Abstract
FGFR1 amplification is recognized as a novel therapy target for non-small-cell lung cancer (NSCLC), especially in squamous cell carcinoma (SCC). However, the association between FGFR1 amplification and the clinicopathological characteristics of NSCLC remains controversial. We performed a meta-analysis of 17 eligible studies to examine the correlation between FGFR1 gene amplification and clinicopathological characteristics. FGFR1 amplification was closely related to these clinicopathological features, including sex (odds ratio [OR] 2.05, 95% confidence interval [CI] 1.50–2.80), smoking (OR 3.31, 95% CI 2.02–5.44), and histology (OR 3.60, 95% CI 2.82–4.59). FGFR1 amplification was associated with shorter overall survival, and no significant heterogeneity existed between studies (I2=3.8%). We should note that publication bias may partly account for these results, but our findings remained significant after the trim-and-fill method (hazard ratio 1.22, 95% CI 1.06–1.40). However, no significant correlation was found with poor disease-free survival (hazard ratio 1.43, 95% CI 0.96–2.12). In conclusion, this study showed that FGFR1 amplification was significantly associated with sex, smoking, and histology. FGFR1 amplification could be a marker of poor prognosis in NSCLC patients, especially in SCC patients.
Keywords: FGFR1, amplification, non-small-cell lung cancer, meta-analysis
Introduction
Lung cancer, which mainly consists of small-cell lung cancer (SCLC) and non-SCLC (NSCLC), is one of the most common malignancies worldwide, and is the leading cause of cancer deaths in the world.1 NSCLC accounts for 75% of all lung cancers and includes two predominant – adenocarcinoma and squamous cell carcinoma (SCC) – which comprise 40% and 25% of NSCLCs, respectively. Despite advances in treatment, the prognosis of lung cancer patients is still poor, and the 5-year overall survival (OS) rate is only 15%. Lung adenocarcinomas with EGFR mutations or EML4–ALK fusions respond effectively to treatment by EGFR and ALK inhibition, respectively.2–4 Unfortunately, these genetic events are rare, and are limited to adenocarcinomas of nonsmoking patients; however, most lung cancer cases are caused by smoking.
FGFR1 has been recognized as one of the promising molecular targets for the treatment of smoking-related lung cancer (SCC) providing a novel therapeutic target for these tumors.5,6 FGFR1 is overexpressed by 10%–20% of lung cancer patients, and is correlated with cigarette-smoking dosage and poor clinical outcomes in resected SCC.7 FGFR1 gene amplification is often associated with FGFR overexpression, which leads to ligand-independent signaling.6
Fluorescence in situ hybridization (FISH) is the standard method for characterizing gene amplification, although it does not have a standard definition of FGFR1 amplification. Differences in FGFR1 copy numbers have been characterized as normal, gain (low amplification), or amplification (high amplification) in some articles, while some studies have used two categories: negative and amplification.8 Other studies have utilized fluorescence quantitative polymerase chain reaction (qPCR), because it is technically less complex, automated, quantitative, and independent of reader interpretation.9,10 The single-nucleotide polymorphism (SNP) array is a useful tool for studying slight variations between whole genomes. As cancer molecular biology progresses, agents targeting the FGFR1 pathway, such as inhibitors or monoclonal antibodies, have been introduced into clinical application.11
Despite a number of individual studies performed in lung cancer patients, the prognostic value of FGFR1-amplification status in a lung cancer patient’s survival remains controversial. Additionally, the clinicopathological features found in those studies varied. Therefore, we performed a systematic review of the literature and conducted a meta-analysis to obtain a more accurate evaluation of the prognostic value of FGFR1 and the clinicopathological features associated with NSCLC. The results of this meta-analysis will help us to design an individualized therapeutic schedule for each patient and to provide closer follow-up care for patients with FGFR1 amplification. Furthermore, based on our understanding of the effect and function of FGFR1 in NSCLC, patients who would potentially profit from FGFR1 inhibitors would be specifically selected for such treatment, which deserves further research for clinical applications.
Materials and methods
Identification and eligibility of relevant studies
The PubMed and ISI Web of Knowledge databases were searched for articles from 1994 to July 2015 relating to FGFR1 and lung cancer. The following Medical Subject Headings keywords and text were used: 1) lung or cancer or tumor or neoplasm or carcinoma; and 2) FGFR1. The references of articles and reviews were also manually searched for additional studies. Eligible studies included in this meta-analysis met the following criteria: 1) the full-text publication should clearly describe studies on the association between FGFR1 gene amplification and lung cancer patient prognosis (OS and/or disease-free survival [DFS]), or 2) directly provide the FGFR1-detection method and present the clinicopathological features of the lung patients. The exclusion criteria were 1) letters, reviews, conference abstracts, and case reports, and 2) overlapping articles, which were also excluded from this meta-analysis, and only the most recent or the most complete study was involved in the analysis, because of the limited data.
Data extraction
Data extraction was performed using a standardized data-extraction form, collecting information on the first author’s name, publication year, median age, patient number, stage, histology, differentiation, detection method, cutoff value, smoking status, risk estimates or data used to calculate risk estimates, confidence intervals (CIs) or data used to calculate CIs, and the rate of FGFR1 amplification. From studies that reported hazard ratios (HRs) in both univariate and multivariate models, we extracted the latter, because these results were more convincing, as there had been adjustment for potential confounders. If only Kaplan–Meier graphs were published, the Kaplan–Meier curves were read by Engauge Digitizer version 4.1 (http://digitizer.sourceforge.net). Two investigators (QQZ and YG) reviewed each eligible study independently and extracted data from all the publications meeting the inclusion criteria. Controversial problems were arbitrated by the third investigator (FJX).
