Abstract
Dongxiang wild rice (Oryza rufipogon Griff.) is the progenitor of cultivated rice (Oryza sativa L.), and is well known for its superior level of tolerance against cold, drought and diseases. To date, however, little is known about the salt-tolerant character of Dongxiang wild rice. To elucidate the molecular genetic mechanisms of salt-stress tolerance in Dongxiang wild rice, the Illumina HiSeq 2000 platform was used to analyze the transcriptome profiles of the leaves and roots at the seedling stage under salt stress compared with those under normal conditions. The analysis results for the sequencing data showed that 6,867 transcripts were differentially expressed in the leaves (2,216 up-regulated and 4,651 down-regulated) and 4,988 transcripts in the roots (3,105 up-regulated and 1,883 down-regulated). Among these differentially expressed genes, the detection of many transcription factor genes demonstrated that multiple regulatory pathways were involved in salt stress tolerance. In addition, the differentially expressed genes were compared with the previous RNA-Seq analysis of salt-stress responses in cultivated rice Nipponbare, indicating the possible specific molecular mechanisms of salt-stress responses for Dongxiang wild rice. A large number of the salt-inducible genes identified in this study were co-localized onto fine-mapped salt-tolerance-related quantitative trait loci, providing candidates for gene cloning and elucidation of molecular mechanisms responsible for salt-stress tolerance in rice.
Introduction
Salt stress is a vital problem for plant growth and agricultural productivity. Rice (Oryza sativa L.) is one of the most important food crops in the world and also a model for genomic research in monocots [1]. However, salinity is one of the most devastating abiotic stresses in rice, and the salt-affected soils currently account for about 20% of the total paddy rice planting area. More seriously, the area of salt-affected irrigated land is expanding and spreading in China [2]. In recent years, many studies have provided valuable insight into the molecular and cellular mechanisms by which rice responds to and tolerate salinity [3–7]. However, the regulatory mechanisms involved in coordinating salt stress tolerance and plant growth are not fully understood.
Dongxiang wild rice (Oryza rufipogon Griff., hereafter referred as DXWR) is the progenitor of cultivated rice (Oryza sativa L.). DXWR, a Chinese type of wild rice grown in Jiangxi Province (28°14’N latitude and 116°30’E longtitude), is considered to be the northernmost region in the world where O. rufipogon is found [8]. DXWR grows in the natural habitats and possesses various characteristics resistant to biotic and abiotic stresses and abundant genetic diversity which have been lost in the cultivated rice. Thus, it is an extremely important resource for providing a valuable gene pool for rice genetic improvement. To date, numerous quantitative trait loci (QTLs) have been mined from the DXWR, which are involved in biotic and abiotic stress tolerances [9–14]. But, the instances of rice improvement in stress tolerances with the help of DXWR were still rather rare. So far, only two genes (OrbHLH001 and OrbHLH2) of DXWR were reported to be involved in the salt stress resistance [15, 16], but the molecular mechanism on the salt stress resistance of DXWR remained unclear.
Gene expression profiling is accelerating our progress toward a comprehensive understanding of the genetic mechanisms that control responses to environmental stress. Microarray analysis and tag-based sequencing approaches have been used to investigate over-all gene expression profiles in various plants [17–21]. However, these technologies have critical limitations, such as low throughput and low sensitivity [22]. Recently, next-generation high-throughput RNA sequencing technology (RNA-Seq) could overcome the drawbacks of array-based technologies. With the high resolution and sensitivity, the RNA-Seq could be used for discovering novel splice junctions, novel transcripts, alternative transcription start sites and rare transcripts [23]. Moreover, RNA-Seq data revealed a high level of reproducibility in both technical and biological replicates [24]. So far, the global gene expression in various plants has been profiled by RNA-Seq [25–29]. In addition, RNA-Seq has been applied to the identification of stress-inducible transcripts in rice [30, 31]. However, little effort is being expended in attempts to investigate stress resistances of DXWR using RNA-Seq.
Transcription factors are integral in linking salt sensory pathways to many tolerance responses. Core sets of transcription factor family genes are differentially expressed in response to elevated external salinity [32], including basic leucine zipper (bZIP) [33], WRKY [34, 35], APETALA2/ETHYLENE RESPONSE FACTOR (AP2/ERF) [36], MYB [37], basic helix-loop-helix (bHLH) [38], and NAC [39] families. These transcription factors, in turn, regulate the expression levels of various genes that may ultimately influence the level of salt tolerance of plants [40]. In rice, recent studies have indicated that a large number of transcription factors were involved in salt stress response, such as OsMYB91 [7], OsbZIP71 [41], OsWRKY42 [42], SERF1 [3], OsTZF1 [43], and OsNAC5 [44].
In rice, important traits such as salt-tolerance, yield and quality are controlled by polygenes or gene complexes that are described as quantitative trait loci (QTL). The recent development of rice molecular maps has facilitated the analysis of QTLs for many factors, such as yield [45], heading date [46], and seed dormancy [47]. The dissection of such a complex trait by means of the QTL mapping-approach will be of great significance for breeding by enhancing the salt tolerance of rice. QTL analyses of salt tolerance have been conducted by several researches [48, 49]. But, the identification of genes underlying QTLs could be time consuming and expensive [50]. To date, few key salt responsive genes were identified by QTLs.
In this study, we used the Illumina sequencing to perform deep transcriptome sequencing to compare the genome-wide differential expression between salt-treatment and normal condition DXWR at the seedling stage. Through identifying potential candidate genes involved in the salt stress response in DXWR, this work may provide useful information and potential genetic resources for the improvement of salt-tolerant characters in rice. Furthermore, this study may provide a starting point for the elucidation of the molecular mechanism underlying the salt stress response in DXWR.
Materials and Methods
Plant material and salt stress treatment
Seeds of Dongxiang wild rice (Oryza rufipogon Griff.; Dongxiang County, Jiangxi Province) and rice Xieqingzao B (O. sativa L. ssp. indica) were immersed in distilled water in the dark, and the uniformly germinated seeds were sown in 96-well plates supported by a plastic container. Seeds were grown in a growth chamber, as previously described [12]. The growth culture solution was renewed every 3 days. After the seedlings had been grown for 14 days, they were transferred on their 96-well plates into containers filled with 200 mM NaCl solution, or with control solution for 12 days. The seedlings were then recovered under normal solution for 3 days, and survival rates were calculated. The experiment was a randomized complete block design with three replications. For RNA-Seq analysis, 14-day-old seedlings of Dongxiang wild rice were grown with or without 200 mM NaCl treatment for 3 days and then the leaves (penultimate leaves) and total roots (separated from the culture solution and washed carefully) of these seedlings were collected and immediately frozen in liquid nitrogen, respectively. For RNA extraction from each treatment group, 10 plants were collected and mixed, to minimize the effect of transcriptome unevenness among plants.
RNA extraction, cDNA library preparation and sequencing
Total RNA was extracted for the three biological replicates from the sampled leaf or root tissues collected from the ten seedlings for each biological replicate using the TRIzol kit following the manufacturer’s instructions (Invitrogen). Total RNA was then purified and concentrated using the RNeasy MinElute cleanup kit (Qiagen). The RNA quality was checked using Bioanalyzer 2100 (Agilent). Equal quantities of total RNA from three biological replicates for each tissue sample were then pooled for the following RNA sequencing. Magnetic beads with Oligo (dT) were used to isolate mRNA from the total RNA. Mixed with the fragmentation buffer, the mRNA is fragmented into short fragments. Then cDNA is synthesized using the mRNA fragments as templates. Short fragments were purified and resolved with EB buffer for end reparation and poly (A) addition. After that, the short fragments were connected with adapters. After agarose gel electrophoresis, the suitable fragments (200 bp) were selected for the PCR amplification as templates. The library was sequenced using the Illumina HiSeq− 2000 platform.
Reads filtration and assessment of differential gene expression
Before assembly, adaptor sequences, empty reads, low quality sequences with ‘N’ percentage over 10% and those containing more than 50% bases with a Q-value < 20 were removed using the Perl program written according to the custom method of Program editing. After filtering, the remaining reads were called clean reads and used for downstream bioinformatics analysis. The retained high-quality reads were mapped to the Nipponbare reference genome [51] by Tophat (-N 2-read-gap-length 3-read-edit-dist 3-read-realign-edit-dist 0-report-secondary-alignments-coverage-search-microexon-search-library-type fr-unstranded-b2-sensitive) [52], and then the resulting aligned reads were used to create a RABT (Reference Annotation Based Transcript) assembly using Cufflinks [53]. The sequence reads were submitted to GenBank GEO database under accession number GSE73181 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73181).
Expression levels for each gene were calculated by quantifying the reads according to the RPKM (reads per kilobase per million reads) method [54]. We used ‘FDR (false discovery rate) ≤ 0.001 and the absolute value of log2 RPKM ratio ≥ 1’ as the threshold to judge the significance of gene expression difference [55].
Gene Ontology (GO) term analysis
Blast2GO program was used to classify unigenes to GO terms based on molecular function, biological processes and cellular components [56] for leaf and root tissues, at p-values < 0.05.
