Abstract
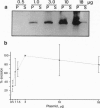
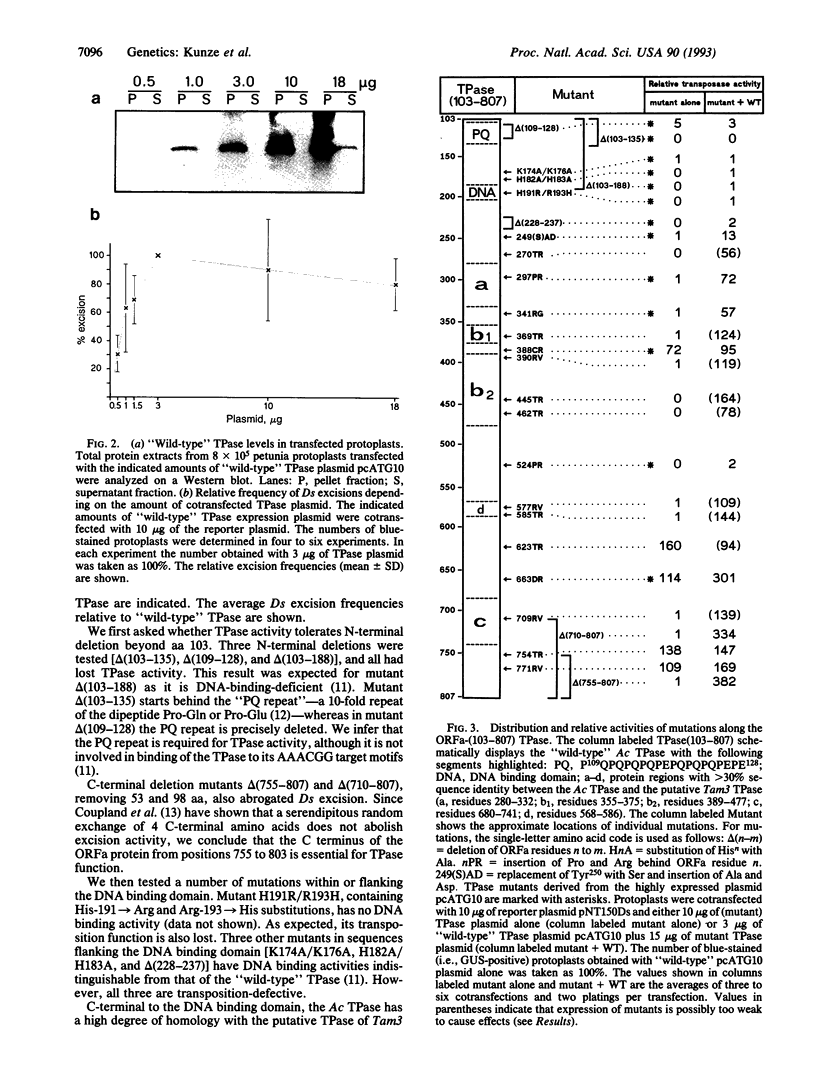
The maize transposable element Activator (Ac) encodes a transposase (TPase) protein, whose DNA-binding domain is located in a basic region around aa 200. The N-terminal 102 aa of the TPase are not required for the transposition reaction. In transfected petunia protoplasts, we analyzed the protein levels of the N-terminally truncated TPase and mutants thereof and the corresponding transposition frequencies. The TPase protein forms large insoluble aggregates at high expression levels. There is no proportionality observed between TPase levels and transposition frequency. Twenty-one mutations (of 26), which are distributed over the whole length of the protein, inactivate the TPase completely. By coexpressing inactive mutant and active truncated TPase, it was found that several mutations have a trans-dominant inhibitory effect. Among those are two DNA-binding-deficient mutants, indicating that inhibition of the active TPase is not caused by competition for the binding sites on the transposon. Accordingly, Ac TPase acts as an oligo- or multimer formed by protein-protein interactions. Peculiarly, two mutants lacking 53 and 98 aa from the C terminus that are themselves transpositionally inactive lead to an increased excision frequency when they are coexpressed with the active truncated TPase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker B., Coupland G., Fedoroff N., Starlinger P., Schell J. Phenotypic assay for excision of the maize controlling element Ac in tobacco. EMBO J. 1987 Jun;6(6):1547–1554. doi: 10.1002/j.1460-2075.1987.tb02399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D., Lütticke R., Li M., Starlinger P. Control of excision frequency of maize transposable element Ds in Petunia protoplasts. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5552–5556. doi: 10.1073/pnas.89.12.5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D. M., Jackson M. S., Kidwell M. G., Dover G. A. KP elements repress P-induced hybrid dysgenesis in Drosophila melanogaster. EMBO J. 1987 Dec 20;6(13):4125–4135. doi: 10.1002/j.1460-2075.1987.tb02758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvi B. R., Hong T. J., Findley S. D., Gelbart W. M. Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell. 1991 Aug 9;66(3):465–471. doi: 10.1016/0092-8674(81)90010-6. [DOI] [PubMed] [Google Scholar]
- Coupland G., Baker B., Schell J., Starlinger P. Characterization of the maize transposable element Ac by internal deletions. EMBO J. 1988 Dec 1;7(12):3653–3659. doi: 10.1002/j.1460-2075.1988.tb03246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmar S., Kunze R. The ORFa protein, the putative transposase of maize transposable element Ac, has a basic DNA binding domain. EMBO J. 1991 Dec;10(13):4003–4010. doi: 10.1002/j.1460-2075.1991.tb04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusswinkel H., Schein S., Courage U., Starlinger P., Kunze R. Detection and abundance of mRNA and protein encoded by transposable element activator (Ac) in maize. Mol Gen Genet. 1991 Feb;225(2):186–192. doi: 10.1007/BF00269846. [DOI] [PubMed] [Google Scholar]
- Hehl R., Nacken W. K., Krause A., Saedler H., Sommer H. Structural analysis of Tam3, a transposable element from Antirrhinum majus, reveals homologies to the Ac element from maize. Plant Mol Biol. 1991 Feb;16(2):369–371. doi: 10.1007/BF00020572. [DOI] [PubMed] [Google Scholar]
