Summary
Variation in amyloid structures profoundly influences a wide array of pathological phenotypes in mammalian protein conformation disorders and dominantly inherited phenotypes in yeast. Here, we describe, for the first time, naturally occurring, self-propagating, structural variants of a prion protein isolated from wild strains of the yeast Saccharomyces cerevisiae. Variants of the [RNQ+] prion propagating in a variety of wild yeast differ biochemically, in their intracellular distributions, and in their ability to promote formation of the [PSI+] prion. [PSI+] is an epigenetic regulator of cellular phenotype and adaptability. Strikingly, we find that most natural [RNQ+] variants induced [PSI+] at high frequencies and the majority of [PSI+] variants elicited strong cellular phenotypes. We hypothesize that the presence of an efficient [RNQ+] template primes the cell for [PSI+] formation in order to induce [PSI+] in conditions where it would be advantageous. These studies utilize naturally occurring structural variants to expand our understanding of the consequences of diverse prion conformations on cellular phenotypes.
Keywords: prion, amyloid, aggregation, variants, yeast, adaptation
Introduction
Prion proteins can adopt multiple infectious conformations from identical amyloidogenic polypeptides (Toyama & Weissman, 2011). Such remarkable structural plasticity can lead to the formation of various amyloid structures, or prion strains, termed conformational variants in yeast. Prion variants often confer diverse pathological phenotypes and transmissibility in mammals (Gambetti et al., 2003) and distinct heritable phenotypes in yeast (Liebman & Chernoff, 2012). Recently, amyloid-forming proteins associated with a variety of neurodegenerative diseases have been characterized as “prion-like” based on their ability to propagate via a seeding mechanism similar to that described for PrP, the mammalian prion protein. Similar to PrPSc, the causative agent of transmissible spongiform encephalopathies (Legname et al., 2006), multiple amyloid structures have been identified for several other amyloidogenic proteins, including Aβ, the proposed causative agent of Alzheimer's disease (Petkova et al., 2005), and α-synuclein (Heise et al., 2005), a major component of the Lewy bodies characteristic of Parkinson's disease. This conformational variation may explain at least some of the diversity in disease progression observed in these disorders.
The [PSI+] prion in Saccharomyces cerevisiae is perhaps the best-characterized example of a protein that is able to adopt a variety of amyloid conformations. [PSI+] is the aggregated form of the translation termination factor Sup35 and is transmitted as a dominantly inherited, epigenetic element to daughter cells (Ter-Avanesyan et al., 1994, Paushkin et al., 1996). In [PSI+] cells, Sup35 is sequestered into self-propagating amyloid, resulting in a loss-of-function and read-through of stop codons. Structural variants of [PSI+] can be distinguished phenotypically using sensitive reporters that depend on the efficiency with which Sup35 is recruited into prion aggregates (Serio & Lindquist, 2000). In contrast to the toxic amyloid associated with human protein misfolding disorders, the prion status of yeast can impact the fitness of the cell and may be advantageous or disadvantageous, depending on the environment (True & Lindquist, 2000, Eaglestone et al., 1999). Moreover, the shift to [PSI+] allows for the generation of phenotypic diversity without any genetic change (Halfmann et al., 2010).
Spontaneous formation of [PSI+] is dependent on the presence of the [RNQ+] prion (Stein & True, 2011). [RNQ+] is formed by ordered aggregation of Rnq1, a protein of unknown function (Sondheimer & Lindquist, 2000, Strawn & True, 2006). In cells harboring [RNQ+], soluble Sup35 is thought to directly interact with prion aggregates of Rnq1 in an inefficient, heterologous templating process that leads to the formation of self-propagating Sup35 aggregates (Derkatch et al., 2000, Derkatch et al., 2004, Liebman & Chernoff, 2012). Thus, this two prion system can regulate cellular phenotypes and adaptability.
Variants of [RNQ+] exhibit different biochemical properties and differ in their abilities to induce [PSI+] (Bradley et al., 2002, Huang et al., 2013, Kalastavadi & True, 2010) suggesting that the [RNQ+] variants represent distinct amyloid structures. While variants of [PSI+] and [RNQ+] have been generated and characterized in the laboratory, to date only variants of the mammalian prion protein, PrPC, are known to exist in natural settings. Naturally occurring prions have been isolated from wild yeast strains (Resende et al., 2003, Halfmann et al., 2012); however, it is unknown if wild populations can form or selectively propagate distinct prion structures. Here, we analyzed the structural variation of the [RNQ+] prion in strains that were previously collected from vineyards, clinical samples and breweries. These strains originated in diverse environments, and have likely evolved or adapted differently based on the stimuli they encountered. The ability to convert to a [PRION+] state, or, more specifically, a variety of [PRION+] states, may have been selected for in specific environments in the wild as a potentially adaptive mechanism. For the first time, we describe a spectrum of prion variants occurring naturally in wild yeast. Interestingly, all of the [RNQ+] variants that we characterized were efficient inducers of [PSI+], but differed in the subset of [PSI+] variants that formed. Further, we found that [RNQ+] structures can be modified after inducing, or interacting with, [PSI+] and in some cases, this interaction resulted in the loss of [RNQ+].
