Abstract
Insulin/IGF-1 signaling (IIS) is a critical regulator of an organism’s most important biological decisions, from growth, development, and metabolism to reproduction and longevity. It primarily does so through the activity of the DAF-16/FOXO transcription factor, whose global targets were identified in C. elegans using whole-worm transcriptional analyses more than a decade ago1. IIS and FOXO also regulate important neuronal and adult behavioral phenotypes, such as the maintenance of memory2 and axon regeneration3 with age, in both mammals4 and C. elegans, but the neuron-specific IIS/FOXO targets that regulate these functions are still unknown. By isolating adult C. elegans neurons for transcriptional profiling, we identified both the wild-type and IIS/FOXO adult neuronal transcriptomes for the first time. IIS/FOXO neuron-specific targets are distinct from canonical IIS/FOXO-regulated longevity and metabolism targets, and are required for IIS/daf-2 mutants’ extended memory. We also discovered that the activity of the forkhead transcription factor FKH-9 in neurons is required for daf-2’s ability to regenerate axons with age, and its activity in non-neuronal tissues is required for daf-2’s long lifespan. Together, neuron-specific and canonical IIS/FOXO-regulated targets enable the coordinated extension of neuronal activities, metabolism, and longevity under low insulin-signaling conditions.
The C. elegans IIS pathway acts both cell autonomously and non-autonomously to control longevity, growth, dauer formation, metabolism, and reproduction5–7 through its regulation of DAF-16/FOXO’s nuclear localization and transcriptional activation. The canonical IIS/FOXO gene set, which identified primarily intestinal and hypodermal targets (Extended Data Fig. 1A–B)1,8,9, has been instructive in our understanding of how insulin signaling regulates a diverse range of activities, including metabolism, autophagy, stress resistance, and proteostasis. However, IIS mutants also exhibit daf-16-dependent neuronal phenotypes, including extended positive olfactory learning2, increased short- and long-term associative memory2, increased thermotaxis learning10, improved neuronal morphology maintenance11,12, and improved axon regeneration3. These phenotypes are unlikely to be regulated by the known intestinal and hypodermal IIS/FOXO targets1,8. Therefore, to understand how IIS/daf-2 animals extend behavioral functionality, we must identify the neuronal targets of FOXO/DAF-16.
We first profiled the expression of daf-16;daf-2 worms with daf-16 rescued in specific tissues6 (Supplementary Table 1). Intestinal daf-16 rescue correlates best with whole-worm profiles (Extended Data Fig. 1A, C). By contrast, neuronal daf-16 rescue profiles are anti-correlated with the intestinal DAF-16 and whole-worm profiles (Extended Data Fig. 1A, C). Surprisingly, many genes induced by neuronal DAF-16 rescue are expressed (Wormbase) or predicted to be expressed in non-neuronal tissues13 (Extended Data Fig. 1D), and have non-neuronal functions (e.g., collagens14; Extended Data Fig. 1B, 1E, Supplementary Table 2). Thus, whole-worm transcriptional analyses of neuronally-rescued DAF-16 failed to reveal targets that account for daf-2 mutants’ daf-16-dependent age-related behaviors. Therefore, we needed to specifically examine transcription in IIS-mutant neurons.
The tough outer cuticle prevents dissociation of adult tissues15, thus the wild-type adult neuronal transcriptome has not been described. To solve this problem, we used rapid, chilled chemomechanical disruption followed immediately by FACS to isolate GFP-marked neurons from wild-type worms, then RNA-sequenced these isolated cells (Fig. 1A–C, Extended Data Fig. 2A–C,F,G, Supplementary Table 3). This method is gentle enough to preserve the integrity of cells and some neurites (Extended Data Fig. 2A), does not involve cell culturing prior to FACS, in contrast to previous methods16, and does not affect transcription (Actinomycin D; Fig. 1B, Extended Data Fig. 2D,E, Supplementary Table 4). Downsampling analysis showed that sufficient sequencing depth was achieved (Extended Data Fig. 2H).
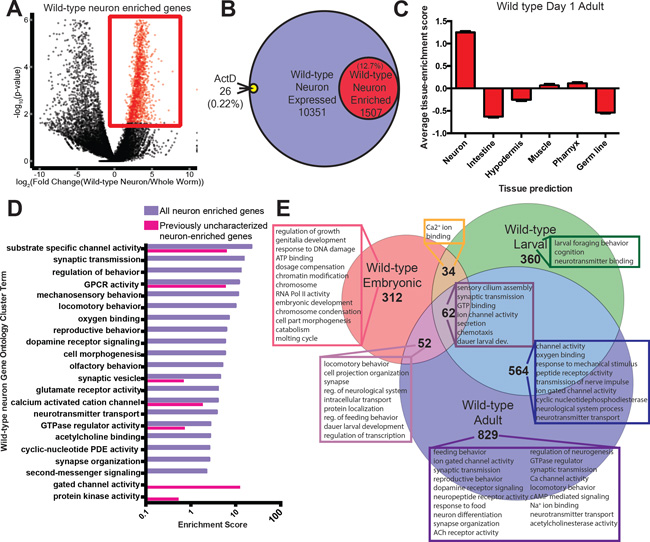
Fig 1. Identification of neuronal IIS/FOXO targets requires neuronal isolation.
A) Volcano plot of neuron-expressed relative to whole worm-expressed genes obtained by neuron-specific RNA sequencing of adult wild-type animals. B) Neuron-expressed and enriched genes are not influenced by cell isolation: treatment with the transcription inhibitor Actinomycin D affected only 0.22% of all neuronal genes (Supplementary Table 4). C) Tissue expression prediction of wild-type adult neuron-enriched genes. Mean ± SEM. D) GO terms highlight the neuronal characteristics of both all and previously uncharacterized neuron-enriched genes. E) Embryonic16, larval16, and adult neuron-enriched genes and significant GO terms transition from developmental to neuronal and behavioral functions (Supplementary Table 5); FDR<10% for all gene sets.
We compared gene expression in isolated wild-type neurons with whole-worm expression to identify genes that are enriched in neurons (Fig. 1A–C). Of the 1507 “neuron enriched” genes (False Discovery Rate (FDR)<0.1; Supplementary Table 3; Fig. 1A,B), only 4% have previously-described expression patterns exclusively in non-neuronal tissues, and “Neuron” is the only significantly-enriched tissue (Fig. 1C, Extended Data Fig. 2F), suggesting the method is highly selective for neuronal transcripts. Gene promoter-gfp tests of previously uncharacterized genes from our “neuron enriched” list confirmed neuronal expression, with no bias for particular neuron types (Extended Data Fig. 3A). We also detected genes previously reported to be expressed only in single neurons or small subsets of neurons, including glr-3 (RIA), ttx-3 (AIY/AIA), and npr-14 (AIY) (Wormbase).
The wild-type neuron-enriched set includes synaptic machinery, ion channels, neurotransmitters, and signaling components (Supplementary Table 3), as well as >700 previously-uncharacterized genes; these genes are predicted to have “neuronal”-like character and function (Fig. 1D). Comparison of the wild-type embryonic and larval neuronal transcriptomes with the adult neuronal transcriptome at the same FDR revealed a shift in functional categories from developmental processes to neuronal function/behavior in the adult neuronal transcriptome (Fig. 1E, Extended Data Fig. 3B,C, Supplementary Table 5), suggesting that previous isolation methods16, either due to early developmental stage isolation or to re-culturing, biased expression toward developmental genes rather than neuronal/behavioral genes.
To identify adult neuronal IIS/FOXO targets, we sequenced RNA from isolated daf-2 and daf-16;daf-2 neurons on Day 1 of adulthood (Fig. 2A, Extended Data Fig. 4, Supplementary Table 6, 8). The IIS/FOXO neuron-isolated gene set is enriched for neuronal expression: 86% and 92% of the up- and down-regulated genes, respectively, are expressed in wild-type neurons. While several of DAF-16’s top Class I targets, including hil-1, sip-1, mtl-1, nnt-1, ins-6, and daf-16 itself, were upregulated in both daf-2 neurons and daf-2 whole worms (Group B; Fig. 2B), most of the IIS/FOXO neuronally-regulated set differs from the canonical whole-worm IIS/FOXOs set1,8 (Fig. 2B). Specifically, in contrast to the metabolism-dominated functions of canonical whole-worm IIS/FOXO targets1,8, the neuronal IIS set GO terms reflect neuron-like functions (Extended Data Fig. 5B): serpentine receptors, GPCRs, syntaxin, globins, kinesins, insulins, ion channels, potassium channels, seven-transmembrane receptors, the NPR-1 neuropeptide receptor, and the SER-3 octopamine receptor are upregulated in daf-2 neurons (Supplementary Table 6). A few genes (fat-3 and crh-1/CREB) are upregulated in daf-2 neurons but downregulated in whole daf-2 animals.