Validation of RNA-Seq by qRT-PCR
qRT-PCR were performed to confirm a set of 32 differentially expressed genes (16 up-regulated and 16 down-regulated genes) randomly selected from each group of salt-inducible genes identified from RNA sequencing using a Chromo 4 real-time PCR detection system. Diluted cDNA was amplified using gene specific primers (S29 Table) and SYBR Green real-time PCR master mix (Toyobo). qRT-PCR was conducted with three biological replicates, and each sample was conducted at least in triplicate and normalized using Tubulin as an internal control [16].
Results and Discussion
Salt stress resistance assessment in the seedling stage of DXWR
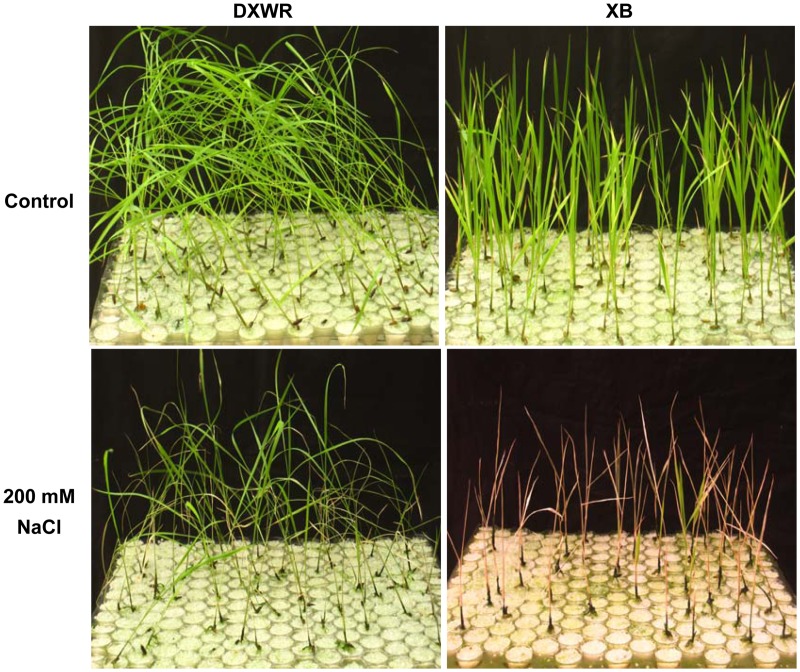
To investigate the character of salt stress resistance in the DXWR seedlings, two-week-old seedlings of DXWR and Xieqingzao B (O. sativa L. ssp. indica, hereafter referred as XB), which is a representative maintainer line in hybrid rice breeding system in China [12] were exposed to the hydroponic solution with 200 mM NaCl for 12 days. When grown under normal, the DXWR and XB seedlings were all green and alive. After exposure to the salt stress, most DXWR seedlings remained green and showed continuous growth, whereas XB seedlings were all dead (Fig 1). The survival rate of the XB seedlings were 0%, but the survival rate of DXWR seedlings reached 80%. These results suggested that DXWR exhibited more salt-stress resistant than the cultivated rice XB at the seedling stage. In this study, 80% of DXWR seedlings were survival under 200 mM NaCl treatment for 12 days, in which the arrest of growth for many varieties of cultivated rice was generally found in previous studies [57–61]. Recent studies have proved that DXWR contained the genes involved in the improvement of salt tolerance in transgenic Arabidopsis [15, 16]. These results indicated that DXWR has the resistant to salt stress compared with the cultivated rice.
Fig 1. The seedlings of DXWR showed stronger salt resistance than the seedlings of XB.
Sequencing statistics
In order to understand the molecular mechanism of salt tolerance of DXWR, RNA-Seq was used to investigate gene expression responding to salt stress at the seedling stage. cDNA libraries were prepared from the leaves and roots of DXWR seedlings and subjected to RNA-Seq analysis on the Illumina HiSeq 2000 platform. A total of 46.8 million, 48.2 million, 43.6 million and 46.7 million high-quality 100-bp paired-end reads were obtained from the leaf and root transcriptome libraries of DXWR seedlings under the normal condition (control) and salt treatment (Table 1). The O. sativa ssp. japonica cv. Nipponbare genome has been completely sequenced through a map-based sequencing strategy [62]. Lu et al. isolated and completely sequenced 1888 putative full-length cDNA clones from wild rice Oryza rufipogon Griff. W1943 for comparative analysis between wild and cultivated rice species. Homology searching of these cDNA sequences revealed that > 96.8% of the wild rice cDNAs were matched to the cultivated rice Nipponbare genome sequence [63]. So in this study, we selected the Nipponbare genome for the references genome to map the reads. The alignment results showed that 70.41–74.63% of the total reads mapped to the reference genome and 58.84–63.34% to the gene regions (including 68.06–73.17% and 36.54–38.57% uniquely matched), respectively (Tables 1 and 2). We found that there was a significant difference in the percentage of reads mapping to the genome and gene regions, especially for the uniquely mapped reads. These results were similar with the previous study that about 72% of the total reads mapped to the genome and 46% to the gene regions (including 68% and 38% uniquely matched) for deep transcriptome sequencing of rhizome and aerial-shoot in Sorghum propinquum [28], suggesting that it might be result from the reads mapping to the intergenic regions and alternative mRNA splicing. The 25.37–29.59% of reads remained unmapped, attributing primarily to the gaps and diversity between DXWR and references genome sequences (Table 1). On average, at least 50% of more than 60% of the mapped genes were covered by the uniquely mapped reads, and only ~15% of the genes had gene coverage of 20% or lower (S1 Fig), indicating a good quality of the transcriptome. Previous studies have revealed that asian cultivated rice (Oryza sativa) was domesticated from the wild species Oryza rufipogon [64] and the sequence of wild rice W1943 had a very high similarity with those of Nipponbare [63]. But, several W1943 cDNAs that did not match to the Nipponbare genomic sequence might be located in the gap of genomic sequence or might be related to wild rice W1943-specific genes [63], suggesting that it has some limitations for using the Nipponbare genome to map the reads of Oryza rufipogon.
Table 1. Summary of Illumina transcriptome reads mapped to the reference genome.
| Reads mapping | Reads number (%) | |||
|---|---|---|---|---|
| LCK | LS | RCK | RS | |
| Total reads | 46,784,432 | 48,221,828 | 43,588,908 | 46,715,548 |
| Total base pairs | 4,678,443,200 | 4,822,182,800 | 4,358,890,800 | 4,671,554,800 |
| Total mapped reads | 34,881,473 (74.56) | 35,987,794 (74.63) | 31,979,139 (73.37) | 32,890,448 (70.41) |
| Perfect match | 25,627,140 (54.78) | 26,067,640 (54.06) | 23,267,595 (53.38) | 23,839,427 (51.03) |
| ≤ 3 bp mismatch | 9,254,333 (19.78) | 9,920,154 (20.57) | 8,711,544 (19.99) | 9,051,021 (19.37) |
| Unique match | 34,230,815 (73.17) | 34,972,876 (72.52) | 31,136,589 (71.43) | 31,795,999 (68.06) |
| Multi-position match | 650,658 (1.39) | 1,014,918 (2.1) | 842,550 (1.93) | 1,094,449 (2.34) |
| Total unmapped reads | 11,902,959 (25.44) | 12,234,034 (25.37) | 11,609,769 (26.63) | 13,825,100 (29.59) |
LCK, LS, RCK, and RS denote the control and salt treatment of the leaves and roots, respectively.
Table 2. Summary of Illumina transcriptome reads mapped to the reference genes.
| Reads mapping | Reads number (%) | |||
|---|---|---|---|---|
| LCK | LS | RCK | RS | |
| Total reads | 46,784,432 | 48,221,828 | 43,588,908 | 46,715,548 |
| Total base pairs | 4,678,443,200 | 4,822,182,800 | 4,358,890,800 | 4,671,554,800 |
| Total mapped reads | 29,634,341 (63.34) | 29,995,403 (62.20) | 26,743,293 (61.35) | 27,488,576 (58.84) |
| Perfect match | 22,232,426 (47.52) | 22,165,310 (45.97) | 19,983,467 (45.85) | 20,422,362 (43.72) |
| ≤ 5 bp mismatch | 7,401,915 (15.82) | 7,830,093 (16.24) | 6,759,826 (15.51) | 7,066,214 (15.13) |
| Unique match | 17,816,293 (38.08) | 17,894,359 (37.11) | 16,812,042 (38.57) | 17,071,704 (36.54) |
| Multi-position match | 11,818,048 (25.26) | 12,101,044 (25.09) | 9,931,251 (22.78) | 10,416,872 (22.30) |
| Total unmapped reads | 17,150,091 (36.66) | 18,226,425 (37.80) | 16,845,615 (38.65) | 19,226,972 (41.16) |
LCK, LS, RCK, and RS denote the control and salt treatment of the leaves and roots, respectively.
