Abstract
Kinase signaling is a major mechanism driving many cancers. While many inhibitors have been developed and are employed in the clinic, resistance due to crosstalk and pathway reprogramming is an emerging problem. High-throughput assays to detect multiple pathway kinases simultaneously could better model these complex relationships and enable drug development to combat this type of resistance. We developed a strategy to take advantage of time-resolved luminescence of Tb3+-chelated phosphotyrosine-containing peptides, which facilitated efficient energy transfer to small molecule fluorophores conjugated to the peptides to produce orthogonally-colored biosensors for two different kinases. This enabled multiplexed detection with high signal to noise in a high-throughput-compatible format. This proof-of-concept study provides a platform that could be applied to other lanthanide metal and fluorophore combinations to achieve even greater multiplexing without the need for phosphospecific antibodies.
Numerous leukemias and lymphomas have been characterized by the clonal expansion of B-lymphocytes due to the deregulation of the B-cell receptor signaling pathway. 1, 2 Tyrosine kinases Lyn, Syk and Btk are the main signal transducers in this pathway, making them popular therapeutic targets for small molecule inhibitors. 3 Despite the identification of this pathway as the cause of disease, effective therapeutic options targeting the B-cell receptor pathway and/or these kinases are still relatively limited. Often these kinase activities are dependent on each other, which can affect the efficacy of inhibitor drugs targeting individual enzymes. There is a need for new detection strategies that offer sensitive and specific detection of multiple kinase activities that can enhance the depth of information obtained in a screening assay, monitoring more than one signal simultaneously and mimicking reconstitution of the relevant pathways.
Förster resonance energy transfer (FRET) based assays have been developed to monitor multiple dynamic cellular processes simultaneously in a single assay. 4–8 However, while useful in some applications, FRET based methods that use organic fluorophores or fluorescent proteins as both the donor and acceptor suffer from limitations including small dynamic ranges, small Stokes shifts/wide emission peaks resulting in spectral bleed through, and the requirement for genetic engineering and expression of protein fluorophores. Lanthanides (Ln3+) have been explored as probes in biological assays for the detection of ligand binding, enzyme activity, and protein-protein interactions due to their unique optical properties. 9–17
Compared to organic fluorophores and fluorescent proteins, Ln3+ have narrow emission bands, large Stokes shifts, and long photoluminescence lifetimes, enabling time-resolved analysis, high sensitivity and specificity of detection due to reduced interference from short-lived background fluorescence. These also allow multiplexed detection via the multiple distinct, well-resolved emission bands that can be exploited for luminescence resonance energy transfer (LRET) to more than one acceptor fluorophore, chosen such that the emission profiles do not overlap (e.g. Fig. 1A). Existing examples of this strategy rely on antibodies for detection, with either the substrate or a substrate-specific antibody tagged with a small molecule fluorophore for emission, and a phosphospecific antibody labeled with a chelated lanthanide for detecting phosphorylation via donation to the small molecule fluorophore.17–20 These strategies are therefore limited to the antibodies available for a given substrate modification, and subject to the costs and handling issues presented by such immunodetection workflows.
Figure 1.
