Abstract
Across bacterial species, metal binding proteins can serve functions in pathogenesis in addition to regulating metal homeostasis. We have compared and contrasted the activities of zinc (Zn2+)-binding lipoproteins AdcA and AdcAII in the Streptococcus pneumoniae TIGR4 background. Exposure to Zn2+-limiting conditions resulted in delayed growth in a strain lacking AdcAII (ΔAdcAII) when compared to wild type bacteria or a mutant lacking AdcA (ΔAdcA). AdcAII failed to interact with the extracellular matrix protein laminin despite homology to laminin-binding proteins of related streptococci. Deletion of AdcA or AdcAII led to significantly increased invasion of A549 human lung epithelial cells and a trend toward increased invasion in vivo. Loss of AdcAII, but not AdcA, was shown to negatively impact early colonization of the nasopharynx. Our findings suggest that expression of AdcAII affects invasiveness of S. pneumoniae in response to available Zn2+ concentrations.
Introduction
Streptococcus pneumoniae normally colonizes the human nasopharynx asymptomatically, but can also cause serious infections, including pneumonia, septicemia, and bacterial meningitis. Thus, the pneumococcus is able to adapt to a wide variety of host environments. An important variable in these diverse niches is the concentration of metals, such as Zn2+. Limitation of Zn2+ results in growth arrest and thus Zn2+ transport is critical to bacterial multiplication and, indirectly, to progression of disease. A key question is whether these metal transporters have additional functions such as directly affecting adherence to host cells or the extracellular matrix.
The LraI (lipoprotein receptor antigen I) family of lipoproteins represents one group of metal transporters. Previously characterized proteins within the LraI family, including the manganese-binding protein PsaA from S. pneumoniae, play roles in both adhesion and metal transport [1, 2]. Adhesion of pneumococci to nasopharyngeal epithelial cells is reduced by antibodies to PsaA, [3] and PsaA has been proven essential for virulence in animal models when inoculated intranasally [4].
AdcAII is homologous to members of the LraI family, several of which have been characterized in other gram-positive pathogenic species. Lmb expressed by Streptococcus agalactiae and Lsp or Lbp expressed by Streptococcus pyogenes are both known to confer laminin-binding properties that promote adhesion and invasion. In S. agalactiae, a mutant lacking Lmb shows decreased adherence to human laminin [5] and reduced invasion into human brain microvascular endothelial cells (HBMEC) [6]. S. pyogenes mutants lacking Lsp are defective in adhesion and invasion of epithelial cells in vitro and are also highly attenuated in a murine subcutaneous ulcer model [7, 8]. This raises the question of what role AdcAII plays in pneumococcal physiology.
Biophysical and structural studies have demonstrated that AdcAII of S. pneumoniae serves as a metal-binding receptor (MBR) for Zn2+, given the presence of a flexible binding loop commonly seen in other Zn2+-binding transporters [9]. Additional studies identified that AdcAII actually serves as a second Zn2+ transporter, along with AdcA of the AdcCBA operon [10], regulating intracellular levels of Zn2+ in pneumococcus [11]. Only recently, have the relative contributions of AdcA and AdcAII been investigated [12]. Crystal structure analysis indicated that both proteins contain a highly homologous Zn2+-binding domain but AdcA has a second Zn2+-binding domain, absent in AdcAII [13]. Both proteins transfer Zn2+ to the AdcBC transporter and have overlapping functions in Zn2+ homeostasis. However, it has been suggested that AdcAII may play a role in other areas of pathogenesis since histidine triad proteins, which are known to be involved in adherence, are encoded directly downstream [14, 15]. AdcAII and pneumococcal histidine triad proteins are upregulated during invasive pneumococcal disease, [16] and expression of adcAII and pht genes is affected by availability of Zn2+ [17, 18]. Additionally, transposon mutagenesis identified AdcAII, AdcB, and AdcC as being involved in colonization [19]. Fluctuations in available Zn2+ concentrations within a host could therefore serve to alter expression of these surface proteins, which in turn could have effects on the pathogen’s virulence.
Given the clinical significance of pneumococcal infections and previous work showing the importance of AdcAII homologs to virulence in other species, we investigated the relative contribution of AdcA and AdcAII to processes including adhesion and invasion. Here we show that AdcA and AdcAII mutants display distinctly different phenotypes during colonization and during growth in zinc-limiting environments.
Materials and Methods
Bacterial strains and growth conditions
Novablue strain E. coli (Merck KGaA, Darmstadt, Germany) was grown at 37°C in Luria-Bertani (LB) broth with agitation or on LB agar supplemented, when appropriate, with spectinomycin (50μg/mL). S. pneumoniae serotype 4 strains TIGR4 and the unencapsulated TIGR4 derivative T4R were grown on tryptic soy agar plates supplemented with 5% defibrinated sheep blood, Kaighn’s medium (F-12K; ATCC 3–2004) with 2mM L-glutamine and 1500mg/L sodium bicarbonate supplemented with 10% FBS, or in Todd-Hewitt broth (BD Biosciences, Sparks MD) supplemented with 0.5% yeast extract [20]. Isogenic mutants of T4R lacking AdcA (ΔAdcA), and AdcAII (ΔAdcAII) were created using splicing by overlap extension (SOE) PCR method using erythromycin and spectinomycin antibiotic cassettes [21] and standard S. pneumoniae transformation procedures. Mutants lacking AdcAII were isolated by selection on blood agar plates supplemented with erythromycin (0.5μg/mL) and ΔAdcA mutants were isolated by selection on blood agar plates supplemented with spectinomycin (500μg/mL). Double mutants (ΔAdcA/ΔAdcAII) were selected for using blood agar plates supplemented with both erythromycin and spectinomycin. All mutants were confirmed by PCR. Primer sequences used for construction of mutants are listed in S1 Table. Complemented mutants of AdcAII (ΔAdcAII+) were created by cloning the adcAII gene from T4R by amplifying the gene by PCR followed by digestion of the product with EcoRI and XmaI restriction enzymes (New England Biolabs). Primers were LMB3 (CGCGATCCCGGGGGTTAAGGAGTTGTTCATGAAGAAAC) and LMB4 (CCGCGGAATTCCTTTCCTCACTTTAATTCTTCTGC) with restriction sites underlined. Following enzymatic digestion, the gene was ligated with the pNE-1 pneumococcal shuttle vector that was similarly digested. The ligation was transformed into E. coli strain DH5α. Purified plasmid was then used to transform ΔAdcAII strains by standard methods and complemented mutants were selected by plating on blood agar plates supplemented with erythromycin (0.5μg/mL) and spectinomycin (500μg/mL). Complementation of AdcAII was confirmed by western blot (S1 Fig).
DNA Manipulation
Chromosomal DNA from TIGR4 S. pneumoniae was purified by standard methods. Plasmid DNA (pNE1) was prepared from E. coli using GenElute HP Plasmid Midiprep kit (Sigma-Aldrich, St. Louis, MO). PCR reactions contained 100-200ng of chromosomal DNA, primers at 200nM, 200μM of each dNTP, indicated buffer, and Takara Taq Ex. After initial incubation at 94°C, reactions completed 30 cycles of incubation at 94°C for 1 minute, 53°C for 1 minute, 72°C for 5 mins, with final incubations at 4°C.