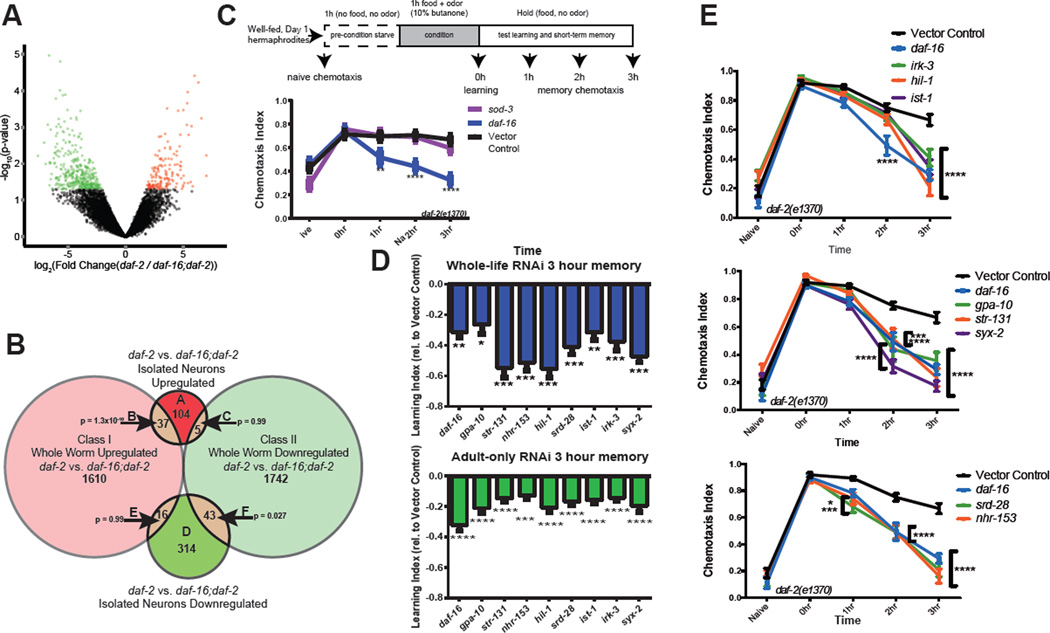
Fig 2. RNA-seq transcriptional profile of isolated neurons reveals IIS/FOXO neuronal transcriptome.
A) Volcano plot of daf-2-regulated, daf-16-dependent up- (red) and down-regulated (green) neuronal genes (p<0.05). B) Comparison of whole-worm (Class I)8 vs neuronal-IIS/FOXO targets. P-values: hypergeometric distributions. C–E) Short-term associative memory (STAM) assays. C) Schematic of STAM assay and chemotaxis profiles of daf-2 treated with (C) sod-3 or (D, E) neuronal IIS/FOXO target gene RNAi. D) Learning indices relative to control RNAi at 3h post-training of daf-2 treated with adult-only (green) or whole life (blue) neuronal IIS/FOXO target gene RNAi. Mean ± SEM, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-way repeated measures ANOVA, Bonferroni post hoc tests.
The IIS/FOXO downregulated set includes serpentine receptors, guanylate cyclases, signaling peptides and receptors (NLPs, FLPs, and NPRs), and the vesicle trafficking G protein rab-28 (Supplementary Table 6). Expression of the sensory neuron cilia protein IFTA-2, which co-localizes with DAF-2 and whose loss increases lifespan17, is downregulated in daf-2 mutants, consistent with the longevity of daf-2 and ciliated sensory neuron mutants18. Similarly, sams-1 (S-adenosyl methionine synthetase), which is downregulated under long-lived Dietary Restriction conditions19, and sma-5 and dbl-1, components of TGF-beta pathways linked with IIS7,20, are downregulated, perhaps coordinating the longevity and reproductive output of these pathways.
Unlike canonical IIS/FOXO targets1, neuronal IIS/FOXO gene promoters are not enriched for the DBE (DAF-16 Binding Element, GTAAAt/cA), but the overlapping, upregulated (Group B) targets’ promoters contain twice as many DBEs (Extended Data Fig. 5A). The overlapping downregulated (Group F) targets are enriched for the PQM-18/DAE motif (CTTATCA1,8; Supplementary Table 7). DAF-16 may regulate neuronal activities indirectly through activation of ~60 IIS/FOXO-upregulated transcription factors (Supplementary Table 6).
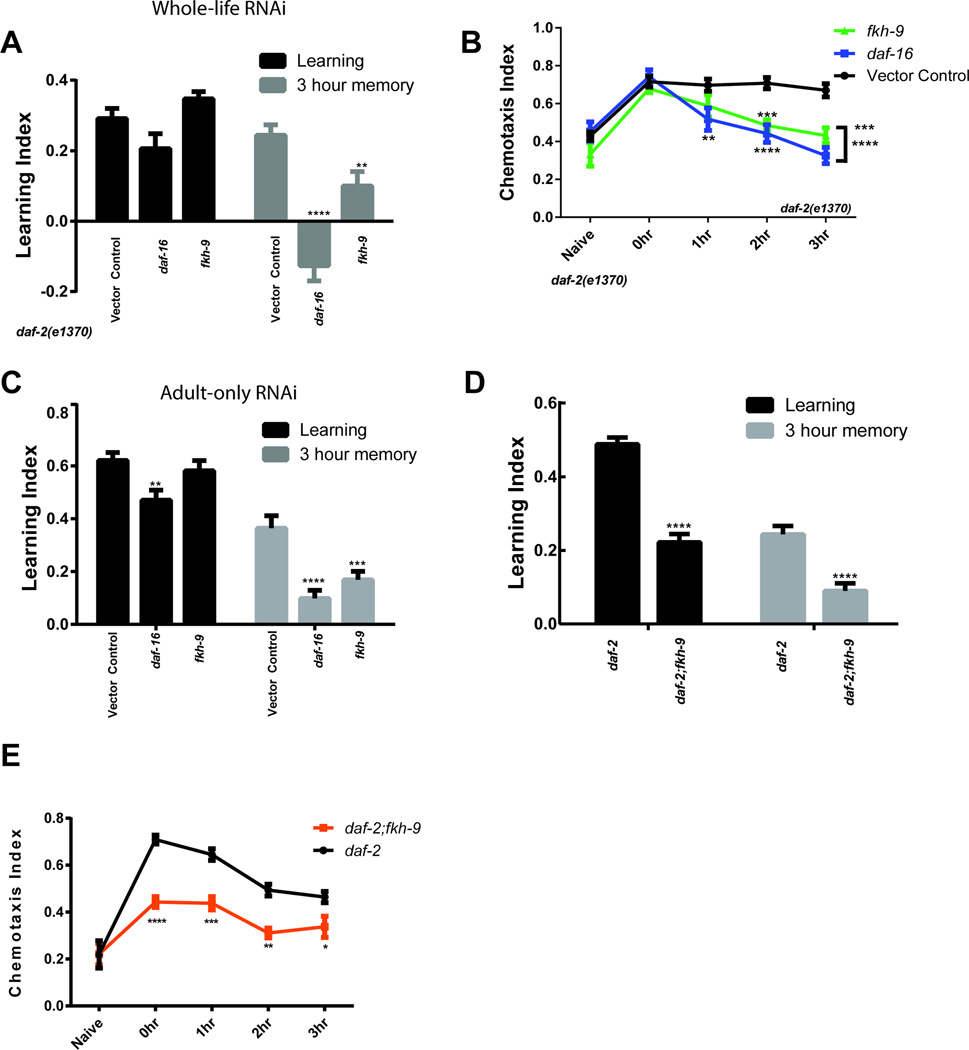
We next tested the roles of top-scoring genes in daf-2-regulated neuronal phenotypes. Long-term and short-term associative memory are both extended in daf-2 mutants in a daf-16-dependent manner2 (Extended Data Fig. 6). The bZIP transcription factor CREB, which is required for long-term memory in many organisms, including C. elegans2, is upregulated by IIS/FOXO in neurons (Supplementary Table 6), correlating with daf-2’s increased long-term memory2,21. However, short-term associative memory (STAM; Fig. 2C) is CREB-independent2, and the genes that enable daf-2’s STAM extension are unknown. While the DAF-16 non-neuronal target sod-3 had no effect on daf-2’s extended STAM (Fig. 2C, Extended Data Fig. 6B–D), knockdown of 8 of the 10 top-ranked, upregulated IIS/FOXO targets significantly decreased daf-2(e1370)’s STAM (Fig. 2D,E), both in whole-life and adult-only RNAi tests. (Neuronal RNAi is effective in learning, STAM, and LTAM tests21.) The variety of genes (ion channels, transcription factors, G-proteins, vesicle fusion proteins) required for daf-2’s extended STAM suggests that decreased insulin signaling affects a broad network of memory extension genes. Several of these genes are also required for wild type’s learning and memory (Extended Data Fig. 6G) suggesting that daf-2 mutants maintain neuronal function, rather than utilizing an alternative short-term memory mechanism.