Statistical analysis
Pooled estimates of the odds ratios (ORs) and their 95% CIs were used to estimate the association between FGFR1 amplification and the clinical parameters of lung cancer, including age, sex, smoking status, histologic type, differentiation, and lymph-node metastasis, as well as stage. Pooled estimates of HRs and their 95% CIs were used to estimate the association between FGFR1 amplification and lung cancer survival. The assumption of statistical heterogeneity among the studies was evaluated using the χ2-based Q-test.12 When I2 was no more than 50%, pooled ORs, relative risks, and 95% CIs were calculated using the Mantel–Haenszel method with fixed-effect models.13 When significant heterogeneity (P<0.1, I2>50%) was detected among the studies, a random-effect model (using the DerSimonian and Laird method) was adopted. If necessary, a sensitivity analysis was also performed to evaluate the influence of individual studies on the final effect. The potential publication bias was assessed by Begg’s funnel plot and Egger’s test.14 We used the trim-and-fill method to evaluate the influence of possible publication bias on the results. A P-value <0.05 was considered significant. All the statistical analyses were performed using Stata package 12.0 (StataCorp LP, College Station, TX, USA).
Results
Search results and characteristics
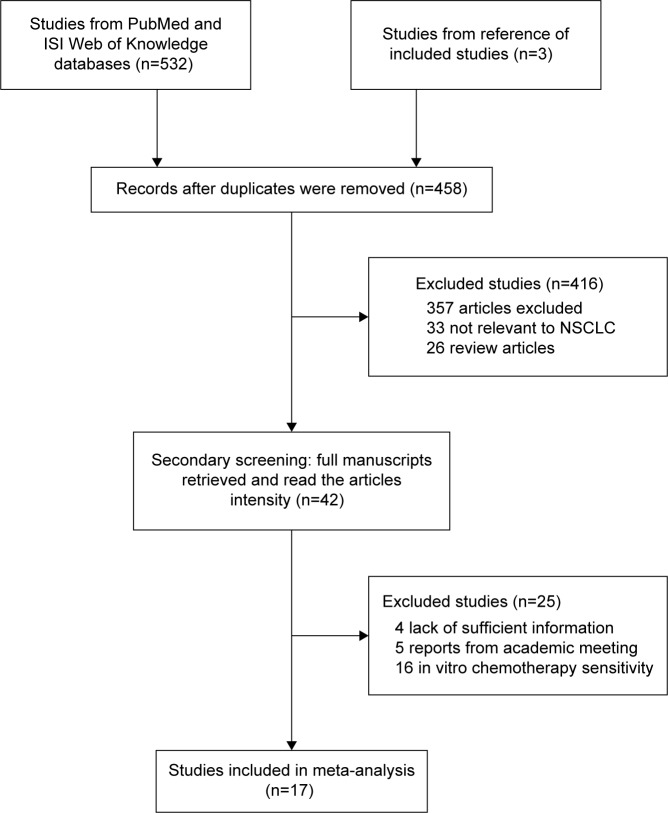
Figure 1 illustrates the process of evaluating articles for inclusion in the review and meta-analysis. Of the 458 abstracts identified, we excluded 416 abstracts and further reviewed 42 full-text articles to determine whether they met our inclusion and exclusion criteria. As a result, 18 eligible studies comprising 4,954 NSCLC cases were included in this meta-analysis.7–10,15–28
Figure 1.
Flowchart of the eligible studies.
Abbreviation: NSCLC, non-small-cell lung cancer.
The main characteristics of the 18 eligible studies are shown in Table 1. Most of the studies investigated FGFR1 amplification by FISH (eleven studies), three studies used qPCR, two articles detected FGFR1 using the SNP-array method, and two studies identified FGFR1 by silver ISH and dual-color ISH. Dutt et al27 clearly summarized patient clinical pathological characteristics of FGFR1 amplification by the Affymetrix 250K SNP array in a previously reported data set.29–33 Among the 17 studies, four studies (900 patients, 18.2%) were performed in Asian populations, and the remaining studies (4,054 patients, 81.8%) involved non-Asian patients.
Table 1.