Results and Discussion
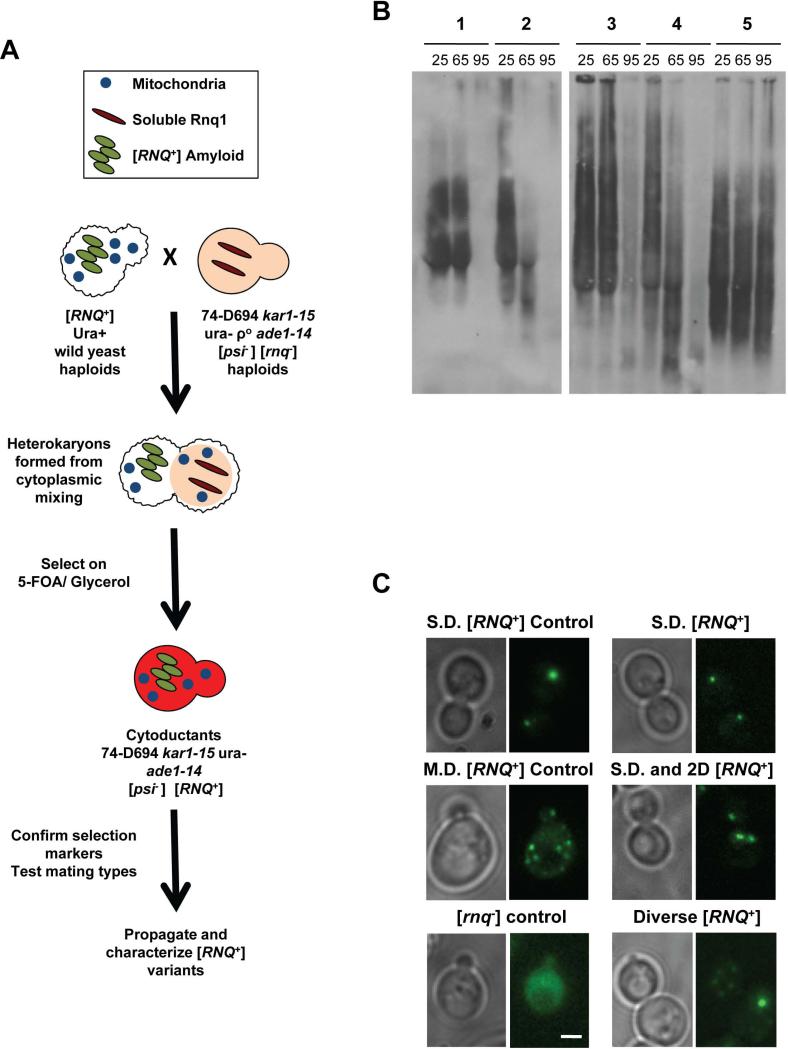
Four different [RNQ+] variants that spontaneously formed in the laboratory environment were previously characterized based on their biochemical, genetic and phenotypic properties (Bagriantsev & Liebman, 2004, Bradley et al., 2002). We used similar criteria to characterize the [RNQ+] prion from a subset of wild yeast originating from a variety of environments (Table 1). We chose eighteen [RNQ+] and four [rnq−] control wild yeast strains and first confirmed them for the presence or absence of [RNQ+] using two biochemical assays: fractionation of the Rnq1 protein to the pellet after high-speed centrifugation, and the presence of Rnq1 aggregates by semi-denaturing detergent agarose gel electrophoresis (SDD-AGE) (Kryndushkin et al., 2003). In order to control for genetic background and to take advantage of a phenotypic reporter allele, we performed cytoplasmic transfer (cytoduction) (Fig. 1a) of wild yeast donors into a [psi-] [rnq-] laboratory yeast strain and confirmed the presence of [RNQ+] by SDD-AGE (Supp. Fig. 1). The recipient strain, 74-D694, contains a Rnq1 sequence that is known to propagate a variety of [RNQ+] variants (Huang et al., 2013) and was used for all further analyses. Notably, in the sequences that we analyzed, we found surprisingly few polymorphisms when we compared the RNQ1 sequence in [RNQ+] haploids generated from the wild strains to that of the S288C reference strain (data not shown).
Table 1.
Wild yeast were isolated from a variety of natural environments
| Strain | Origin | Source |
|---|---|---|
| RM9-1B | vineyard | Mortimer |
| YJM326 | clinical isolate | McCusker/Fay |
| YSP163α | oak exudate (Pennsylvania) | Sniegowski/Fay |
| CLIB382 | super-attenuated beer, brewery (Ireland) | Kruglyak/Fay |
| RM5-1B | vineyard (Italy) | Mortimer |
| 12447-1-3a | unknown | Mortimer |
| FL100 | laboratory | Lacroute/Fay |
| YJM653 | clinical isolate | McCusker |
| Y-2209 | leaves from Lepidopterous (California) | USDA |
| F1643 | clinical wound | Fink |
| T73 | vineyard | Perez/Fay |
| Y-399 | fruit (cherries) | USDA |
| F1640 | clinical wound | Fink |
| YJM237 | laboratory S288c | McCusker |
| YJM431 | hybrid between Y55 and S288c lab strains | McCusker |
| RM7-1A | vineyard | Mortimer |
| CLIB324 | baker's yeast (Saigon, Vietnam) | Kruglyak/Fay |
| Y-27806 | clinical isolate | USDA |
| F1632 | clinical isolate | Fink |
| F1653 | clinical isolate | Fink |
| UCD939 | grapes, vineyard (Italy) | Bisson |
| UCD824 | commercial wine | Bisson |
Figure 1. [RNQ+] variants can be distinguished by differences in temperature sensitivity and by cellular aggregate distribution.
A. Schematic depicting cytoplasmic transfer of [RNQ+] from yeast of wild origin into the [psi−] [rnq−] strain 74-D694, as detailed in Materials and Methods. B. Cell lysates from 74-D694 yeast cytoduced with wild [RNQ+] prions were incubated for 7 minutes at 25°C, 65°C or 95°C. Lysates were then subjected to SDD-AGE and western blotting using an anti-Rnq1 antibody. Example yeast strains containing aggregates with different temperature sensitivities are shown and are listed as follows: 1) no breakdown at 65°C, eliminated at 95°C, 2) breakdown at 65°C, eliminated at 95°C, 3) no breakdown at 65°C, remain at 95°C, 4) 74-D694 Multi-Dot High [RNQ+] control: breakdown at 65°C, eliminated at 95°C, 5) 74-D694 Single-Dot High [RNQ+] control: no breakdown at 65°C, remain at 95°C. C. 74-D694 yeast that were cytoduced with wild [RNQ+] prions were transformed with the Rnq-GFP plasmid and imaged as detailed in Materials and Methods. Two 74-D694 cytoductants for each wild [RNQ+] strain were examined in three independent experiments. More than 100 cells were analyzed for each wild strain. Photos are Z stacks of a representative subset of cells. M.D.= multiple fluorescent puncta, 2.D.= two fluorescent puncta, S.D.= single fluorescent puncta. Scale bar in the lower right corner of the [rnq−] control= 2μm and applies to all panels.