Identification of differentially expressed genes (DEGs) by RNA-Seq
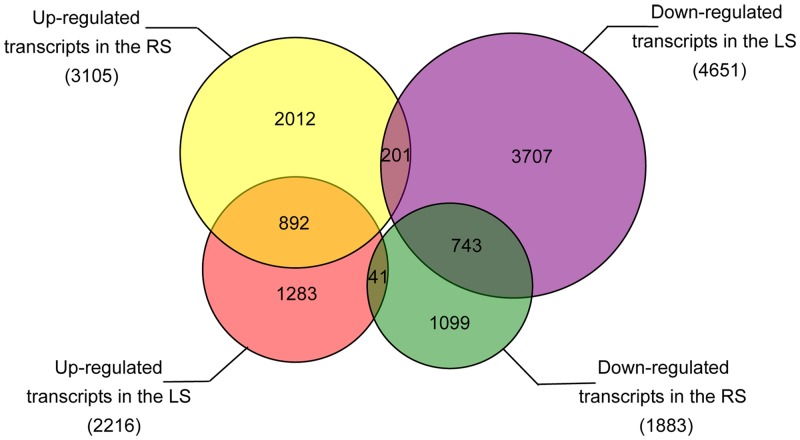
Gene expression levels can be estimated from Illumina sequencing data based on the number of raw reads [65]. Putative DEGs from the treatment vs. control (LS vs. LCK and RS vs. RCK) were identified and a total of 2,216 transcripts showed up-regulation (S1 Table) and 4,651 transcripts showed down-regulation (S2 Table) in LS vs. LCK, whereas 3,105 transcripts showed up-regulation (S3 Table) and 1,883 transcripts showed down-regulation (S4 Table) in RS vs. RCK (Fig 2). Among the DEGs, 892 transcripts were up-regulated and 743 transcripts were down-regulated in both the LS vs. LCK and RS vs. RCK. Interestingly, 41 transcripts were up-regulated in LS vs. LCK but down-regulated in RS vs. RCK, and 201 transcripts were down-regulated in LS vs. LCK but up-regulated in RS vs. RCK. In these 201 transcripts, we detected that the expression of seven genes were dramatically changed (the absolute value of log2 RPKM ratio > 3), including LOC_Os03g37290, LOC_Os06g31800, LOC_Os09g13440, LOC_Os09g19229, LOC_Os10g13430, LOC_Os10g41040, and LOC_Os12g28177. But, so far little is known about their functions.
Fig 2. The number of up- and down-regulated transcripts in the LS and RS compared with the LCK and RCK.
LS, leaves with salt treatment; RS, roots with salt treatment; LCK, leaves without salt treatment; RCK, roots without salt treatment.
Among the DEGs, we found that a number of genes have been proved to be involved in responding to salt stress in the previous studies (Table 3). We also found that most of these genes are transcription factors (TFs), suggesting that TFs play critical roles in responding to salt stress via transcriptional regulation of the downstream genes responsible for plant tolerance to salt challenges.
Table 3. List of published salt resistant genes among the DEGs detected by RNA-Seq.
| Gene name | Gene ID | Up or down (Log2ratio) | References | |
|---|---|---|---|---|
| LS vs. LCK | RS vs. RCK | |||
| ZFP179 | LOC_Os01g62190 | up (3.67) | none | [61] |
| ZFP182 | LOC_Os03g60560 | up (5.65) | up (2.68) | [66] |
| ZFP252 | LOC_Os12g39400 | up (2.48) | none | [67] |
| SNAC1 | LOC_Os03g60080 | up (1.46) | up (1.72) | [68] |
| SNAC2/OsNAC6 | LOC_Os01g66120 | up (2.28) | up (1.21) | [69, 70] |
| OsNAC5 | LOC_Os11g08210 | up (1.26) | up (1.15) | [44] |
| ONAC045 | LOC_Os11g03370 | none | up (1.01) | [71] |
| OsACA6 | LOC_Os04g51610 | up (1.39) | none | [72] |
| OsMYB2 | LOC_Os03g20090 | up (1.39) | up (1.64) | [57] |
| OsbZIP23 | LOC_Os02g52780 | up (1.72) | none | [73] |
| OsBIERF3 | LOC_Os02g43790 | down (-1.13) | up (1.36) | [74] |
| OsHKT1 | LOC_Os06g48810 | none | down (-2.64) | [75] |
| OsLEA3-2 | LOC_Os06g21910 | up (12.63) | none | [76] |
| RSOsPR10 | LOC_Os12g36830 | up (1.91) | up (4.08) | [77] |
| OsTZF1 | LOC_Os05g10670 | up (1.15) | up (1.35) | [43] |
Besides ZFP179, ZFP182 and ZFP252 which have been described for responding to salt stress by the previous studies, other zinc finger proteins (ZFPs) were also detected. For example, two C2H2-type ZFP genes, ZOS3-12 (LOC_Os03g32230) which has been reported to be induced by nitrogen-starvation in rice [78] and ZOS5-08 (LOC_Os05g37190) which has been proved to respond to grazing defoliation [79], indicating that ZFPs could play important roles in regulating various stress tolerance. A genome-wide annotation analysis of the CCCH-type ZFPs identified 67 genes in rice [80]. Apart from OsTZF1 [43], we also detected other many CCCH-type ZFP genes (S5 Table), indicating that these CCCH-type ZFPs could be functional redundancy in salt-stress tolerance.
NAC (NAM, ATAF, and CUC) proteins are a class of plant-specific TFs, which contain a highly conserved N-terminal domain known as the NAC domain. Ooka et al. reported that 75 and 105 NAC genes were predicted in rice and Arabidopsis genomes, respectively [81]. Overexpression of four NAC domain genes, SNAC1 [68], SNAC2 [70], OsNAC5 [82], and ONAC045 [71], in rice conferred tolerance to salt stress. Besides these four genes described by previous studies, another 16 NAC domain genes were also found in our study (S6 Table). Our data combined with the previously determined functions of the NAC domain genes strongly suggested their important roles in salt-stress tolerance. Additionally, the 9 of 16 genes were down-regulated by salt stress in the leaves and/or roots of seedlings, and 6 of 16 genes were up-regulated by salt stress in the leaves or roots of seedlings. Interestingly, we also observed one gene, ONAC066 (LOC_Os03g56580) that was down-regulated in the leaves, but up-regulated in the roots by salt stress. These results suggested that a number of NAC domain genes were involved in salinity tolerance, but there were different potential mechanisms of salt stress responses regulated by NAC domain genes.
MYB-type TFs play a diverse role in plant development and response to abiotic stress. MYB proteins contain one, two, or three imperfect repeats (51–53 amino acids) in their DNA-binding domain, and they are further classified into three subfamilies, type MYBR2R3, MYB-related, and type MYBR1R2R3, depending on the number of repeats in their MYB domains [83]. Recently, two R2R3-type MYB proteins (OsMYB2 [57] and OsMYB91 [7]) and one R1R2R3-type MYB protein (OsMYB3R-2) [84] were identified to be involved in response to salt stress. We found 62 MYB-type genes which were induced by salt treatment in the leaves and/or roots of DXWR seedlings besides OsMYB2 (S7 Table). Among these genes, 47 genes were R2R3-type, 13 genes were MYB-related type, and 2 genes were R1R2R3-type. This result was consistent with that among the MYB proteins in plants, the MYB family with the two-repeat (R2R3) was the most common one and a large number of them were involved in responses of plants to environmental stress [85]. Moreover, a few MYB-related type genes were detected by our RNA-Seq analyses, and most of them (9 of 13) were up-regulated by salt stress, but so far, little is known about that the MYB-related type proteins are related with the response to salt stress. Our data indicated that the MYB-related type TFs could also participate in the response to salt stress, or there was another different potential mechanism of salt stress responses regulated by the MYB-related type TFs in DXWR seedlings compared with cultivated rice.
In plants, like other TF genes, members of the bZIP (basic leucine zipper) TF family also have been identified to regulate diverse plant-specific phenomena, including seed maturation and germination, floral induction and development, light signaling, and abiotic stress tolerance. 89 genes encoding bZIP TFs have been identified and characterized in the rice genome and they were classified into 10 subfamilies [86]. Recent studies have reported that OsbZIP05 (OSBZ8) [87] and OsbZIP23 [73] conferred salinity tolerance in rice. Besides OsbZIP23, we also found 27 salt-induced bZIP TFs distributed in 6 subfamilies (I, IV, V, VI, VII, and IX) (S8 Table). OsbZIP38 (LIP19), OsbZIP52 (RISBZ5), OsbZIP87 (OsOBF1), OsbZIP12, OsbZIP16, and OsbZIP45 have been identified to confer abiotic stress tolerance [88–92]. According to the previous studies and our RNA-Seq analyses, we observed that all these stress-inducible bZIP TFs were included in the above 6 subfamilies, suggesting that the bZIP TFs of these 6 subfamilies may play more important roles in responding to abiotic stress compared with the other subfamilies. Furthermore, recent studies have proved that OsbZIP12 [93], OsbZIP23 [73], and OsbZIP45 [92] were involved in not only drought tolerance but also salinity tolerance. OsbZIP16 also has been identified to positively regulate drought resistance in rice [91]. OsbZIP71 was shown to play an important role in ABA-mediated drought and salt tolerance in rice [41]. Our data combined with the previous studies strongly suggested that there could be the similar potential mechanisms regulated by the bZIP TFs between the salt and drought stress responses.