Main characteristics and results of the eligible studies
| First author (references) | Year | Country | Cancer type | Stage | Patient number | Median age, years (range) | Detection method | Cutoff value | Positive rate (%) | Clinicopathological features | HR estimation | HR for overall survival (95% CI) | HR for disease-free survival (95% CI) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seo et al28 | 2014 | Korea | NSCLC | I–III | 369 (AC 230, SCC 139) | 65 (21–84) | FISH | FGFR1 gene copy number ≥6.2 | 32/369 (8.7%) | G, C, LN, S, H | HR | SCC: 1.79 (0.83–3.87) | SCC: 1.63 (0.87–3.07) |
| Cihoric et al8 | 2014 | Switzerland | NSCLC | I–II | 329 (SCC 169, AC 137, LCC 23) | 66.9 (42–83) | FISH | FGFR1/CEP8 signal ratio ≥2.0 | 41/329 (12.5%) | G, C, T, H | HR | NSCLC: 2.06 (1.05–4.05); SCC: 1.05 (0.57–1.93) | NSCLC: 1.46 (0.76–2.81); SCC 1.12 (0.48–2.58) |
| Wynes et al15 | 2014 | Poland | NSCLC | I–IV | 189 (AC 55, SCC 103, LCC 5, Other 26) | 64 (37–85) | SISH | FGFR1 gene copy number ≥4, or FGFR1:CEP8 ratio ≥2 | 14/182 (8%) | A, G, C, S, D, H | HR | 0.99 (0.50–1.96) | NA |
| Russell et al16 | 2014 | Australia | NSCLC | I–IV | 338 (AC 99, SCC 178, LCC 41, Other 20) | 69 (19–87) | FISH | High FGFR1 amplification: FGFR1/centromere 8 (CEN8) ≥2, or the tumor cell percentage with ≥15 FGFR1 signals ≥10%, and the average number of FGFR1 signals/tumor cell nucleus ≥6; Low FGFR1: tumor cell percentage with ≥5 FGFR1 signals ≥50% | 49/352 (13.9%) | H | HR | NSCLC: 1.09 (0.72–1.66); SCC: 1.01 (0.65–1.58) | SCC: 1.04 (0.67–1.60) |
| Toschi et al17 | 2014 | Italy | NSCLC | I–IV | 447 (AC 244, SCC 138, Other 65) | 66 (33–86) | FISH | Gene copy gain: ≥4 gene copies/cell; | Amplification: | G, C, S, H# | Survival curve | 0.99 (0.70–1.40) | NA |
| Serizawa et al18 | 2014 | Japan | AC | I–IV | 411 | 68 (29–89) | qPCR | Amplification: presence of gene clusters | 37/445 (8.3%); Copy-number gain 37/445 (8.3%) | ||||
| Pros et al19 | 2013 | Spain | NSCLC | I–IV | 265 (AC 86, SCC 150, LCC 26, Other 3) | NA | FISH | The ratio of the normalized quantity of FGFR1/COL8A1 ≥2 | 2/411 (0.05%) | C | – | NA | NA |
| Gadgeel et al9 | 2013 | US | NSCLC | I–IV | qPCR | FGFR1 copy-number >12 or presence of gene clusters | 17/265 (6%) | G, C, H | – | NA | NA | ||
| (Training cohort) | 203 (AC 98, SCC 79, LCC 15, Other 11) | 66.2 (35.0–83.8) | FGFR1 exon 15 copy-number value >3.50 | G, H | HR | ||||||||
| (Validation cohort) | 142 (AC 71, SCC 57, LCC 13, Other 1) | 65.2 (25.8–81.9) | 12/203 (5.9%) | 2.19 (1.02–4.75) | NA | ||||||||
| Craddock et al20 | 2013 | Canada | SCC | I–IV | 135 | 69.2 (44.0–83.9) | FISH | 5/142 (3.5%) | 2.91 (1.14–7.41) | NA | |||
| Tran et al21 | 2013 | Australia | NSCLC | I–III | 264 (AC 115, SCC 101, LCC 44, Other 4) | 66.5 (57.8–75.2) | Dual-color FISH | FGFR1 copy number ≥5.0 | 22/121 (18.2%) | G, C, S | HR | 1.33 (0.67–2.62) | 1.15 (0.59–2.25) |
| Kim et al7§ | 2013 | Korea | SCC | I–III | 262 | 66 (36–81) | FISH | Amplification: FGFR1/CEP8 ≥2.0, or mean FGFR1 signals per tumor cell ≥6.0, or percentage of tumor cells or containing FGFR1 clusters ≥10%; FGFR1 copy-number gain: the mean of FGFR1 signals was between 4 and 6 or at least 50% of counted cells contained ≥4 FGFR1 signals | Amplification: 37/264 (14%); Copy-number gain 12/264 (4.5%) | G, C, S, D, H# | Survival curve | 1.29 (0.85–1.95) | NA |
| Heist et al22 | 2012 | US | SCC | I–IV | 226 | 69 (38–91) | FISH | High amplification: FGFR1/CEP8 ≥9.0; | High amplification: | G, C, LN, S, H | HR | 1.83 (1.15–2.89) | 2.24 (1.45–3.45) |
| Kohler et al23 | 2012 | Germany | NSCLC | I–IV | 236 (AC 64, SCC 133, LCC 4, Other 35) | NA | FISH | Low amplification: FGFR1/CEP8 >2 and <9 | 34/262 (13.0%); Low amplification: 105/262 (40.1%) | ||||
| Schildhaus et al24 | 2012 | Germany | NSCLC | NA | 420 (AC 100, SCC 307, Other 13) | NA | FISH | FGFR1/CEP8 ≥2.2 | 37/226 (16%) | G, C, S | Survival curve | 0.84 (0.53–1.33) | NA |
| Zhang et al25 | 2012 | People’s Republic of China | NSCLC | I–IV | 127 (AC 76, SCC 48, Other 3) | NA | FISH | FGFR1 copy-number ≥4 | 14/133 (10.5%) | G, H | – | SCC: 2.64 (1.43–4.86) | NA |
| Sasaki et al10 | 2012 | Japan | SCC | I–IV | 100 | NA (29–86) | qPCR | FGFR1/CEN8 ≥2.0 or FGFR1 signals/cell nucleus ≥6 or the percentage of tumor cells containing ≥15 FGFR1 signals or large clusters is ≥10% or the percentage of tumor cells containing ≥5 FGFR1 signals is ≥50% | 58/290 (20%) for SCC, 0/97 (0%) for AC, 2/13 (15.4%) for others | H | – | NA | NA |
| Weiss et al26 | 2010 | US and Switzerland | NSCLC | NA | 232 (AC 77, SCC 155) | NA | SNP array | FGFR1/CEP8 ≥2.0 or cluster signals ≥10% of tumor cells | 11/127 (8.7%) | G, C, S, LN, H | – | NA | NA |
| Dutt et al27 | 2011 | US | NSCLC | I–IV | 628 (AC 555, SCC 46, Other 27) | NA | SNP array | FGFR1 gene copy number >4 | 32/100 (32%) | G, C, S, D, LN, H | – | 1.48 (0.57–3.86) | NA |
| Chromosome 8p12 that included FGFR1 ≥4 copies | AC, 1/77 (1.3%); SCC, 15/115 (9.7%) | C, H | Survival curve | 1.19 (0.78–1.81)*,† | NA | ||||||||
| Log2 ratio >0.7 or 3.25 normalized DNA copies | 32/628 (5.96%) | A, S, D, H | – | NA | NA |
Notes:
FGFR1–positive (included FGFR1 amplification and copy-number gain);
high amplification vs not high amplification;
HR FGFR1 copy number >9 vs copy number =2;
only includes SCC patients.
