C3 botulin toxin substrate 1/p21-activated kinase 1 pathway drive myosin light chain phosphorylation in frontal cortex in schizophrenia. Biol Psychiatry 71: 906–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MD, Wood K, Haroutunian V, Meador-Woodruff JH (2013). Dysfunction of the ubiquitin proteasome and ubiquitin-like systems in schizophrenia. Neuropsychopharmacology 38: 1910–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AR, Mele M, Vaz SH, Kellermayer B, Grimaldi M, Colino-Oliveira M et al (2015). Differential role of the proteasome in the early and late phases of BDNF-induced facilitation of LTP. J Neurosci 35: 3319–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet RA, Henteleff RA, Zhang W, Sampson AR, Lewis DA (2009). Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology 34: 374–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K (2013). The proteasome: From basic mechanisms to emerging roles. Keio J Med 62: 1–12. [DOI] [PubMed] [Google Scholar]
- Tucholski J, Simmons MS, Pinner AL, Haroutunian V, Meador-Woodruff JH (2013. a). Abnormal N-linked glycosylation of cortical AMPA receptor subunits in schizophrenia. Schizophr Res 146: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucholski J, Simmons MS, Pinner AL, McMillan LD, Haroutunian V, Meador-Woodruff JH (2013. b). N-linked glycosylation of cortical N-methyl-D-aspartate and kainite receptor subunits in schizophrenia. Neuroreport 24: 688–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N et al (2002). The structure of the mammalian 20S proteasome at 2.75A resolution. Structure 10: 609–618. [DOI] [PubMed] [Google Scholar]
- Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH 3rd, Donovan DM et al (2001). Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull 55: 641–650. [DOI] [PubMed] [Google Scholar]
- Voges D, Zwickl P, Baumeister W (1999). The 26S proteasome: A molecular machine designed for controlled proteolysis. Annu Rev Biochem 68: 1015–1068. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang S, Viollet B, Zou MH (2012). Regulation of the proteasome by AMPK in endothelial cells: the role of O-GlcNAc transferase (OGT). PLoS ONE 7: e36717. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhang XY, Liang J, Chen da C, Xiu MH, Yang FD, Kosten TA et al (2012). Low BDNF is associated with cognitive impairment in chronic patients with schizophrenia. Psychopharmacology 222: 277–284. [DOI] [PubMed] [Google Scholar]
- Zemoura K, Benke D (2014). Proteasomal degradation of γ-aminobutyric acidB receptors is mediated by the interaction of the GABAB2 C terminus with the proteasomal ATPase Rtp6 and regulated and neuronal activity. J Biol Chem 289: 7738–7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl P, Grziwa A, Pühler G, Dahlmann B, Lottspeich F, Baumeister W (1992). Primary structure of the Thermoplasma proteasome and its implications for the structure, function, and evolution of the multicatalytic proteinase. Biochemistry 31: 964–972. [DOI] [PubMed] [Google Scholar]
- Zwickl P, Kleinz J, Baumeister W (1994). Critical elements in proteasome assembly. Nat Struct Biol 1: 765–770. [DOI] [PubMed] [Google Scholar]