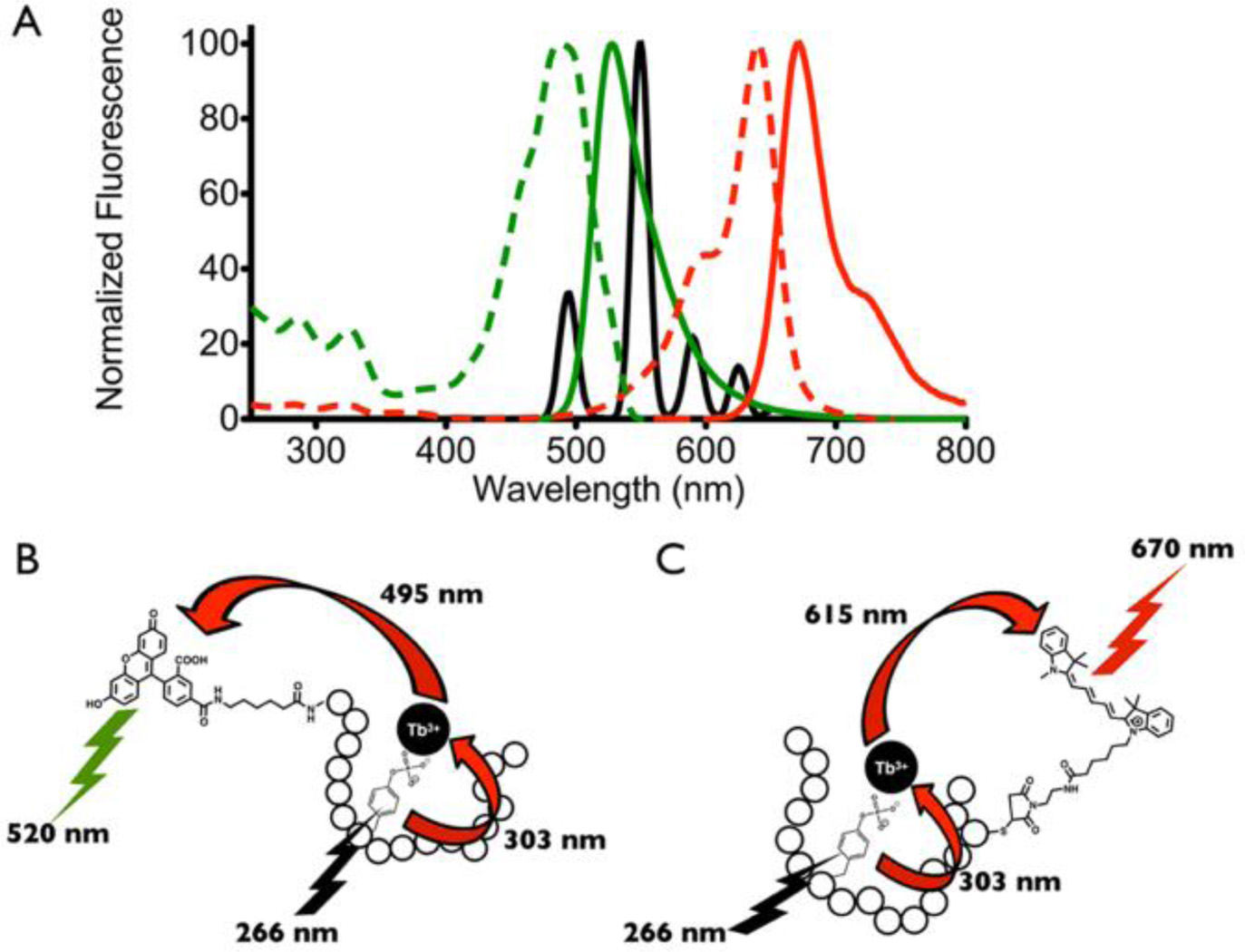
Multiplexed detection using time-resolved lanthanide-based resonance energy transfer (TR-LRET) and fluorophore conjugated peptide biosensors. (A) Emission spectrum of phosphopeptide-Tb3+ complex (black), excitation (dashed lines) and emission (solid lines) spectra of the two acceptor fluorophores 5-FAM (green) and Cy5 (red). Schematic illustrating TR-LRET detection of Lyn (B) and Syk (C) tyrosine kinase activities using the 5-FAM-SFAStide-A (5-FAM-Ahx-GGEEDEDIYEELDEPGGKbiotinGG) and SAStide-Cy5 (GGDEEDYEEPDEPGGCCy5GG) biosensors respectively.