Abbreviations: HR, hazard ratio; CI, confidence interval; NSCLC, non-small-cell lung cancer; AC, adenocarcinoma; SCC, squamous cell carcinoma; LCC, large cell carcinoma; FISH, fluorescence in situ hybridization; SISH, silver ISH; qPCR, quantitative polymerase chain reaction; SNP, single-nucleotide polymorphism; G, sex; C, smoking status; S, stage; D, histologic differentiation; H, histology; LN, lymph-node metastasis; P, performance status; NA, not available; T, tumor size; A, age.
FGFR1 amplification and clinicopathologic features
Table 2 presents the results of the meta-analysis in NSCLC patients. Overall, there was no association between age, lymph-node metastasis, differentiation, tumor size or stage, and FGFR1 amplification (P>0.05). The OR (95% CI) was 1.19 (0.51–2.75) for age (≥60 years vs <60 years), 1.20 (0.79–1.83) for lymph-node metastasis (yes vs no), and 0.40 (0.11–1.41) for differentiation (good vs moderate or poor). However, positive FGFR1 amplification was associated with sex, smoking status, and histology (SCC vs non-SCC) in lung cancer patients (P<0.05). The OR (95% CI) was 2.32 (1.71–3.14) for sex (male vs female), 3.31 (2.02–5.44) for smoking status (smoking vs no smoking), and 3.60 (2.82–4.59) for histology (SCC vs non-SCC).
Table 2.
FGFR1 amplification and clinicopathological features for NSCLC
| Patient characteristics | Included studies | Heterogeneity test
|
Meta-analysis
|
Outcomes
|
||
|---|---|---|---|---|---|---|
| I2 (%) | P-value | Model | OR (95% CI) | P-value | ||
| NSCLC | ||||||
| Sex (male vs female) | 12 | 16.3 | 0.284 | Fixed | 2.32 (1.71–3.14) | <0.001 |
| Age (≥60 years vs <60 years) | 2 | 0 | 0.454 | Fixed | 1.19 (0.51–2.75) | 0.687 |
| Smoking vs no smoking | 12 | 33.9 | 0.119 | Fixed | 3.84 (2.29–6.43) | <0.001 |
| Histology (SCC vs non-SCC) | 13 | 42.3 | 0.054 | Fixed | 3.60 (2.82–4.59) | <0.001 |
| Lymph-node metastasis (yes vs no) | 4 | 30.6 | 0.229 | Fixed | 1.20 (0.79–1.83) | 0.384 |
| Differentiation (good vs moderate or poor) | 5 | 54.9 | 0.064 | Random | 0.40 (0.11–1.41) | 0.154 |
| Tumor size (T3 + T4 vs T1 + T2) | 4 | 0 | 0.898 | Fixed | 1.53 (0.94–2.47) | 0.081 |
| Stage (III–IV vs I–II) | 8 | 0 | 0.965 | Fixed | 0.97 (0.73–1.29) | 0.853 |
| SCC only | ||||||
| Sex (male vs female) | 5 | 0 | 0.733 | Fixed | 2.35 (1.41–3.92) | 0.001 |
| Smoking vs no smoking | 5 | 61.7 | 0.034 | Random | 2.57 (0.56–11.76) | 0.225 |
| Lymph-node metastasis (yes vs no) | 2 | 27.5 | 0.252 | Fixed | 1.13 (0.70–1.83) | 0.632 |
| Differentiation (good vs moderate or poor) | 2 | 46.9 | 0.17 | Random | 0.91 (0.13–6.15) | 0.921 |
| Tumor size (T3 + T4 vs T1 + T2) | 2 | 0 | 0.406 | Fixed | 1.75 (0.93–3.27) | 0.079 |
| Stage (III–IV vs I–II) | 4 | 0 | 0.775 | Fixed | 0.87 (0.55–1.37) | 0.543 |
Abbreviations: NSCLC, non-small-cell lung cancer; OR, odds ratio; CI, confidence interval; SCC, squamous cell carcinoma.
When pooling SCC studies only, positive FGFR1 amplification was associated with sex (male vs female, pooled OR 2.41, 95% CI 1.43–4.08; P=0.001). Although the pooled OR values were greater than 1.0, we did not find a significant association between patient smoking status (smoking vs no smoking, pooled OR 3.86, 95% CI 0.61–24.48; P>0.05). In accordance with NSCLC patients’ pathological features, there was no significant association between FGFR1 amplification and lymph-node metastasis, differentiation, tumor size, or stage. The pooled ORs (95% CI) were 0.94 (0.40–2.23), 0.91 (0.13–6.15), 1.36 (0.58–3.22), and 0.84 (0.50–1.39), respectively.
