Abstract
Purpose: The direct transmission of avian influenza viruses to human and increasing drug resisted strains posing new threats for public health. Therefore, development of efficient vaccines is needed to generate protective and persistent immunity to the viruses.
Methods: Three motifs of Mx protein sequence in human, mouse and poultry located in interferon induced (GTP ase) domain were candidate as biologic adjuvant for enhancing the immune responses against influenza virus. Chimera proteins composed with the conserved HA2 subunit of influenza virus and the Mx motifs named HA2/Mx were modeled and evaluated by in silico analysis includes bioinformatics algorithms in order to explore biological characteristics of these peptides.
Results: Amongst the predicted models, HA2/Mx1 peptide showed the better results following protein structures prediction, antigenic epitopes determination and model quality evaluation. Comparative homology modeling was performed with Swiss Model and the model was validated using ProSA. Epitope predictions revealed the construct could induce both B and T cell epitopes that expect a high immune response.
Conclusion: Taken together, these data indicate that the HA2/Mx1 chimera peptide can be potentiated for developing an adjuvant-fused influenza vaccine capable of stimulating effective immune response.
Keywords: Influenza virus, Peptid vaccine, HA2 subunit, Mx
Introduction
Influenza virus subtypes cause a wide spectrum of symptoms ranging from mild illness to fatal disease in bird and mammalian species. The genetic variations of the viruses emerge and re-emerge of new antigenic variants, and direct transmission of avian viruses to human raise a concern in recurring health problem.1-4 Control and treatment of influenza depends mainly on vaccination and chemoprophylaxis with the approved antiviral drugs. The emergence of drug resistant strains of influenza viruses under drug selective pressure highlights the need to development of new influenza therapies.5,6 The current influenza vaccines are designed to elicit B-cell responses so, development of gene vaccines have been interested because significant potential for induction of both humoral and cell-mediated immunity through the priming of CD4 and CD8 cells. Induction of immune responses against influenza viruses have been considered by targeting various viral proteins.7-10 Hemagglutinin (HA) plays a vital role in the attachment and activation of membrane fusion for entry of the virus into host cells and comprises major neutralizing epitopes has additional considerations as highly immunogene.11,12 Viral HA precursor is cleaved by host trypsin-like proteolytic enzymes into HA1 and HA2 subunits. However development HA-based vaccine is often difficult to attain due to emergence variants that have undergone sufficient antigenic drift in HA1 globular head to evade existing antibody responses.13,14 It has been shown that HA2 is highly conserved as compared to HA1 subunit and antibodies recognizing stalk domain of the subunit neutralize the virus and provide sufficiently protection against infection and do cross-react with the HA of other subtypes.15-17
Generally enhanced and directed immune responses to viral vaccine can be achieved by using adjuvants. The molecules act by different mechanisms include prolonging the exposure time of antigen to the immune system, enhancing the delivery of antigen, directing antigen presentation by the MHC, or providing immune stimulatory signals that potentiate the immune response.18-20 The ability of molecular adjutants such as cytokines, bacterial derivatives and immune system regulator proteins to improve the immunogenicity of vaccines as a novel strategy is under evaluation.21-26 Here we explore the idea of using host cellular protein to elicit immune responses against influenza virus and choose Mx protein which belongs to the class of dynamin-like large guanosine triphosphatases (GTPases) and involves in interferon induction and immune system regulation.26,27 We focus on details of viral HA2 and cellular Mx interactions for possibly induction immunity against influenza infection using computational chemistry and bioinformatics tools. Prediction of structures, properties, functions and solvent accessibility of proteins plays a crucial role in determining antigenicity and immunogeneicity. Such results are used in a wide range of applications in molecular biology, medical science and drug and vaccine design.23,28,29 The in silico study was designed based on comparative modeling techniques to predict the structure, properties and functions of HA2/Mx chimera protein constructs to candidate an efficient gene vaccine against influenza infection.
Materials and Methods
HA2 Influenza A virus sequence data collection
Datasets of HA2 deduced amino acid sequences based on the circulated H9N2 influenza subtypes from their emergence to 2013 were derived from GeneBank. All of the sequences were aligned using ClustalW program with default parameters. The conserved HA2 peptide encoded 189 amino acids in length were determined using BioEdit.
Mx sequence data collection
Datasets of Mx peptide sequences from Homo sapiens, Mus musculus, and Gallus gallus were derived from the Uniprot. All of the sequences were aligned and three conserved motifs in GTPase domain (interferon induced domain) determined.