Previously, we demonstrated development of peptide biosensors capable of detecting tyrosine kinase activity through phosphorylation-enhanced terbium (Tb3+) luminescence.21–23 Here we show extension to a multiplexed detection platform for simultaneous monitoring of multiple tyrosine kinase activities (Lyn and Syk) via SFAStide-A and SAStide substrates (sequences given in Table 1).21, 22 Multi-colored detection was achieved through time-resolved luminescence energy transfer (TR-LRET) by employing the phosphopeptide-Tb3+ complexes as the energy donors and the conjugated fluorophores cyanine 5 (Cy5) and 5-carboxyfluorescein (5-FAM) respectively, as the energy acceptors (Figure 1A).
Table 1.
| Name | Kinase | Sequence |
|---|---|---|
| 5-FAM-SFAStide-A | Src-family | 5-FAM-Ahx-GGEEDEDIYEELDEPGGKbGG |
| SAStide-Cy5 | Syk | GGDEEDYEEPDEPGGCCy5GG |
5-FAM=5-carboxyfluorescein; Ahx=6-aminohexanoic acid; Kb=biotinyl-L-lysine; CCy5=cysteine thiol conjugated with Cy5.
Sequence segments represented in bold are the core kinase recognition/Tb3+-chelation residues of the biosensor.
5-FAM was selected as the acceptor to couple with the pSFAStide-A-Tb3+ complex because its broad excitation peak at 495 nm matches well with the 5D4 → 7F6 emission band of Tb3+ centered at 495 nm. Sensitized excitation of the phosphorylated 5-FAM-SFAStide-A-Tb3+ complex through phosphotyrosine triggers energy transfer to 5-FAM, giving emission from 5-FAM at its characteristic wavelength (~520nm), which falls in a relatively “empty” region of the Tb3+ emission spectrum (Figure 1B). Similarly, detection of pSAStide-Cy5-Tb3+ complex is achieved based on the overlap of the Cy5 excitation band with the 5D4 → 7F4 and 5D4 → 7F3 emission bands of Tb3+ centered at 595 nm and 620 nm, giving Cy5 emission at its characteristic wavelength (~670 nm) which is also free of interference from Tb3+ emission (Figure 1C).
Phosphorylated and unphosphorylated forms of SAStide-Cy5 and 5-FAM-SFAStide-A were synthesized as controls. As we characterized in our previous work, phosphorylation of the peptide substrates resulted in physiochemical and photophysical changes in the peptide-Tb3+ complex that enable detection of kinase activity. These changes include enhancing the Tb3+ binding affinity, reducing the Tb3+ chelate hydration number, increasing the Tb3+ luminescence lifetime, and shifting the excitation wavelength of tyrosine.21–23
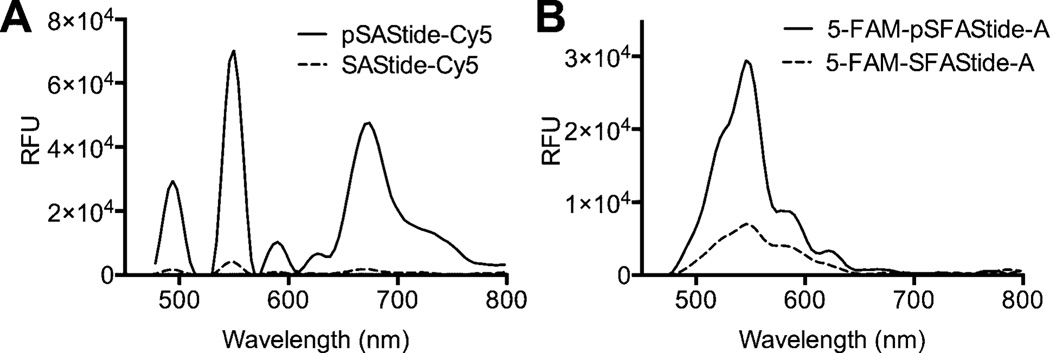
Time-resolved analysis of each peptide biosensor in the presence of Tb3+ gave the four characteristic luminescence emission peaks from Tb3+ as well as the fluorescence emission peak from the conjugated fluorophore label (Figure 2A, B). Quantitative comparison of the emission spectra between the phosphorylated and unphosphorylated biosensors showed a 25-fold increase in intensity at the Cy5 emission maximum (λ670) for pSAStide-Cy5 (Figure 2A), and a 3.9-fold increase in intensity at the 5-FAM emission maximum (λ520) for 5-FAM-pSFAStide-A (Figure 2B). Control experiments in the presence and absence of Tb3+ showed that excitation of pSAStide-Cy5 at 266 nm was Tb3+- and therefore LRET-dependent rather than arising from direct excitation of the fluorophore. Excitation of 5-FAM-pSFAStide-A at 266 nm was also Tb3+/LRET-dependent, but also showed some low-level background excitation at ~330 nm even in the absence of Tb3+ (Supporting Information