The aspartic protease from N. gracilis was crystallized in complex with the inhibitor pepstatin A using a newly formulated low-pH screen. X-ray diffraction data showed that the crystals diffracted to 2.8 Å resolution.
Keywords: aspartic proteases, nepenthesins, Nepenthes gracilis, low-pH crystallization screen
Abstract
Nepenthesins are aspartic proteases secreted by carnivorous pitcher plants of the genus Nepenthes. They significantly differ in sequence from other plant aspartic proteases. This difference, which provides more cysteine residues in the structure of nepenthesins, may contribute to their unique stability profile. Recombinantly produced nepenthesin 1 (rNep1) from N. gracilis in complex with pepstatin A was crystallized under two different crystallization conditions using a newly formulated low-pH crystallization screen. The diffraction data were processed to 2.9 and 2.8 Å resolution, respectively. The crystals belonged to space group P212121, with unit-cell parameters a = 86.63, b = 95.90, c = 105.40 Å, α = β = γ = 90° and a = 86.28, b = 97.22, c = 103.78 Å, α = β = γ = 90°, respectively. Matthews coefficient and solvent-content calculations suggest the presence of two molecules of rNep1 in the asymmetric unit. Here, the details of the crystallization experiment and analysis of the X-ray data are reported.
1. Introduction
Aspartic proteases (APs; EC 3.4.23) are a relatively small group of proteolytic enzymes that can be found across all forms of life (Davies, 1990 ▸; Dunn, 2002 ▸). These enzymes are mostly expressed as zymogens capable of auto-activation in an acidic environment (Dunn, 1997 ▸; Khan & James, 1998 ▸). Aspartic proteases are systematically classified by the MEROPS database into one family, which is further divided into several subfamilies. The two major subfamilies are pepsin-like proteases (A1) and retroviral proteases (A2) (Rawlings et al., 2013 ▸).
Crystallographic analysis of various pepsin-like proteases revealed that they are mostly composed of β-sheet secondary structures. Their three-dimensional structure consists of two lobes with a very similar fold. The active site is located in the cleft between the two lobes and contains two catalytic aspartate residues, both of which occur in a conserved sequence: Asp-Thr/Ser-Gly. In porcine pepsin and endothiapepsin these residues have been identified as Asp32 and Asp215 (Coates et al., 2001 ▸; Veerapandian et al., 1992 ▸). The majority of aspartic proteases also have a flap structure made up of a β-hairpin that completes their active site (Madala et al., 2010 ▸) and participates in substrate binding.
Within the A1 MEROPS subfamily, nepenthesins represent a distinct group. These enzymes are produced by the carnivorous plants of the genus Nepenthes (Takahashi et al., 2008 ▸), where they function as a component of the pitcher fluid. In contrast to other plant aspartic proteases, nepenthesins lack the typical plant-specific insert. However, they have another insertion called the nepenthesin-type aspartic protease-specific insert. This sequence, which is not present in classical animal pepsin-like proteases, provides additional cysteine residues to their primary structure (Athauda et al., 2004 ▸; Takahashi et al., 2005 ▸). The formation of additional disulfide bridges by the higher number of cysteine residues is considered to be the major cause of the unusual stability of nepenthesins over a wide range of pH and temperature. On the other hand, it probably causes their high susceptibility to denaturing and reducing agents. The cleavage properties of nepenthesins also differ from those typical of other aspartic proteases (Rey et al., 2013 ▸; Kadek, Mrazek et al., 2014 ▸; Yang et al., 2015 ▸), making nepenthesins an effective alternative to porcine pepsin A in the hydrogen/deuterium exchange–mass spectrometry workflow.
In a previous study, we described a convenient way to obtain high amounts of nepenthesin 1 from N. gracilis using heterologous production in Escherichia coli. The recombinant enzyme has the enzymatic and physicochemical properties expected for this protease. Recombinantly produced nepenthesin 1 is active in a highly acidic environment, with a pH optimum in the range of 2.2–3.1. At slightly acidic to neutral pH the protease is inactive (Kadek, Tretyachenko et al., 2014 ▸). Given the specific stability and cleavage features of nepenthesin 1, we decided to study the structure of the enzyme in a form close to the native and functional form, i.e. at a pH close to the optimum.
Crystallization of proteins at extremely low pH requires specific approaches. Standard crystallization screens typically do not include pH extremes and do not allow the screening of various combinations of precipitants at low pH (see Table 1 ▸).