Chimeric HA2/Mx constructs design and characterization
To design a single peptide construction the C-terminus of HA2 fragment were fused to each of Mx motif using a repeat of hydrophobic amino acid linkers (EAAAK). The kozak sequence was introduced to increase the efficiency of translational initiation. The bioinformatics analyses were ran on the three HA2/Mx constructs. The physicochemical properties, hydrophobicity, hydrophilicity, surface accessibility and electrostatic potential of the HA2/Mx construct proteins were identified using Prot-Param (http://expasy.org/tools/protparam.html). These peptides sequences were inverted to nucleotide sequences and were consistent to mouse practical codons by GeneScript. Then codon adaption index (CAI) score and the average GC content were estimated.
Proteins structures prediction
To aid alignment correction and loop modeling, secondary structures of HA2/Mx chimera proteins were predicted by using PSIPRED tool (http://bioinf.cs.ucl.ac.uk/psipred). Protein structure and three dimensional (3D) models or tertiary structure of the chimeric constructs were predicted by Phyre (http://www.sbg.bio.ic.ac.uk/phyre2/html/). The 3D model is visualized in different representation patterns by the Swiss-Pdb Viewer (http://spdbv.vital_it.ch/).
Homology modeling and model quality and validation
The 3D models were constructed from the sequence alignment between the constructs and the template proteins using SWISSMODEL30 with parameters of energy minimization value. The energy minimization was computed with the GROMOS96 implantation of the software.
In order to assess the reliability of the modeled structure of HA2/Mx, the root mean square deviation (RMSD) was calculated by superimposing it on the template structure using a 3D structural superposition. The backbone conformation of the modeled structure was calculated by analyzing the phi (Φ) and psi (ψ) torsion angles using RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). Finally, the quality of the consistency between the template and the modeled HA2/Mx was evaluated using validated by ProSA (https://prosa.services.came.sbg.ac.at/prosa.php) which gives the overall model quality based on the Cα positions.
Prediction of post-translational modifications
Certain post-translational modifications may occur in the eukaryote protein sequences such as NetNGly, NetOGly and YingOYang. For more information on the correct folding of the HA2/Mx peptides, N-glycosylation of NXS/T amino acids sequences (where X is any amino acids except prolin) and O-(beta)-GlcNAc were evaluated at http://www.cbs.dtu.dk.
Potential antigenic sites prediction
The amino acid sequences were predicted for linear B-cell epitopes using Immune Epitope Database (IEDB) server (http://tools.immuneepitope.org/tools/bcell/tutorial.jsp). The antigenic sites in the chimeric models were determined using Kolaskar and Tongaonkar antigenicity prediction method based on physicochemical properties of amino acid residues (i.e. hydrophilicity, accessibility and flexibility) with about 75% accuracy. Solvent accessible scale for delineating hydrophobic and hydrophilic characteristics of the chimera protein sequences was predicted using Vadar. Prediction of T-cell epitopes in the protein sequences was performed based on integrating the peptide major histocompatibility (MHC) class I binding, proteasomal C terminal cleavage and transporters associated with antigen processing efficiency by using the NetCTL tool in the server and SYFPEITHI (http://www.syfpeithi.de/Scripts/MHCServer.dll/EpitopePrediction.htm).
Results
Chimeric HA2/Mx physicochemical properties
Each of the Mx1 13SGKSSVLEALSGVALPR30, Mx2 103VPDLTLIDLPGITRVAV120 and Mx3 152NVDIATTEALSMAQEVD169 motif was selected and fused by repeats of EAAAK sequence to HA2 peptide sequence named HA2/Mx1, HA2/Mx2 and HA2/Mx3. The physicochemical properties of the constructs are shown in Table 1. The physicochemical properties were similar except that the negatively charged residue of the HA2/Mx3 was slightly higher than other constructs. The calculated instability indexes suggest HA2/Mxs are stable proteins. The negative GRAVY indexes of -0.378, -0.461, and -0.438 are indicative of a hydrophilic and soluble proteins. By optimizing Codons to proposed constructs, CAI score achieved 1 and the average GC content was > 50 percent for these models.