The previously characterized laboratory [RNQ+] variants showed varying sensitivities to increased temperature (Bagriantsev & Liebman, 2004, Huang et al., 2013). To see if Rnq1 aggregates from wild yeast could be similarly differentiated, we assessed the stability of Rnq1 aggregates generated in wild yeast upon treatment with increasing temperature. We subjected lysates from 74-D694 yeast containing [RNQ+] from wild strains to three temperatures and assessed the breakdown and distribution of SDS-resistant Rnq1 aggregates by SDD-AGE (Supp. Fig. 2). We found that wild [RNQ+] variants can be distinguished by differences in sensitivity to increased temperature and segregated into one of three groups: 1) breakdown at 65°C, gone at 95°C, 2) no breakdown at 65°C, gone at 95°C, 3) no breakdown at 65°C, aggregates remain at 95°C. (Fig. 1b, Table 2). These results demonstrate that, similarly to [RNQ+] variants that formed spontaneously in the laboratory, Rnq1 aggregates from wild [RNQ+] yeast show different sensitivities to high temperatures.
Table 2.
[RNQ+] variants isolated from wild yeast exhibit diverse biochemical properties and aggregate distribution
| Strain | Temperature sensitivity of Rnq1 aggregates | Rnq1-GFP distribution |
|---|---|---|
| RM9-1B | [rnq] | diffuse |
| YJM326 | [rnq] | diffuse |
| YSP163a | [rnq] | diffuse |
| C1:0382 | [rnq] | diffuse |
| RM5-1B | Breakdown at 65°C, gone at 95°C | SD |
| 12447-1-3a | Breakdown at 65°C, gone at 95°C | SD |
| FL100 | Breakdown at 65°C, gone at 95°C | SD |
| YJM653 | Breakdown at 65°C, gone at 95°C | SD |
| Y-2209 | Breakdown at 65°C, gone at 95°C | SD |
| F1643 | Breakdown at 65°C, gone at 95°C | SD |
| T73 | Breakdown at 65°C, gone at 95°C | SD and 2D |
| Y-399 | Breakdown at 65°C, gone at 95°C | SD and 2D |
| F1640 | Breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| YJM237 | Breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| YJM431 | Breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| RM7-1A | Breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| CLIB324 | Breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| Y-27806 | No breakdown at 65°C, gone at 95°C | SD |
| F1632 | No breakdown at 65°C, gone at 95°C | SD |
| F1653 | No breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| UCD939 | No breakdown at 65°C, gone at 95°C | diverse: SD,2D,MD |
| UCD824 | No breakdown at 65°C, aggregates remain at 95°C | SD |
Lysates from 74-D694 yeast cells harboring the wild [RNQ+] variants from the yeast strains listed in column 1 were incubated at 25°C, 65°C or 95°C, and analyzed by SDD-AGE as detailed in Materials and Methods and Figure 1. Variants were categorized as listed in column 2. Rnq1-GFP was expressed in 74-D694 yeast cells harboring the wild [RNQ+] variants and live cells were analyzed by fluorescence microscopy. Variants were categorized according to the number of fluorescent puncta contained per cell in column 3. MD= multiple fluorescent puncta, 2D= two fluorescent puncta, SD= single fluorescent puncta. Four [rnq-] control 74-D694 strains showed only monomer when analyzed by SDD-AGE and had diffuse GFP fluorescence.
Another distinguishing feature of the [RNQ+] variants is variant-specific fluorescent distribution when a fusion of Rnq1 and GFP (Rnq-GFP) was expressed in cells containing [RNQ+] prions (Bradley & Liebman, 2003). Upon co-expression with Rnq-GFP, the laboratory-generated [RNQ+] variants used in this study consistently conform to either a ‘single dot’ (S.D.) pattern, containing one large focus, or a ‘multi-dot’ (M.D.) distribution, in which several small puncta are distributed throughout the cell (Fig. 1C). We sought to determine whether the putative structural differences of the wild [RNQ+] variants, based on their thermal stabilities, would correlate to differences in cellular Rnq1 aggregate distribution when co-expressed with the Rnq-GFP protein. We found that cells containing the various wild [RNQ+] prions exhibited a spectrum of aggregate distributions (Fig. 1C, Table 2). Moreover, these patterns deviated from the previously characterized S.D. and M.D. structures. Additionally, when we compared the temperature sensitivity of the Rnq1 aggregates to the Rnq-GFP distributions, we noted that these properties did not always correlate (Table 2). For example, 13 of the 18 of the wild [RNQ+] variants were sensitive to temperatures of 65°C and completely broken down after treatment at 95°C. However, these variants differed in the pattern of fluorescent puncta in the cell. These data suggest that wild yeast harbor an array of [RNQ+] variants and propagate a range of prion structures.
[PSI+] alters the efficiency of translation termination and can influence cellular adaptation and fitness. As such, naturally occurring [PSI+] might act as a mechanism to modify cellular phenotypes when they would be beneficial (True & Lindquist, 2000, Eaglestone et al., 1999, Halfmann et al., 2010). A rigorous analysis of spontaneous prion formation in wild yeast revealed that a minority of strains stably propagated the [PSI+] prion (Halfmann et al., 2012). Interestingly, all of these strains contained the [RNQ+] prion, indicating that [RNQ+] likely acts as a naturally occurring [PSI+] inducer in wild yeast.