Table 1. Low-pH extremes in frequently used commercially available screens.
| Screen | Lowest pH available | No. of conditions in which the pH is 4.0 or lower |
|---|---|---|
| Morpheus (Molecular Dimensions) | 6.5 | 0 |
| JBScreen Classic (Jena Bioscience) | 4.6 | 0 |
| JBScreen JCSG++ (Jena Bioscience) | 4.0 | 2 |
| Index (Hampton Research) | 3.5 | 3 |
| Crystal Screen (Hampton Research) | 4.6 | 0 |
| SaltRx (Hampton Research) | 4.6 | 0 |
| PEGRx (Hampton Research) | 3.5 | 8 |
In the current report, we describe successful crystallization with the use of a newly formulated low-pH screen and a preliminary X-ray analysis of nepenthesin 1 in complex with pepstatin A.
2. Materials and methods
2.1. Macromolecule production
Recombinant nepenthesin 1 (rNep1) was produced and purified according to a previously published protocol (Kadek, Tretyachenko et al. 2014 ▸). The production procedure is summarized in Table 2 ▸. Briefly, E. coli C41 (DE3) cells were transformed with pET-21a/Nep1 vector. After expression, the cells were lysed by sonication and the protein was isolated in the form of inclusion bodies, which were resolubilized into a denaturing buffer consisting of 8 M urea, 1 mM EDTA, 1 mM glycine, 500 mM NaCl, 300 mM β-mercaptoethanol, 50 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) pH 10.5. Following dissolution, Nep1 was refolded by stepwise dialyses into 50 mM Tris–HCl pH 11, 50 mM Tris–HCl pH 7.5 and then into PBS buffer pH 7.5. Finally, the protein solution was concentrated by pressure ultrafiltration using a 30 kDa membrane (Millipore, USA), cleared by centrifugation (24 000g, 30 min, 277 K), acidified by the addition of 1 M glycine buffer pH 2.5 and kept overnight at 277 K to let the protease auto-activate. As a last step, the active protease was concentrated using an Amicon 10 kDa ultrafiltration device (Millipore, USA) and diluted into 50 mM glycine buffer pH 2.5.
Table 2. Details of the expression of recombinant nepenthesin 1.
| Source organism | N. gracilis |
| Forward primer | CATATGACGTCAAGAACAGCTC |
| Reverse primer | AAGCTTTCACGACGCACCACATTG |
| Cloning vector | pBSSK− (Invitrogen, USA) |
| Expression vector | pET-21a (Invitrogen, USA) |
| Expression host | E. coli C41 (DE3) (Lucigen, USA) |
| Complete amino-acid sequence of the construct produced† | TSRTALNHRHEAKVTGFQIMLEHVDSGKNLTKFQLLERAIERGSRRLQRLEAMLNGPSGVETSVYAGDGEYLMNLSIGTPAQPFSAIMDTGSDLIWTQCQPCTQCFNQSTPIFNPQGSSSFSTLPCSSQLCQALSSPTCSNNFCQYTYGYGDGSETQGSMGTETLTFGSVSIPNITFGCGENNQGFGQGNGAGLVGMGRGPLSLPSQLDVTKFSYCMTPIGSSTPSNLLLGSLANSVTAGSPNTTLIQSSQIPTFYYITLNGLSVGSTRLPIDPSAFALNSNNGTGGIIIDSGTTLTYFVNNAYQSVRQEFISQINLPVVNGSSSGFDLCFQTPSDPSNLQIPTFVMHFDGGDLELPSENYFISPSNGLICLAMGSSSQGMSIFGNIQQQNMLVVYDTGNSVVSFASAQCGAS |
The propeptide sequence is underlined.
2.2. Crystallization
In order to map the catalytic site and to avoid autodigestion of the protein, a complex of rNep1 with the inhibitor pepstatin A was prepared. The solution of rNep1 was incubated at 277 K with a 2.5 molar excess of pepstatin A. Since pepstatin A is insoluble in water, DMSO/acetic acid [9:1(v:v)] was used to prepare the concentrated inhibitor solution. The final DMSO concentration in the rNep1–pepstatin complex solution was around 3%(v/v). The sample was concentrated using a Nanosep centrifugal device with an Omega membrane to a final protein concentration of 10 mg ml−1 in 50 mM glycine pH 2.5.