Table 1. Physicochemical properties of the HA2/Mx chimera peptides .
| Physicochemical properties | HA2/Mx | HA2/Mx | HA2/Mx |
| MW (kDa) | 23955.3 | 23970.1 | 23834.1 |
| Negatively charged residue (Asp+Glu) | 5.73 | 5.24 | 6.33 |
| Positively charged residue (Arg+Lys) | 31 | 33 | 30 |
| Theoretical pI | 28 | 27 | 29 |
| Instability index | 36.14 | 36.06 | 36.14 |
| Aliphatic Index | 90.14 | 84.17 | 85.55 |
| GRAVY (Grad Average of Hydropathicity) | -0.378 | -0.461 | -0.438 |
Protein structures prediction
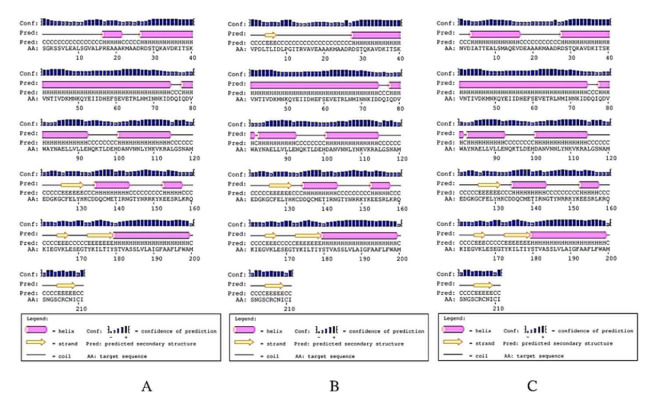
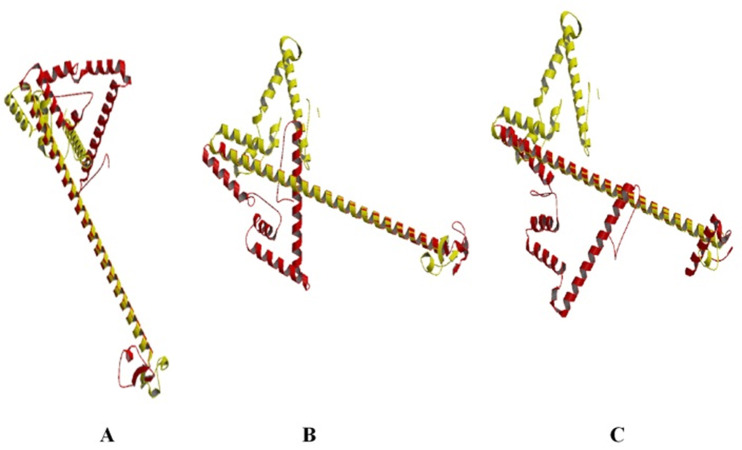
Analysis of the chimeric protein secondary structure (Figure 1) indicated 35.07% random coils, 55.45% α-helixes and 9.47% β-sheets for HA2/Mx1 construct. The HA2/Mx2 showed lower random coils (25.59%) and higher α-helixes (64.45%) and β-sheet (11.37%). The 31.75% random coils, 58.29% α-helixes and 9.95% β-sheets for HA2/Mx3 was detected. The 3D structures were modeled (Figure 2) and evaluated for choosing the best model.
Figure 1.
PSIPRED graphical result of HA2/Mx chimera peptide secondary structure prediction. A) HA2/Mx1 construct, B) HA2/Mx2 construct, and C) HA2/Mx3 construct.
Figure 2.
Three dimensional structure of the HA2/Mx chimera peptides modeled by the Swiss-Pdb Viewer software. A) HA2/Mx1 construct, B) HA2/Mx2 construct, and C) HA2/Mx3 construct.