Quantitative Real-time PCR
S. pneumoniae T4R was grown in Todd-Hewitt medium supplemented with 0.5% yeast extract to O.D.600 of 0.6 prior to the addition of Zn2+-chelating agent TPEN (30μM) (Sigma-Aldrich P4413) or ZnS04 (100μM). After addition of TPEN or ZnS04, bacteria were incubated at 37°C for 5 mins. At the 5 min time point 2mL of bacterial culture was added to 4mL of RNAprotect (Qiagen). Samples were incubated at room temperature for 5 mins. Two mL of bacterial culture in RNAprotect was pelleted for 5 mins at 13,000rpm, pellet was resuspended in 1mL of cold RNase free PBS, and samples were centrifuged again for 5 mins at 13,000rpm. Supernatant was decanted and pellet was resuspended in 400μL RLT buffer (Qiagen) with 2-Mercaptoethanol (Sigma-Aldrich M6250). Samples were sonicated three times (15 seconds), 600μL of RLT buffer was added to each sample, and sample was transferred to 500μL of 0.7mm Zirconia beads. Samples were bead beat for 5 mins using Mini-BeadBeater 16 (BioSpec Products). Lysate was centrifuged on tabletop centrifuge for 1 min. 700μL of sample was placed over Qiashredder (Qiagen) per manufacturer’s instructions. 100% Ethanol was added at 0.6 volume of the sample. RNA was then purified using a Qiagen RNeasy Mini Kit (Qiagen), optional on-column DNase treatment was performed, RNA was quantitated using a Qubit, and 50ng from each sample was used to synthesize cDNA using a Maxima First Strand cDNA synthesis kit (Thermo Scientific). cDNA products were diluted 1:100 and 1μL of cDNA was used as template for real-time PCR using (Luminaris Color HiGreen High ROX qPCR Master Mix (Thermo Scientific) per manufacturer’s instructions. Oligonucleotide sequences for adcAII were 5’ TCGGATGATTCAGTCAAGTAGTG and 5’ CAGACTTCCTGCCCAAGATT. Oligonucleotide sequences for adcA were 5’ GATAAGGCTTACGCAGAAGGT and 5’ GTCCATAGTCCAAGGCAAGATAG. Gyrase A served as a control gene and oligonucleotide sequences were 5’ ATGACCTCTTGGCTCTGATTG and 5’ CAACTCTGTACGGCGCTTAT. Relative fold changes were determined by ΔΔCt analysis comparing samples treated with TPEN or ZnSO4 to untreated controls, using GyrA as an internal housekeeping control gene.
Adhesion assay and invasion assay
Adhesion and invasion assays were performed in the A549 cell line. Cells were seeded to 80–90% confluence in a 24-well plate (Invitrogen). Cells were incubated with 1 x 106 CFU/mL bacteria at 37°C in 5% CO2. After 30 mins, cells were washed with D-PBS with Calcium and Magnesium (D-PBS +Ca/Mg) three times, trypsinized, and diluted 1:10 in PBS. 10μL was plated on blood agar and incubated at 37°C overnight. For invasion assays, cells were prepared in the same manner as for the adhesion assays. Cells were incubated with 1 x 106 CFU/mL bacteria for 2 hrs. Cells were then washed three times with D-PBS +Ca/Mg, and incubated with appropriate cell culture media with penicillin/gentamycin (10μg/mL and 250μg/mL, respectively) for 1 hr. Cells were again washed with D-PBS +Ca/Mg, trypsinized, and lysed with Triton X-100 (0.0125% final conc.). Serial dilutions were plated on TSA supplemented with 5% sheep blood and incubated overnight at 37°C in 5% CO2. Some experiments were performed in the same manner with the exception that bacteria were inoculated into media containing ZnS04 (200μM) prior to being seeded onto A549 monolayers. Each experimental condition was performed in three replicate wells and was repeated at least three times.
Fluorescence Microscopy
Invasion assays were performed as described above with the exception that A549 cells were seeded to 80% confluency in 2 well chamber slides (Nalge Nunc Int, Rochester, NY). After 2 hrs of contact with bacteria followed by 1 hr in penicillin/gentamycin supplemented media, cells were washed with D-PBS +Ca/Mg and fixed with 4% paraformaldehyde (in PBS) at 25°C for 10 mins. Cells were then rinsed three times with PBS and blocked with PBS plus 0.5% BSA, 4% mouse serum, and 0.5% Triton-X 100 for 15–30 mins. Cells were incubated with anti-TEPC-15 primary antibody (Sigma-Aldrich M-1421, specific for phosphorylcholine at 1:40) overnight at 4°C, rinsed two times with PBS for 5 mins each, prior to being incubated with goat anti-mouse IgA conjugated to rhodamine (Southern Biotech 1040–03 Birmingham, AL, at 1:100) for 1 hr. Cells were then rinsed two times with PBS for 5 mins each then 0.5mL of 300nM DAPI was added and incubated for 20 mins at room temperature. Cells were then rinsed three times with PBS before the chambers were removed, then 10μL of VECTASHIELD Mounting Medium (Vector H-1000) was added, and a cover slip was applied. Cells were visualized by fluorescence microscopy using a Zeiss Axioskop 2 Plus.
Mouse challenges
All experiments involving animals were planned and conducted in accordance with guidelines of the MSU Institutional Animal Care and Use Committee. The committee specifically approved procedures performed in the study presented here. C57BL/6 mice were maintained at Biosafety Level 1 and 2 facilities in the Mississippi State University Animal Facility. Six-week old female mice were anesthetized with isoflurane and challenged intranasally with either 20μL of PBS containing 7x103 (for colonization) or 100μl of PBS containing 3.6x104 (for invasion) of TIGR4, ΔAdcA, or ΔAdcAII. At 24 hrs and 48 hrs post-infection, mice were humanely euthanized by deep isoflurane inhalation followed by cervical dislocation and confirmed by incubation for 5 min under CO2. During the time course of the infections, no mice became severely ill or required early/humane endpoint sacrifices. This was determined by monitoring physical or behavioral changes including, weight loss, dehydration, loss of balance, or becoming moribund. Nasal washes, nasal tissues, and blood samples were collected. Bacterial titers were determined by plating serial dilutions on tryptic soy agar plates supplemented with 5% defibrinated sheep blood with neomycin (20μg/mL), followed by a16 hr incubation at 37°C.