Before crystallization, the sample monodispersity was checked by dynamic light scattering (Xtal Concepts SpectroLight 600, R h = 4.2 nm, polydispersity = 12.3%). Screening to obtain initial crystallization conditions was performed by the sitting-drop vapour-diffusion method using the commercially available crystallization screening kits Index, Crystal Screen 2 and PEGRx2 from Hampton Research. As rNep1 is most active in highly acidic environment, an acidic pH screen containing common precipitants was used in addition to the commercially available screening kits. At low pH, the selection of biologically compatible buffers is limited. The buffer systems in the screen were chosen according to our previous experience to be compatible with commonly used salts and polymers. The composition of the low-pH screen is summarized in Table 3 ▸.
Table 3. Composition of the low-pH screen.
| Condition | Salt | Polymer | Second precipitant | Buffer |
|---|---|---|---|---|
| 1 | 2 M ammonium sulfate | 0.1 M KCl/HCl pH 1.5 | ||
| 2 | 2 M ammonium sulfate | 0.1 M glycine pH 2.5 | ||
| 3 | 2 M ammonium sulfate | 0.1 M citric acid/sodium citrate pH 3 | ||
| 4 | 25%(w/v) PEG 3350 | 0.1 M KCl/HCl pH 1.5 | ||
| 5 | 25%(w/v) PEG 3350 | 0.1 M glycine pH 2.5 | ||
| 6 | 25%(w/v) PEG 3350 | 0.1 M citric acid/sodium citrate pH 3 | ||
| 7 | 0.2 M MgCl2 | 20%(w/v) PEG 3350 | 0.1 M KCl/HCl pH 1.5 | |
| 8 | 0.2 M MgCl2 | 20%(w/v) PEG 3350 | 0.1 M glycine pH 2.5 | |
| 9 | 0.2 M MgCl2 | 20%(w/v) PEG 3350 | 0.1 M citric acid/sodium citrate pH 3 | |
| 10 | 0.2 M ammonium acetate | 40% MPD | 0.1 M KCl/HCl pH 1.5 | |
| 11 | 0.2 M ammonium acetate | 40% MPD | 0.1 M glycine pH 2.5 | |
| 12 | 0.2 M ammonium acetate | 40% MPD | 0.1 M acid/sodium citrate pH 3 | |
| 13 | 10%(v/v) PEG 400 | 10%(v/v) ethylene glycol | 0.1 M KCl/HCl pH 1.5 | |
| 14 | 10%(v/v) PEG 400 | 10%(v/v) ethylene glycol | 0.1 M glycine pH 2.5 | |
| 15 | 10%(v/v) PEG 400 | 10%(v/v) ethylene glycol | 0.1 M acid/sodium citrate pH 3 |
Droplets consisting of 0.3 µl protein and 0.3 µl reservoir solution were equilibrated against 50 µl reservoir solution in CrystalQuick 96-well sitting-drop plates (Greiner). The protein concentration was 11 mg ml−1 for apo rNep1 and 10 mg ml−1 for the rNep1–pepstatin A complex.
The screening was performed at 291 and 283 K for the apo form and at 291 K for the complex with the inhibitor. The only optimizable crystallization hits were those for rNep1–pepstatin A in the low-pH screen. All other conditions did not provide results apart from crystalline precipitate, which could not be optimized. No hits were observed for the apo form or at a pH higher than 3.5.
The most promising conditions producing microcrystals were optimized using the hanging-drop vapour-diffusion method in EasyXtal 15-well plates (Qiagen) by varying the amount of precipitant and the ratio of protein and precipitant in the drops, by the use of additives and by exchanging PEG 3350 for PEG 1500, PEG 5000 MME or PEG 10 000 at concentrations between 10 and 30%(w/v). The final crystallization conditions consisted of (i) 25%(w/v) PEG 3350, 0.1 M glycine pH 2.5, with a 2:1 volume ratio of protein and precipitant in the drop, and (ii) 2 M ammonium sulfate, 0.1 M glycine pH 2.5, 0.025%(v/v) dichloromethane, with a 1:1 volume ratio of protein and precipitant in the drop (Table 4 ▸). Crystals of the rNep1–pepstatin A complex were grown at 291 K in two weeks. The approximate dimensions of the crystals were 70 × 50 × 5 µm.