Homology modeling and model quality evaluation
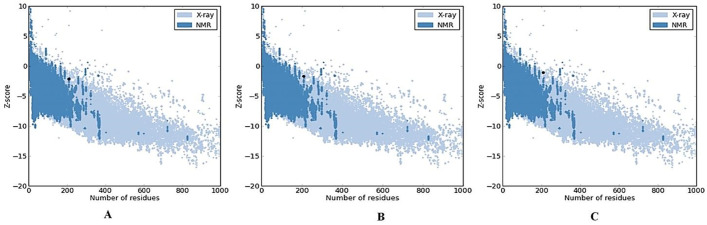
Homology modeling was used to determine the 3D structure of HA2/Mx. SWISSMODEL search with default parameters was performed to find suitable templates for homology modeling The HA2/Mx1 construct was modeled with 93.0% confidence and 74% identity between query and template protein sequence by the single highest scoring template. The other constructs were modeled with lower confidence percentages (92.86 and 88.16) and the same identity. To select the best chimera protein model, the generated 3D HA2/Mx models were compared together. The models were qualifies by model geometry RMSD, energy minimization, and the Ramachandran plot. The peptide structures were superimposed for the calculation of RMSD. The value for HA2/Mx1 construct was estimated 1.93 while the similar lower values were obtained for other constructs (1.42 and 1.48). The predicted HA2/Mx1-3 models were subjected to energy minimization by implementation of the Swiss-PDB viewer (-1,231.092, -1,179.402, and -1,138.913 kcal/mol for three models respectively) and analysis of the results indicating that the constructs had acceptable stability. The predicted structure was further validated for its reliability and structural quality based on the Ramachandran plot. The plots for the three HA2/Mx peptides were generated to assess the quality of the structures built using homology modeling. The plots showed φ and ψ torsion angles for all residues in these models and clustered around secondary structure regions that define the backbone conformation. Results of RAMPAGE for HA2/Mx1 showed 97.2% of residues are located in the favored region, 1.4% in the allowed and only 1.4% in the outlier regions (Figure 3). The percent of residues placed in allowed region was lower for HA2/Mx2 (96.2) and HA2/Mx3 (94.3). The overall quality of these models was validated and the energy criteria for the modeled structure were compared with the potential mean force obtained from a large set of known protein structures. The ProSA Z-scores were obtained -2.12, -1.65, and -1.08 for these constructs, respectively (Figure 4) and -2.63 for the template’s 3D structure.
Figure 3.
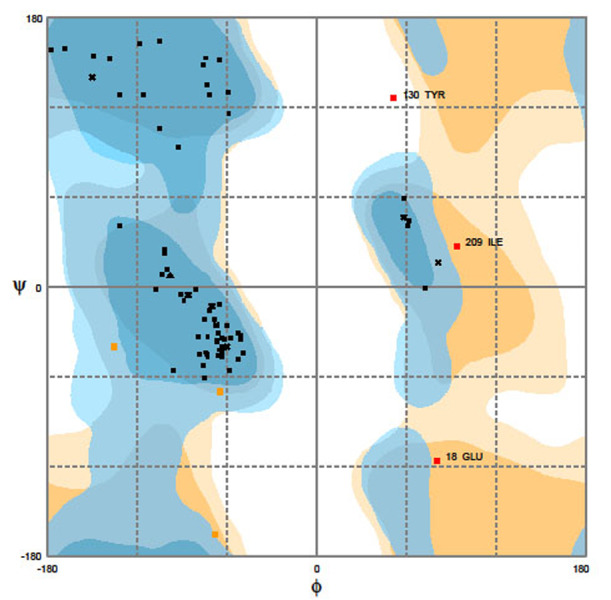
Evaluation of HA2/Mx1 quality by Ramachandran plot. Percentage of the residue was 97.2% in the favored region, 1.4% in allowed region, and 1.4% in outlier region. The φ and ψ torsion angles of amino acid residues in the proteins were reasonably accurate.
Figure 4.
ProSA model quality validation of HA2/Mx chimera peptides. A) HA2/Mx1: Z-score -2.12; B) HA2/Mx2: -1.65; C) HA2/Mx3: -1.08.
Prediction of post-translational modifications
Two N-glycosylation sites at positions 139 and 148 and two O-(beta)-GlcNAc sites at positions 24 and 101 in all of the HA2/Mx constructs were determined which were similar to the HA2 peptide of H9N2 influenza viruses.