The [RNQ+] amyloid structure impacts both the frequency of de novo [PSI+] formation and the specific [PSI+] variants that are formed in laboratory conditions (Bradley & Liebman, 2003, Sharma & Liebman, 2013). Furthermore, [RNQ+] variants in laboratory strains that show similar biochemical properties and cellular aggregate distributions are easily distinguished from each other by their abilities to induce [PSI+] (Bradley et al., 2002, Bradley & Liebman, 2003). Therefore, we sought to determine whether the naturally occurring wild [RNQ+] variants showed different [PSI+]-inducing capabilities.
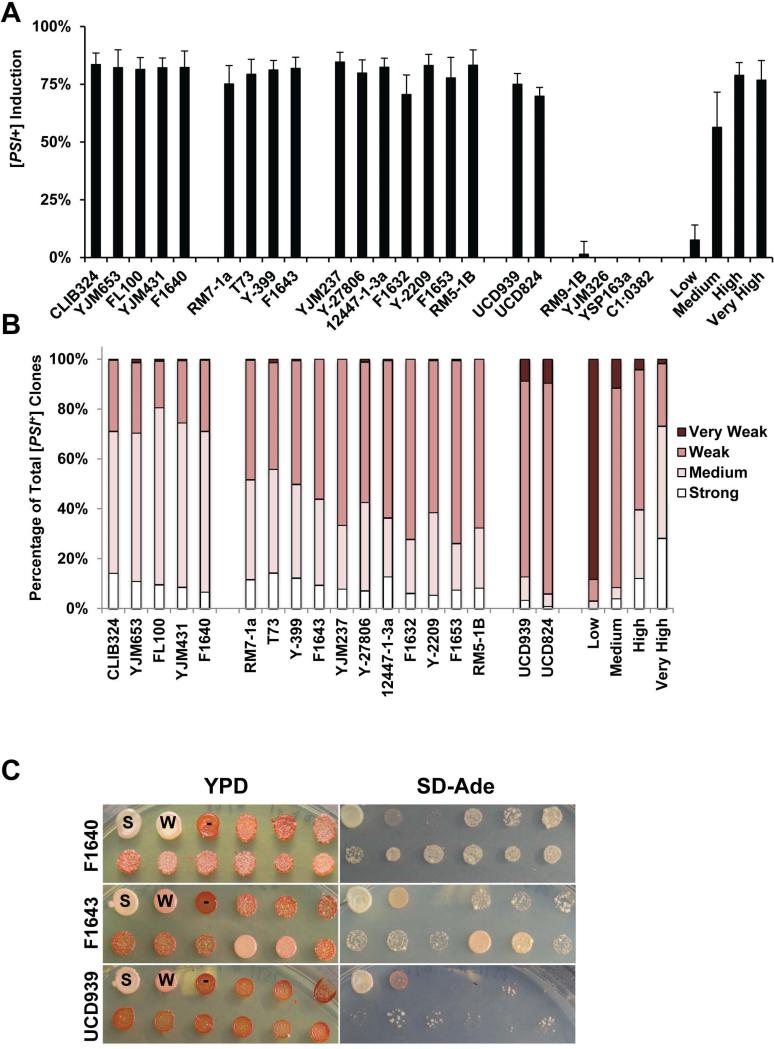
Full-length Sup35 was over-expressed in 74-D694 yeast containing [RNQ+] from the wild, and cells were analyzed for [PSI+] induction utilizing a nonsense mutation in the ADE1 gene (Liebman & Derkatch, 1999). This reporter is used to phenotypically monitor the [PSI+] state using a colorimetric assay and the ability to grow on media lacking adenine (see Materials and Methods). Interestingly, we found that all of the [RNQ+] prions from the wild yeast showed similar levels of [PSI+] induction of 75-90% upon Sup35 over-expression (O.E.) (Fig. 2A). The consistency in [PSI+] induction was unexpected considering the different biochemical properties and aggregate distribution characteristics we observed (Fig. 1). This high level of [PSI+] induction was similar to the ‘high’ and ‘very high’ [RNQ+] variants previously characterized, indicating that the majority of wild yeast harbor an efficient template for the cross-seeding of Sup35. The various wild donor strains likely evolved differentially based on their extracellular environments and stressors they encounter. We hypothesize that an effective [RNQ+] template may have been selected for as an evolutionarily adaptive mechanism in order to more readily promote de novo [PSI+] formation in conditions where it would be beneficial.
Figure 2. [RNQ+] prions from wild yeast induce [PSI+] at levels similar to a previously-characterized high [RNQ+] variant.
A. For each wild strain donor, Sup35 was O.E. in two different 74-D694 [RNQ+] cytoductants to induce [PSI+]. Colonies were scored as [PSI+] as detailed in Materials and Methods. Experiments were repeated at least three times and at least 300 colonies were scored for each clone, per experiment. Four wild [rnq−] control cytoductant strains, which lacked Rnq1 aggregates by SDD-AGE (Supplemental Figure 2), never induced [PSI+], indicating that the cytoduction protocol did not readily generate spontaneous [RNQ+]. B. [PSI+] colonies were isolated and re-grown on YPD, SD-Ade, and YPD + 3mM GdnHCl solid media, for verification and scoring of [PSI+] variants. Between 200 and 600 [PSI+] colonies from 3 separate experiments were isolated for each wild strain donor and scored for the strength of the [PSI+] variant. C. Example spotting plates of yeast with one representative strain from each of the three groups. Controls are the first three spots of the top row: strong [PSI+] (S), weak [PSI+] (W) and [psi−] (-).