Table 4. Crystallization conditions of rNep1–pepstatin optimized from conditions 2 and 5 of the low-pH crystallization screen.
| Condition (i) | Condition (ii) | |
|---|---|---|
| Method | Hanging-drop vapour diffusion | Hanging-drop vapour diffusion |
| Plate type | EasyXtal 15-well tool (Qiagen) | EasyXtal 15-well tool (Qiagen) |
| Temperature (K) | 291 | 291 |
| Protein concentration (mg ml−1) | 10 | 10 |
| Buffer composition of protein solution | 0.05 M glycine pH 2.5 | 0.05 M glycine pH 2.5 |
| Composition of reservoir solution | 25%(w/v) PEG 3350, 0.1 M glycine pH 2.5 | 2 M ammonium sulfate, 0.1 M glycine pH 2.5, 0.025%(v/v) dichloromethane |
| Volume and ratio of drop (protein:reservoir solution) | 1.5 µl (2:1) | 1 µl (1:1) |
| Volume of reservoir (µl) | 500 | 500 |
2.3. Data collection and processing
For diffraction data collection, single crystals of the rNep1–pepstatin A complex were flash-cooled after soaking in cryosolution consisting of 15%(v/v) PEG 400, 25%(w/v) PEG 3350, 0.1 M glycine pH 2.5 for crystals grown in condition (i) and 20%(v/v) glycerol, 2 M ammonium sulfate, 0.1 M glycine pH 2.5 for crystals grown in condition (ii). Data sets were collected under cryogenic conditions (100 K) on beamline 14.1 at BESSY II HZB, Berlin, Germany (Mueller et al., 2015 ▸). For both types of crystals, the wavelength of the radiation was set to 0.91841 Å and a Pilatus 6M detector was used to record X-ray diffraction intensities in shutterless mode. For crystal A, grown in the final crystallization condition (i), an oscillation range of 0.3° per frame was used and the crystal-to-detector distance was set to 615.7 mm. The crystal diffracted to 2.9 Å resolution. For crystal B, grown in the final crystallization condition (ii), an oscillation range of 0.1° per frame was used and the crystal-to-detector distance was set to 495.2 mm. The crystal diffracted to 2.8 Å resolution. All data were indexed, merged and processed using the XDS package (Kabsch, 2010 ▸). The data statistics are summarized in Table 5 ▸.
Table 5. Data collection and processing.
Values in parentheses are for the outer shell.
| Crystal A | Crystal B | |
|---|---|---|
| Beamline | 14.1, BESSY II | 14.1, BESSY II |
| Wavelength (Å) | 0.91841 | 0.91841 |
| Temperature (K) | 100 | 100 |
| Detector | Pilatus 6M | Pilatus 6M |
| Crystal-to-detector distance (mm) | 615.7 | 495.2 |
| Rotation range per image (°) | 0.3 | 0.1 |
| Total rotation range (°) | 240 | 180 |
| Exposure time per image (s) | 1.5 | 0.5 |
| Space group | P212121 | P212121 |
| a, b, c (Å) | 86.63, 95.90, 105.40 | 86.28, 97.22, 103.78 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Mosaicity (°) | 0.34 | 0.24 |
| Resolution range (Å) | 47.95–2.90 (3.07–2.90) | 48.61–2.81 (2.96–2.81) |
| Total No. of reflections | 172087 (28189) | 142752 (21048) |
| No. of unique reflections | 20123 (3179) | 21937 (3147) |
| Completeness (%) | 99.9 (99.8) | 99.9 (99.7) |
| Multiplicity | 8.6 (8.9) | 6.5 (6.7) |
| 〈I/σ(I)〉 | 5.9 (2.0) | 4.8 (2.2) |
| R r.i.m. | 0.292 (0.981) | 0.382 (0.958) |
| Overall B factor from Wilson plot (Å2) | 41.3 | 13.4 |
The diffraction data obtained were highly anisotropic; the recommended resolution limits along a*, b* and c*, according to the Anisotropy Diffraction Server (Strong et al., 2006 ▸), are 2.9, 2.9 and 3.9 Å, respectively, for crystal A and 2.8, 2.8 and 3.4 Å, respectively, for crystal B.