Antigenicity evaluation
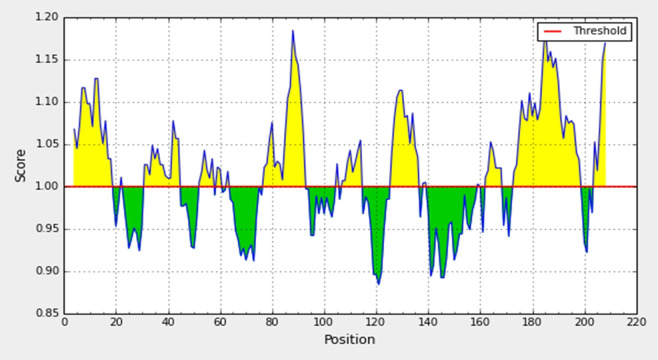
The ultimate results of the potential B cell epitopes of these peptides were shown in Table 2. In contract to HA2/Mx1, similar predicted B cell epitopes (60, 74, 95, 118, 143, and 167) were predicted for HA2/Mx2 and HA2/Mx3 constructs. Figure 5 shows the antigenic determinant plot for HA2/Mx1; x-axis shows amino acid position and y-axis shows antigenic propensity score. Average antigenic for HA2/Mx1 chimera protein is 1.016; all residues above 1.0 are potentially antigenic in Kolaskar and Tongaonkar algorithm. There are five antigenic determinants in sequence with the highest pick at position 85-95 amino acid residue and at the end of sequence. Antigenic index directly showed the epitope forming capacity of HA2/Mx1 chimera peptide. Estimation of the solvent accessible scale was shown that the chimera peptide is hydrophobic and contains segments of low complexity and high-predicted flexibility. For HA2/Mx2 and HA2/Mx3, the average antigenic propensities were calculated -0.104 and -0.048 respectively. According to the algorithm these constructs may not be elicit an antibody response. The vaccine-formulated adjuvant can be greatly modulated the T helper responses. NetCTL tool was used to identify the T cell epitope of HA2/Mx chimera peptides. The high scores predicted regions of the peptides were shown in Table 3. As a result, HA2/Mx1 was predicted to have an additional high score T cell epitope sequence at position 5.
Table 2. Prediction of linear B- cell epitopes in HA2/Mx chimer peptides .
| Construct | Position | Amino acid sequence |
| HA2/Mx1 | 4 | SSVLEALSGVALPRE |
| 31 | QKAVDKITSKVNTI | |
| 52 | YEIIDH | |
| 77 | IQDVWAYNAELLVLLE | |
| 107 | LYNKVKRA | |
| 126 | CFELYHKCDDQ | |
| 162 | IEGVKLE | |
| 173 | YKILTIYSTVASSLVLAIGFAAFLFW | |
| HA2/Mx2 | 22 | KMAADRDSTQKAVDK |
| 60 | S | |
| 74 | DDQ | |
| 95 | KTLDEHDAN | |
| 118 | NAMEDGK | |
| 143 | NGTYNRRKYKEESRL | |
| 167 | LESE | |
| HA2/Mx3 | 4 | IA |
| 15 | EVDEAAAKMAADRDSTQKAVDK | |
| 60 | S | |
| 74 | DDQ | |
| 95 | KTLDEHDAN | |
| 118 | NAMEDGK | |
| 143 | NGTYNRRKYKEESRL | |
| 167 | LESE |
Figure 5.
Antigenicity prediction plot of HA2/ Mx1 chimera peptide an average 1.016 using Kolaskar-Tongaonkar method.
Table 3. Prediction of high score T- cell epitopes in HA2/Mx chimer peptides .
| Position | HA2/Mx1 | Position | HA2/Mx2 | Position | HA2/Mx3 |
| 89 | VLLENQKTL | 89 | VLLENQKTL | 89 | VLLENQKTL |
| 5 | SVLEALSGV | 36 | KITSKVNTI | 36 | KITSKVNTI |
| 36 | KITSKVNTI | 72 | KIDDQIQDV | 72 | KIDDQIQDV |
| 72 | KIDDQIQDV | 96 | TLDEHDANV | 96 | TLDEHDANV |
| 96 | TLDEHDANV | 83 | YNAELLVLL | 83 | YNAELLVLL |
| 83 | YNAELLVLL | 174 | KILTTYSTV | 174 | KILTTYSTV |
| 174 | KILTTYSTV | 188 | LAIGFAAFL | 188 | LAIGFAAFL |
| 188 | LAIGFAAFL | 54 | IIDNEFSEV | 54 | IIDNEFSEV |
| 54 | IIDNEFSEV | 65 | RLNMINNKI | 65 | RLNMINNKI |
| 65 | RLNMINNKI | 29 | STQKAVDKI | 29 | STQKAVDKI |
| 29 | STQKAVDKI | 33 | AVDKITSKV | 33 | AVDKITSKV |
| 33 | AVDKITSKV | 182 | VASSLVLAI | 182 | VASSLVLAI |
| 182 | VASSLVLAI | 185 | SLVLAIGFA | 185 | SLVLAIGFA |
| 185 | SLVLAIGFA | 79 | DVWAYNAEL | 79 | DVWAYNAEL |
| 79 | DVWAYNAEL | 103 | KVNNLYNKV | 103 | KVNNLYNKV |
| 103 | KVNNLYNKV | 106 | NLYNKVKRA | 106 | NLYNKVKRA |
| 106 | NLYNKVKRA | 167 | LESEGTYKI | 167 | LESEGTYKI |
| 167 | LESEGTYKI | 47 | KMNKQYEII | 47 | KMNKQYEII |
| 47 | KMNKQYEII | 82 | AYNAELLVL | 82 | AYNAELLVL |