[PSI+] variants differ in the efficiency with which they recruit soluble Sup35 into aggregates, which modulates the read-through of stop codons in messenger RNA (Cox et al., 1988, Uptain et al., 2001, Patino et al., 1996). “Strong” prion variants recruit monomeric protein more effectively into the aggregated state than “weak” variants, and thereby generate a correspondingly stronger nonsense suppression phenotype. We suggest that selective formation and propagation of specific [PSI+] variants could act as a cellular mechanism to regulate translational efficiency, so we sought to characterize the effect of wild [RNQ+] variants on the variants of [PSI+] induced. We selected newly induced [PSI+] colonies and scored them for strength of the [PSI+] variant (Figure 2B). Although many of the wild [RNQ+] variants induced [PSI+] at similar frequencies, we found differences in the [PSI+] variants formed, and this differentiated the wild [RNQ+] variants from each other (Fig. 2B). These variants generally fell into one of 3 groups (Fig. 2C). [RNQ+] from wild strains YJM431, CLIB324, YJM653, FL100 and F1640 induced a range of [PSI+] variants with generally stronger phenotypes. Another group of wild [RNQ+] variants (RM7-1a, T73, F1643, Y-399, 12447-1-3a, Y-27806, F1632, Y-2209, YJM237, F1653, and RM5-1B) induced [PSI+] variants of intermediate strength, where strong [PSI+] was rare, and approximately 50% or more of the clones analyzed were characterized as weak or very weak [PSI+]. In the final group, which contained [RNQ+] variants from strains UCD939 and UCD824, approximately 90% of the [PSI+] variants that formed were weak or very weak. These results indicate that the distribution and strength of newly-induced [PSI+] variants more effectively distinguishes [RNQ+] variants than the level of [PSI+] induction.
In addition to the wild strains that naturally harbor [RNQ+], but not [PSI+], we acquired wild strains that were previously characterized as [RNQ+] [PSI+] (Halfmann et al., 2012). In two of these strains, [PSI+] was extremely unstable and, upon cytoduction of UCD939 and UCD824 into our 74-D694 lab strain recipient we failed to isolate pink colonies or clones that were [PSI+] when analyzed using our biochemical assays. Importantly, all of the cytoductants we isolated were [RNQ+] by SDD-AGE analysis, confirming successful cytoplasmic transfer (Supp. Fig. 1). In performing the same analysis as above, we found that, upon Sup35 O.E., the newly induced [PSI+] variants were mostly weak [PSI+] (Fig. 2B,C). Weak [PSI+] variants show decreased mitotic stability as compared to stronger [PSI+] variants (Derkatch et al., 1997, Derkatch et al., 1996). Therefore, the [PSI+] variants that were initially identified in these strains (Halfmann et al., 2012) were also likely very weak, and thus readily lost. These results suggest that the [PSI+] variants that we induced by transient O.E. of Sup35 likely correlate with the naturally existing [PSI+] variants.
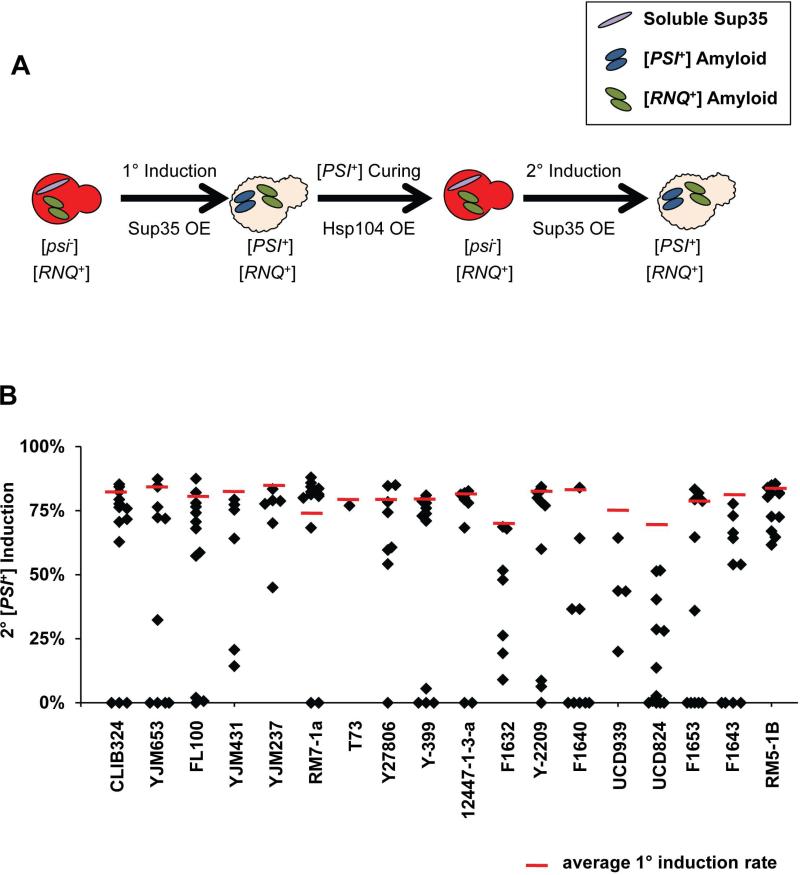
[PSI+] induction is proposed to occur through a templating process whereby soluble Sup35 protein interacts with Rnq1 aggregates in [RNQ+] cells to form [PSI+] (Stein & True, 2011). Previously, incompatibilities between select variants of [PSI+] and [RNQ+] have been reported, indicating that the prion proteins likely interact both before and after the conversion into the prion state (Bradley & Liebman, 2003). We were interested in the impact of the putative [RNQ+]-[PSI+] interaction on the wild [RNQ+] variant template. Because the ability to induce [PSI+] is the most sensitive way to differentiate between structurally-distinct [RNQ+] variants, we characterized the post-[PSI+] induction [RNQ+] variants by first curing [PSI+], followed by [PSI+] re-induction (Materials and Methods, Fig. 3A).
Figure 3. Secondary [PSI] induction can be modified by prior [PSI+] induction.
A. Schematic depicting the protocol for the 2° induction of [PSI+], as detailed in Materials and Methods. B. After curing [PSI+], [PSI+] was re-induced by O.E. of Sup35. Cells were scored for the [PSI+] variant using the colorimetric readout described in Materials and Methods. The dashed red line represents the level of primary [PSI+] induction. Each marker represents the [PSI+] induction value of one re-induced clone.