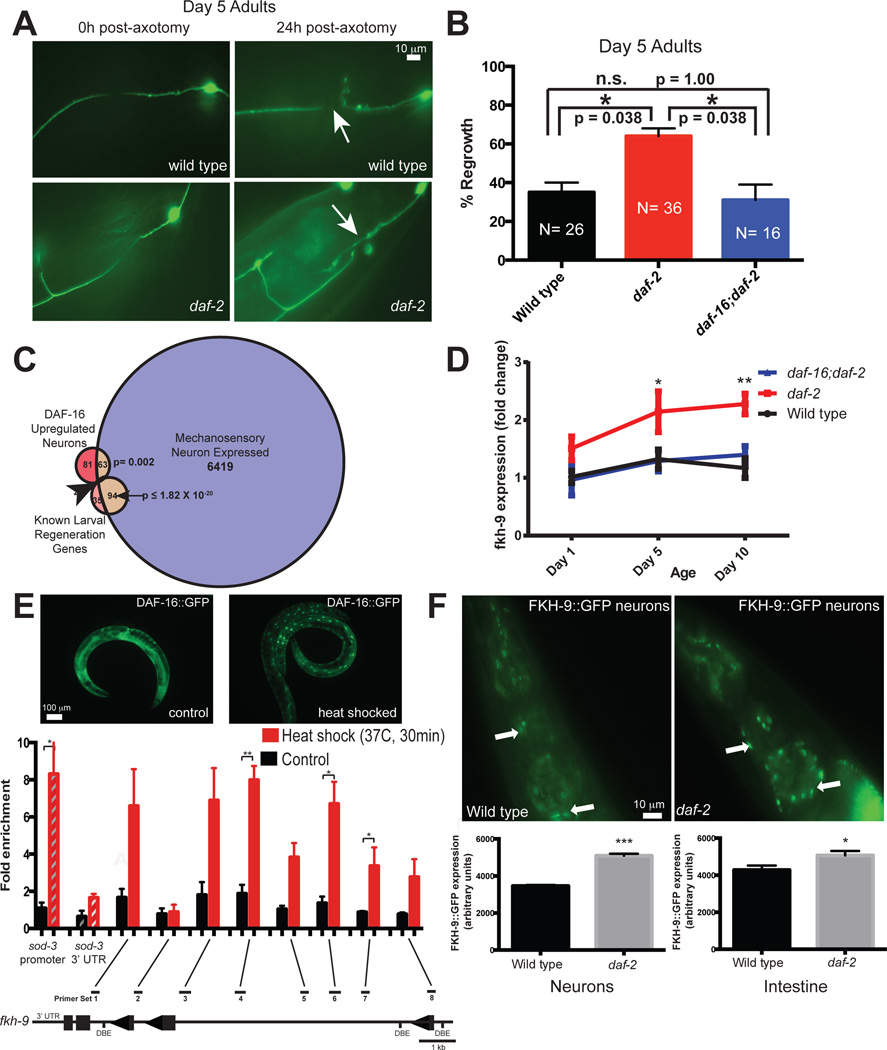
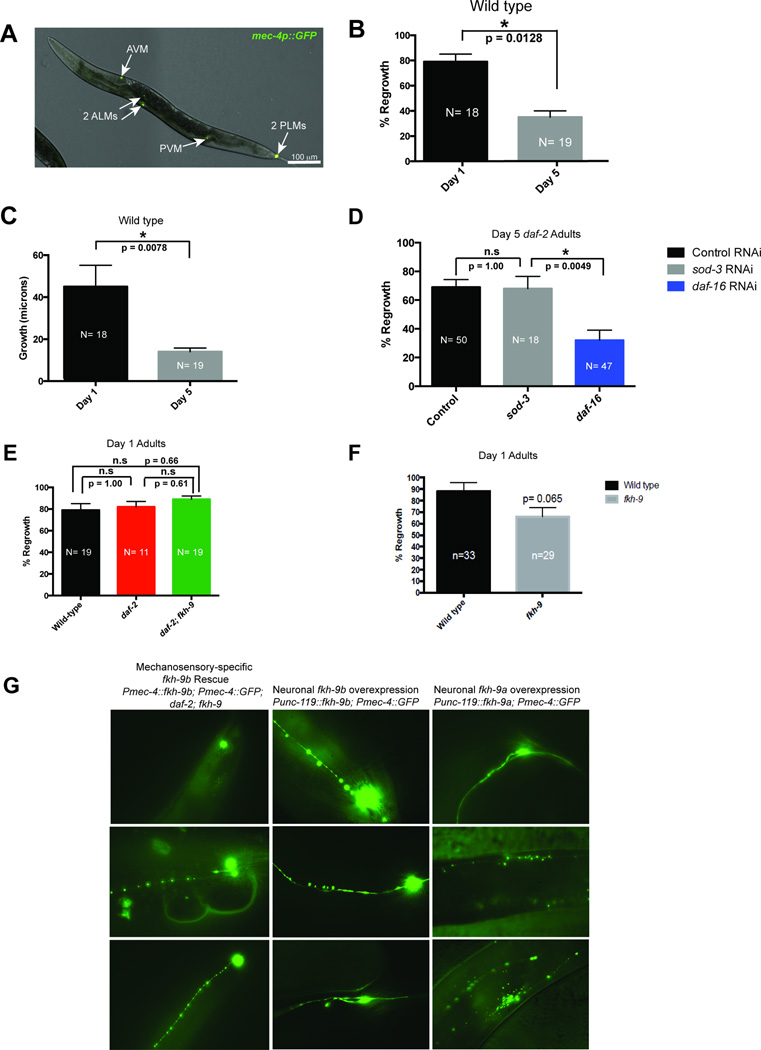
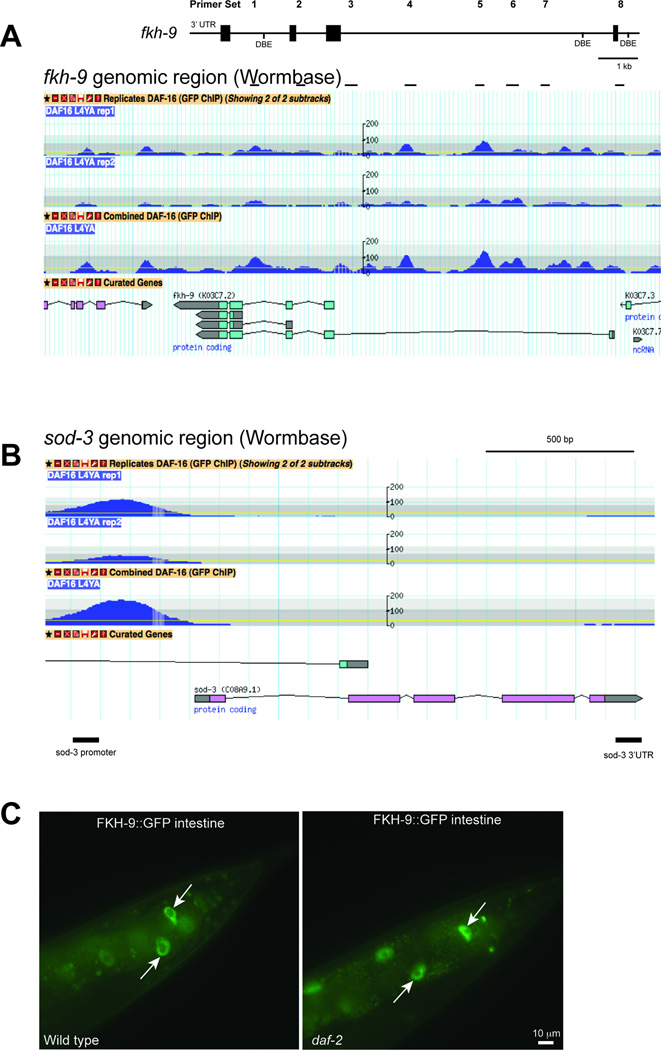
daf-2 mutants also maintain motor neuron axon regeneration ability with age in a daf-16-dependent manner3, and we found this is also true for mechanosensory neurons (Fig. 3A,B, Extended Data Fig. 7A–D). To identify factors that enable axon regeneration with age, we isolated and RNA-sequenced six adult mechanosensory neurons (Fig. 3C, Supplementary Table 9); this set includes 94 known larval regeneration genes from limited candidate screens22 (p≤ 1.82 × 10−20). To find daf-2/daf-16-dependent axon regeneration candidates, we identified mechanosensory genes that are also regulated by neuronal IIS/FOXO (Fig. 3C, Supplementary Table 9; p<0.002). The forkhead transcription factor FKH-9 is a neuronal IIS/FOXO target (Supplementary Table 6) and a canonical Class I target1, and is expressed in mechanosensory neurons (Supplementary Table 9). fkh-9’s promoter is occupied by DAF-16, which we confirmed by chIP-qPCR (Fig. 3E, Extended Data Fig. 8A, B). FKH-9::GFP localized to nuclei, and neurons were the primary site of differential FKH-9::GFP levels in daf-2 mutants (Fig. 3F, Extended Data Fig. 8C), all suggesting a role for FKH-9 in daf-2/daf-16-mediated neuronal function.