| 82 | AYNAELLVL | 157 | LKRQKIEGV | 157 | LKRQKIEGV |
| 157 | LKRQKIEGV | 187 | VLAIGFAAF | 187 | VLAIGFAAF |
| 187 | VLAIGFAAF | - | - | - | - |
Discussion
Point mutations and coexistence of different influenza subtypes in the same susceptible population are likely to generate appropriate conditions for the emergence of new antigenic variants and or novel reassortment strains. Circulation and coexistence of these variants highlight the need to develop an efficacious vaccine that can provide robust protection against influenza infection. Herein, we describe the construction of a novel chimera protein comprising the conserved influenza HA2 and different motifs of Mx protein as biological adjuvant. Previous studies have been shown that vaccination with the stalk domain of HA (HA2) elicited immune sera with broader reactivity and provided full protection against death and partial protection against disease following lethal viral challenge.16,17,31 Due to simultaneously point mutations in the HA1 and the absence of these changes in HA2, recent studies on the production of peptide vaccine have been focused on HA2. Generally the protective potential of an immunogene could be increased by optimization of the delivery and immunogenicity using selection of suitable and safe adjuvant. Recently many approaches have been applied to introduce a proper adjuvant for enhancing immunity against influenza.21-24 However, some post vaccination adverse reactions frequently occurs in recipients. This study was based on the idea of introducing a biological adjuvant to reduce the frequency of the reactions by using in silico prediction, which can save the expense of synthetic peptides and the working time. The Mx sequence homology analysis revealed that three conserved motifs are placed in GTPase domain of mammalian and bird species. Each of the motifs was ligated to the C-teriminus of HA2 peptide by EAAAK linker that provide structural flexibility, improving protein stability and increasing biological activity.23,28 The ability of the chimera proteins in induction of immune responses against influenza infection was evaluated subsequently. Prediction of secondary structure of the peptides (Figure 1) was shown the high percentage of helices in their structure makes HA2/Mxs more flexible for folding, which might increase protein interactions.
Analysis of 3D structures will help in the identification of binding sites and may lead to the designing of new vaccines.28,29 Several structure assessment methods including Ramachandran plot RMSD, and Z-scores were used to check reliability of the predicted 3D models. PROCHECK displayed a higher percentage (97.2) of residues in the most favoured regions of HA2/Mx1 (Figure 4). This indicated that the backbone dihedral angles, Φ and ψ, in the HA2/Mx1 3D model, were reasonably accurate. The RMSD value indicates the degree to which two 3D structures are similar. The RMSD value obtained from superimposition of HA2/Mx2 and HA2/Mx3 was found to be lower than HA2/Mx1, suggesting the more similarity between the structures. The Z-score was used to check whether the chimera peptides placed within the range of scores typically found for native HA2s of similar size. Among them the Z-score of modelled HA2/Mx1 (-2.12) is within the range of score typically found for the template’s 3D structure (-2.63) and confirm the quality of the homology model.