Impact of FGFR1 amplification on overall survival in NSCLC patients
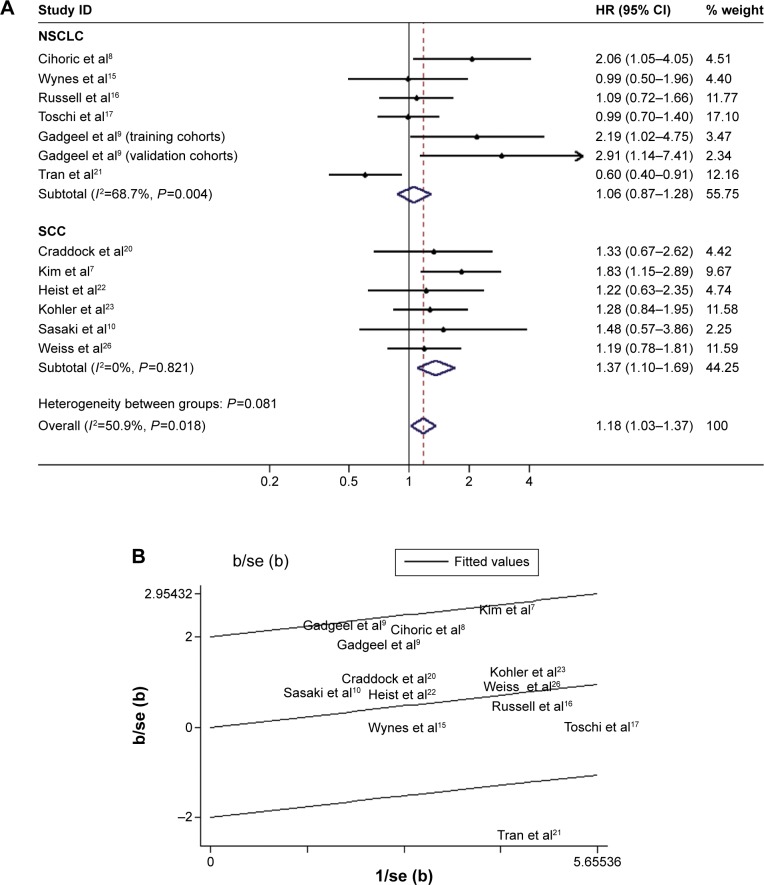
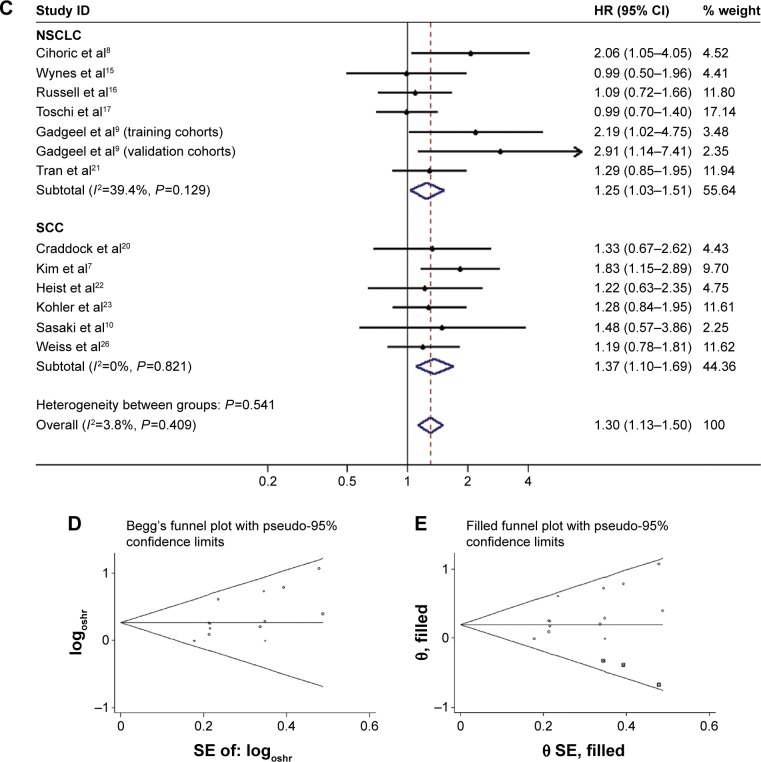
The combined HR for 13 studies evaluating FGFR1 amplification on OS was 1.35 (95% CI 1.05–1.73), suggesting that FGFR1 amplification was an indicator of poor prognosis in NSCLC patients (Figure 2A). However, significant heterogeneity was observed among the studies (I2=50.9%, P=0.018). In Figure 2B, one article is identified in the Galbraith plot as an outlier.21 This study investigated “FGFR1 positive” status, and included whether the FGFR1 ISH expressed FGFR1 amplification or copy-number gain.21 According to Kim et al, there is no significant difference in messenger RNA-expression levels between low FGFR1 gene amplification and disomy,7 so the HRs of FGFR1 amplified vs nonamplified extracted data from the Kaplan–Meier curves were more suitable for further analyses. Indeed, the adjusted association of FGFR1 amplification and NSCLC patients’ OS had lower heterogeneity (I2=3.8%, P=0.41) and predicted a worse prognosis (fixed-effect model, HR 1.30, 95% CI 1.13–1.50; P<0.001; Figure 2C).
Figure 2.
Forest plot and Begg’s funnel plot of the association between FGFR1 amplification and NSCLC patient OS.
Notes: Studies are sorted in order of publication year. (A) Forest plot of HR for the association of FGFR1 amplification with OS in primary studies (random-effect model); (B) Galbraith plot of association between FGFR1 amplification and NSCLC with overall survival; (C) forest plot of HR for the association of FGFR1 amplification with OS, with adjusted values (fixed-effect model); (D) Egger’s publication showed obvious publication bias (P<0.05) for studies regarding FGFR1 amplification and OS in the meta-analysis; (E) adjusted funnel plot for publication bias.