We found that previously being [PSI+] induced striking changes in the ability of [RNQ+] to promote re-induction of [PSI+]. In fact, only three [RNQ+] variants (YJM237, RM5-1B, and T73) always retained the ability to induce [PSI+] (Fig. 3B). Additionally, in several of the strains, a subset of clones exhibited lower frequencies of [PSI+] induction than those seen during the primary induction, or failed to induce [PSI+] at all. We hypothesize that the interaction of Rnq1 aggregates with Sup35 during [PSI+] induction modified the [RNQ+] template, resulting in altered [PSI+]-induction capabilities in a subset of clones. A transient, or more permanent, alteration to [RNQ+] may allow for a greater level of control by [RNQ+] on [PSI+]. Moreover, differential effects on [RNQ+] structures may increase cellular fitness and broaden [PSI+]-mediated adaptatability. Thus, this two-prion system provides for a greater range of diversity of prion structures in the cellular population, which may act to control a variety of phenotypes.
Naturally occurring yeast prion variants have not been described to date (Wickner et al., 2009). Our current results demonstrate, for the first time, that variants of the [RNQ+] prion naturally exist in the wild. Moreover, we show that the majority of [RNQ+] variants effectively and differentially induce [PSI+]. Although the function of the Rnq1 protein has not been defined, it is clear that the [RNQ+] prion serves as a critical regulator of [PSI+] formation. As [PSI+] formation modulates translation termination, these effects can dramatically influence the expression of a variety of proteins with diverse cellular functions. It would be reasonable to predict that [PSI+]-induced changes to global translation termination would be detrimental to cells (McGlinchey et al., 2011). Interestingly, the presence of Sup35 aggregates can be either advantageous or disadvantageous, depending on the cellular environment (Eaglestone et al., 1999, Halfmann et al., 2012, True et al., 2004, True & Lindquist, 2000). In fact, certain stressful growth conditions actually increase prion formation (Chernoff, 2007, Derkatch et al., 2001, Newnam et al., 2011, Tyedmers et al., 2008). Moreover, prions provide an example of how changes in protein conformation can alter cellular phenotypes and subsequently generate phenotypic diversity, without any permanent genetic change. Prion variants serve as an additional level of epigenetic cellular control, offering a gradient of phenotypes, rather than simply a [PRION+] or [prion−] state. The resulting continuum of structural and phenotypic variation might impact cellular fitness in response to different environmental stressors to generate a range of adaptive phenotypes to facilitate evolutionarily advantageous change.
Experimental Procedures
Strains, Plasmids and Cultivation Procedures
Yeast cells were grown and manipulated using established techniques (Guthrie & Fink, 1991). The strain 74-D694 (MATA ade1-14 trp1-289 his3Δ-200 ura3-52 leu2-3,112) of Saccharomyces cerevisiae was used as a control for all experiments (Chernoff et al., 1995). Isogenic 74-D694 strains harboring the low, medium, high and very high [RNQ+] variants were kindly provided by Dr. Susan Liebman (Bradley et al., 2002). Wild yeast were received from a variety of sources (see Table 1). The strain 74-D694 (MATA or alpha ade1-14 trp1-289 his3Δ-200 ura3-52 leu2-3,112 kar1-15 ρ) was utilized as a recipient for cytoplasmic transfer from wild yeast.
Untransformed cells were grown in rich media, which is composed of 1% yeast extract/ 1% peptone/ 2% dextrose (YPD). For [PSI+] induction experiments, we utilized the pEMBL-SUP35 plasmid (pSup2) which contains a URA3 marker and SUP35 under the control of its native promoter (Ter-Avanesyan et al., 1993). A plasmid with a URA3 selection marker containing the coding region of HSP104 under the GPD promoter (pGPD-104) was used to cure [PSI+] cells (Mumberg et al., 1995). A plasmid containing the coding region of Rnq1 under the CUP1 promoter (pCUP-GFP RNQ) with a URA3 selection marker was used to label RNQ1 aggregates (Sondheimer & Lindquist, 2000). Cells transformed with either pSup2, pGPD-104 or pRS316-CUP1 GFP-RNQ1 were grown in synthetic media lacking uracil (SD-Ura) and containing 2% glucose, to maintain selection of the plasmid. YPD media supplemented with 3 mM guanidine hydrochloride (YPD + GdnHCl) was used to inhibit Hsp104 activity and cure both [PSI+] and [RNQ+] prions (Tuite et al., 1981). In order to assess the [PSI+] phenotype (nonsense suppression of ade1-14), cells were grown on solid synthetic media lacking adenine (SD-Ade) and containing 2% glucose.
Cytoduction
Transfer of [RNQ+] into the 74-D694 laboratory yeast strain using cytoplasmic mixing experiments (cytoduction) was performed as previously described (Halfmann et al., 2012). Haploids of wild yeast donor strains were generated by disrupting the HO locus with a KanMX4 antibiotic-resistance marker, in diploid wild yeast. Diploids were sporulated, and haploids in which HO was disrupted were selected for by plating cells on YPD plates containing Geneticin and confirmed to be haploids by mating type tests. Haploid wild yeast clones (donors) were mixed with 74-D694 ura- respiration deficient (ρ), kar1-15, [rnq−] cells (recipients) on solid YPD medium and grown for 6 hours at 30°C. A swipe of the cell mixture was re-plated onto solid media containing 5-Fluororotic acid (5-FOA), which inhibits the growth of cells expressing URA3 or URA5 (Sikorski & Boeke, 1991), and placed at 30°C for ~40 hours. Cells that grew on 5-FOA media were presumed to contain the recipient nuclei and were struck on YP-Glycerol plates to select for cells containing functional mitochondria, acquired from the cytoplasm of the wild donors. Cells that grew on YPD, 5-FOA and YP-Glycerol solid media but did not grow on SD-Ura were tested for the presence of [RNQ+] using SDD-AGE. Two independent [RNQ+] clones for each donor strain were selected and both were tested in all further assays.