In plant kingdom, AP2/ERF (APETALA2/ethylene response factor) is a large family of TFs. To date, 163 AP2/ERF-type TFs were identified in rice and divided into four subfamilies [94]. The AP2/ERF-type TFs have been shown to be highly involved in the salt stress tolerance mechanisms [3, 74, 95–97]. Our RNA-Seq analyses showed that 41 AP2/ERF-type TF genes were up-regulated or down-regulated by salt stress in the leaves and/or roots of seedlings. Among these genes, eight genes belonged to the AP2 subfamily; twelve genes belonged to the DREB subfamily, seventeen genes belonged to the ERF subfamily, and four genes belonged to the RAV subfamily (S9 Table). Although a few AP2/ERF genes were identified to be related with the response to salt stress, they all belonged to the DREB and ERF subfamilies and little is known about whether the TFs of AP2 and RAV subfamilies participate in the salt-stress response. We found eight AP2 genes and four RAV genes, suggesting that there could be some unknown molecular mechanisms of salt stress responses regulated by the TFs of AP2 and RAV subfamilies in rice or some special mechanism of salt stress responses regulated by AP2/ERF-type TFs in DXWR seedlings compared with cultivated rice. Additionally, we found all eight AP2 genes were down-regulated by salt stress and except of one gene (LOC_Os11g03540) was down-regulated in both the leaves and roots, all the other genes were down-regulated only in leaves. These results suggested that the AP2-type TFs could mainly negatively regulate salt stress responses in the leaves of DXWR seedlings.
To further identify the novel TFs involved in the salt stress tolerance in our study, the striking up-regulated TF genes were selected (the value of log2 RPKM ratio > 3). In the LS vs. LCK, we detected 20 TF genes, including two bHLH (basic helix-loop-helix) genes, seven ZFP genes, and two WRKY genes (S10 Table). In the RS vs. RCK, we found 26 TF genes, including nine ZFP genes, five WRKY genes, and four bHLH genes (S11 Table). Among these genes, four genes were up-regulated both in the LS vs. LCK and the RS vs. RCK, including OsWRKY42 (LOC_Os02g26430), two Homeodomain-leucine zipper TF genes (LOC_Os02g43330 and LOC_Os04g45810), and one GRAS family TF gene (LOC_Os11g04400). To date, expect OsWRKY42 [42] and OsbHLH133 (LOC_Os12g32400) [98] which have been studied for their functions, little was known about the functions for the other genes in rice. Research on salt regulatory TFs has mainly focused on single factors and linear pathways. However, emerging findings increasingly suggest integration of the TFs in dynamic network hubs as well as interaction and competition of pathways manifesting complexity of molecular links in stress adaptation. After salt stress, the plant hormone biosynthesis was changed dynamically and then Ca2+, ROS (reactive oxygen species), and hormone signaling cascades were activated. At the next step, the global transcriptional profile of the plant (including TF families, such as bZIP, NAC, AP2/ERF, WRKY, bHLH and MYB) could be altered and regulated many salt stress responsive genes expression. Ultimately, these early signaling pathways result in expression and activation of cellular detoxification mechanisms, including HKT, NHX, and the SOS Na+ transport mechanisms as well as osmotic protection strategies [40]. Furthermore, within this hierarchical network, cellular stress responses might be fine tuned by interaction and competition of TFs that regulate sub-clusters of the stress transcriptome. In the hierarchical network of TFs for stress adaptation, bZIP TFs could be in the upstream position and other TFs might have functions in regulating sub-networks of adaptation to salt stress [32].
Besides a large number of TFs, two HKT (high-affinity potassium transporter) genes, OsHKT1 and OsHKT7 were detected by our RNA-Seq analyses. Consisting with the observation that the OsHKT1 transcript was significantly down-regulated in salt-tolerant rice under NaCl stress [75], we observed that OsHKT1 was down-regulated by salt stress in the roots of DXWR seedlings. There is little known about that OsHKT7 contributes to salt tolerance in rice, but its homologue, TmHKT7 in durum wheat (Triticum turgidum subsp. durum) was identified to be involved in salt stress response [99]. We found that OsHKT7 was down-regulated by salt stress both in the leaves and roots, suggesting that OsHKT1 and OsHKT7 could act redundantly to negatively regulate salt stress response.
To confirm the validity of the DEG data, quantitative RT-PCR was performed to investigate the expression patterns of 32 randomly selected genes under the same conditions. We compared the results obtained from quantitative RT-PCR with the gene expression profiles from RNA-Seq analysis. Expression trends were consistent for all transcripts in both analyses, with a correlation coefficient of R2 = 0.8961 (S2 Fig).
Functional classification by Gene Ontology (GO)
Web Gene Ontology Annotation Plot (WEGO) software [100] was used to perform the GO classifications and to draw the GO tree to classify the up- and down-regulated transcripts into putative functional groups for the LS vs. LCK and RS vs. RCK. A total of 15,636 and 17,937 transcripts were assigned GO terms for the DEGs in the LS vs. LCK and RS vs. RCK, respectively. Among the 15,636 transcripts from the LS vs. LCK (Fig 3A), there were 5,633 transcripts at the cellular level, 5,068 transcripts at the molecular level and 4,935 transcripts at the biological level. Among the 17,937 transcripts from the RS vs. RCK (Fig 3B), there were 6,302 transcripts at the cellular level, 5,907 transcripts at the molecular level and 5,728 transcripts at the biological level. In the biological process category, cellular process and metabolic process were the most highly represented groups, suggesting that extensive metabolic activities were taking place both in the leaves and roots of DXWR seedlings with salt treatment. Within the cellular component category, transcripts that corresponded to cell, cell parts and cell organelles were the most abundant. Binding and catalytic activities were the most abundant groups within the molecular functional category.
Fig 3. Gene ontology (GO) classification of the unigenes from the LS-vs-LCK (A) and RS-vs-RCK (B).
We further identified GO terms in the biological process category that were over-represented (P < 0.05) in DEGs of LS vs. LCK and RS vs. RCK, respectively (S12 and S13 Tables). These GO terms served as indicators of significantly different biological processes underlying salt-stress responses in the leaves and roots. GO terms such as metabolic process, catabolic process, and response to stimulus (Table 4) were enriched in both sets of transcripts from the two tissues, suggesting that the same biological processes were required to maintain the tissues activities during salt treatment. However, some striking differences were found between the two sets of enriched GO terms. In particular, GO terms related to cell cycle process and DNA reproductive process were highly enriched in the DEGs of LS vs. LCK, suggesting that salt stress may affect the reproduction of the plant cell and the plant may utilize this effect to resist the salt stress in the leaves of seedlings. For the RS vs. RCK, there were a large number of DEGs involved in transport, response to metal ion, response to abiotic stimulus, and regulation of reactive oxygen species metabolic process. This result indicated that the roots could be the main tissue for the plant to resist the salt stress and maintaining a low cytosolic Na+ concentration could be the most important way to improve salt-stress tolerance for the plant [40]. We also further identified GO terms in the cellular component and molecular function categories that were over-represented (P < 0.05) in DEGs of LS vs. LCK and RS vs. RCK, respectively (S14–S17 Tables). In the cellular component category, cell periphery and extracellular region were enriched in both sets of transcripts from the two tissues (S18 Table). This indicated that that salt stress affected the cell-cell interactions. The cytoplasmic membrane-bounded vesicle was also enriched in the two tissues, indicating an involvement of endosomal transport proteins, including NHXs, in plant salt tolerance, by transporting K+ into vacuoles to control organelle pH and ion homeostasis [101]. For the molecular function category, endopeptidase regulator activity was enriched in the two tissues (S19 Table). This suggested that some stress responsive genes expression might be regulated by posttranslational modification to cope with salinity stress. The peptidase regulator activity and carbohydrate derivative binding were enriched and this was consistent with the previous studies that the accumulation of organic osmolytes, such as sugar alcohols, proline, and polyamines, plays a key role in maintaining the low intracellular osmotic potential of plants and in preventing the harmful effects of salt stress [102, 103].