Fig 3. FKH-9 is a direct target of DAF-16 and is expressed in mechanosensory neurons.
A, B) daf-16 is required for daf-2’s enhanced Day 5 axon regeneration, Mean ± SEM, *p<0.05, Fisher’s exact test. C) Known larval regeneration genes are significantly enriched in the Day 1 adult mechanosensory transcriptome. 63 genes are both DAF-16 targets and expressed in mechanosensory neurons (<5%FDR). D) fkh-9 mRNA levels are higher in aged daf-2 compared to wild type in a daf-16-dependent manner. N=4 biological replicates, two-way ANOVA, Bonferroni post hoc tests. E) Chromatin immunoprecipitation of DAF-16::GFP worms with and without heat shock, which mobilizes DAF-16 into the nucleus. DAF-16 binds to the sod-3 promoter but not its 3’ UTR, and to the fkh-9 promoter at multiple locations (Extended Data Figure 8). Fold enrichment relative to wild-type (not expressing DAF-16::GFP) is shown (mean ± SEM, two-tailed t-test, N=3 biological replicates). F) Neuronal FKH-9::GFP (fkh-9p::fkh-9::gfp) expression in daf-2 compared to wild type. N=25 animals. Mean ± SEM, two-tailed t-test. D–F) *p<0.05, **p<0.01, ***p<0.001.
While there is no effect on the first day of adulthood (Extended Data Fig. 7E,F), loss of fkh-9 severely impairs daf-2’s axon regeneration ability in aged (Day 5) worms (Fig. 4A), correlating with an increased difference in fkh-9 expression levels between wild-type and daf-2 (Fig. 3D). Pan-neuronal fkh-9 expression rescues the ability of Day 5 daf-2;fkh-9 worms to regenerate PLM axons (Fig. 4B,C). fkh-9 levels are critical for neuron morphology, as fkh-9 neuronal overexpression causes axonal defects (Extended Data Fig. 7G)
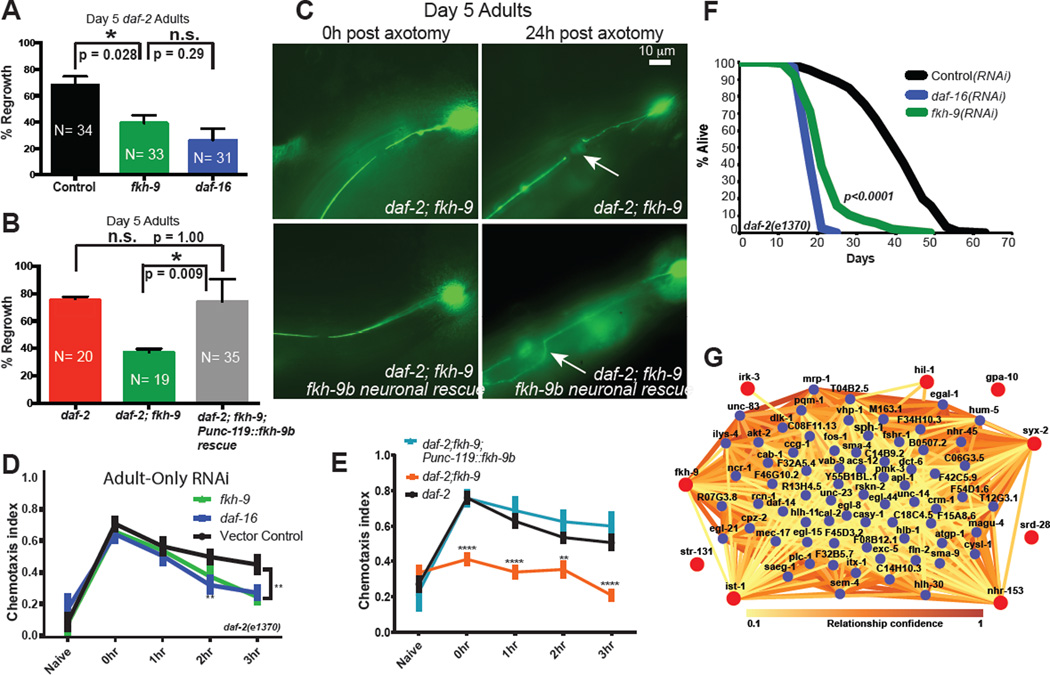
Fig 4. FKH-9 is required for daf-2's improved axon regeneration, short-term associative memory, and lifespan.
A) fkh-9 knockdown reduces axon regeneration of Day 5 daf-2 mutants, as does daf-16 knockdown. B, C) Neuronally-expressed fkh-9 rescues Day 5 axon regeneration in daf-2;fkh-9 mutants. Mean ± SEM, *p<0.05, Fisher’s exact test. D) fkh-9 is required for daf-2’s enhanced memory in adult-only RNAi-treated worms. E) Neuronally-expressed fkh-9 rescues extended STAM in daf-2;fkh-9 mutants with defective learning and memory. Mean ± SEM, **p<0.01, ***p<0.001, ****p<0.0001, two-way repeated measures ANOVA, Bonferroni post hoc tests. F) Adult-specific fkh-9 RNAi treatment reduces daf-2 mutant lifespan. Median lifespan: control RNAi 42 days, fkh-9 RNAi 21 days, daf-16 RNAi 21 days. p<0.0001 for control RNAi vs. daf-16 RNAi and control vs. fkh-9 RNAi, log-rank test. N=144 worms per strain. G) IMP network analysis of DAF-16 neuronal target genes with STAM phenotypes (red circles).
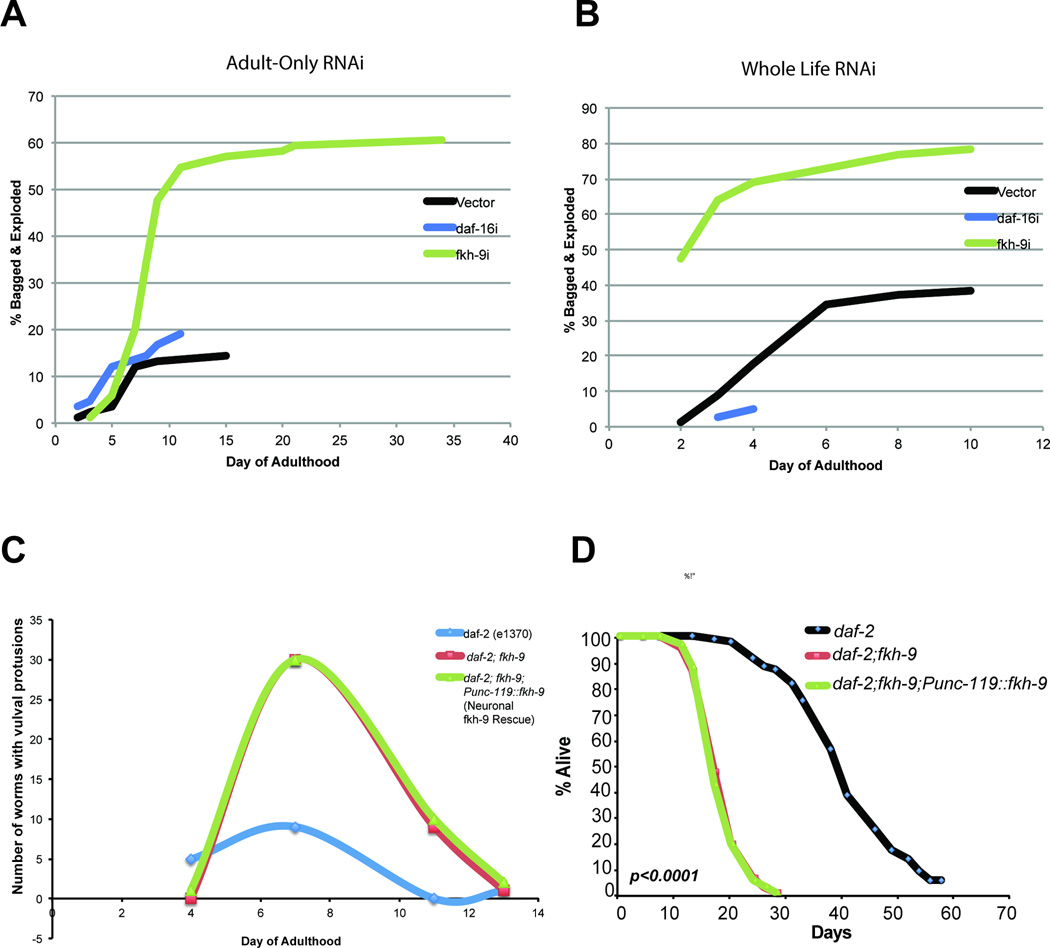
Adult-specific and whole-life reduction of fkh-9 also severely impair daf-2’s extended STAM (Fig. 4D, Extended Data Fig. 9). daf-2;fkh-9 double mutants are defective in both STAM and learning, and neuronal fkh-9 expression rescues these defects (Fig. 4E, Extended Data Fig. 9D,E), suggesting fkh-9 is required for daf-2’s extended memory and normal neuronal development. Day 1 and 5 fkh-9 expression levels correlate with STAM and axon regeneration (Fig 3D). fkh-9 reduction delays development, and reduction during adulthood causes severe matricide (Extended Data Fig. 10A–C). fkh-9 knockdown in adult daf-2 worms treated with FUdR to block matricide20 significantly shortens lifespan (40–50%; Fig 4F). Pan-neuronal fkh-9 expression does not rescue lifespan (Extended Data Fig. 10D), suggesting that FKH-9 acts in non-neuronal tissues to regulate lifespan. Thus, IIS/FOXO-regulated FKH-9 function is important for both neuronal and non-neuronal growth and development, as well as adult memory and axon regeneration. Interestingly, FKH-9’s mammalian homolog FoxG1 is required for axon outgrowth23 and is the most highly-induced gene in spinal cords treated with radial glial cell transplant following spinal cord injury24.