Microarray analysis
To determine if loss of AdcAII has an effect on global gene expression in S. pneumoniae, microarray analysis was performed to compare transcript levels between T4R and its isogenic mutant ΔAdcAII. T4R and the ΔAdcAII knockout strain were cultured in C+Y media. Bacterial RNA was harvested from mid-log-phase cultures by the Qiagen RNeasy Mini Kit (Qiagen). Microarray experiments were performed as described previously [16]. Whole genome S. pneumoniae oligonucleotide microarrays obtained from the PFGRC at the J. Craig Venter Institute (http://pfgrc.jcvi.org/) were used for microarray experiments (lot numbers 6QSP11062007A and 6QSP11012007A). The array consisted of 70-mer oligonucleotide probes representing 2060 open reading frames from S. pneumoniae strain TIGR4 and 457 from strains G54 and R6, and each probe sequence was spotted at least five times on the array. Data have been submitted to NCBI under accession GSM358422, GSM358242, GSM358241, GSM358231, GSM358229 and GSM358101. To analyze the microarray data, a series of filtration algorithms was applied to eliminate spots with poor-quality data. Spots flagged (as bad, absent or not found) by the image analysis software GenePix 6.0 (Axon Corp., Union City, CA) and spots having a signal-to-noise ratio less than 1.5 or a background-corrected signal reading less than 20 in both Cy3and Cy5 channels were excluded from analysis. To correct the intensity bias, loess normalization was applied to each microarray. Of at least five replicate spots on each array, only genes whose corresponding spots passed the filtering criterion at least 50% of the times were reported in further analysis. Of the 11960 spots dedicated to TIGR4, approximately 16.8% were eliminated because of poor quality or low signal intensity. These were typically gene specific, with all five spots failing to give adequate signal intensity, indicative of either poor hybridization to the oligonucleotide probe or low abundance of the specific transcript. For each experiment, results were summarized from three independent microarray replicates, which included one dye flip experiment for adjusting the dye bias. The average of the fold changes from at least two replicated experiments for each gene was calculated, and those with a minimum two-fold change were listed in respective tables. All genes included in the table had a P-value less than 0.05, using a t-distribution test.
Western Blotting
Bacterial cultures were grown in Todd-Hewitt medium supplemented with 0.5% yeast extract to an O.D.600 of 0.8. Cultures were centrifuged and pellets were lysed in 100μL Pneumococcal lysis buffer (0.01% SDS, 0.1% DOC, and 0.015M Na Citrate). Equal amounts of protein were determined using a BCA Protein Assay (Pierce) loaded in Mini-Protean gel (BIO-RAD), transferred using a semi-dry transfer method (BIO-RAD Trans Blot SD), blocked with 5% non-fat milk, and probed with anti-Sp0463 (pilus) as the primary antibody, and goat-anti mouse conjugated to HRP as the secondary antibody. For western blot confirmation of AdcAII complementation, blotted lysates were probed with polyclonal rabbit antiserum against AdcAII (Rockland labs).
Production of recombinant AdcAII constructs
The AdcAII gene lacking the signal sequence was amplified from TIGR4 chromosomal DNA using primers BMLMBF2 and BMLMBR containing EcoRI and NdeI restriction sites, respectively. Primer sequences listed in S1 Table. Restriction digests included 100-200ng of PCR product or plasmid DNA, enzymes, indicated buffer, and bovine serum albumin (BSA) when indicated by manufacturer (New England Biolabs). Digests were incubated for at least 6 hrs at 37°C and DNA was isolated using QIAquick PCR Purification Kit (Qiagen, Valencia, CA) for gene inserts or QIAquick Gel Extraction kit (Qiagen) for plasmids according to manufacturer’s instructions. Restricted PCR products were ligated into previously digested expression vector pET-28 (Invitrogen) prior to transformation into E. coli using standard molecular biology techniques. Recombinant histidine-tagged protein was purified using HIS-Select Nickel Affinity Gel (Sigma-Aldrich)
Dot blot
Interaction between AdcAII and human matrix proteins was determined by dot blot analysis as previously described [8]. Using a BIO-DOT vacuum apparatus (Bio-Rad), 2μg, 5μg, and 10μg of collagen (Sigma-Aldrich), fibronectin, and laminin (Sigma-Aldrich) were transferred in 20μL spots onto PVDF membrane (Millipore, Bedford, MA) previously soaked with 100% methanol and rinsed in transfer buffer (4.5% Tris, 22% glycine, 5% methanol). Anti-AdcAII polyclonal antiserum served as a control for binding to the membrane. The blot was blocked for 30 min in PBST containing 2% BSA (Sigma-Aldrich Inc.) prior to incubation with biotinylated AdcAII protein (10μg/mL) in PBST-2% BSA for 2 hrs at room temperature. AdcAII was biotin labeled using the Biotin Labeling Kit (Roche, Indianapolis, IN) per manufacturer’s instructions. After incubation with biotinylated AdcAII, the blot was washed with PBST and incubated with horseradish peroxidase strepavidin (1:5000) (Vector Laboratories, Burlingame, CA) prior to another wash and development using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL). For a laminin-binding control, recombinant 37/67kDa laminin receptor protein was used as a probe and developed using anti-LR mAb (1:500) (Abcam, Cambridge, MA) and HRP-conjugated goat anti-mouse secondary (1:10000) (Bio-Rad).
Biophysical Characterization
Circular dichroism (CD) experiments were performed on an Olis DSM 20 spectropolarimeter. Protein concentrations were estimated using an extinction coefficient at 280 nm of 24,410 M-1 cm-1 [22]. AdcAII was measured in a 0.1 mm pathlength cell at a concentration of 42 μM at 25°C in 25 mM Tris buffer pH 7.5. Data were collected at intervals of 0.5 nm over the far-UV range of 260 to 190 nm. After data collection, spectra were smoothed using the Savitsky-Golay algorithm over a window size of 25 points [23]. The processed data were analyzed using the CDSSTR program to predict the secondary structure content [24, 25]. To facilitate analysis, a convenient Python program was developed, streamlining data analysis for the CDSSTR core program. This program runs on Mac, Linux, and Windows computers and is available at http://fitzkee.chemistry.msstate.edu/software/. The crystal structure of AdcAII (PDB ID 3CX3, chain A) was used for comparison with CD-based secondary structure predictions [9]. Backbone secondary structure in the crystal structure was assigned using DSSP [26].
Dynamic light scattering (DLS) measurements were collected on a Wyatt DynaPro NanoStar instrument under identical conditions as CD measurements. Samples were filtered using an Anotop 10 syringe filter with a 0.02 μm pore size (GE Healthcare Life Sciences, New Brunswick, NJ). Sample loss during filtration was determined to be minimal (< 10%) based on absorbance measurements. Cumulant and regularization analysis were both applied to the DLS data and agreed to within 2 Å with low polydispersity (< 20%). The native state hydrodynamic radius (RH) was estimated from the crystal structure (PDB 3CX3) by dividing the largest Cα-Cα distance by two (RH ≈ 30.5 Å).
Statistics
In instances where values from three or more experiments were compared, we used a two-tailed Student’s t-test to test for statistical significance. A P value less than 0.05 was considered significant.