[PSI+] Induction Frequency
74-D694 yeast containing [RNQ+] prions from wild yeast and 74-D694 yeast cells containing four previously characterized [RNQ+] variants (Bradley et al., 2002) were transformed with pSup2, spread on solid SD-Ura medium and incubated for 4 days at 30°C. Approximately 5-10 Ura+ colonies were used to inoculate 4mls of SD-Ura selection media, and cultures were grown for 16 hours at 30°C with rotation. 300μl of diluted culture (1:8,000 in water) were spread using glass beads onto 150mm YPD solid media. Plates were incubated at 30°C for 5 days, and then moved to 4°C overnight for color development. [PSI+] induction was quantified as previously described (Huang et al., 2013). In each experiment, [PSI+] status was assessed in at least 300 colonies for each strain, using a colorimetic assay which depends on the read-through of the ade1-14 mutant allele. This [PSI+]-mediated nonsense suppression prevents the accumulation of a red pigment, resulting in colonies to be white or pink. Petite yeast are beige and easily distinguishable from [PSI+] or [psi−] yeast with functional mitochondria, and were excluded from analyses and further characterization. Each experiment was repeated at least 3 times.
Scoring of [PSI+] Phenotypes
[PSI+] colonies that were completely white or pink, or contained white or pink sectors, were spotted onto YPD, SD-Ade and YPD + 3mM GdnHCl using a pinning device. YPD and YPD + 3mM GdnHCl plates were grown for 3 days, and SD-Ade plates were grown for 6 days, at 30°C. Plates were then moved to 4°C overnight for development of the red pigment. Clones that were completely red on YPD + 3mM GdnHCl were scored for the strength of the [PSI+] phenotype primarily using the degree of cell growth on SD-Ade plates. Non-[PSI+] nonsense suppressors are not readily cured by GdnHCl. These clones represented less than 1% of the total, were readily distinguishable and were not included in the data analyses.
SDD-AGE
74-D694 yeast containing the wild [RNQ+] variants, or one of the four [RNQ+] variants that were previously characterized (Bradley et al., 2002), were grown overnight at 30° with agitation in 10mls rich media (YPD). Cells were lysed, using glass beads, in a buffer containing containing 25mM Tris (pH 7.5), 100mM NaCl, 10mM MgCl2, 1mM EDTA, 0.5mM DTT, 3mM phenylmethanesulfonylfluoride, 40mM N-Ethylmaleimide, and complete mini protease inhibitor (Roche). Cell lysates were normalized for the total amount of protein using the Bradford Assay. Equal amounts of total protein were incubated for 7 minutes at the indicated temperature in loading buffer (180mM Tris-HCl pH 6.8, 15% Glycerol, 6% SDS and bromophenol blue) and subsequently loaded onto 1.5% agarose gels containing 0.1% SDS. Prion aggregates were separated using SDD-AGE and subjected to western blotting as previously described (Kryndushkin et al., 2003). Rnq1 protein was detected with a polyclonal anti-Rnq1 antibody.
Secondary [PSI+] Induction
Newly induced [PSI+] colonies were spotted from solid YPD to solid media containing YPD, SD-Ade and YPD + 3mM GdnHCl. [PSI+] cells that were curable on YPD + 3mM GdnHCl and harboring an array of [PSI+] variants, were selected from YPD, resuspended in water and re-plated on media containing 5-FOA, to select for cells that had lost the pSup2 plasmid. Colonies were selected from 5-FOA and plated onto YPD and SD-Ura solid medium. Uracil auxotrophs were presumed to have lost the pSup2 plasmid. Clones that remained pink on YPD were selected, grown overnight in liquid YPD media, and transformed with pGPD-104 to cure [PSI+]. Colonies that were transformed with pGPD-104 were selected from SD-Ura solid media and struck onto YPD. Red colonies were selected from YPD and re-grown on 5-FOA solid media to select against cells containing pGPD-104. Red colonies were selected from 5-FOA and re-struck on YPD to confirm that they remained red and SD-Ura to confirm that they had lost the pGPD-104 plasmid. Clones that grew on 5-FOA, failed to grow on SD-Ura and that were red on YPD were transformed with pSup2 to re-induce [PSI+]. 5-10 Ura+ colonies were used to inoculate a 4ml culture of YPD and grown overnight at 30°C with rotation. Cultures were diluted 1:8,000 in sterile water, 300ul was spread onto 15cm solid YPD plates, plates were placed in a 30°C incubator for 5 days and then moved to 4°C for color development. Colonies containing pink or white were scored as [PSI+].
Microscopy
In order to visualize Rnq1 aggregates, yeast were transformed with pRS316 Rnq1 (153-405)-GFP under control of the CUP1 copper inducible promoter. Rnq1-GFP was previously shown to ‘decorate’ the Rnq1 aggregates in cells (Sondheimer & Lindquist, 2000), generating foci that can be visualized using fluorescence microscopy. Cells were grown overnight in 4mls of SDUra at 30°C with rotation for 16 hours. Cells were spun down, normalized to an OD600=0.2 and returned to 30°C. After 3 hours of growth, CuSO4 was added to a final concentration of 50μM and cells were placed back on the rotator at 30°C for ~2.5 hours prior to imaging. Cell cultures were spun down, and cell pellets were washed once with water and resuspended in phosphate buffered saline. 2μl of cells were placed directly onto VWR 3in × 1in, 1mm-thick slides, covered with fisherbrand No.#1.5 microscope coverslips and secured with nail polish. Live yeast cells were imaged at room temperature with a Zeiss Axiovert 200 Inverted Fluorescent Microscope outfitted with a Zeiss 100x/1.4 NA oil objective and collected using a Cascade 512B EMCCD camera (Photometrics. Tucson, AZ). Z-stacks of several focal planes were collected using an excitation of 488nm or phase filter, and each Z-stack was analyzed and digitally deconvoluted (using no neighbors setting) with Slidebook 5.0 (Intelligent Imaging Innovations).