The HA protein can undergo posttranslational glycosylation modification which is essential for HA folding and transport. The glycosylation sites are involved in HA biological functions, including receptor binding activity and evasion of host immunity. Alternation in the process may affect host innate immune system recognition and the ability of the HA to induce adaptive immune response.31 Seven glycosylation sites are determined in HA H9N2 viruses and two of them located at HA2.2 Prediction of posttranslational modifications of HA2/Mx chimera peptides showed that the number and location of glycosylation sites of the constructs were not differed from other HA2 deposited in GenBank. This suggests that ligation of Mx motif to HA2 did not affect the glycosylation pattern of the peptide. Finally HA2/Mx 3D structures constructed by a homology modelling method were employed to compare immunogenic future of the peptides. The majority of available in silico methods for predicting B-cell immunogenic regions focus on linear epitopes and are based on several amino acid-based propensity scales, including hydrophilicity, solvent accessibility, secondary structure, flexibility, and antigeneicity.23,29,32 The total residues lying in HA2/Mx1 B cell epitopes were differ from other constructs. Nearly half of amino acid residues in each predicted B cell epitope belonging to different antigenic regions were hydrophobic and the common residues were L, E, A, and V. Since such amino acid residues do not directly contribute to the interaction with antibodies, the surface structures of antigenic sites that are accessible for antibodies were detected. The antigenic peptides of HA2/Mx1 located in solvent accessible regions and contain low complexity and high-predicted flexibility. Data from solvent accessible surface was confirmed by an extended structure of the chimera peptide with more than 55% of α-helices. NetCTL and SYFPEITHI were used to predict the T cell epitopes in HA2/Mx peptides. All of the predicted epitopes were located in HA2 peptide fragment and only one additional potential sequences was detected in Mx motif of HA2/Mx1. It seems that the additional epitope can be role in stimulating and enhancing cellular immune response against influenza infection.
Conclusion
The three dimensional structure and functional properties of the chimera peptide constructed by HA2 as viral immunogenic and cellular Mx motifs as adjuvant were computed in silico. The applicability of these proteins was analyzed to candidate a sufficient potent vaccine against influenza. Our results suggest that the HA2/Mx1 chimera peptide potentiates to induce humoral and cellular immune responses against influenza virus. The protective immunity of the HA2/Mx1 protein against influenza viruses and the immunostimulatory effect of Mx on the chimeric HA2 protein need to be investigated in animal models.
Acknowledgments
This work was supported by Razi Vaccine & Serum Research Institute, Karaj, Iran.
Ethical Issues
Not applicable.
Conflict of Interest
This study is a part of PhD thesis that conceived and designed by S.Shahsavandi. All of the authors declare that they have no conflict of interest.
References
- 1.Russell CJ, Webster RG. The genesis of a pandemic influenza virus. Cell. 2005;123(3):368–71. doi: 10.1016/j.cell.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 2.Shahsavandi S, Salmanian AH, Ghorashi SA, Masoudi S, Ebrahimi MM. Evolutionary characterization of hemagglutinin gene of h9n2 influenza viruses isolated from asia. Res Vet Sci. 2012;93(1):234–9. doi: 10.1016/j.rvsc.2011.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A. et al. Avian-to-human transmission of h9n2 subtype influenza a viruses: Relationship between h9n2 and h5n1 human isolates. Proc Natl Acad Sci U S A. 2000;97(17):9654–8. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM. et al. Human infection with an avian h9n2 influenza a virus in hong kong in 2003. J Clin Microbiol. 2005;43(11):5760–7. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang SM, Song JM, Compans RW. Novel vaccines against influenza viruses. Virus Res. 2011;162(1-2):31–8. doi: 10.1016/j.virusres.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee YT, Kim KH, Ko EJ, Lee YN, Kim MC, Kwon YM. et al. New vaccines against influenza virus. Clin Exp Vaccine Res. 2014;3(1):12–28. doi: 10.7774/cevr.2014.3.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H, Wang L, Compans RW, Wang BZ. Universal influenza vaccines, a dream to be realized soon. Viruses. 2014;6(5):1974–91. doi: 10.3390/v6051974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo J, Zheng D, Zhang W, Fang F, Wang H, Sun Y. et al. Induction of cross-protection against influenza a virus by DNA prime-intranasal protein boost strategy based on nucleoprotein. Virol J. 2012;9:286. doi: 10.1186/1743-422X-9-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pica N, Palese P. Toward a universal influenza virus vaccine: Prospects and challenges. Annu Rev Med. 2013;64:189–202. doi: 10.1146/annurev-med-120611-145115. [DOI] [PubMed] [Google Scholar]