Network analysis using fkh-9 and the other 8 neuronal DAF-16 STAM genes (Fig. 4G, Supplementary Table 10) identified casy-1, which is required for several forms of associative learning and memory2,25–27, apl-1, the C. elegans ortholog of amyloid precursor protein (APP) that can disrupt sensory plasticity28, and dlk-1, the only previously known regulator of age-dependent axon regeneration3,29. Additionally, genes involved in neuronal degeneration (mec-17), neuronal development (egl-44, sem-4), neuronal function (egl-21, rnc-1, vab-9, cysl-1), synaptic regulation and function (cab-1, hlb-1, magu-4, sph-1, unc-64), and axon outgrowth (unc-14) and regeneration (egl-8, fos-1, pmk-3), were connected to the STAM genes. PQM-18, whose motif (DAE) is overrepresented in neuronal IIS target promoters, and other IIS (akt-2, dct-6, hlh-30), TGF-β (daf-14, sma-4, crm-1, sma-9, sma-1, sta-1), and MAPK pathway (vhp-1, pmk-3) components emerged in the network. Transcriptional regulation by IIS/FOXO and its targets may lead to broader, indirect transcriptional and non-transcriptional regulation of genes with important neuronal functions.
Plasticity in development, reproduction, and longevity allows organisms to respond appropriately to nutrient availability and changes in their environment. The IIS pathway is a critical mediator of these decisions, with FOXO selecting transcriptional targets to execute specific biochemical functions in each tissue, including factors that maintain cognitive function with age. daf-2 worms maintain neuronal behaviors with age by utilizing a set of transcriptional targets that are distinct from previously-identified metabolic and stress resistance targets expressed in other tissues. These genes may regulate additional neuronal targets through non-transcriptional mechanisms (Fig. 4G). The regulation of tissue-specific transcriptional programs is important to coordinate phenotypic responses, extending neuronal abilities in concert with daf-2’s extended longevity and reproductive span.
Methods
Adult cell isolation
Day 1 adult neuronally GFP-labeled worms (Punc119::GFP or Pmec-4::GFP) were prepared for cell isolation as previously described15 with modifications (Extended Data Fig. 2). Synchronized adult worms were washed with M9 buffer to remove excess bacteria. The pellet (~250 µl) was washed with 500 µl lysis buffer (200 mM DTT, 0.25% SDS, 20 mM Hepes pH 8.0, 3% sucrose) and resuspended in 1000 µl lysis buffer. Worms were incubated in lysis buffer with gentle rocking for 6.5 minutes at room temperature. The pellet was washed 6× with M9 and resuspended in 20 mg/ml pronase from Streptomyces griseus (Sigma-Aldrich). Worms were incubated at room temperature (<20 minutes) with periodic mechanical disruption by pipetting every 2 min. When most worm bodies were dissociated, leaving only small debris and eggs, ice-cold PBS buffer containing 2% fetal bovine serum (Gibco) was added. RNA from FACS-sorted neurons was prepared for RNA-seq and subsequent analysis (see Extended Data for details).
Short-term associative memory assay
Memory assays were performed as described2.
Axon Regeneration Assays
In vivo laser axotomy of PLM neurons was performed as described22.
Extended Data
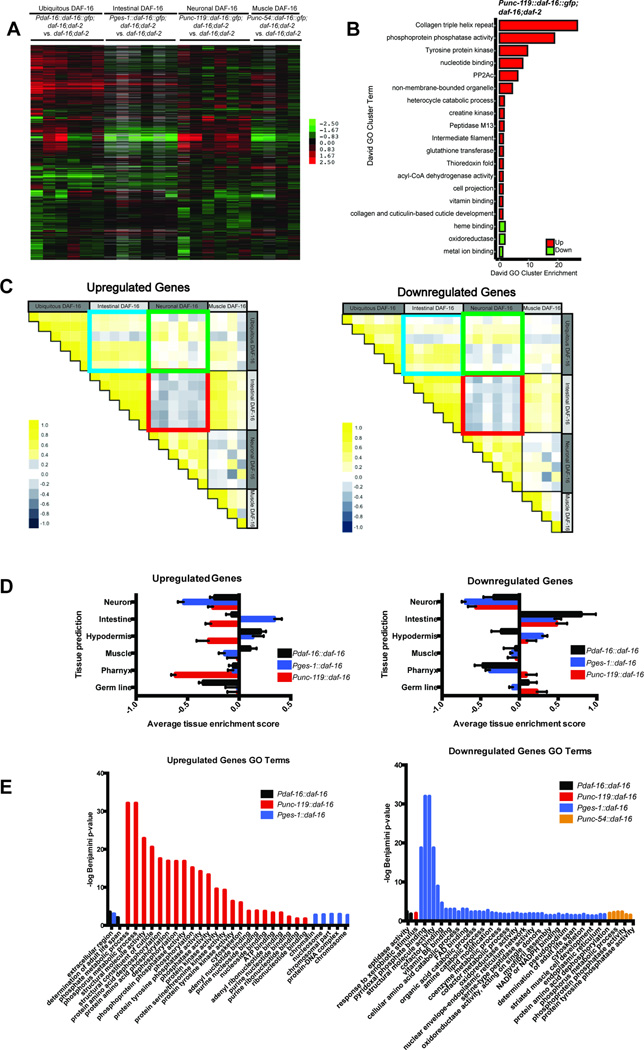
Extended Data Fig 1.
A) DAF-16 tissue-specific transgenics; heat map of all genes with expression differences ≥1.5-fold in ≥3 arrays. B) Significant Gene Ontology (GO) cluster terms from Punc-119::daf-16-regulated up- and down-regulated genes (Enrichment score >1). C) Pairwise Pearson correlations between arrays of DAF-16-up-regulated or down-regulated targets. The red box highlights the negative correlation between neuronal DAF-16 rescued targets (Punc-119::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2) and intestinal DAF-16 targets (Pges-1::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2), while the blue box shows the positive correlation between intestinal DAF-16 targets (Pges-1::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2) and whole worm DAF-16 targets (Pdaf-16::daf-16::gfp; daf-16;daf-2 vs daf-16;daf-2). The green box shows the weak correlation between neuronal rescued and whole worm DAF-16 targets. D) Tissue enrichment analysis (Mean ± SEM) of significant DAF-16-rescued up- and down-regulated genes (Supplementary Table 1) (FDR < 0.5). E) Significant Gene Ontology (GO) terms (adjusted p-value < 0.05) for DAF-16 up-regulated and down-regulated genes from whole worm, intestine-, neuron-, and muscle-rescued DAF-16 strains. Genes used for GO analysis (Supplementary Table 2) were derived from SAM analysis of the microarrays in (A) and Supplementary Table 1.