Results
Relative importance of AdcA and AdcAII during Zn2+ limitation
Previous work in the laboratory-adapted strains D39 and R6 have shown that expression of both adcA and adcAII is important for growth in low Zn2+ conditions [12, 27]. Using derivatives of the meningitis isolate TIGR4, wild type (T4R) and isogenic mutants ΔAdcA, ΔAdcAII, and ΔAdcA/AdcAII were grown in THY media containing various concentrations of the Zn2+-specific chelator, N,N,N',N'-Tetrakis-(2-pyridylmethyl) ethylenediamine (TPEN) [12]. In the absence of TPEN, only the double mutant ΔAdcA/AdcAII displayed a growth defect (Fig 1A). Titration of the TPEN concentration indicated that ΔAdcAII demonstrated delayed growth compared to wild type T4R at concentrations where growth of ΔAdcA was virtually unaffected (Fig 1B). Supplementation of TPEN-containing media with ZnSO4 complemented the growth defect of ΔAdcAII, confirming the effect was Zn2+-dependent (data not shown). Based on these results, AdcAII appears to be more important for growth in Zn2+limiting conditions than AdcA.
Fig 1. Effects of zinc concentration on growth of Adc mutants.

A) Representative growth curve from three runs of T4R wild type and Adc mutants grown in THY media (pH6.5) in the absence (A) or presence (B) of the Zn-specific chelator TPEN (21μM).
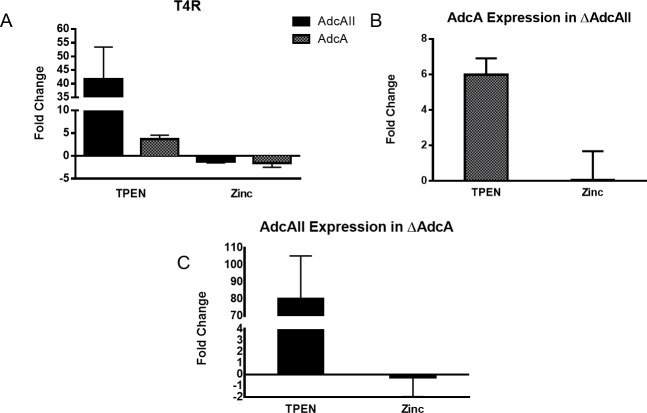
We also assessed the relative expression of adcA and adcAII under Zn2+-limiting and sufficient conditions. Addition of the Zn2+ chelator to wild type T4R resulted in 42-fold upregulation of adcAII but only 4-fold upregulation of adcA (Fig 2A). In the ΔAdcAII strain, adcA upregulation in the presence of the chelator was still only modest at 6-fold (Fig 2B); conversely, in the ΔAdcA strain, adcAII was strongly upregulated 80-fold in the presence of TPEN (Fig 2C). Thus, it appears that adcAII expression is more tightly coupled to Zn2+ concentration in the environment than adcA.
Fig 2. Effects of zinc on expression of adcA and adcAII in zinc rich (100μM ZnSO4) or low zinc environment (30μM TPEN).
A) Expression of adcA (grey) and adcAII [28] in wild type T4R as measured by qRT-PCR ΔΔCT analysis. B) Expression of adcA in ΔAdcAII. C) Expression of adcAII in ΔAdcA.
Contribution of AdcA and AdcAII to colonization and invasion
Homologs of AdcAII have been shown in related bacterial species to play a role in adhesion and invasion of host cells [6, 8, 29]. To determine if AdcAII serves a similar function for pneumococcus, we performed adhesion and invasion assays in the A549 human lung epithelial cell line. Interestingly, the loss of AdcAII led to a slight decrease in the number of bacterial cells adherent to A549 cells, while loss of AdcA led to no change in ability to adhere. Loss of both AdcA and AdcAII led to a significant decrease in adherence (Fig 3A; **p<0.0005). However, both ΔAdcA and ΔAdcAII strains were significantly more invasive in comparison to wild type T4R (Fig 3B; *p<0.005). The hyperinvasive phenotype of ΔAdcAII and ΔAdcA appears to be due to insufficient zinc acquisition. When zinc is supplemented at 200μM into media prior to invasion assays, the invasive phenotype of the mutants mimic that of the wild type (Fig 3D). Invasion data is not shown for the ΔAdcA/ΔAdcAII strain due to a growth defect of the strain during the two hr incubation required for invasion assays. We believe this to be a result of the mutant’s inability to acquire sufficient zinc since supplementation of 100μM ZnSO4 restored growth of the ΔAdcA/ΔAdcAII mutant (data not shown). Increased invasion of ΔAdcA and ΔAdcAII as compared to wild type T4R was also confirmed using fluorescent microscopy of A549 cells. The invasive phenotype of the complemented ΔAdcAII strain (ΔAdcAII+) returned closely to that of wild type T4R (Fig 3C). Western blot confirmed expression of AdcAII in our complemented ΔAdcAII+ strain, but expression was not equal to that of wild type T4R (S1 Fig). This could explain why invasion levels, though decreased, are not decreased to the same extent as that of wild type T4R.
Fig 3. Effect of AdcA and AdcAII on adhesion and invasion in A549 cell line.
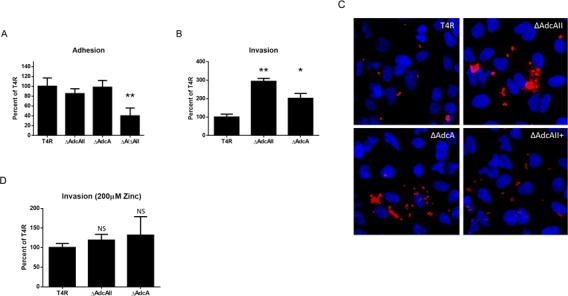
A) Adhesion relative to T4R (100% representing approximately 0.9% of inoculum adhered). Bacteria were incubated with A549 cells for 30 mins in 24-well plates and adherent bacteria were quantitated. ** = P<0.0005. B) Invasion relative to T4R (100% representing approximately 0.03% of inoculum invaded). Bacteria were incubated for 2 hrs with A549 cells, followed by 1 hr in the presence of antibiotics to kill extracellular bacteria; cells were lysed, and intracellular bacteria were quantitated. * = P<0.005 and ** = P<0.0005. C) Fluorescent images of A549 cells incubated with T4R, ΔAdcA ΔAdcAII, ΔAdcAII+ as in B. Cells were washed, permeabilized, and bacteria were stained with anti-TEPC-15 antibody (in red). Cell nuclei are stained with DAPI (blue). D) Invasion relative to T4R (%). Bacteria were incubated for 2 hrs with A549 cells with 200μM ZnSO4, followed by 1 hr in the presence of antibiotics to kill extracellular bacteria; cells were lysed, and intracellular bacteria were quantitated. All experiments were conducted at least three times. Each experiment consisted of triplicate sample wells.
Unencapsulated T4R from the TIGR4 strain background was used for these studies since encapsulated strains are known to poorly invade in vitro, and our goal was to specifically understand the role of surface proteins. Increased invasion was not due to growth differences between the strains as determined by comparing growth in F-12K (A549) media (data not shown).