Abbreviations: NSCLC, non-small-cell lung cancer; SCC, squamous cell carcinoma; OS, overall survival; HR, hazard ratio; CI, confidence interval; SE, standard error; oshr, overall survival hazard ratio.
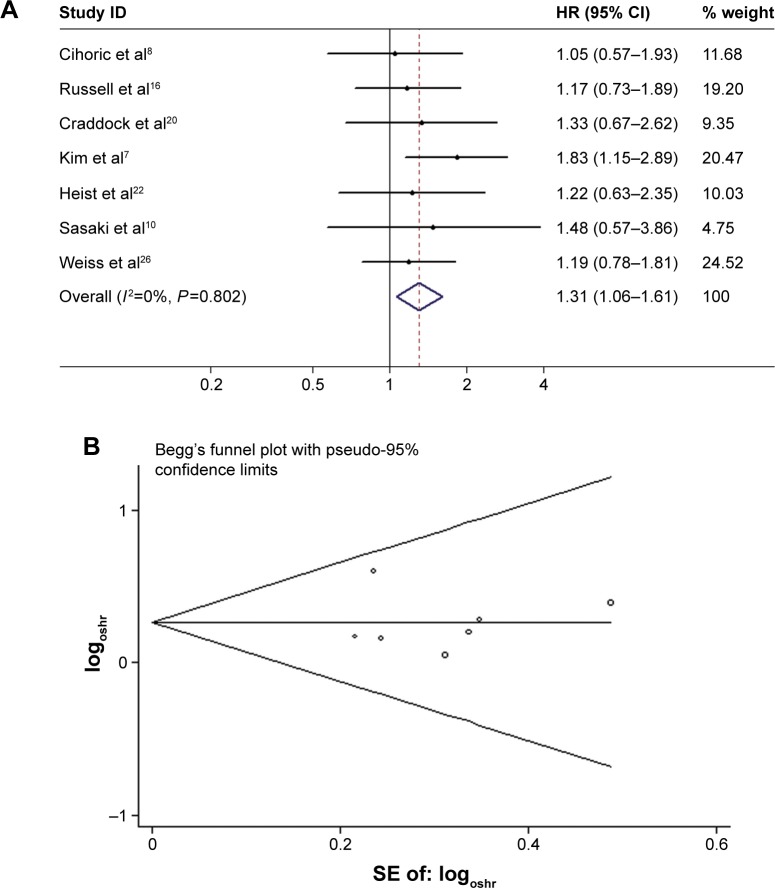
We also separately analyzed the studies that included the histological type of SCC patients only. After pooling the seven studies, the combined HR was 1.31 (95% CI 1.06–1.61, P<0.05), suggesting that FGFR1 amplification had a significant impact on SCC patients’ OS (Figure 3A). When we limited the analysis to the studies using qPCR, the pooled HR was 1.04 (95% CI 0.75–1.44, P>0.05). When we included the study with the largest sample size (sample size >300), the combined HR was 1.27 (95% CI 1.01–1.60, P<0.05). For subgroup analyses based on ethnicity (Asian or non-Asian) and large study, both results suggested that FGFR1 amplification had a significant negative impact on survival (Table 3).
Figure 3.
Forest plot (A) and Begg’s funnel plot (B) of the association between FGFR1 amplification and SCC patient OS.
Abbreviations: SCC, squamous cell carcinoma; OS, overall survival; HR, hazard ratio; CI, confidence interval; SE, standard error; oshr, overall survival hazard ratio.
Table 3.
Meta-analysis of FGFR1 and prognosis in NSCLC patients
| Categories | Study population (reference) | Meta-analysis model | HR (95% CI) | I2 (%) | Ph | P-value |
|---|---|---|---|---|---|---|
| Overall survival | ||||||
| Overall | 13 (7–10,15–17,20–23,26,28) | Fixed | 1.30 (1.13–1.50) | 3.8 | 0.409 | <0.001 |
| NSCLC only | 7 (8,9,15–17,21,28) | Fixed | 1.25 (1.03–1.51) | 39.4 | 0.129 | 0.024 |
| SCC only | 7 (7,8,10,16,20,22,26) | Fixed | 1.31 (1.06–1.61) | 0 | 0.802 | 0.012 |
| Asian | 2 (18,28) | Fixed | 1.76 (1.16–2.66) | 0 | 0.695 | 0.008 |
| Non-Asian | 11 (7–10,15–17,20,22,23,26) | Fixed | 1.25 (1.07–1.45) | 0 | 0.44 | 0.005 |
| FISH analysis | 9 (7,8,15–17,20,22,23,28) | Fixed | 1.37 (1.16–1.61) | 0 | 0.621 | <0.001 |
| qPCR analysis | 2 (9,10) | Fixed | 1.04 (0.75–1.44) | 0 | 0.439 | 0.621 |
| Large study (n.300) | Fixed | 1.27 (1.01–1.60) | 0 | 0.052 | 0.042 | |
| Disease-free survival | ||||||
| Overall | 4 (7,8,20,28) | Random | 1.43 (0.96–2.12) | 54.3 | 0.087 | 0.075 |
| NSCLC only | 1 (8) | – | 1.46 (0.76–2.81) | – | – | .0.05 |
| SCC only | 4 (7,8,16,28) | Random | 1.37 (0.89–2.09) | 56.5 | 0.075 | 0.152 |
Note: P-value for heterogeneity, based on Q-test.