S5), which we speculate can be attributed to delayed fluorescence or phosphorescence of the fluorescein, possibly from peptide adsorbed onto the surface of the well since such longer lived emission (up to the ms range) has been previously observed for fluorescein particularly when adsorbed onto surfaces or in solid state environments.24–26 However this background excitation of 5-FAM would not substantially affect the LRET readout for the assay since excitation is performed at 266 nm, which did not show any signal for excitation in the absence of Tb3+. Accordingly, the relevant changes in the intensity of the fluorophore signals upon phosphorylation of their respective peptides provide sensor-specific spectral features that can be monitored to determine phosphorylation of the sensors and consequently kinase activity.
Figure 2.
Time-Resolved Lanthanide-based Resonance Energy Transfer (TR-LRET) detection of phosphorylation-dependent signals and fluorescence cross-interference. (A) Time-resolved luminescence emission spectra for SAStide-Cy5 (dashed line) and pSAStide-Cy5 (solid line). (B) 5-FAM-SFAStide-A (dashed line) and 5-FAM-pSFAStide-A (solid line). Spectra were collected from 15 µM peptide in the presence of 100 µM Tb3+ in 10 mM HEPES, 100 mM NaCl, pH 7.5, λex = 266 nm, in 50 µL total volume, 1 ms collection time, 50 µs delay time, and sensitivity 180. Data represent the average of experiments performed in triplicate.
In order to achieve multiplex detection in the same sample, the reaction and detection conditions needed to be optimized to have limited cross-interference between sensors. Cross-interference was evaluated by analyzing the fluorophore signal from an unphosphorylated sensor in the presence of the other phosphorylated biosensor. To accomplish this, the concentrations of the biosensors and Tb3+, as well as the delay time, were varied and TR-LRET spectra collected. Quantification was accomplished by Gaussian fitting of the fluorophore emission peaks and integrating the resulting curves for each peak (Supporting Information S6). Under the optimized conditions, the TR-LRET spectra for each phosphorylated biosensor displayed minimal signal from cross-interfering fluorophore, while giving significantly stronger signal for the desired fluorophore (Supporting Information S7). TR-LRET distance parameters were also characterized (Supporting Information S8 and Table 1).
Next, a calibration curve was plotted to show the quantitative relationship between sensor phosphorylation and its corresponding TR-LRET signal for each sensor (Supporting Information S9). Experiments were performed in the presence of the unphosphorylated form of the other biosensor and the kinase reaction buffer (to best mimic the conditions of a multiplexed kinase reaction). Proportion of phosphorylated peptide was quantitatively determined by integrating the signal centered at 520 nm for 5-FAM and 670 nm for Cy5. The high signal to noise ratio observed in the initial control experiments was maintained in the presence of the reaction buffer with 7.6:1 for SAStide-Cy5 and 5.8:1 for 5-FAM-SFAStide-A. Z’-factor and signal window (SW) values were calculated and shown to be appropriate for HTS with Z’-factor values of 0.72 and 0.78, and SW of 13.27 and 12.65, for SAStide-Cy5 and 5-FAM-SFAStide-A, respectively. Details of these calculations are provided in the supporting information.