Table 4. The significant GO terms of DEGs for the biological process category both in the LS vs. LCK and RS vs. RCK.
| GO term | GO term annotation |
|---|---|
| GO:0006022 | aminoglycan metabolic process |
| GO:0006026 | aminoglycan catabolic process |
| GO:0006030 | chitin metabolic process |
| GO:0006032 | chitin catabolic process |
| GO:0010035 | response to inorganic substance |
| GO:0010466 | negative regulation of peptidase activity |
| GO:0022607 | cellular component assembly |
| GO:0034622 | cellular macromolecular complex assembly |
| GO:0043933 | macromolecular complex subunit organization |
| GO:0046348 | amino sugar catabolic process |
| GO:0050896 | response to stimulus |
| GO:0051346 | negative regulation of hydrolase activity |
| GO:0052547 | regulation of peptidase activity |
| GO:0065003 | macromolecular complex assembly |
| GO:1901072 | glucosamine-containing compound catabolic process |
Especially, we detected some enriched GO terms for the biological process category in LS vs. LCK, including chromatin assembly or disassembly, nucleosome assembly, histone phosphorylation and histone H3-K9 methylation, which belonged to epigenetic processes and were related to transcription. Efficiency of gene expression is highly influenced by chromatin structure that might be modulated epigenetically by processes as DNA methylation and posttranslational modifications of histones. The histone-mediated structure of nucleosomes in the chromatin might be modified and thus influence nucleosome density, binding efficiency of TFs, and transcriptional activity [104]. Several studies have shown that chromatin modification contribute to plant environmental adaptation [105, 106]. However, in contrast to the detailed knowledge on influences of epigenetic mechanisms on developmental processes, information on epigenetic regulation of abiotic stress resistance is still rare.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway mapping
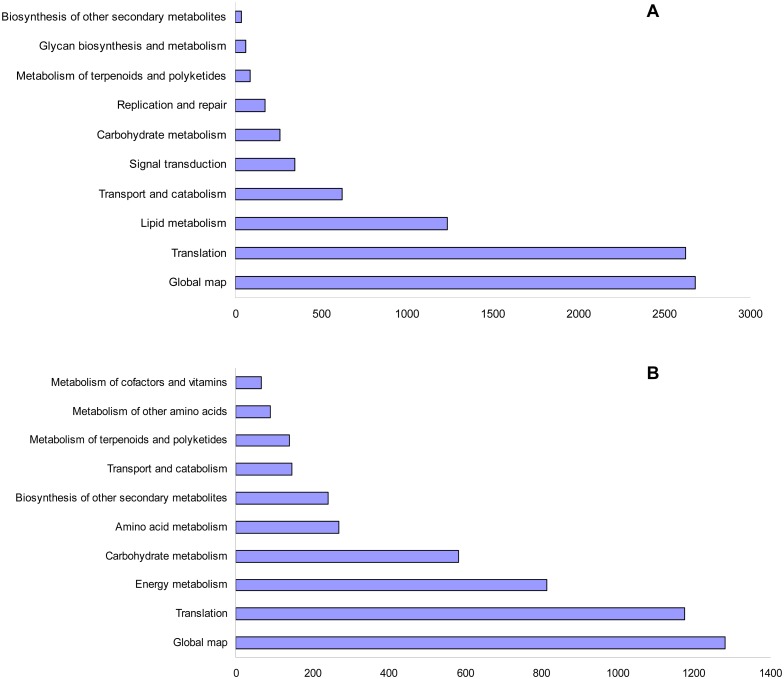
To identify metabolic pathways in which DEGs were involved and enriched, pathway-based analysis was performed using the KEGG pathway database [107]. As a result, 4,131 of 6,876 in the LS vs. LCK and 3,069of 4,988 in the RS vs. RCK were classified into 20 functional categories. Then they were classified into 125 and 126 subcategories, respectively. We further identified over-represented KEGG Orthology (KO) terms (Q-value < 0.05), and classified them into 10 categories, respectively (Fig 4). As shown in Fig 4, these transcripts belonged mainly to the following KEGG pathways both in the LS vs. LCK and RS vs. RCK: Biosynthesis of other secondary metabolites, carbohydrate metabolism, global map, metabolism of terpenoids and polyketides, translation, and transport and catabolism. Then they were further classified into 19 and 27 subcategories (S20 and S21 Tables). Among these subcategories, we found that five subcategories, metabolic pathways, biosynthesis of secondary metabolites, amino sugar and nucleotide sugar metabolism, RNA transport, and mRNA surveillance pathway were over-represented both in the LS vs. LCK and RS vs. RCK (S3 Fig). Furthermore, RNA transport and mRNA surveillance pathways were related to transcription, indicating that these pathways might have a modulating impact on the regulation of salt-inducible gene expression. In contrast, KO terms related to endocytosis, ether lipid metabolism, and glycerophospholipid metabolism were exclusively enriched in the LS vs. LCK. This result suggested that there could be considerable differences of biochemical and physiological processes between the leaves and roots of seedlings during salt stress. These annotations provide a valuable resource for investigating specific processes, functions, and pathways of salt-stress responses in different tissues.
Fig 4. KEGG pathway assignments in the LS vs. LCK (A) and RS vs. RCK (B).
The represented categories (Q-value≤ 0.05) and the number of transcripts predicted to belong to each category are shown.
Comparative analysis of the DEGs in our RNA-Seq with those previously identified by RNA-Seq in cultivated rice Nipponbare
Cultivated rice is considered to have been domesticated from wild rice thousands of years ago. However, in the long-term domestication, cultivated rice lost various valuable traits with regard to tolerance to cold, drought and salinity which derived from wild rice [9]. To investigate the similarities and differences of salt stress responsive mechanisms between cultivated rice and wild rice, we compared our RNA-Seq data with the data of the previous study which used RNA-Seq to investigate the DEGs in the shoots and roots of Nipponbare seedlings with 1 h of salinity stress compared with normal rice shoots and roots [30]. According to the above criteria: false discovery rate (FDR) ≤ 0.001 and the absolute value of log2 RPKM ratio ≥ 1, the previous study showed that a total of 665 genes were up-regulation (S22 Table) and 296 genes were down-regulation (S23 Table) in shoots, whereas 1,365 genes were up-regulation (S24 Table) and 1,425 genes were down-regulation (S25 Table) in roots. Among the DEGs, 352 genes were up-regulated and 76 genes were down-regulated in both the shoots and roots.
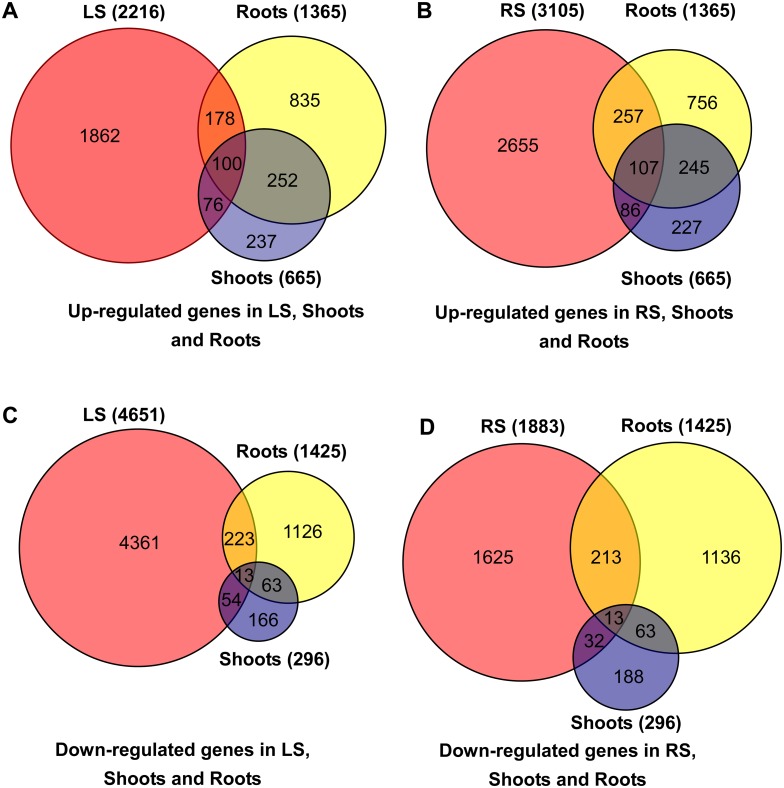
Comparing our data with the previous study, we found that 176 genes were up-regulated in both the LS vs. LCK (for convenient description, LS vs. LCK was named as LS) and shoots, 278 genes were up-regulated in both the LS and roots, and 100 genes were all up-regulated in the LS, shoots and roots (Fig 5A). We also found that 67 genes were down-regulated in both the LS and shoots, 236 genes were down-regulated in both the LS and roots, and 13 genes were all down-regulated in the LS, shoots and roots (Fig 5C). We observed that 193 genes were up-regulated in both the RS vs. RCK (for convenient description, RS vs. RCK was named as RS) and shoots, 364 genes were up-regulated in both RS and roots, and 107 genes were all up-regulated in RS, shoots and roots (Fig 5B). We also observed that 45 genes were down-regulated in both the RS and shoots, 226 genes were down-regulated in both the RS and roots, and 13 genes were all down-regulated in the RS, shoots and roots (Fig 5D).
Fig 5. The number of up- and down-regulated genes in the LS (LS vs. LCK), RS (RS vs. RCK), shoots, and roots.
(A), up-regulated genes in the LS, shoots, and roots; (B), up-regulated genes in the RS, shoots, and roots; (C), down-regulated genes in the LS, shoots, and roots; (D), down-regulated genes in the RS, shoots, and roots.