Abbreviations: NSCLC, non-small-cell lung cancer; HR, hazard ratio; CI, confidence interval; SCC, squamous cell carcinoma; FISH, fluorescence in situ hybridization; qPCR, quantitative polymerase chain reaction.
The impact of FGFR1 amplification on disease-free survival in NSCLC patients
Four studies reported the relationship between FGFR1 amplification and DFS in NSCLC patients. Pooled data from all four studies showed that FGFR1 amplification was not significantly correlated with poor DFS, with a pooled-estimate HR of 1.43 (95% CI 0.96–2.12, P=0.075) and significant heterogeneity in the data (I2=54.3%, P=0.087). The combined HR was 1.37 (95% CI 0.89–2.09) based on the four studies of SCC, which also demonstrated a nonsignificant association between FGFR1 amplification and SCC patients’ DFS (Figure 3B).
Publication bias
We constructed funnel plots and performed Egger’s tests to assess publication bias. As a result, we observed evidence for publication bias (P=0.044 for Begg’s test) for OS in all NSCLC patients, and the funnel plot was not symmetrical (Figure 2D). This might be a limitation for our analysis, because studies with negative findings, especially those with small sample sizes, were less likely to be published. By using the trim-and-fill method, we showed that if the publication bias were the only source of the funnel-plot asymmetry, we required three more studies to balance the funnel plot (Figure 2E). The adjusted risk estimate was attenuated, but the remaining HR was significant (1.22, 95% CI 1.06–1.40; P<0.05), indicating the stability of our results. However, no obvious publication bias was detected by either Begg’s test (P=0.55) or Egger’s test (P=0.91) for OS in SCC patients. For progression-free survival PFS survival, Egger’s test indicated that there was no evidence of significant publication bias after assessing the funnel plot for the studies included in our meta-analysis (P=0.91 for Begg’s test, Figure 3B).
Discussion
In the present study, we collected all available, published articles and performed a meta-analysis to examine the association between FGFR1 amplification and clinicopathological characteristics. Eighteen studies were critically reviewed to clarify the controversial results from previous reports. Our meta-analysis showed that FGFR1 amplification was enriched in males, smokers, and SCC patients. FGFR1 amplification was significantly associated with poor OS in NSCLC patients (HR 1.30, 95% CI 1.13–1.50; P<0.001), and the association did not vary by ethnicity. Despite higher HR values for DFS in FGFR1-amplification patients, the difference was not significant (HR 1.43, 95% CI 0.96–2.12; P>0.05). These findings might be important for the prognosis and treatment of NSCLC patients, in addition to improving the understanding of FGFR1 molecular biology in NSCLC patients.
As a member of the FGFR family, FGFR1 has been studied in many human tumors, and has been found to be amplified or overexpressed in clinical tumor samples from NSCLC patients, especially SCC patients.27,34 Accumulating evidence suggests that FGFR1 plays an essential and active role in tumor-cell proliferation, angiogenesis, migration, and survival, and increased FGFR1 amplification is currently recognized as the predictive biomarker for preselected patients with SCCs for entry into clinical trials of the FGFR-specific tyrosine-kinase inhibitors.15 Our findings provide a clue as to how to select suitable patients with NSCLC for anti-FGFR therapy, and more suitable and cost-effective detection methods should be established.
In an article by Yang et al, FGFR1 amplification did not significantly influence the prognosis of SCC patients, even though the subgroup analysis found poor NSCLC prognoses among Asian patients.35 The authors also found that although FGFR1 amplification was significantly more prevalent in SCC patients, it was not a prognosis marker in NSCLC patients. Compared with this previous meta-analysis, the present study is much larger, and includes over four times as many cancer cases as the earlier study. Our analysis also comprehensively reviewed clinicopathologic features. In addition, several subgroup analyses were conducted to identify potential sources of heterogeneity. Heterogeneity for the HRs in NSCLC was observed among the studies. This heterogeneity may be due to various factors, such as diversity in the population characteristics, differences in the detection methods, and differences in the cutoff levels to determine FGFR1.
There were also several limitations to our study. First, HRs calculated from data or extracted from survival curves might be less reliable than a direct analysis of variance. Second, a standardized reading-and-evaluation strategy and evaluation criteria for amplification must be established. Third, the sample size in PFS studies was not sufficient to detect a significant difference between FGFR1 amplification and PFS. Third, we only enrolled suitable English-language literature reports, potentially introducing bias due to the language criteria.
The present meta-analysis shows that FGFR1 amplification is significantly associated with sex, smoking, and histology. FGFR1 amplification patients had poorer OS, indicating that FGFR1 may be a marker for poor prognosis in NSCLC patients, and is a promising therapeutic target. Large, well-designed prospective studies are required to investigate the precise prognostic significance and clinicopathologic differences of FGFR1 amplification.