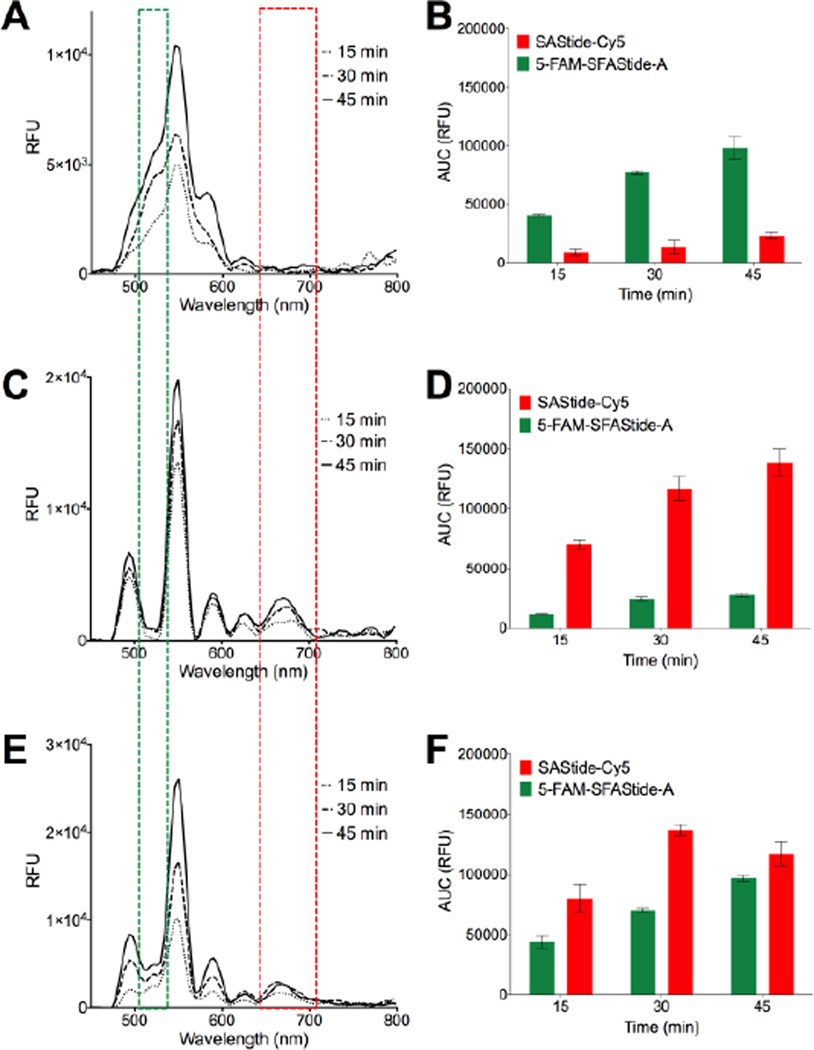
After establishing the relationship between sensor phosphorylation and TR-LRET signal, we employed the two biosensors in a kinase assay. Analysis of Syk and Lyn activities in vitro was accomplished using the purified kinases with the kinase reaction buffer and detection conditions described in the supporting information. Briefly, after pre-incubation of the kinases with the reaction buffer for 10 minutes, the reaction was initiated by the addition of the biosensor(s). Aliquots were removed from the reaction, quenched with urea, treated with Tb3+, and brought to a volume of 100 µL. In the presence of only one or the other of the kinases, TR-LRET emission spectra for each respective biosensor displayed an increase in the conjugated dye’s fluorescence signal (with minimal bleed through or background interference from the fluorophore attached to the other biosensor) over the time course of the reaction (Figure 3A–D). These results confirmed the relative specificity of each biosensor for its individual kinase, in agreement with previously reported results from our laboratory for SAStide and a separate assay using ELISA-based chemifluorescence detection for SFAStide-A (Supporting Information S10).27 Finally, to demonstrate multiplex detection, both biosensors were incubated with both kinases in a single reaction. A simultaneous increase in intensity for both fluorophores was seen over the time course, indicating an increase in phosphorylation of both peptides (Figure 3E).