Furthermore, we observed that 57 genes and 8 genes were all up-regulated and down-regulated in the LS, RS, shoots, and roots, respectively (S26 and S27 Tables). Among the 57 up-regulated genes, one LEA (late embryogenesis abundant) gene (LOC_Os01g21250) was found and the previous studies have proved that the LEA genes, OsLEA3-2 [76] and JcLEA [108] conferred salinity tolerance in the transgenic Arabidopsis plants, respectively. Our data combined with the previous studies strongly suggested that the plants could have a common molecular mechanism regulated by LEA proteins to respond to salt stress in different tissues. We also found four WRKY TFs, WRKY24, WRKY28, WRKY70, and WRKY113 among the 57 up-regulated genes. This result is consistent with the previous observations that WRKY-type TFs play important roles in the adaptation to abiotic stresses [109–111]. Two salt-responsive genes (LOC_Os01g64360 and LOC_Os02g52670) which belonged to MYB TF family and AP2/ERF TF family respectively were also observed not only in DXWR but also in cultivated rice Nipponbare, indicating that the salt-stress response pathway regulated by MYB and AP2/ERF TFs in cultivated rice may originate from wild rice.
Plants respond to abiotoc stress through a complexity of signaling pathways, and the dephosphorylation mediated by protein phosphatase (PP) is an important event in this process. In the rice genome, 90 PP2C were identified and classified into 11 groups [112]. Recent studies have reported that two PP2C genes, OsPP18 [113] and OsPP108 [114] were essential for plant tolerance to several abiotic stresses, such as drought, salt, mannitol, and oxidative stresses. Among the 57 up-regulated genes, we also found two PP2C genes (LOC_Os03g16170 and LOC_Os09g15670). These results suggested that many PP2C genes could act redundantly in the phosphorylation cycles to make the phosphorylated proteins to switch rapidly from one state to another, which allows rice to respond to stress stimuli rapidly and accurately. 13 TPP (trehalose-6-phosphate phosphatase) genes and 14 TPS (trehalose-6-phosphate synthase) have been identified in the rice and one TPP gene, OsTPP1 [115] and one TPS gene, OsTPS1 [116] have been found that their overexpression in rice enhanced tolerance to abiotic stress by activating stress tolerance related genes. Besides OsTPP5 (LOC_Os03g26910) which was found in the 57 up-regulated genes, we also found three TPP genes (OsTPP6, OsTPP7, and OsTPP8), and four TPS genes (OsTPS4, OsTPS6, OsTPS9, and OsTPS10) in DXWR and Nipponbare, but not all in the LS, RS, shoots, and roots. Moreover, one TPS gene, OsTPS8 was only found in DXWR but not in Nipponbare. These results suggested that TPP genes and TPS genes could act together to confer stress tolerance through activating stress responsive genes, but different tissues had their own specific salt-stress response pathway regulated by TPP and TPS genes.
Two RLCK (receptor-like cytoplasmic kinase) genes, OsRLCK167 (LOC_Os04g56060) and OsRLCK253 (LOC_Os08g28710) were observed in the 8 down-regulated genes and 57 up-regulated genes, respectively. This result was consistent with the previous study that OsRLCK167 was down-regulated and OsRLCK253 was up-regulated under salt stress in rice seedlings [117]. To date, however, the molecular mechanism of salt-stress responses regulated by RLCK genes remained unknown. According to our RNA-Seq analyses and previous researches, we supposed that these two genes could play antagonistic roles in responding to salt stress. Taken together, the number of salt-inducible genes in DXWR is much more than that of cultivated rice Nipponbare, whether it was the number of up-regulated genes or down-regulated genes, suggesting that DXWR may have more complex molecular mechanisms and signaling pathways than the cultivated rice Nipponbare to preferably adapt to salt stress. Meanwhile, we should realize that there could be limitations in comparison of RNA-Seq results, due to technical differences, such as differences in depth of RNA-Seq experiments and differences in read lengths and mapping. Therefore, the differences between DXWR and Nipponbare in salt-stress responses need to be further analyzed in future.
DEGs mapped to the previously identified salt stress related QTL intervals
Map-based cloning is one approach that may be used for the identification of genes underlying QTLs. Several QTLs associated with yield [118], cold tolerance [13], brown planthopper resistance [11], and drought tolerance [9] have been identified in DXWR. Based on the Gramene QTL database (www.gramene.org/db/), a total of 17 QTLs related to salt tolerance in rice have been identified. We located 276 genes differentially regulated by salt stress on 7 of these identified QTL intervals. Among them, the QTLs qST1, qSDS-6, and qRNC-9 had the greatest number of co-localized DEGs with 82, 41, and 41 genes, respectively (S28 Table).
The QTL qST1 is the most important QTL related to salt stress tolerance in rice at seedling stage [119]. 82 DEGs were co-localized on the qST1 interval. Among them, many TFs were found, including two NAC genes, ONAC048 and ONAC068; three WRKY genes, WRKY22, WRKY24, and WRKY108; three C2H2-type ZFP genes, ZOS1-14, ZOS1-15, and ZOS1-17; three bZIP genes, OsbZIP10, OsbZIP11, and OsbZIP12; one MYB gene (LOC_Os01g64360); one C3HC4-type ZFP gene ((LOC_Os01g64620); one AP2/ERF gene (LOC_Os01g64790). We also found one PP2C gene ((LOC_Os01g62760) and the P5CS1 gene which encodes for a bifunctional enzyme that catalyzes the rate limiting reaction in proline biosynthesis in living organisms and has been proved to be involved in abiotic stress tolerance [120].
The major QTL associated with survival days of seedlings under salt stress, designated as qSDS-6 [121], was co-localized with 41 DEGs, including one GATA-type ZFP gene (LOC_Os06g37450) and a VQ gene (LOC_Os06g40090) which has been reported to be involved in response to abiotic stresses [122]. 41DEGs co-localized on a major QTL qRNC-9, which included four TF genes, two bHLH (basic helix-loop-helix) genes (LOC_Os09g24490 and LOC_Os09g28210), one WRKY gene WRKY62, and one C3HC4-type ZFP gene (LOC_Os09g29310). In addition, we also found one annexin gene (LOC_Os09g27990) and the rice annexin genes have been found to be regulated in seedlings stage by various abiotic stressors including salinity, drought, heat and cold [123]. By combining further functional identification and QTL fine mapping, the co-localized DEGs detected in this RAN-Seq analysis may provide the basic for gene cloning and elucidation of the common molecular mechanisms responsible for salt tolerance between in the DXWR and rice.
Conclusions
DXWR has several desired agronomic traits, and therefore it is considered as an important genetic resource for rice breeding. In this study, we used RNA-Seq platform to systematically investigate the salinity stress-inducible transcripts of leaves and roots at the seedling stage of DXWR. Identified DEGs with contrasting expression patterns between leaves and roots in response to salt stress are excellent targets for further functional studies to understand more specific molecular mechanisms of salt tolerance. Furthermore, this data set, compared with the previous RNA-Seq analysis and QTLs for salt tolerance in rice, provides clues for candidate transcripts and more complete information that are essential for future studies into the specific molecular mechanisms of salt-stress responses in DXWR. Further experiments are in progress towards functional validation of putative candidate genes to provide genetic resources for the improvement of salt-stress tolerance in rice.
Supporting Information
(PDF)
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
(PDF)
(XLS)
(PDF)
Acknowledgments
We thank Prof. Jieyun Zhuang and Dr. Junyu Chen for technical support and excellent discussions, and the associate editor and anonymous reviewers for their valuable suggestions.
Abbreviations
- AP2
APETALA2
- bHLH
basic helix-loop-helix
- bZIP
basic leucine zipper
- DEGs
differentially expressed genes
- DXWR
Dongxiang wild rice
- ERF
ethylene response factor
- FDR
false discovery rate
- GO
Gene Ontology
- HKT
high-affinity potassium transporter;
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KO
KEGG Orthology
- LEA
late embryogenesis abundant
- NAC
NAM, ATAF, and CUC
- PP
protein phosphatase
- RPKM
reads per kilobase per million reads
- QTLs
quantitative trait loci
- RLCK
receptor-like cytoplasmic kinase
- TFs
transcription factors
- TPP
trehalose-6-phosphate phosphatase
- WEGO
Web Gene Ontology Annotation Plot
- XB
Xieqingzao B
- ZFPs
zinc finger proteins
Data Availability
The sequence reads were submitted to GenBank GEO database under accession number GSE73181 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73181).