Acknowledgments
This study was supported by the China Postdoctoral Science Foundation (2012M521189), the Zhejiang Provincial Postdoctoral Science Foundation (Bsh1202064), the National Natural Science Foundation of China (81172081), and the Zhejiang Provincial Medical and Health Science Technology Project (2015119812). The abstract of this paper was presented at the 2015 ASCO Annual Meeting as a poster presentation with interim findings. The poster’s abstract was published in “Poster abstracts” in the Journal of Clinical Oncology (2015;33 Suppl:e22188; http://meetinglibrary.asco.org/content/149309-156). The actual paper, however, has never been published.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Liu Z, Sun Y, et al. Discovery and identification of new non-ATP competitive FGFR1 inhibitors with therapeutic potential on non-small-cell lung cancer. Cancer Lett. 2014;344:82–89. doi: 10.1016/j.canlet.2013.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Malchers F, Dietlein F, Schöttle J, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 2014;4:246–257. doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 7.Kim HR, Kim DJ, Kang DR, et al. Fibroblast growth factor receptor 1 gene amplification is associated with poor survival and cigarette smoking dosage in patients with resected squamous cell lung cancer. J Clin Oncol. 2013;31:731–737. doi: 10.1200/JCO.2012.43.8622. [DOI] [PubMed] [Google Scholar]
- 8.Cihoric N, Savic S, Schneider S, et al. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer. 2014;110:2914–2922. doi: 10.1038/bjc.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gadgeel SM, Chen W, Cote ML, et al. Fibroblast growth factor receptor 1 amplification in non-small cell lung cancer by quantitative real-time PCR. PLoS One. 2013;8:e79820. doi: 10.1371/journal.pone.0079820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasaki H, Shitara M, Yokota K, et al. Increased FGFR1 copy number in lung squamous cell carcinomas. Mol Med Rep. 2012;5:725–728. doi: 10.3892/mmr.2011.715. [DOI] [PubMed] [Google Scholar]
- 11.André F, Bachelot T, Campone M, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 12.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 14.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res. 2014;20:3299–3309. doi: 10.1158/1078-0432.CCR-13-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russell PA, Yu Y, Young RJ, et al. Prevalence, morphology, and natural history of FGFR1-amplified lung cancer, including squamous cell carcinoma, detected by FISH and SISH. Mod Pathol. 2014;27:1621–1631. doi: 10.1038/modpathol.2014.71. [DOI] [PubMed] [Google Scholar]
- 17.Toschi L, Finocchiaro G, Nguyen TT, et al. Increased SOX2 gene copy number is associated with FGFR1 and PIK3CA gene gain in non-small cell lung cancer and predicts improved survival in early stage disease. PLoS One. 2014;9:e95303. doi: 10.1371/journal.pone.0095303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serizawa M, Koh Y, Kenmotsu H, et al. Assessment of mutational profile of Japanese lung adenocarcinoma patients by multitarget assays: a prospective, single-institute study. Cancer. 2014;120:1471–1481. doi: 10.1002/cncr.28604. [DOI] [PubMed] [Google Scholar]
- 19.Pros E, Lantuejoul S, Sanchez-Verde L, et al. Determining the profiles and parameters for gene amplification testing of growth factor receptors in lung cancer. Int J Cancer. 2013;133:898–907. doi: 10.1002/ijc.28090. [DOI] [PubMed] [Google Scholar]
- 20.Craddock KJ, Ludkovski O, Sykes J, Shepherd FA, Tsao MS. Prognostic value of fibroblast growth factor receptor 1 gene locus amplification in resected lung squamous cell carcinoma. J Thorac Oncol. 2013;8:1371–1377. doi: 10.1097/JTO.0b013e3182a46fe9. [DOI] [PubMed] [Google Scholar]
- 21.Tran TN, Selinger CI, Kohonen-Corish MR, et al. Fibroblast growth factor receptor 1 (FGFR1) copy number is an independent prognostic factor in non-small cell lung cancer. Lung Cancer. 2013;81:462–467. doi: 10.1016/j.lungcan.2013.05.015. [DOI] [PubMed] [Google Scholar]
- 22.Heist RS, Mino-Kenudson M, Sequist LV, et al. FGFR1 amplification in squamous cell carcinoma of the lung. J Thorac Oncol. 2012;7:1775–1780. doi: 10.1097/JTO.0b013e31826aed28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohler LH, Mireskandari M, Knösel T, et al. FGFR1 expression and gene copy numbers in human lung cancer. Virchows Arch. 2012;461:49–57. doi: 10.1007/s00428-012-1250-y. [DOI] [PubMed] [Google Scholar]
- 24.Schildhaus HU, Heukamp LC, Merkelbach-Bruse S, et al. Definition of a fluorescence in-situ hybridization score identifies high- and low-level FGFR1 amplification types in squamous cell lung cancer. Mod Pathol. 2012;25:1473–1480. doi: 10.1038/modpathol.2012.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–6667. doi: 10.1158/1078-0432.CCR-12-2694. [DOI] [PubMed] [Google Scholar]
- 26.Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutt A, Ramos AH, Hammerman PS, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS One. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seo AN, Jin Y, Lee HJ, et al. FGFR1 amplification is associated with poor prognosis and smoking in non-small-cell lung cancer. Virchows Arch. 2014;65:547–558. doi: 10.1007/s00428-014-1634-2. [DOI] [PubMed] [Google Scholar]
- 29.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos AH, Dutt A, Mermel C, et al. Amplification of chromosomal segment 4q12 in non-small cell lung cancer. Cancer Biol Ther. 2009;8:2042–2050. doi: 10.4161/cbt.8.21.9764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sos ML, Michel K, Zander T, et al. Predicting drug susceptibility of non-small cell lung cancers based on genetic lesions. J Clin Invest. 2009;119:1727–1740. doi: 10.1172/JCI37127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beroukhim R, Mermel CH, Porter D, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Göke F, Franzen A, Menon R, et al. Rationale for treatment of metastatic squamous cell carcinoma of the lung using fibroblast growth factor receptor inhibitors. Chest. 2012;142:1020–1026. doi: 10.1378/chest.11-2943. [DOI] [PubMed] [Google Scholar]
- 35.Yang W, Yao YW, Zeng JL, et al. Prognostic value of FGFR1 gene copy number in patients with non-small cell lung cancer: a meta-analysis. J Thorac Dis. 2014;6:803–809. doi: 10.3978/j.issn.2072-1439.2014.05.02. [DOI] [PMC free article] [PubMed] [Google Scholar]