Figure 3.
Simultaneous multiplexed in vitro detection of Syk and Lyn kinase activities. (A) In vitro Lyn assay luminescence emission spectra in the presence of both 5-FAM-SFAStide-A and SAStide-Cy5. (C) In vitro Syk assay luminescence emission spectra in the presence of both 5-FAM-SFAStide-A and SAStide-Cy5. (E) In vitro Lyn and Syk assay luminescence emission spectra the presence of both 5-FAM-SFAStide-A and SAStide-Cy5. (B, D, F) Quantification of 5-FAM-SFAStide-A signal and SAStide-Cy5 signal for each assay. The green and red boxes represent the approximate spectral regions represented in the Gaussian fitted curves used to integrate the signal (see supporting information S6 for more detail). Assays were performed in the presence of 2.5 µM 5-FAM-SFAStide-A, 12.5 µM SAStide-Cy5, Lyn, Syk or both kinases (15 nM), 100 µM ATP, 10 mM MgCl2 and 0.2 ng/µL BSA.
In summary, we have presented the development of a platform for detection of kinase activity that leverages the overlap of the multiple distinct emission bands of Tb3+ with orthogonal fluorescently labeled peptide substrates that are capable of phosphorylation-enhanced Tb3+ luminescence. Multiplexed kinase activity detection has remained a challenge in the field, with only a few examples of successful implementation. The Lawrence group accomplished dual kinase detection using the environmentally sensitive fluorophores oxazine and cascade yellow conjugated to peptide substrates for the Lyn and Abl kinases, respectively. 28 Unfortunately, most environmentally-sensitive fluorophores are limited in their application in more complex or higher throughput systems by small dynamic ranges and problems with background fluorescence.
A key point is that the approach presented here circumvents some of the limitations of antibody-based TR-FRET/LRET approaches and complements the previous strategies, enabling direct sensing of phosphate incorporation to the biosensors—avoiding the need for antibody labels and giving high signal-to-noise, streamlining the path from enzyme reaction to assay read-out. This strategy should be compatible with other kinases and fluorophores to increase the number of activities monitored in a single reaction, setting the stage for pathway-based drug screening to target signaling pathway reprogramming in inhibitor resistance. Future application to real-time activity monitoring would further extend the utility of this strategy.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by a National Cancer Institute K99/R00 Pathway to Independence award and R01 to L.L.P (CA127161 and CA182543) and a grant from the Purdue Center for Cancer Research (P30CA023168).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Detailed experimental conditions, peptide characterization, TR-LRET parameters (e.g. lifetimes, etc.) and additional supporting data as referenced in the text are provided. This material is available free of charge via the Internet at http://pubs.acs.org.
Conflict of Interest Disclosure
Dr. Parker and Dr. Lipchik own equity in and serve on the Scientific Advisory Board of KinaSense. Dr. Parker's relationship with KinaSense has been reviewed and managed by the University of Minnesota in accordance with its conflict of interest polices.