3. Results and discussion
The suitability of protein samples for crystallization was verified by DLS measurements. rNep1 at pH 2.5 appeared as a monodisperse solution of particles with an R h of 4.2 nm (corresponding to a monomer) and was suitable for crystallization. Although extreme pH values do not contribute significantly to successful crystallization for most proteins (Kantardjieff & Rupp, 2004 ▸), crystallization screening of the rNep1–pepstatin A complex gave several hits at low-pH extremes (pH 1.5–3.5): conditions 1–9 yielded high amounts of plate-like microcrystals, with the largest sized microcrystals in conditions 2, 5 and 6. No crystallization hits were observed at a pH higher than 3.5. The pI of rNep1, as calculated by the EMBOSS software suite (Rice et al., 2000 ▸), is 3.56. The identified low-pH crystallization conditions are in accordance with the observation of Kirkwood et al. (2015 ▸) that 85% of proteins crystallize within two pH units of their pI. However, as opposed to the previously reported results (Kirkwood et al., 2015 ▸; Kantardjieff & Rupp, 2004 ▸), rNep1 does not tend to crystallize at a more neutral pH, i.e. above its pI. Our observation illustrates the importance of screening at low pH when preliminary screens do not produce any crystallization hits or during optimization, especially for proteins that have very low pI values.
Optimization of crystallization conditions 2 and 5 yielded plate-like crystals (Fig. 1 ▸). Even though the two crystals of the rNep1–pepstatin A complex belonged to the same space group P212121, they possessed somewhat different unit cells (see Table 4 ▸).
Figure 1.
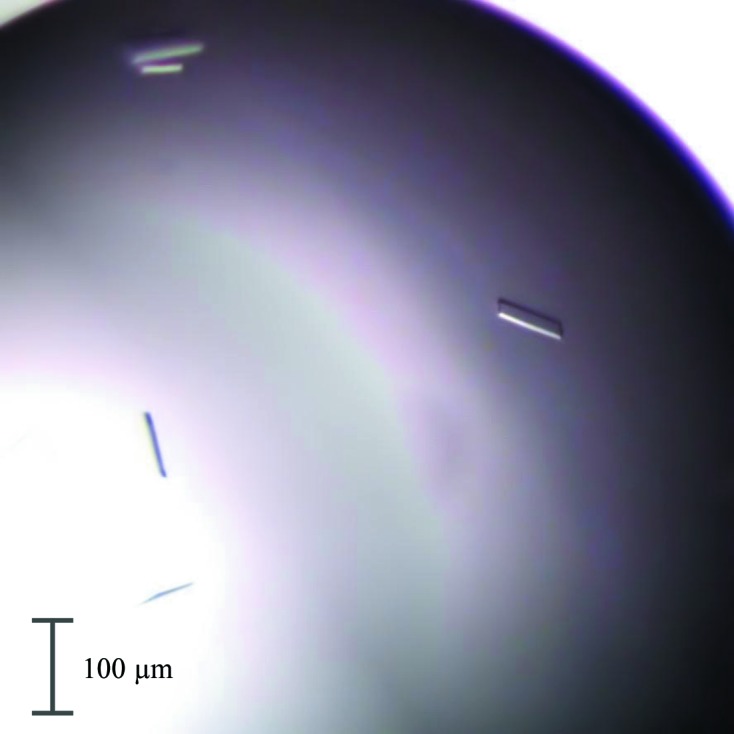
Crystals of the rNep1–pepstatin A complex grown in condition (i). The largest dimension is approximately 70 µm.
Based on the Matthews coefficient, we propose that the asymmetric unit contains two rNep1 molecules. In this case, the Matthews coefficients for the two data sets are 2.93 and 2.91 Å3 Da−1, corresponding to solvent contents of 58.0 and 57.8%, respectively. The self-rotation function for these data sets calculated by MOLREP (Vagin & Teplyakov, 2010 ▸) does not reveal the existence of a noncrystallographic twofold axis, suggesting that the putative noncrystallographic axis is probably parallel to one of the crystallographic axes.
Structure-solution attempts by molecular replacement using a distant homology model (human progastricin; PDB entry 1htr; 27% sequence identity; Moore et al., 1995 ▸) did not yield a satisfying solution. Experimental phasing and other molecular-replacement attempts are under way. Structure determination of rNep1 will help to explain the observed differences in the stability and cleavage properties of nepenthesin 1 in comparison with other pepsin-like proteases.
Acknowledgments
This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (grant Nos. EE2.3.30.0029 and LG14009), the Grant Agency of the Czech Republic (grant P206/12/0503), BIOCEV CZ.1.05/1.1.00/02.0109 from the ERDF and by institutional support from IBT CAS, v.v.i. (RVO 86652036). AK also acknowledges funding by Charles University project UNCE_204025/2012. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement No. 283570).