Supplementary Material
Acknowledgements
This work was supported by the National Institutes of Health grant GM072778 (to H.L.T.) and by the Institutional Training Grant T32-AI007172 (L.W.). We thank members of the True lab for helpful discussions. We appreciate Kevin Stein and Kathryn Keefer's critical reading of the manuscript. We thank Dr. S. Liebman, Dr. Y. Chernoff, Dr. R. Halfmann, Dr. D. Jarosz, Dr. J. Fay, Dr. R. Mortimer, Dr. J. McCusker, Dr. P. Sniegowski, Dr. L. Kruglyak, Dr. J. Perez, Dr. F. Lacroute Dr. L. Bisson and Dr. G. Fink for yeast strains and plasmids.
Footnotes
Authors' contributions
LW carried out all of the experiments. LW and HLT conceived and designed the study and drafted the manuscript. Both authors read and approved the final manuscript.
Disclosures
The authors disclose that they have no competing financial interests. The authors declare that they have no conflicts of interest.
References
- Bagriantsev S, Liebman SW. Specificity of prion assembly in vivo. [PSI+] and [PIN+] form separate structures in yeast. J Biol Chem. 2004;279:51042–51048. doi: 10.1074/jbc.M410611200. [DOI] [PubMed] [Google Scholar]
- Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;30:30. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley ME, Liebman SW. Destabilizing interactions among [PSI(+)] and [PIN(+)] yeast prion variants. Genetics. 2003;165:1675–1685. doi: 10.1093/genetics/165.4.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO. Stress and prions: lessons from the yeast model. FEBS Lett. 2007;581:3695–3701. doi: 10.1016/j.febslet.2007.04.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast. 1988;4:159–178. doi: 10.1002/yea.320040302. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? Embo J. 2000;19:1942–1952. doi: 10.1093/emboj/19.9.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. Embo J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetti P, Kong Q, Zou W, Parchi P, Chen SG. Sporadic and familial CJD: classification and characterisation. Br Med Bull. 2003;66:213–239. doi: 10.1093/bmb/66.1.213. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. Academic Press, Inc.; San Diego: 1991. [Google Scholar]
- Halfmann R, Alberti S, Lindquist S. Prions, protein homeostasis, and phenotypic diversity. Trends Cell Biol. 2010;20:125–133. doi: 10.1016/j.tcb.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci U S A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang VJ, Stein KC, True HL. Spontaneous Variants of the [RNQ+] Prion in Yeast Demonstrate the Extensive Conformational Diversity Possible with Prion Proteins. PLoS One. 2013;8:e79582. doi: 10.1371/journal.pone.0079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalastavadi T, True HL. Analysis of the [RNQ+] prion reveals stability of amyloid fibers as the key determinant of yeast prion variant propagation. The Journal of biological chemistry. 2010;285:20748–20755. doi: 10.1074/jbc.M110.115303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J Biol Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Legname G, Nguyen HO, Peretz D, Cohen FE, DeArmond SJ, Prusiner SB. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc Natl Acad Sci U S A. 2006;103:19105–19110. doi: 10.1073/pnas.0608970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebman SW, Derkatch IL. The yeast [PSI+] prion: making sense of nonsense. J Biol Chem. 1999;274:1181–1184. doi: 10.1074/jbc.274.3.1181. [DOI] [PubMed] [Google Scholar]
- McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci U S A. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Newnam GP, Birchmore JL, Chernoff YO. Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol. 2011;408:432–448. doi: 10.1016/j.jmb.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science. 1996;273:622–626. doi: 10.1126/science.273.5275.622. [DOI] [PubMed] [Google Scholar]
- Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau WM, Mattson MP, Tycko R. Self-propagating, molecular-level polymorphism in Alzheimer's beta-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol. 2003;49:1005–1017. doi: 10.1046/j.1365-2958.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- Serio TR, Lindquist SL. Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol. 2000;10:98–105. doi: 10.1016/s0962-8924(99)01711-0. [DOI] [PubMed] [Google Scholar]
- Sharma J, Liebman SW. Exploring the Basis of [PIN(+)] Variant Differences in [PSI(+)] Induction. Journal of molecular biology. 2013;425:3046–3059. doi: 10.1016/j.jmb.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boeke JD. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Stein KC, True HL. The [RNQ+] prion: a model of both functional and pathological amyloid. Prion. 2011;5:291–298. doi: 10.4161/pri.5.4.18213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, True HL. Deletion of RNQ1 gene reveals novel functional relationship between divergently transcribed Bik1p/CLIP-170 and Sfi1p in spindle pole body separation. Curr Genet. 2006;50:347–366. doi: 10.1007/s00294-006-0098-6. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Dagkesamanskaya AR, Kushnirov VV, Smirnov VN. The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics. 1994;137:671–676. doi: 10.1093/genetics/137.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan MD, Kushnirov VV, Dagkesamanskaya AR, Didichenko SA, Chernoff YO, Inge-Vechtomov SG, Smirnov VN. Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol. 1993;7:683–692. doi: 10.1111/j.1365-2958.1993.tb01159.x. [DOI] [PubMed] [Google Scholar]
- Toyama BH, Weissman JS. Amyloid structure: conformational diversity and consequences. Annu Rev Biochem. 2011;80:557–585. doi: 10.1146/annurev-biochem-090908-120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. Epub 2004 Aug 2015. [DOI] [PubMed] [Google Scholar]
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi−] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI(+)] are distinguished by their efficiencies of prion- mediated conformational conversion. Embo J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner RB, Edskes HK, Shewmaker F, Kryndushkin D, Nemecek J. Prion variants, species barriers, generation and propagation. J Biol. 2009;8:47. doi: 10.1186/jbiol148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.