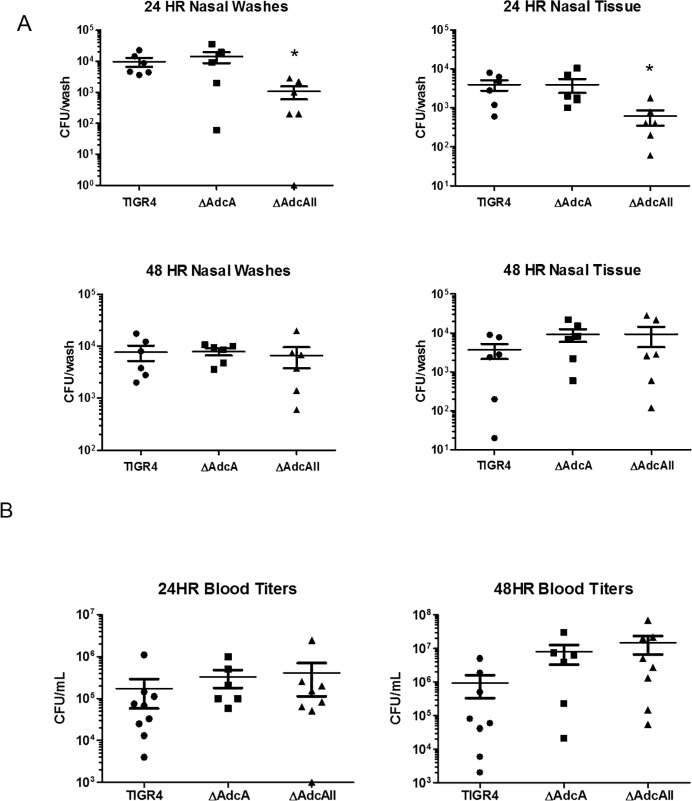
To assess the functions of AdcA and AdcAII in vivo, mice were challenged intranasally with S. pneumoniae TIGR4, T4ΔAdcA, and T4ΔAdcAII. Nasal washes, nasal tissues, and blood titers were collected at 24 and 48 hrs post-infection. The T4ΔAdcAII strain was found in significantly less abundance in nasal washes and nasal tissues taken 24 hrs post-inoculation; however, no significant differences were seen with T4ΔAdcA. However, at 48 hrs post-challenge, no significant differences were seen between TIGR4 and the mutants in either nasal washes or nasal tissues (Fig 4A). Thus, a modest colonization defect was seen only for T4ΔAdcAII and only early after challenge. Assessment of invasion as determined by blood bacterial counts demonstrated a trend for increased invasion for both T4ΔAdcA and T4ΔAdcAII, though these data did not reach statistical significance (Fig 4B). Similarly to the in vitro assays, the T4ΔAdcA/ΔAdcAII strain was not used during animal challenges due to growth deficiencies.
Fig 4. Effect of AdcAII on in vivo colonization and invasion.
A) For colonization studies, mice were inoculated intranasally with bacteria (7x103 CFU). At 24 and 48 hrs, mice were sacrificed and 200μL of PBS was passed from the trachea through the nose. Nasal passages were also collected, homogenized, and serial dilutions were plated. B) For invasion studies, mice were inoculated intranasally with bacteria (3.6x104 CFU). Serial dilutions of blood samples were plated at 24 and 48 hrs. Results have been pooled from two independent experiments.
Zinc-dependent gene expression
Microarray analysis was used to compare differential gene expression between TIGR4 and the ΔAdcAII strain. Ten genes were upregulated and twelve genes were downregulated in ΔAdcAII. Some of the genes upregulated in the ΔAdcAII strain include phtD and a v-type ATP synthase. Some of the genes that were the most strongly downregulated include neuraminidase B, a 50s ribosomal protein, and a sialidase B precursor (S2 Table). While the gene encoding phtD (SP1003) is directly downstream of the gene for adcAII (SP1002), upregulation was not due to polar effects since supplementation with zinc decreased the upregulation (S2 Fig). In order to investigate the role PhtD may have in the hyperinvasive phenotype of ΔAdcAII, we performed invasion assays as previously described. Interestingly, we saw that ΔPhtD had a similar phenotype to ΔAdcAII as it was approximately three times more invasive than wild type T4R (S3 Fig) (p<0.05). Thus indicating to us that though phtD is upregulated in ΔAdcAII, it is not responsible for the hyperinvasive phenotype.
The two-component systems CbpR/S and HK/RR03 and the metalloregulators MerR and PsaR are known to down-regulate expression of pilus [30]. Mutations in these loci result in hyper-invasion of epithelial cells similar to that seen with ΔAdcAII [30]. This similarity suggested a potential role for AdcAII in regulating pilus gene expression. While microarray analysis did not identify changes in pilus gene expression in ΔAdcAII, western blot was used to assess expression of pilus in wild type T4R, ΔAdcA, and ΔAdcAII; however, no significant differences were seen (Fig 5).
Fig 5. Western blot assessing pilus SP0463 expression.
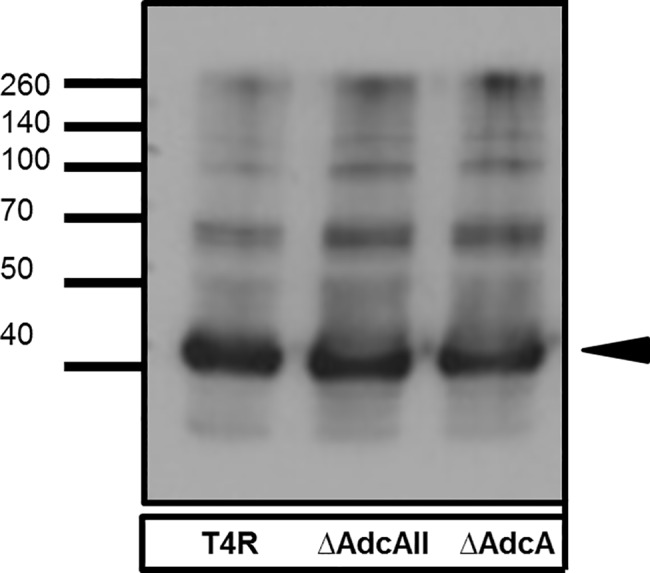
Lysates from T4R, ΔAdcA, and ΔAdcAII strains were blotted and probed with anti-SP0463 antibody and developed. Black arrow indicates SP0463 pilus monomer.
Assessment of potential laminin binding
AdcAII homologs, Lmb of S. agalactiae and Lbp or Lsp of S. pyogenes, have been shown to interact with the extracellular matrix protein laminin, increasing adhesion to host cells [5, 8, 29]. Given the degree of homology between AdcAII and these proteins (>60% identity), we investigated a potential role for AdcAII in interaction with extracellular matrix proteins. For these studies the adcAII gene from S. pneumoniae TIGR4 was expressed as a recombinant protein and purified by affinity chromatography. Using a dot blot technique, AdcAII failed to interact with human laminin (Fig 6) or the extracellular matrix proteins collagen or fibronectin (data not shown). Additionally, interaction of laminin and recombinant AdcAII by co-immunoprecipitation or ELISA could not be demonstrated (data not shown). Together, these data indicate that while highly homologous to laminin-binding proteins of related streptococci, AdcAII does not share an affinity for laminin.
Fig 6. Dot blot demonstrating lack of laminin binding by AdcAII.