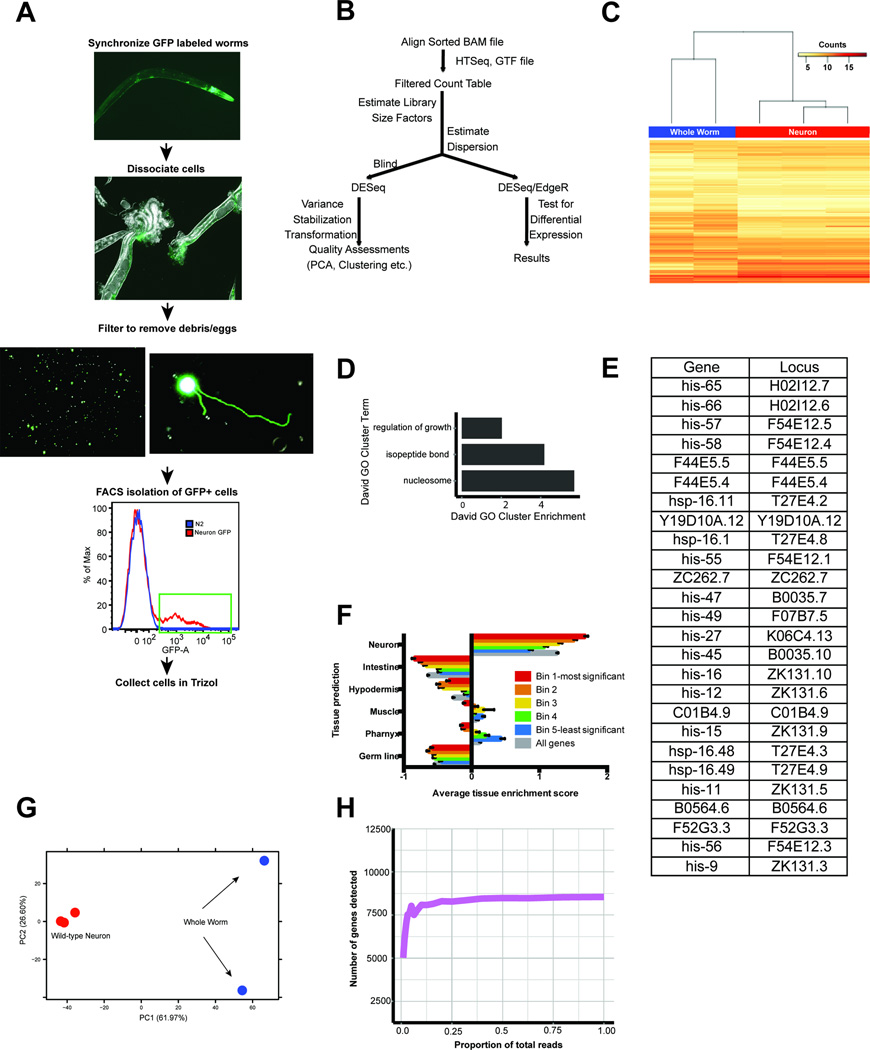
Extended Data Fig 2.
A) Pipeline for isolation of adult cells for FACS and RNA sequencing. B) Workflow for RNAseq data analysis of isolated neurons. C) Heat map of wild type neuron-expressed relative to whole worm-expressed genes. D) Actinomycin D (transcription inhibitor) treatment (100 µg/ml) during the cell isolation process demonstrates that the neuron isolation technique induces minimal transcriptional changes in wild type animals. Gene Ontology (GO) Terms represent genes up-regulated in the absence of Actinomycin D (Fig 1B, Supplementary Table 4). E) The 26 differentially expressed genes from Actinomycin D treatment are listed. F) C. elegans tissue gene expression prediction confirms neuronal character of adult wild-type neuron-enriched genes. Neuron-enriched genes were divided among equal bins according to p-value. Bin 1: FDR<0.003%; Bin 2: 0.003%–0.03%; Bin 3: 0.03%–1.3%; Bin 4: 1.3%–4%; Bin 5: 4–10%. G) Principal component analysis (PCA) shows a clear separation between wild-type adult neuronal and whole-worm samples. H) Down-sampling of wild-type neuron sequencing reads demonstrates sufficient sampling depth. The number of genes detected at the 3 counts per million threshold (for expressed genes) with different proportions of total sequencing depth analyzed.
Extended Data Fig 3.
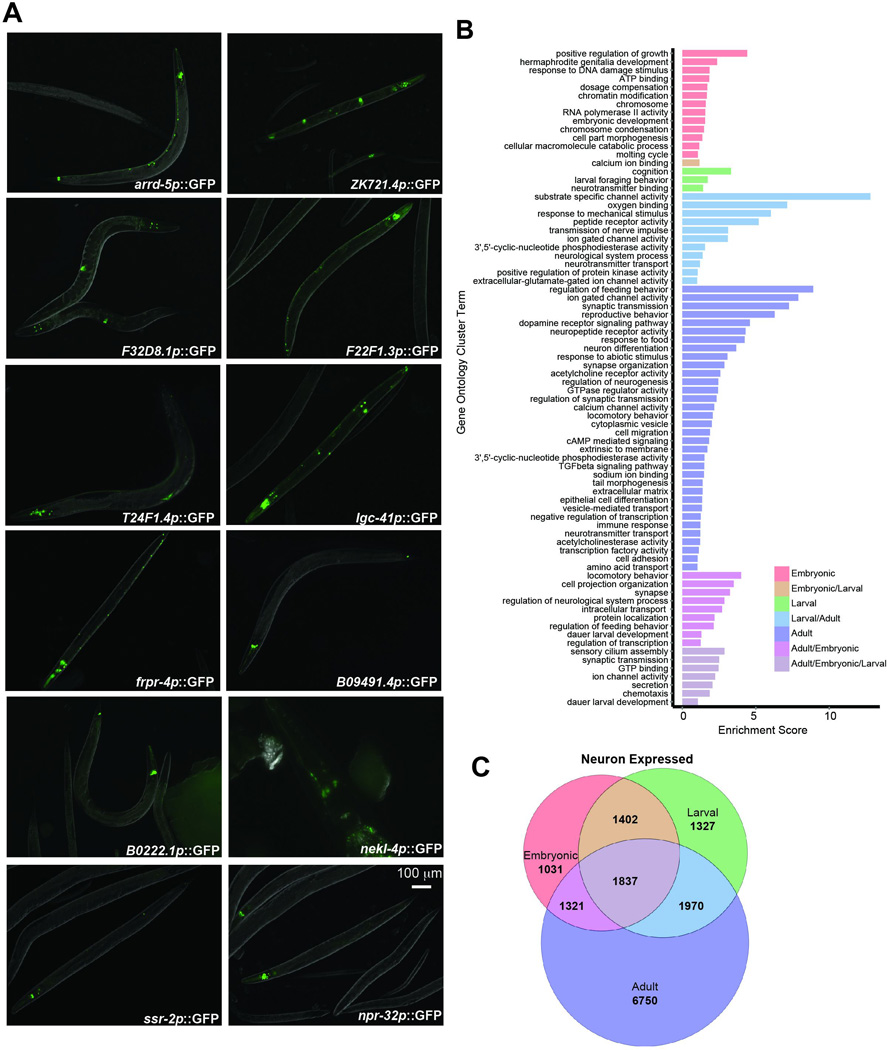
A) Promoter::GFP transcriptional fusions of candidate uncharacterized neuronal genes (Day 1 of adulthood). B) Gene Ontology clusters were generated from the categories in Fig 1E. Non-overlapping GO Terms suggest a transition from development-related processes in embryonic and larval animals to neuronal processes involved in behavior in adults (Supplementary Table 5). C) Venn diagram depicting the overlap between genes classified as “expressed” among embryonic and larval neurons16 and adult neurons from our RNA-seq analysis (Supplementary Table 5).