References
- Athauda, S. P., Matsumoto, K., Rajapakshe, S., Kuribayashi, M., Kojima, M., Kubomura-Yoshida, N., Iwamatsu, A., Shibata, C., Inoue, H. & Takahashi, K. (2004). Biochem. J. 381, 295–306. [DOI] [PMC free article] [PubMed]
- Coates, L., Erskine, P. T., Wood, S. P., Myles, D. A. A. & Cooper, J. B. (2001). Biochemistry, 40, 13149–13157. [DOI] [PubMed]
- Davies, D. R. (1990). Annu. Rev. Biophys. Biophys. Chem. 19, 189–215. [DOI] [PubMed]
- Dunn, B. M. (1997). Nature Struct. Mol. Biol. 4, 969–972. [DOI] [PubMed]
- Dunn, B. M. (2002). Chem. Rev. 102, 4431–4458. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kadek, A., Mrazek, H., Halada, P., Rey, M., Schriemer, D. C. & Man, P. (2014). Anal. Chem. 86, 4287–4294. [DOI] [PubMed]
- Kadek, A., Tretyachenko, V., Mrazek, H., Ivanova, L., Halada, P., Rey, M., Schriemer, D. C. & Man, P. (2014). Protein Expr. Purif. 95, 121–128. [DOI] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2004). Bioinformatics, 20, 2162–2168. [DOI] [PubMed]
- Khan, A. R. & James, M. N. G. (1998). Protein Sci. 7, 815–836. [DOI] [PMC free article] [PubMed]
- Kirkwood, J., Hargreaves, D., O’Keefe, S. & Wilson, J. (2015). Bioinformatics, 31, 1444–1451. [DOI] [PMC free article] [PubMed]
- Madala, P. K., Tyndall, J. D. A., Nall, T. & Fairlie, D. P. (2010). Chem. Rev. 110, PR1–PR31. [DOI] [PubMed]
- Moore, S. A., Sielecki, A. R., Chernaia, M. M., Tarasova, N. I. & James, M. N. G. (1995). J. Mol. Biol. 247, 466–485. [DOI] [PubMed]
- Mueller, U., Förster, R., Hellmig, M., Huschmann, F. U., Kastner, A., Malecki, P., Pühringer, S., Röwer, M., Sparta, K., Steffien, M., Ühlein, M., Wilk, P. & Weiss, M. S. (2015). Eur. Phys. J. Plus, 130, 141.
- Rawlings, N. D., Waller, M., Barrett, A. J. & Bateman, A. (2013). Nucleic Acids Res. 42, D503–D509. [DOI] [PMC free article] [PubMed]
- Rey, M., Yang, M., Burns, K. M., Yu, Y., Lees-Miller, S. P. & Schriemer, D. C. (2013). Mol. Cell. Proteomics, 12, 464–472. [DOI] [PMC free article] [PubMed]
- Rice, P., Longden, I. & Bleasby, A. (2000). Trends Genet. 16, 276–277. [DOI] [PubMed]
- Strong, M., Sawaya, M. R., Wang, S., Phillips, M., Cascio, D. & Eisenberg, D. (2006). Proc. Natl Acad. Sci. USA, 103, 8060–8065. [DOI] [PMC free article] [PubMed]
- Takahashi, K., Athauda, S., Matsumoto, K., Rajapakshe, S., Kuribayashi, M., Kojima, M., Kubomura-Yoshida, N., Iwamatsu, A., Shibata, C. & Inoue, H. (2005). Curr. Protein Pept. Sci. 6, 513–525. [DOI] [PubMed]
- Takahashi, K., Niwa, H., Yokota, N., Kubota, K. & Inoue, H. (2008). Plant Physiol. Biochem. 46, 724–729. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Veerapandian, B., Cooper, J. B., Šali, A., Blundell, T. L., Rosati, R. L., Dominy, B. W., Damon, D. B. & Hoover, D. J. (1992). Protein Sci. 1, 322–328. [DOI] [PMC free article] [PubMed]
- Yang, M., Hoeppner, M., Rey, M., Kadek, A., Man, P. & Schriemer, D. C. (2015). Anal. Chem. 87, 6681–6687. [DOI] [PubMed]