- Houba-Hérin N., Becker D., Post A., Larondelle Y., Starlinger P. Excision of a Ds-like maize transposable element (Ac delta) in a transient assay in Petunia is enhanced by a truncated coding region of the transposable element Ac. Mol Gen Genet. 1990 Oct;224(1):17–23. doi: 10.1007/BF00259446. [DOI] [PubMed] [Google Scholar]
- Isberg R. R., Lazaar A. L., Syvanen M. Regulation of Tn5 by the right-repeat proteins: control at the level of the transposition reaction? Cell. 1982 Oct;30(3):883–892. doi: 10.1016/0092-8674(82)90293-8. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Yin J. C., Reznikoff W. S. Control of Tn5 transposition in Escherichia coli is mediated by protein from the right repeat. Cell. 1982 Oct;30(3):873–882. doi: 10.1016/0092-8674(82)90292-6. [DOI] [PubMed] [Google Scholar]
- Kunze R., Stochaj U., Laufs J., Starlinger P. Transcription of transposable element Activator (Ac) of Zea mays L. EMBO J. 1987 Jun;6(6):1555–1563. doi: 10.1002/j.1460-2075.1987.tb02400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. G., Starlinger P. Mutational analysis of the N terminus of the protein of maize transposable element Ac. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6044–6048. doi: 10.1073/pnas.87.16.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCLINTOCK B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Misra S., Rio D. C. Cytotype control of Drosophila P element transposition: the 66 kd protein is a repressor of transposase activity. Cell. 1990 Jul 27;62(2):269–284. doi: 10.1016/0092-8674(90)90365-l. [DOI] [PubMed] [Google Scholar]
- Nitasaka E., Mukai T., Yamazaki T. Repressor of P elements in Drosophila melanogaster: Cytotype determination by a defective P element carrying only open reading frames 0 through 2. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7605–7608. doi: 10.1073/pnas.84.21.7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio D. C., Laski F. A., Rubin G. M. Identification and immunochemical analysis of biologically active Drosophila P element transposase. Cell. 1986 Jan 17;44(1):21–32. doi: 10.1016/0092-8674(86)90481-2. [DOI] [PubMed] [Google Scholar]
- Rio D. C. Molecular mechanisms regulating Drosophila P element transposition. Annu Rev Genet. 1990;24:543–578. doi: 10.1146/annurev.ge.24.120190.002551. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Engels W. R. Modified P elements that mimic the P cytotype in Drosophila melanogaster. Genetics. 1989 Dec;123(4):815–824. doi: 10.1093/genetics/123.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler H., Nevers P. Transposition in plants: a molecular model. EMBO J. 1985 Mar;4(3):585–590. doi: 10.1002/j.1460-2075.1985.tb03670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield S. R., Harrison K., Nurrish S. J., Jones J. D. Promoter fusions to the Activator transposase gene cause distinct patterns of Dissociation excision in tobacco cotyledons. Plant Cell. 1992 May;4(5):573–582. doi: 10.1105/tpc.4.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y., Ohtsubo E. Frameshifting is required for production of the transposase encoded by insertion sequence 1. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4609–4613. doi: 10.1073/pnas.86.12.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streck R. D., Macgaffey J. E., Beckendorf S. K. The structure of hobo transposable elements and their insertion sites. EMBO J. 1986 Dec 20;5(13):3615–3623. doi: 10.1002/j.1460-2075.1986.tb04690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]