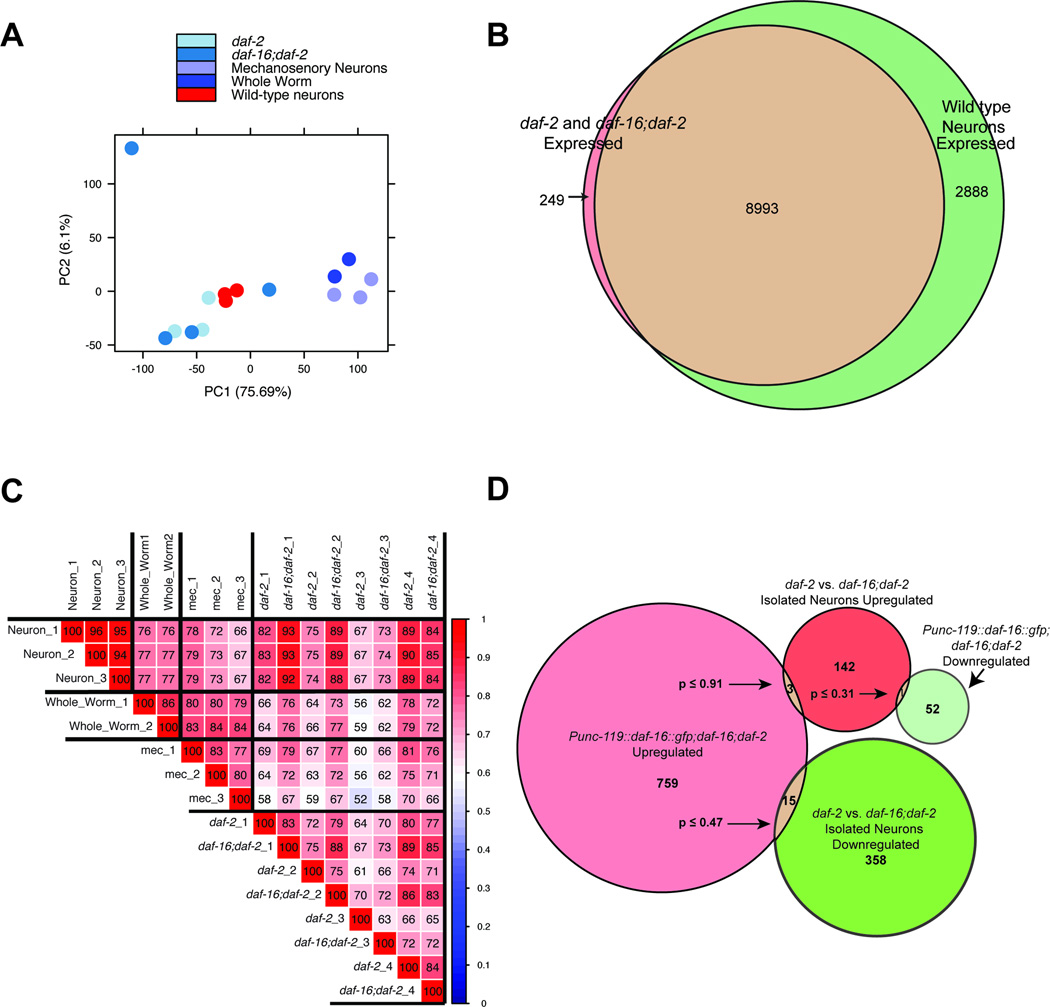
Extended Data Fig 4.
A) Principal Components Analysis of the whole worm and isolated adult neuron samples obtained for this study. B) Venn diagram depicting the overlap of daf-2- and daf-16;daf-2-expressed genes with those expressed in wild-type adult neurons. C) Spearman correlation of whole-worm and isolated adult neuron samples. D) The DAF-16 cell-autonomous and cell non-autonomous targets are distinct. The number of genes that overlap between neuronal DAF-16-rescued whole-worm targets (Punc-119::daf-16::gfp;daf-16;daf-2 vs daf-16;daf-2) and isolated neuron IIS targets (daf-2 vs daf-16;daf-2) is shown (Supplementary Table 8). Hypergeometric distribution analysis (p-values) shows that the extent of overlap between the gene categories is not significant.
Extended Data Fig 5.
A) The different classes of neuronal IIS/FOXO genes shown in Figure 2B were analyzed for DBE and DAE sequences in the 1kb upstream promoter regions. The genome-wide % of DBE and DAE occurrences across the 1kb promoters of all gene-encoding regions is reported. Comparison of whole-worm (Class I)8 vs neuronal-IIS/FOXO-regulated targets. P-values: hypergeometric distributions. B) GO terms of Class I whole worm8 vs. neuronal-IIS up-regulated genes (left) and Class II whole worm8 vs neuronal-IIS down-regulated genes (right) (Supplementary Table 5).
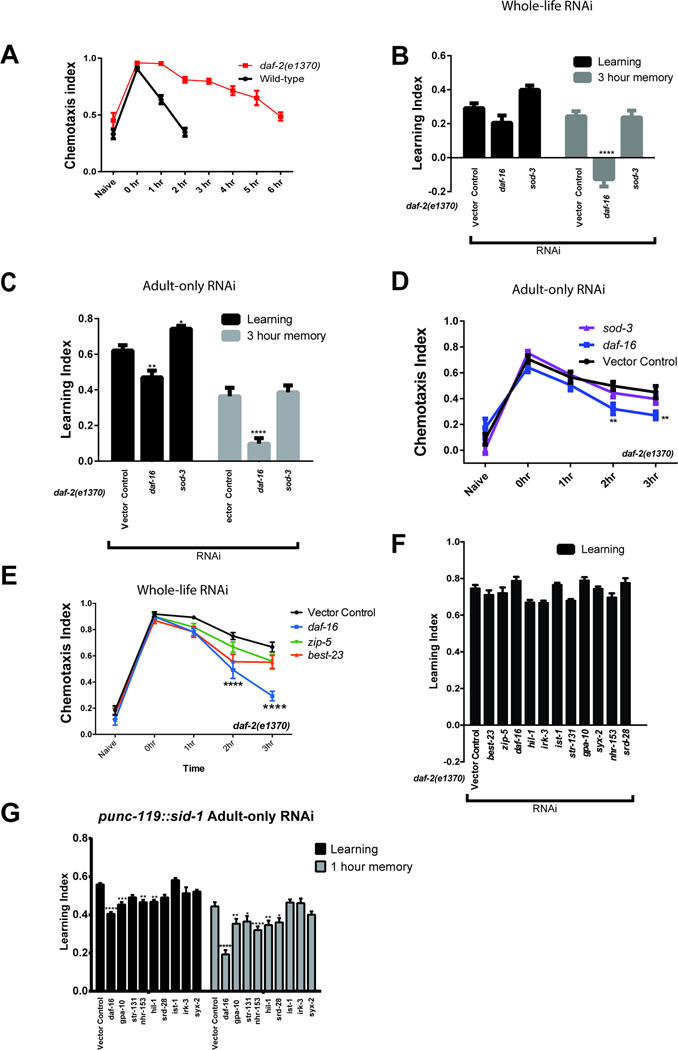
Extended Data Fig 6.
daf-2 is required for various forms of C. elegans associative learning2,27,30–33. daf-16 is required for daf-2’s improvements and extensions of abilities with age2. daf-2 mutants are defective for salt chemotaxis learning27,30,31, and daf-16 is not involved in salt chemotaxis learning27,30,31. Furthermore, salt learning utilizes a unique daf-2c isoform27 in a daf-16-independent manner30, suggesting a learning mechanism distinct from the associative memory paradigms studied here. We are specifically interested in understanding how activation of DAF-16 results in the improved and extended abilities of daf-2 mutants to carry out olfactory associative learning2, short-term associative memory2,33, and long-term associative memory2, all of which require daf-16. A) Chemotaxis index profile of wild type (N2) and daf-2 animals at time points following memory training. B) RNAi knockdown of sod-3, a non-neuronal DAF-16-regulated target that influences lifespan, has no effect on the extended short-term associative memory (STAM) of daf-2 mutants when treated with RNAi-feeding bacteria throughout the whole life (B) or only the post-developmental (adult-only) period (C, D) of the animal. daf-2 worms treated with daf-16 RNAi have defective STAM, as previously reported2. E) Knockdown of the neuronal IIS candidate genes zip-5 and best-23 does not affect STAM. Time-courses showing the chemotaxis index for each time point are shown in D and E. Learning indices are shown in B, C, F, and G. B–E) Two-way repeated measures ANOVA, Bonferroni post hoc tests. F) Treatment of daf-2 worms with neuronal DAF-16 target RNAi does not affect short-term associative learning. G) Neuronal-RNAi sensitive worms (Punc-119::sid-1) in a wild-type background were treated only during adulthood with RNAi targeted against the neuronal DAF-16 target genes. (0h) Learning and 1 h short-term associative memory time points are shown. A–G) Mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Extended Data Fig 7.
A) Six adult mechanosensory neurons labeled by mec-4p::GFP were isolated for RNA-seq. B) Axon length from the cell body to the site of injury was measured in µm immediately after axotomy and 24 hours later. Regenerative capacity of wild-type PLM axons declines from day 1 to day 5 of adulthood. C) Day 5 wild-type animals regrow axons that are significantly shorter than in Day 1 animals. D) Axotomies of daf-2 mutants grown on vector control, sod-3, or daf-16 RNAi demonstrate that sod-3, a lifespan-regulating DAF-16 target, does not influence the axon regeneration capacity of daf-2 worms at Day 5 of adulthood. E) fkh-9 does not affect the regenerative capacity of daf-2 axons on Day 1 of adulthood. F) fkh-9 is not required for axon regeneration in Day 1 adults. B–F) Mean ± SEM, Fisher’s exact test, *p < 0.05. G) Overexpression of the a and b isoforms of fkh-9 in wild-type animals causes axonal structural defects. Rescuing fkh-9 activity in the mechanosensory neurons of daf-2;fkh-9 mutants results in severe beading and degeneration of axons.