- 10.Watanabe T, Watanabe S, Kim JH, Hatta M, Kawaoka Y. Novel approach to the development of effective h5n1 influenza a virus vaccines: Use of m2 cytoplasmic tail mutants. J Virol. 2008;82(5):2486–92. doi: 10.1128/JVI.01899-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285(37):28403–9. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao Y, Koh X, Dong L, Du X, Wu A, Ding X. et al. Rapid estimation of binding activity of influenza virus hemagglutinin to human and avian receptors. PLoS One. 2011;6(4):e18664. doi: 10.1371/journal.pone.0018664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okumura Y, Takahashi E, Yano M, Ohuchi M, Daidoji T, Nakaya T. et al. Novel type ii transmembrane serine proteases, mspl and tmprss13, proteolytically activate membrane fusion activity of the hemagglutinin of highly pathogenic avian influenza viruses and induce their multicycle replication. J Virol. 2010;84(10):5089–96. doi: 10.1128/JVI.02605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zarrin Lebas N, Shahsavandi S, Mohammadi A, Ebrahimi MM, Bakhshesh M. Replication efficiency of influenza A virus H9N2: A comparative analysis between different origin cell types. Jundishapur J Microbiol. 2013;6(9):e8584. doi: 10.5812/jjm.8584. [DOI] [Google Scholar]
- 15.Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza a virus: Where do we stand? Microbes Infect. 2008;10(9):1024–9. doi: 10.1016/j.micinf.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khanna M, Sharma S, Kumar B, Rajput R. Protective immunity based on the conserved hemagglutinin stalk domain and its prospects for universal influenza vaccine development. Biomed Res Int. 2014;2014:546274. doi: 10.1155/2014/546274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K. et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010;1(1) doi: 10.1128/mBio.00018-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Singh M. Selection of adjuvants for enhanced vaccine potency. World J Vaccines. 2011;1(2):33–78. doi: 10.4236/wjv.2011.12007. [DOI] [Google Scholar]
- 19.Awate S, Babiuk LA, Mutwiri G. Mechanisms of action of adjuvants. Front Immunol. 2013;4:114. doi: 10.3389/fimmu.2013.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Even-Or O, Samira S, Ellis R, Kedar E, Barenholz Y. Adjuvanted influenza vaccines. Expert Rev Vaccines. 2013;12(9):1095–108. doi: 10.1586/14760584.2013.825445. [DOI] [PubMed] [Google Scholar]
- 21.Fan X, Hashem AM, Chen Z, Li C, Doyle T, Zhang Y. et al. Targeting the ha2 subunit of influenza a virus hemagglutinin via cd40l provides universal protection against diverse subtypes. Mucosal Immunol. 2015;8(1):211–20. doi: 10.1038/mi.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liniger M, Summerfield A, Ruggli N. Mda5 can be exploited as efficacious genetic adjuvant for DNA vaccination against lethal h5n1 influenza virus infection in chickens. PLoS One. 2012;7(12):e49952. doi: 10.1371/journal.pone.0049952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shahsavandi S, Ebrahimi MM, Sadeghi K, Mahravani H. Design of a heterosubtypic epitope-based peptide vaccine fused with hemokinin-1 against influenza viruses. Virol Sin. 2015 doi: 10.1007/s12250-014-3504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riedl K, Riedl R, von Gabain A, Nagy E, Lingnau K. The novel adjuvant ic31 strongly improves influenza vaccine-specific cellular and humoral immune responses in young adult and aged mice. Vaccine. 2008;26(27-28):3461–8. doi: 10.1016/j.vaccine.2008.04.029. [DOI] [PubMed] [Google Scholar]
- 25.Sadeghi K, Shahsavandi S, Ebrahimi MM, Mahravani H, Fazel H. Hemokinin-1 molecular adjuvant: an approach to enhance the efficacy of influenza vaccine. Arak Med Univ J. 2014;17(11):62–9. [Google Scholar]
- 26.Soleimani S, Shahsavandi S, Maddadgar O, Mahravani H, Lotfi M. Mx bio adjuvant for enhancing immune responses against influenza virus. Tehran Univ Med J. 2015;73:192–201. [Google Scholar]
- 27.Haller O, Stertz S, Kochs G. The mx gtpase family of interferon-induced antiviral proteins. Microbes Infect. 2007;9(14-15):1636–43. doi: 10.1016/j.micinf.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Tomar N, De RK. Immunoinformatics: An integrated scenario. Immunology. 2010;131(2):153–68. doi: 10.1111/j.1365-2567.2010.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, Wu W, Negre NN, White KP, Li C, Shah PK. Determinants of antigenicity and specificity in immune response for protein sequences. BMC Bioinformatics. 2011;12:251. doi: 10.1186/1471-2105-12-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arnold K, Bordoli L, Kopp J, Schwede T. The swiss-model workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 31.Vigerust DJ, Shepherd VL. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007;15(5):211–8. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ben-Yedidia T, Arnon R. Epitope-based vaccine against influenza. Expert Rev Vaccines. 2007;6(6):939–48. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]