Funding Statement
This study was supported by the grants from the National Natural Science Foundation of China (31360327 and 31500259), the Natural Science Foundation of Jiangxi Province, China (20142BAB214012 and 20151BAB204006), and the Key Projects of Jiangxi Education Department (KJLD12059). The funders had no rule in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot 2007. November 01; 100(5): 959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang J, Qu Y, Yang C, Ma X, Cao G, Zhao Z, et al. Identification of QTLs associated with salt or alkaline tolerance at the seedling stage in rice under salt or alkaline stress. Euphytica 2015. February 01; 201(3): 441–52. [Google Scholar]
- 3.Schmidt R, Mieulet D, Hubberten HM, Obata T, Hoefgen R, Fernie AR, et al. Salt-responsive ERF1 regulates reactive oxygen species-dependent signaling during the initial response to salt stress in rice. Plant Cell 2013. June 01; 25(6): 2115–31. 10.1105/tpc.113.113068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh A, Pareek A, Sopory SK, Singla-Pareek SL. A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J 2014. October 01; 80(1): 93–105. 10.1111/tpj.12621 [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Uddin MI, Tanaka K, Yin L, Shi Z, Qi Y, et al. Maintenance of Chloroplast Structure and Function by Overexpression of the Rice MONOGALACTOSYLDIACYLGLYCEROL SYNTHASE Gene Leads to Enhanced Salt Tolerance in Tobacco. Plant Physiol 2014. May 19; 165(3): 1144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou H, Lin H, Chen S, Becker K, Yang Y, Zhao J, et al. Inhibition of the Arabidopsis Salt Overly Sensitive Pathway by 14-3-3 Proteins. Plant Cell 2014. March 21; 26(3): 1166–82. 10.1105/tpc.113.117069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu N, Cheng S, Liu X, Du H, Dai M, Zhou D, et al. The R2R3-type MYB gene OsMYB91 has a function in coordinating plant growth and salt stress tolerance in rice. Plant Sci 2015. July 01; 236(0):146–56. [DOI] [PubMed] [Google Scholar]
- 8.Xie J, Agrama HA, Kong D, Zhuang J, Hu B, Wan Y, et al. Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon). Genet Resour Crop Ev 2010. April 01; 57(4): 597–609. [Google Scholar]
- 9.Zhang X, Zhou S, Fu Y, Su Z, Wang X, Sun C. Identification of a drought tolerant introgression line derived from Dongxiang common wild rice (O. rufipogon Griff.). Plant Mol Biol 2006. September 01; 62(1–2): 247–59. [DOI] [PubMed] [Google Scholar]
- 10.Xia H, Lu BR, Su J, Chen R, Rong J, Song Z, et al. Normal expression of insect-resistant transgene in progeny of common wild rice crossed with genetically modified rice: its implication in ecological biosafety assessment. Theor Appl Genet 2009. August 01; 119(4): 635–44. 10.1007/s00122-009-1075-5 [DOI] [PubMed] [Google Scholar]
- 11.Huang D, Chen J, Lai F, Liu G, Zhuang J. Analysis of Quantitative Trait Loci for Resistance to Brown Planthopper in Dongxiang Wild Rice (Oryza rufipogon Griff.). Acta Agronomica Sinica 2012. February 01; 38(2): 210–4. [Google Scholar]
- 12.Zhang F, Cui F, Zhang L, Wen X, Luo X, Zhou Y, et al. Development and identification of a introgression line with strong drought resistance at seedling stage derived from Oryza sativa L. mating with Oryza rufipogon Griff. Euphytica 2014. November 01; 200(1): 1–7. [Google Scholar]
- 13.Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, et al. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet 2015. April 11; 128(7): 1359–71. 10.1007/s00122-015-2511-3 [DOI] [PubMed] [Google Scholar]
- 14.Xiao N, Huang WN, Li AH, Gao Y, Li YH, Pan CH, et al. Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor Appl Genet 2015. January 01; 128(1): 173–85. 10.1007/s00122-014-2420-x [DOI] [PubMed] [Google Scholar]
- 15.Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol 2009. August 15; 166(12): 1296–306. 10.1016/j.jplph.2009.02.007 [DOI] [PubMed] [Google Scholar]
- 16.Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep 2010. September 01; 29(9): 977–86. 10.1007/s00299-010-0883-z [DOI] [PubMed] [Google Scholar]
- 17.Pratt LH, Liang C, Shah M, Sun F, Wang H, Reid SP, et al. Sorghum expressed sequence tags identify signature genes for drought, pathogenesis, and skotomorphogenesis from a milestone set of 16,801 unique transcripts. Plant Physiol 2005. October 01; 139(2): 869–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SJ, Huang Y, Ayoubi P. Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 2006. April 01; 223(5): 932–47. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Tian L, Lee HS, Wei NE, Jiang H, Watson B, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 2006. January 01; 172(1): 507–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge X, Chen W, Song S, Wang W, Hu S, Yu J. Transcriptomic profiling of mature embryo from an elite super-hybrid rice LYP9 and its parental lines. BMC Plant Biol 2008. January 20; 8:114 10.1186/1471-2229-8-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiemann A, Fu J, Schrag TA, Melchinger AE, Frisch M, Scholten S. Correlation between parental transcriptome and field data for the characterization of heterosis in Zea mays L. Theor Appl Genet 2010. January 01; 120(2): 401–13. 10.1007/s00122-009-1189-9 [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Jenssen TK, Trimarchi J, Punzo C, Cepko CL, Ohno-Machado L, et al. Comparison of hybridization-based and sequencing-based gene expression technologies on biological replicates. BMC Genomics 2007January 20; 8: 153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 2009. January 01; 10(1): 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.T H P, Friedlander MR, Almlof J, Sammeth M, Pulyakhina I, Anvar SY, et al. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol 2013. November 01; 31(11): 1015–22. 10.1038/nbt.2702 [DOI] [PubMed] [Google Scholar]
- 25.Filichkin SA, Priest HD, Givan SA, Shen R, Bryant DW, Fox SE, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res 2010. January 01; 20(1): 45–58. 10.1101/gr.093302.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kakumanu A, Ambavaram MM, Klumas C, Krishnan A, Batlang U, Myers E, et al. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol 2012. October 01; 160(2): 846–67. 10.1104/pp.112.200444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koenig D, Jimenez-Gomez JM, Kimura S, Fulop D, Chitwood DH, Headland LR, et al. Comparative transcriptomics reveals patterns of selection in domesticated and wild tomato. Proc Natl Acad Sci U S A. 2013. July 09; 110(28): E2655–62. 10.1073/pnas.1309606110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang T, Zhao X, Wang W, Huang L, Liu X, Zong Y, et al. Deep transcriptome sequencing of rhizome and aerial-shoot in Sorghum propinquum. Plant Mol Biol 2014. February 01; 84(3): 315–27. 10.1007/s11103-013-0135-z [DOI] [PubMed] [Google Scholar]
- 29.Liao JL, Zhou HW, Peng Q, Zhong PA, Zhang HY, He C, et al. Transcriptome changes in rice (Oryza sativa L.) in response to high night temperature stress at the early milky stage. BMC Genomics 2015. January 20; 16: 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizuno H, Kawahara Y, Sakai H, Kanamori H, Wakimoto H, Yamagata H, et al. Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 2010. January 20; 11: 683 10.1186/1471-2164-11-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oono Y, Kawahara Y, Kanamori H, Mizuno H, Yamagata H, Yamamoto M, et al. mRNA-Seq Reveals a Comprehensive Transcriptome Profile of Rice under Phosphate Stress. Rice 2011. June 01; 4(2): 50–65. [Google Scholar]
- 32.Golldack D, Luking I, Yang O. Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep 2011. August 01; 30(8): 1383–91. 10.1007/s00299-011-1068-0 [DOI] [PubMed] [Google Scholar]
- 33.Yang O, Popova OV, Suthoff U, Luking I, Dietz KJ, Golldack D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009. May 01; 436(1–2): 45–55. 10.1016/j.gene.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 34.Jiang Y, Deyholos MK. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol Biol 2009. January 01; 69(1–2): 91–105. 10.1007/s11103-008-9408-3 [DOI] [PubMed] [Google Scholar]
- 35.Wang F, Chen HW, Li QT, Wei W, Li W, Zhang WK, et al. GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J 2015. July 01; 83(2): 224–36. 10.1111/tpj.12879 [DOI] [PubMed] [Google Scholar]
- 36.Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, et al. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 2010. June 01; 457(1–2): 1–12. 10.1016/j.gene.2010.02.011 [DOI] [PubMed] [Google Scholar]
- 37.Cui MH, Yoo KS, Hyoung S, Nguyen HT, Kim YY, Kim HJ, et al. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. Febs Lett 2013. June 19; 587(12): 1773–8. 10.1016/j.febslet.2013.04.028 [DOI] [PubMed] [Google Scholar]
- 38.Liu W, Tai H, Li S, Gao W, Zhao M, Xie C, et al. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol 2014. March 01; 201(4): 1192–1204. 10.1111/nph.12607 [DOI] [PubMed] [Google Scholar]
- 39.Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004. September 01; 16(9): 2481–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deinlein U, Stephan AB, Horie T, Luo W, Xu G, Schroeder JI. Plant salt-tolerance mechanisms. Trends Plant Sci 2014. June 01; 19(6): 371–79. 10.1016/j.tplants.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu C, Mao B, Ou S, Wang W, Liu L, Wu Y, et al. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol Biol 2014. October 01; 84(1–2): 19–36. 10.1007/s11103-013-0115-3 [DOI] [PubMed] [Google Scholar]