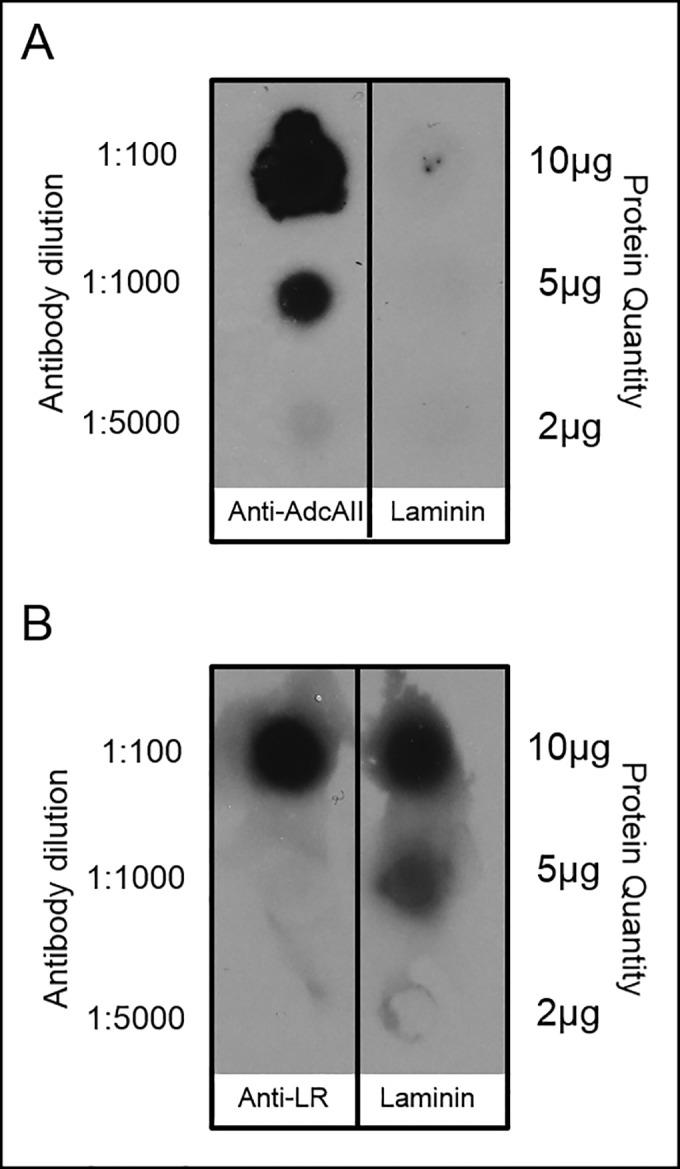
A) Laminin (2 μg, 5 μg, and 10 μg spots) or anti-AdcAII antibody (1:100, 1:1000, 1:5000 spots) as control were probed with biotinylated recombinant AdcAII protein. B) To demonstrate laminin could interact with ligands using this method laminin protein (2 μg, 5 μg, and 10 μg spots) or anti-laminin receptor antibody (1:100, 1:1000, 1:5000 spots) as a control probed with recombinant laminin receptor and developed with anti-LR antibody.
Biophysical Characterization of AdcAII
Circular dichroism (CD) spectroscopy and dynamic light scattering (DLS) were used to confirm the folded conformation of AdcAII. The far-UV CD spectrum of AdcAII suggests a folded structure (S4A Fig), and singular value decomposition (SVD) of CD data predict an α-helical content of 46% and a β-strand content of 7%. These values are in good agreement with the crystal structure of AdcAII (38% α-helix and 11% β-strand) [9]. The DLS results also suggest a compact, globular structure for purified AdcAII (S4B Fig). The observed hydrodynamic radius (RH) from DLS is 27.5 ± 0.4 Å, and this is also in good agreement with the radius predicted using the crystal structure (30.5 Å). These data strongly suggest that the protein in our studies is in its native confirmation; therefore, the lack of detectable binding to ECM components is not a result of misfolded or aggregated protein, but rather an intrinsic property of the protein itself.
Discussion
The goal of the current study was to expand the possible functional roles of the S. pneumoniae Zn2+-binding lipoproteins AdcAII and AdcA from metal transport to virulence. Proteins homologous to AdcAII have been found to interact with the extracellular matrix protein laminin and contribute to virulence by participating in adhesion [5]. AdcA failed to show any significant phenotype in these activities and thus appears to be limited to metal transport. In contrast, AdcAII showed extensive roles in activities beyond metal transport. AdcAII did not associate with laminin as measured by affinity chromatography, dot blot analysis, and co-immunoprecipitation. However, loss of AdcAII attenuated adhesion and strongly increased invasion of host cells. This data combined with our data showing gene expression of adcA, adcAII, and phtD are zinc-dependent leads us to believe that the hyper-invasiveness of ΔAdcA and ΔAdcAII is a result of the zinc limitation. This is underlined by the fact that invasion assays demonstrate that the mutants are equally as invasive as wild type when zinc is supplemented to the media.
Over the course of progression from nasopharyngeal colonization to invasive disease the pneumococcus must respond to a broad range of Zn2+ concentrations [31]. The availability of Zn2+ is known to affect both colonization and development of invasive pneumococcal disease in mice [32, 33]. Zn2+ levels are generally thought to be higher in the nasopharynx and the lungs compared to levels found in the blood [34, 35]. Within the lungs, Zn2+ has been shown to be found at higher concentrations at the apical surface of the epithelium [36]. This suggests that the bacteria encounter higher levels of Zn2+ at the cellular surface prior to invasion into the blood stream [36]. Also, Plumptre et al. have shown that Zn2+ levels increase significantly in niches containing S. pneumoniae as early as 48 hrs post infection. Increased Zn2+ concentrations would decrease expression of the adcA and adcAII genes, and based on our results, may limit invasive potential of the bacteria. Though Plumptre et al. demonstrated this in the D39 background, our data support these findings in the TIGR4 background of S. pneumoniae as well [12].
Expression of AdcAII, the AdcCBA Zn2+ transporter operon, and all of the histidine triad proteins of pneumococcus are regulated by the Zn2+-dependent repressor AdcR [10, 17, 37]. This is consistent with AdcA and AdcAII serving as Zn2+ transporters and with their expression being reduced when Zn2+ levels are high. Bayle et al. have proposed that AdcA and AdcAII serve redundant roles in Zn2+ transport with one capable of substituting for the other when Zn2+ is low [11]. However, our results show a distinct effect of low Zn2+ on growth of mutants lacking only AdcAII. The differences between our findings and those of Plumptre et al, could be explained by differences in culture media between their group and ours. Alternatively, there could be AdcR-dependent differences in the strains R6 and T4R, though the sequences are nearly identical as determined through ClustalW alignment. Also, their studies identified that mutation of both AdcA and AdcAII resulted in complete loss of virulence in animal models of infection. This result is likely due to a complete inability of the bacteria to regulate intracellular Zn2+ concentrations in vivo. In contrast, our results with strains lacking AdcA or AdcAII alone demonstrate at least a trend for increased invasion both in vitro and in vivo. Plumptre et al. have performed challenge experiments with ΔAdcAII and ΔAdcA mutants in the D39 background and found similar results to ours with regards to colonization; however, their AdcAII mutant displayed a trend for decreased invasion at 24 hrs post-infection that then rebounded by 48 hrs to that of wild type[12]. In our model, the difference in invasion between ΔAdcAII and TIGR4 increased from 24 hrs to 48 hrs, indicating the relative importance of AdcA and AdcAII may be strain dependent.