Extended Data Fig 8.
Wormbase (www.wormbase.org) gene models for (A) fkh-9 and (B) sod-3 are shown with modENCODE data for DAF-16 ChIP-seq experiments. A) Primer sets for ChIP-qPCR are depicted. C) Posterior intestinal FKH-9::GFP expression is only modestly increased in daf-2 compared to wild-type animals expressing fkh-9p::fkh-9::gfp. N= 25 animals.
Extended Data Fig 9.
A, B) Whole-life RNAi of fkh-9 reduces daf-2 STAM. C) RNAi knockdown of fkh-9 exclusively during adulthood results in reduced daf-2 STAM comparable to daf-16 RNAi-treatment. D, E) daf-2;fkh-9 mutants have reduced learning (tested immediately following STAM training) and STAM compared to daf-2. Mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Time-courses showing the chemotaxis index for each time point are shown in B and E. Learning indices are shown in A, C, and D.
Extended Data Fig 10.
Adult-only (A) or whole-life (B) fkh-9 RNAi treatment increases matricide in daf-2 worms. The cumulative % of animals dead as a result of bagging and/or exploding was recorded every other day. Two biological replicates were performed, with a representative experiment shown. C) Neuronal rescue of fkh-9 in daf-2;fkh-9 animals does not diminish the rate of vulval protrusions with age. N ≥ 60 per conditions for each experiment. D) Neuronal rescue of fkh-9 does not restore longevity of the daf-2;fkh-9 double mutant. daf-2 median lifespan: 41 days, daf-2;fkh-9 20 days, daf-2;fkh-9;Punc-119::fkh-9 20 days. p < 0.0001 for daf-2 vs. both daf-2;fkh-9 and daf-2;fkh-9;Punc-119::fkh-9. N=112 worms per strain. Censor rate for daf-2 19%, daf-2;fkh-9 51%, daf-2;fkh-9;Punc-119::fkh-9 56%.
Supplementary Material
Acknowledgments
We thank the C. elegans Genetics Center for strains and Wormbase (WS250); L. Parsons for RNA-seq data support; J. Wiggins and the Lewis-Sigler High Throughput Sequencing Core Facility for RNA-seq support; C. DeCoste and the Flow Cytometry Facility for assistance; V. Yao for tissue prediction analysis; R. DiLoreto for chemotaxis assay analysis; Z. Gitai, the Murphy lab, and W. Mair for discussion. Funding was provided by a Keck Scholars Program fellowship (CTM), NIH Cognitive Aging R01 (CTM), and NRSA (RK), NRSA (RA), NSF (JL), and NJCBIR (VL) fellowships.
Footnotes
Author Contributions. CTM, RK, VL, and JL designed experiments. RK, VL, RA, JL, JA, and CTM performed experiments and analyzed data. RK, VL, and JL performed tissue isolation and RNA-seq. AW, RK, VL, and JL performed bioinformatics analysis. RK, VL, and RA performed short-term memory experiments. VL performed axon regeneration experiments. RK, VL, and CTM wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Murphy CT, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 2.Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne AB, et al. Insulin/IGF1 signaling inhibits age-dependent axon regeneration. Neuron. 2014;81:561–573. doi: 10.1016/j.neuron.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sjöberg J, Kanje M. Insulin-like growth factor (IGF-1) as a stimulator of regeneration in the freeze-injured rat sciatic nerve. Brain Res. 1989;485:102–108. doi: 10.1016/0006-8993(89)90671-9. [DOI] [PubMed] [Google Scholar]
- 5.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 6.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 7.Luo S, Kleemann GA, Ashraf JM, Shaw WM, Murphy CT. TGF-β and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell. 2010;143:299–312. doi: 10.1016/j.cell.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tepper RG, et al. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell. 2013;154:676–690. doi: 10.1016/j.cell.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang P, Judy M, Lee S-J, Kenyon C. Direct and indirect gene regulation by a life-extending FOXO protein in C. elegans: roles for GATA factors and lipid gene regulators. Cell Metab. 2013;17:85–100. doi: 10.1016/j.cmet.2012.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murakami H, Bessinger K, Hellmann J, Murakami S. Aging-dependent and -independent modulation of associative learning behavior by insulin/insulin-like growth factor-1 signal in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci. 2005;25:10894–10904. doi: 10.1523/JNEUROSCI.3600-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tank EMH, Rodgers KE, Kenyon C. Spontaneous age-related neurite branching in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci. 2011;31:9279–9288. doi: 10.1523/JNEUROSCI.6606-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth ML, et al. Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. J. Neurosci. Off. J. Soc. Neurosci. 2012;32:8778–8790. doi: 10.1523/JNEUROSCI.1494-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comput. Biol. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ewald CY, Landis JN, Abate JP, Murphy CT, Blackwell TK. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. 2014 doi: 10.1038/nature14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S, Banerjee D, Kuhn JR. Isolation and culture of larval cells from C. elegans. PloS One. 2011;6:e19505. doi: 10.1371/journal.pone.0019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spencer WC, et al. Isolation of specific neurons from C. elegans larvae for gene expression profiling. PloS One. 2014;9:e112102. doi: 10.1371/journal.pone.0112102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schafer JC, et al. IFTA-2 is a conserved cilia protein involved in pathways regulating longevity and dauer formation in Caenorhabditis elegans. J. Cell Sci. 2006;119:4088–4100. doi: 10.1242/jcs.03187. [DOI] [PubMed] [Google Scholar]
- 18.Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- 19.Hansen M, Hsu A-L, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr. Biol. CB. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lakhina V, et al. Genome-wide Functional Analysis of CREB/Long-Term Memory-Dependent Transcription Reveals Distinct Basal and Memory Gene Expression Programs. Neuron. 2015;85:330–345. doi: 10.1016/j.neuron.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen L, et al. Axon regeneration pathways identified by systematic genetic screening in C. elegans. Neuron. 2011;71:1043–1057. doi: 10.1016/j.neuron.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian NM, Pratt T, Price DJ. Foxg1 regulates retinal axon pathfinding by repressing an ipsilateral program in nasal retina and by causing optic chiasm cells to exert a net axonal growth-promoting activity. Dev. Camb. Engl. 2008;135:4081–4089. doi: 10.1242/dev.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang Y-W, et al. Rapid induction of genes associated with tissue protection and neural development in contused adult spinal cord after radial glial cell transplantation. J. Neurotrauma. 2009;26:979–993. doi: 10.1089/neu.2008.0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda DD, et al. CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 2008;105:5260–5265. doi: 10.1073/pnas.0711894105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoerndli FJ, et al. A conserved function of C. elegans CASY-1 calsyntenin in associative learning. PloS One. 2009;4:e4880. doi: 10.1371/journal.pone.0004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno H, et al. Role of synaptic phosphatidylinositol 3-kinase in a behavioral learning response in C. elegans. Science. 2014;345:313–317. doi: 10.1126/science.1250709. [DOI] [PubMed] [Google Scholar]
- 28.Ewald CY, et al. Pan-neuronal expression of APL-1, an APP-related protein, disrupts olfactory, gustatory, and touch plasticity in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci. 2012;32:10156–10169. doi: 10.1523/JNEUROSCI.0495-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hammarlund M, Nix P, Hauth L, Jorgensen EM, Bastiani M. Axon regeneration requires a conserved MAP kinase pathway. Science. 2009;323:802–806. doi: 10.1126/science.1165527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Extended Data References
- 30.Tomioka M, et al. The insulin/PI 3-kinase pathway regulates salt chemotaxis learning in Caenorhabditis elegans. Neuron. 2006;51:613–625. doi: 10.1016/j.neuron.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 31.Kodama E, et al. Insulin-like signaling and the neural circuit for integrative behavior in C. elegans. Genes Dev. 2006;20:2955–2960. doi: 10.1101/gad.1479906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami H, Bessinger K, Hellmann J, Murakami S. Aging-dependent and -independent modulation of associative learning behavior by insulin/insulin-like growth factor-1 signal in Caenorhabditis elegans. J. Neurosci. Off. J. Soc. Neurosci. 2005;25:10894–10904. doi: 10.1523/JNEUROSCI.3600-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein GM, Murphy CT. C. elegans positive olfactory associative memory is a molecularly conserved behavioral paradigm. Neurobiol. Learn. Mem. 2014;115:86–94. doi: 10.1016/j.nlm.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.