REFERENCES
- 1.Kuppers R. Nat Rev Cancer. 2005;5:251–262. doi: 10.1038/nrc1589. [DOI] [PubMed] [Google Scholar]
- 2.Nogai H, Dorken B, Lenz G. J Clin Oncol. 2011;29:1803–1811. doi: 10.1200/JCO.2010.33.3252. [DOI] [PubMed] [Google Scholar]
- 3.Mahadevan D, Fisher RI. J Clin Oncol. 2011;29:1876–1884. doi: 10.1200/JCO.2010.32.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peyker A, Rocks O, Bastiaens PI. Chembiochem. 2005;6:78–85. doi: 10.1002/cbic.200400280. [DOI] [PubMed] [Google Scholar]
- 5.Galperin E, Verkhusha VV, Sorkin A. Nat Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 6.Kienzler A, Flehr R, Kramer RA, Gehne S, Kumke MU, Bannwarth W. Bioconjug Chem. 2011;22:1852–1863. doi: 10.1021/bc2002659. [DOI] [PubMed] [Google Scholar]
- 7.Piljic A, Schultz C. ACS Chem Biol. 2008;3:156–160. doi: 10.1021/cb700247q. [DOI] [PubMed] [Google Scholar]
- 8.Ding Y, Ai HW, Hoi H, Campbell RE. Anal Chem. 2011;83:9687–9693. doi: 10.1021/ac202595g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermanson SB, Carlson CB, Riddle SM, Zhao J, Vogel KW, Nichols RJ, Bi K. PLoS One. 2012;7:e43580. doi: 10.1371/journal.pone.0043580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeyakumar M, Webb P, Baxter JD, Scanlan TS, Katzenellenbogen JA. Biochemistry. 2008;47:7465–7476. doi: 10.1021/bi800393u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeyakumar M, Katzenellenbogen JA. Anal Biochem. 2009;386:73–78. doi: 10.1016/j.ab.2008.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajapakse HE, Gahlaut N, Mohandessi S, Yu D, Turner JR, Miller LW. Proc Natl Acad Sci U S A. 2010;107:13582–13587. doi: 10.1073/pnas.1002025107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sculimbrene BR, Imperiali B. J Am Chem Soc. 2006;128:7346–7352. doi: 10.1021/ja061188a. [DOI] [PubMed] [Google Scholar]
- 14.Vuojola J, Syrjanpaa M, Lamminmaki U, Soukka T. Anal Chem. 2013;85:1367–1373. doi: 10.1021/ac302030q. [DOI] [PubMed] [Google Scholar]
- 15.Weitz EA, Chang JY, Rosenfield AH, Pierre VC. J Am Chem Soc. 2012;134:16099–16102. doi: 10.1021/ja304373u. [DOI] [PubMed] [Google Scholar]
- 16.Yapici E, Reddy DR, Miller LW. Chembiochem. 2012;13:553–558. 489. doi: 10.1002/cbic.201100710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrandt N, Wegner KD, Algar WR. Coord Chem Rev. 2014;273:125–138. [Google Scholar]
- 18.Kim SH, Gunther JR, Katzenellenbogen JA. J Am Chem Soc. 2010;132:4685–4692. doi: 10.1021/ja100248q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton RA, Vogel KW. J Biomol Screen. 2010;15:1008–1015. doi: 10.1177/1087057110368993. [DOI] [PubMed] [Google Scholar]
- 20.Kupcho KR, Stafslien DK, DeRosier T, Hallis TM, Ozers MS, Vogel KW. J Am Chem Soc. 2007;129:13372–13373. doi: 10.1021/ja074791h. [DOI] [PubMed] [Google Scholar]
- 21.Lipchik AM, Parker LL. Anal Chem. 2013;85:2582–2588. doi: 10.1021/ac3023422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipchik AM, Perez M, Bolton S, Dumrongprechachan V, Ouellette SB, Cui W, Parker LL. J Am Chem Soc. 2015;137:2484–2494. doi: 10.1021/ja507164a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui W, Parker LL. Chem Commun (Camb) 2015;51:362–365. doi: 10.1039/c4cc07453j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmichael I, Helman WP, Hug GL. J Phys Chem Ref Data. 1987;16:239–260. [Google Scholar]
- 25.Roalstad S, Rue C, LeMaster CB, Lasko C. J Chem Educ. 1997;74:853–855. [Google Scholar]
- 26.Lam SK, Lo D. Chem Phys Lett. 1997;281:35–43. [Google Scholar]
- 27.Lipchik AM, Killins RL, Geahlen RL, Parker LL. Biochemistry. 2012;51:7515–7524. doi: 10.1021/bi300970h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Zimmerman EI, Toutchkine A, Martin TD, Graves LM, Lawrence DS. ACS Chem Biol. 2010;5:887–895. doi: 10.1021/cb100099h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.