In conclusion, we sought to characterize differences between AdcA and AdcAII in zinc homeostasis and contribution to virulence. This study has provided a link between expression of AdcAII, AdcA, and invasive potential of S. pneumoniae. It was determined that S. pneumoniae strains lacking either AdcAII or AdcA are more invasive than wild type strains. Additionally, AdcAII may be more important for both Zn2+ homeostasis and the regulation of pneumococcal virulence.
Supporting Information
Lysates from T4R, ΔAdcAII, and ΔAdcAII+ strains were blotted and probed with anti-LMB antibody and developed. Black arrow indicates AdcAII.
(TIF)
A) Basal expression of phtD in wild type T4R vs ΔAdcAII as measured by qRT-PCR ΔΔCT analysis. B) Expression of phtD in wild type T4R vs ΔAdcAII when supplemented with 100μM ZnSO4.
(TIF)
Invasion relative to T4R (%). Bacteria were incubated for 2 hrs with A549 cells, followed by 1 hr in the presence of antibiotics to kill extracellular bacteria; cells were lysed, and intracellular bacteria were quantitated. * = P<0.05. All experiments were conducted at least three times. Each experiment consisted of triplicate sample wells.
(TIF)
A) Far-UV Circular Dichroism (CD) spectrum of 40 μM AdcAII at 25°C in 25mM Tris buffer pH 7.5 [28]. The reconstructed spectrum from singular-value decomposition (SVD) is also shown (red) to demonstrate good agreement between the observed data and secondary structure predictions. The data have been processed as described in materials and methods. B) Regularization fit results from dynamic light scattering (DLS) for AdcAII under identical conditions to (A). The average observed hydrodynamic radius (RH) is 27.5 ± 0.4 Å. This compares favorably with the crystal structure value of 30.5 Å.
(TIF)
(TIF)
Bacterial RNA was harvested at OD600 0.5 and used to synthesize cDNA for hybridization to pneumococcal microarray.
(TIF)
Acknowledgments
We would like to thank Granger Ridout and Yong-Dong Wang for assistance with microarray analysis.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by National Institutes of Health grant number R03AI103294 (JAT), National Institutes of Health grant number R01AI27913 (EIT), and the American Lebanese Syrian Associated Charities (www.nih.gov). The lab of JAT was supported by grant #P20GM103646 (Center for Biomedical Research Excellence in Pathogen Host Interactions) from the National Institute for General Medical Sciences. NCF is supported by NIH grant R15GM113152.
References
- 1.Sampson JS, O'Connor SP, Stinson AR, Tharpe JA, Russell H. Cloning and nucleotide sequence analysis of psaA, the Streptococcus pneumoniae gene encoding a 37-kilodalton protein homologous to previously reported Streptococcus sp. adhesins. Infect Immun. 1994;62(1):319–24. Epub 1994/01/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell H, Tharpe JA, Wells DE, White EH, Johnson JE. Monoclonal antibody recognizing a species-specific protein from Streptococcus pneumoniae. J Clin Microbiol. 1990;28(10):2191–5. Epub 1990/10/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero-Steiner S, Pilishvili T, Sampson JS, Johnson SE, Stinson A, Carlone GM, et al. Inhibition of pneumococcal adherence to human nasopharyngeal epithelial cells by anti-PsaA antibodies. Clin Diagn Lab Immunol. 2003;10(2):246–51. Epub 2003/03/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry AM, Paton JC. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64(12):5255–62. Epub 1996/12/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spellerberg B, Rozdzinski E, Martin S, Weber-Heynemann J, Schnitzler N, Lutticken R, et al. Lmb, a protein with similarities to the LraI adhesin family, mediates attachment of Streptococcus agalactiae to human laminin. Infect Immun. 1999;67(2):871–8. Epub 1999/01/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenenbaum T, Spellerberg B, Adam R, Vogel M, Kim KS, Schroten H. Streptococcus agalactiae invasion of human brain microvascular endothelial cells is promoted by the laminin-binding protein Lmb. Microbes Infect. 2007;9(6):714–20. Epub 2007/04/03. S1286-4579(07)00096-2 [pii] 10.1016/j.micinf.2007.02.015 . [DOI] [PubMed] [Google Scholar]
- 7.Weston BF, Brenot A, Caparon MG. The metal homeostasis protein, Lsp, of Streptococcus pyogenes is necessary for acquisition of zinc and virulence. Infect Immun. 2009;77(7):2840–8. Epub 2009/04/29. IAI.01299-08 [pii] 10.1128/IAI.01299-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsner A, Kreikemeyer B, Braun-Kiewnick A, Spellerberg B, Buttaro BA, Podbielski A. Involvement of Lsp, a member of the LraI-lipoprotein family in Streptococcus pyogenes, in eukaryotic cell adhesion and internalization. Infect Immun. 2002;70(9):4859–69. Epub 2002/08/17. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loisel E, Jacquamet L, Serre L, Bauvois C, Ferrer JL, Vernet T, et al. AdcAII, a new pneumococcal Zn-binding protein homologous with ABC transporters: biochemical and structural analysis. J Mol Biol. 2008;381(3):594–606. Epub 2008/07/18. S0022-2836(08)00660-8 [pii] 10.1016/j.jmb.2008.05.068 . [DOI] [PubMed] [Google Scholar]
- 10.Dintilhac A, Claverys JP. The adc locus, which affects competence for genetic transformation in Streptococcus pneumoniae, encodes an ABC transporter with a putative lipoprotein homologous to a family of streptococcal adhesins. Res Microbiol. 1997;148(2):119–31. Epub 1997/02/01. S0923-2508(97)87643-7 [pii] 10.1016/S0923-2508(97)87643-7 . [DOI] [PubMed] [Google Scholar]
- 11.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol. 2011. Epub 2011/10/26. 10.1111/j.1365-2958.2011.07862.x . [DOI] [PubMed] [Google Scholar]
- 12.Plumptre CD, Eijkelkamp BA, Morey JR, Behr F, Counago RM, Ogunniyi AD, et al. AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Molecular microbiology. 2014;91(4):834–51. 10.1111/mmi.12504 . [DOI] [PubMed] [Google Scholar]
- 13.Loisel E, Chimalapati S, Bougault C, Imberty A, Gallet B, Di Guilmi AM, et al. Biochemical characterization of the histidine triad protein PhtD as a cell surface zinc-binding protein of pneumococcus. Biochemistry. 2011;50(17):3551–8. Epub 2011/03/24. 10.1021/bi200012f . [DOI] [PubMed] [Google Scholar]
- 14.Adamou JE, Heinrichs JH, Erwin AL, Walsh W, Gayle T, Dormitzer M, et al. Identification and characterization of a novel family of pneumococcal proteins that are protective against sepsis. Infect Immun. 2001;69(2):949–58. Epub 2001/02/13. 10.1128/IAI.69.2.949-958.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan MN, Pichichero ME. Vaccine candidates PhtD and PhtE of Streptococcus pneumoniae are adhesins that elicit functional antibodies in humans. Vaccine. 2012;30(18):2900–7. 10.1016/j.vaccine.2012.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orihuela CJ, Radin JN, Sublett JE, Gao G, Kaushal D, Tuomanen EI. Microarray analysis of pneumococcal gene expression during invasive disease. Infect Immun. 2004;72(10):5582–96. Epub 2004/09/24. 10.1128/IAI.72.10.5582-5596.2004 72/10/5582 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, et al. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J. 2009;23(3):731–8. Epub 2008/10/31. fj.08-119537 [pii] 10.1096/fj.08-119537 . [DOI] [PubMed] [Google Scholar]
- 18.Shafeeq S, Kloosterman TG, Kuipers OP. Transcriptional response of Streptococcus pneumoniae to Zn2+) limitation and the repressor/activator function of AdcR. Metallomics. 2011;3(6):609–18. Epub 2011/05/24. 10.1039/c1mt00030f . [DOI] [PubMed] [Google Scholar]
- 19.van Opijnen T, Camilli A. A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome research. 2012;22(12):2541–51. 10.1101/gr.137430.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laurance WF, Useche DC, Rendeiro J, Kalka M, Bradshaw CJ, Sloan SP, et al. Averting biodiversity collapse in tropical forest protected areas. Nature. 2012;489(7415):290–4. 10.1038/nature11318 . [DOI] [PubMed] [Google Scholar]
- 21.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77(1):51–9. Epub 1989/04/15. 0378-1119(89)90358-2 [pii]. . [DOI] [PubMed] [Google Scholar]
- 22.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Science. 1995;4(11):2411–23. 10.1002/pro.5560041120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protocols. 2007;1(6):2876–90. http://www.nature.com/nprot/journal/v1/n6/suppinfo/nprot.2006.202_S1.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennessey JP, Johnson WC. Information content in the circular dichroism of proteins. Biochemistry. 1981;20(5):1085–94. 10.1021/bi00508a007 [DOI] [PubMed] [Google Scholar]
- 25.Sreerama N, Woody RW. A Self-Consistent Method for the Analysis of Protein Secondary Structure from Circular Dichroism. Analytical Biochemistry. 1993;209(1):32–44. 10.1006/abio.1993.1079. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22(12):2577–637. 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- 27.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Molecular microbiology. 2011;82(4):904–16. Epub 2011/10/26. 10.1111/j.1365-2958.2011.07862.x . [DOI] [PubMed] [Google Scholar]
- 28.Nguyen AI, Blackmore KJ, Carter SM, Zarkesh RA, Heyduk AF. One- and two-electron reactivity of a tantalum(V) complex with a redox-active tris(amido) ligand. J Am Chem Soc. 2009;131(9):3307–16. Epub 2009/02/18. 10.1021/ja808542j 10.1021/ja808542j [pii]. . [DOI] [PubMed] [Google Scholar]
- 29.Terao Y, Kawabata S, Kunitomo E, Nakagawa I, Hamada S. Novel laminin-binding protein of Streptococcus pyogenes, Lbp, is involved in adhesion to epithelial cells. Infect Immun. 2002;70(2):993–7. Epub 2002/01/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosch JW, Mann B, Thornton J, Sublett J, Tuomanen E. Convergence of regulatory networks on the pilus locus of Streptococcus pneumoniae. Infect Immun. 2008;76(7):3187–96. Epub 2008/04/30. IAI.00054-08 [pii] 10.1128/IAI.00054-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Honsa ES, Johnson MDL, Rosch JW. The roles of transition metals in the physiology and pathogenesis of Streptococcus pneumoniae. Frontiers in Cellular and Infection Microbiology. 2013;3:92 10.3389/fcimb.2013.00092 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strand TA, Briles DE, Gjessing HK, Maage A, Bhan MK, Sommerfelt H. Pneumococcal pulmonary infection, septicaemia and survival in young zinc-depleted mice. Br J Nutr. 2001;86(2):301–6. Epub 2001/08/15. S0007114501001726 [pii]. . [DOI] [PubMed] [Google Scholar]
- 33.Strand TA, Hollingshead SK, Julshamn K, Briles DE, Blomberg B, Sommerfelt H. Effects of zinc deficiency and pneumococcal surface protein a immunization on zinc status and the risk of severe infection in mice. Infect Immun. 2003;71(4):2009–13. Epub 2003/03/26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJ, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Molecular microbiology. 2007;65(4):1049–63. Epub 2007/07/21. MMI5849 [pii] 10.1111/j.1365-2958.2007.05849.x . [DOI] [PubMed] [Google Scholar]
- 35.Versieck J. Trace elements in human body fluids and tissues. Crit Rev Clin Lab Sci. 1985;22(2):97–184. Epub 1985/01/01. 10.3109/10408368509165788 . [DOI] [PubMed] [Google Scholar]
- 36.Carter JE, Truong-Tran AQ, Grosser D, Ho L, Ruffin RE, Zalewski PD. Involvement of redox events in caspase activation in zinc-depleted airway epithelial cells. Biochem Biophys Res Commun. 2002;297(4):1062–70. Epub 2002/10/03. S0006291X02022921 [pii]. . [DOI] [PubMed] [Google Scholar]
- 37.Reyes-Caballero H, Guerra AJ, Jacobsen FE, Kazmierczak KM, Cowart D, Koppolu UM, et al. The metalloregulatory zinc site in Streptococcus pneumoniae AdcR, a zinc-activated MarR family repressor. J Mol Biol. 2010;403(2):197–216. Epub 2010/09/02. S0022-2836(10)00892-2 [pii] 10.1016/j.jmb.2010.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lysates from T4R, ΔAdcAII, and ΔAdcAII+ strains were blotted and probed with anti-LMB antibody and developed. Black arrow indicates AdcAII.
(TIF)
A) Basal expression of phtD in wild type T4R vs ΔAdcAII as measured by qRT-PCR ΔΔCT analysis. B) Expression of phtD in wild type T4R vs ΔAdcAII when supplemented with 100μM ZnSO4.
(TIF)
Invasion relative to T4R (%). Bacteria were incubated for 2 hrs with A549 cells, followed by 1 hr in the presence of antibiotics to kill extracellular bacteria; cells were lysed, and intracellular bacteria were quantitated. * = P<0.05. All experiments were conducted at least three times. Each experiment consisted of triplicate sample wells.
(TIF)
A) Far-UV Circular Dichroism (CD) spectrum of 40 μM AdcAII at 25°C in 25mM Tris buffer pH 7.5 [28]. The reconstructed spectrum from singular-value decomposition (SVD) is also shown (red) to demonstrate good agreement between the observed data and secondary structure predictions. The data have been processed as described in materials and methods. B) Regularization fit results from dynamic light scattering (DLS) for AdcAII under identical conditions to (A). The average observed hydrodynamic radius (RH) is 27.5 ± 0.4 Å. This compares favorably with the crystal structure value of 30.5 Å.
(TIF)
(TIF)
Bacterial RNA was harvested at OD600 0.5 and used to synthesize cDNA for hybridization to pneumococcal microarray.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.