- 42.Han M, Kim CY, Lee J, Lee SK, Jeon JS. OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol Cells 2014. July 01; 37(7): 532–9. 10.14348/molcells.2014.0128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jan A, Maruyama K, Todaka D, Kidokoro S, Abo M, Yoshimura E, et al. OsTZF1, a CCCH-tandem zinc finger protein, confers delayed senescence and stress tolerance in rice by regulating stress-related genes. Plant Physiol 2013. March 01; 161(3): 1202–16. 10.1104/pp.112.205385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song SY, Chen Y, Chen J, Dai XY, Zhang WH. Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 2011. August 01; 234(2): 331–45. 10.1007/s00425-011-1403-2 [DOI] [PubMed] [Google Scholar]
- 45.Zhuang JY, Fan YY, Rao ZM, Wu JL, Xia YW, Zheng KL. Analysis on additive effects and additive-by-additive epistatic effects of QTLs for yield traits in a recombinant inbred line population of rice. Theor Appl Genet 2002. December 01; 105(8): 1137–45. [DOI] [PubMed] [Google Scholar]
- 46.Zhang ZH, Wang K, Guo L, Zhu YJ, Fan YY, Cheng SH, et al. Pleiotropism of the photoperiod-insensitive allele of Hd1 on heading date, plant height and yield traits in rice. PLoS One 2012. January 20; 7(12): e52538 10.1371/journal.pone.0052538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W, Xu L, Bai X, Xing Y. Quantitative trait loci for seed dormancy in rice. Euphytica 2011. April 01; 178(3): 427–35. [Google Scholar]
- 48.Koyama ML, Levesley A, Koebner RM, Flowers TJ, Yeo AR. Quantitative trait loci for component physiological traits determining salt tolerance in rice. Plant Physiol 2001. January 01; 125(1): 406–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong J, He P, Qian Q, Shen L, Zhu L, Chen S. Identification of salt-tolerance QTL in rice (Oryza sativa L.). Chinese Sci Bull 1999. January 01; 44(1): 68–71. [Google Scholar]
- 50.Price AH. Believe it or not, QTLs are accurate!. Trends Plant Sci 2006. May 01; 11(5): 213–6. [DOI] [PubMed] [Google Scholar]
- 51.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011. January 20; 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009. May 01; 25(9): 1105–11. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010. May 01; 28(5): 511–5. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008. July 01; 5(7): 621–8. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- 55.Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. The Annals of Statistics 2001. August 01; 29(4): 1165–88. [Google Scholar]
- 56.Conesa A, Gotz S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008. January 20; 2008: 619832 10.1155/2008/619832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang A, Dai X, Zhang WH. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 2012. April 01; 63(7): 2541–56. 10.1093/jxb/err431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen LJ, Wuriyanghan H, Zhang YQ, Duan KX, Chen HW, Li QT, et al. An S-Domain Receptor-Like Kinase, OsSIK2, Confers Abiotic Stress Tolerance and Delays Dark-Induced Leaf Senescence in Rice. Plant Physiol 2013. December 01; 163(4): 1752–65. 10.1104/pp.113.224881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J, Sun S, Xu D, Lan H, Sun H, Wang Z, et al. A TFIIIA-type zinc finger protein confers multiple abiotic stress tolerances in transgenic rice (Oryza sativa L.). Plant Mol Biol 2012. October 01; 80(3): 337–50. 10.1007/s11103-012-9955-5 [DOI] [PubMed] [Google Scholar]
- 60.Mallikarjuna G, Mallikarjuna K, Reddy MK, Kaul T. Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol Lett 2011. August 01; 33(8): 1689–97. 10.1007/s10529-011-0620-x [DOI] [PubMed] [Google Scholar]
- 61.Sun SJ, Guo SQ, Yang X, Bao YM, Tang HJ, Sun H, et al. Functional analysis of a novel Cys2/His2-type zinc finger protein involved in salt tolerance in rice. J Exp Bot 2010. June 01; 61(10): 2807–18. 10.1093/jxb/erq120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sequencing PRG. The map-based sequence of the rice genome. 2005. August 11; 436(7052): 793–800. [DOI] [PubMed] [Google Scholar]
- 63.Lu T, Yu S, Fan D, Mu J, Shangguan Y, Wang Z, et al. Collection and comparative analysis of 1888 full-length cDNAs from wild rice Oryza rufipogon Griff. W1943. Dna Res 2008. October 01; 15(5): 285–95. 10.1093/dnares/dsn018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 2012. October 25; 490(7421): 497–501. 10.1038/nature11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bloom JS, Khan Z, Kruglyak L, Singh M, Caudy AA. Measuring differential gene expression by short read sequencing: quantitative comparison to 2-channel gene expression microarrays. BMC Genomics 2009. January 20; 10: 221 10.1186/1471-2164-10-221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Huang J, Yang X, Wang MM, Tang HJ, Ding LY, Shen Y, et al. A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta 2007. April 01; 1769(4): 220–7. [DOI] [PubMed] [Google Scholar]
- 67.Xu DQ, Huang J, Guo SQ, Yang X, Bao YM, Tang HJ, et al. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.). Febs Lett 2008. April 02; 582(7): 1037–43. 10.1016/j.febslet.2008.02.052 [DOI] [PubMed] [Google Scholar]
- 68.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, et al. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A 2006. August 29; 103(35): 12987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, et al. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 2007. August 01; 51(4): 617–30. [DOI] [PubMed] [Google Scholar]
- 70.Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 2008. May 01; 67(1–2): 169–81. 10.1007/s11103-008-9309-5 [DOI] [PubMed] [Google Scholar]
- 71.Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Bioph Res Co 2009. February 20; 379(4): 985–9. [DOI] [PubMed] [Google Scholar]
- 72.Huda KM, Banu MS, Garg B, Tula S, Tuteja R, Tuteja N. OsACA6, a P-type IIB Ca(2+) ATPase promotes salinity and drought stress tolerance in tobacco by ROS scavenging and enhancing the expression of stress-responsive genes. Plant J 2013. December 01 76(6): 997–1015. 10.1111/tpj.12352 [DOI] [PubMed] [Google Scholar]
- 73.Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 2008. December 01; 148(4): 1938–52. 10.1104/pp.108.128199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao Y, Wu Y, Zheng Z, Song F. Overexpression of the rice EREBP-like gene OsBIERF3 enhances disease resistance and salt tolerance in transgenic tobacco. Physiol Mol Plant P 2005. September 01; 67(3–5): 202–11. [Google Scholar]
- 75.Kader MA, Seidel T, Golldack D, Lindberg S. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. J Exp Bot 2006. January 20; 57(15): 4257–68. [DOI] [PubMed] [Google Scholar]
- 76.Duan J, Cai W. OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS One 2012. January 20; 7(9): e45117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T. A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol 2004. May 01; 45(5): 550–9. [DOI] [PubMed] [Google Scholar]
- 78.Yang W, Yoon J, Choi H, Fan Y, Chen R, An G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol 2015. January 20; 15: 31 10.1186/s12870-015-0425-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen S, Li XQ, Zhao A, Wang L, Li X, Shi Q, et al. Genes and pathways induced in early response to defoliation in rice seedlings. Curr Issues Mol Biol 2009. January 20; 11(2): 81–100. [PubMed] [Google Scholar]
- 80.Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 2008. January 20; 9: 44 10.1186/1471-2164-9-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. Dna Res 2003. December 31; 10(6): 239–47. [DOI] [PubMed] [Google Scholar]
- 82.Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, et al. The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 2010. September 01; 284(3): 173–83. 10.1007/s00438-010-0557-0 [DOI] [PubMed] [Google Scholar]
- 83.Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 2006. January 01; 60(1): 107–24. [DOI] [PubMed] [Google Scholar]
- 84.Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, et al. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiol 2007. April 01; 143(4): 1739–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001. October 01; 4(5): 447–56. [DOI] [PubMed] [Google Scholar]
- 86.Nijhawan A, Jain M, Tyagi AK, Khurana JP. Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 2008. February 01; 146(2): 333–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mukherjee K, Choudhury AR, Gupta B, Gupta S, Sengupta DN. An ABRE-binding factor, OSBZ8, is highly expressed in salt tolerant cultivars than in salt sensitive cultivars of indica rice. BMC Plant Biol 2006. January 20; 6: 18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, et al. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 2005. October 01; 46(10): 1623–34. [DOI] [PubMed] [Google Scholar]
- 89.Liu C, Wu Y, Wang X. bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 2012. June 01; 235(6): 1157–69. 10.1007/s00425-011-1564-z [DOI] [PubMed] [Google Scholar]
- 90.Zhang X, Rerksiri W, Liu A, Zhou X, Xiong H, Xiang J, et al. Transcriptome profile reveals heat response mechanism at molecular and metabolic levels in rice flag leaf. Gene 2013. November 10; 530(2): 185–92. 10.1016/j.gene.2013.08.048 [DOI] [PubMed] [Google Scholar]
- 91.Chen H, Chen W, Zhou J, He H, Chen L, Chen H, et al. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci 2012. September 01; 193–194: 8–17. 10.1016/j.plantsci.2012.05.003 [DOI] [PubMed] [Google Scholar]
- 92.Park S, Jeong JS, Lee KH, Kim YS, Choi YD, Kim J. OsbZIP23 and OsbZIP45, members of the rice basic leucine zipper transcription factor family, are involved in drought tolerance. Plant Biotechnology Reports 2015March 01; 9(2): 89–96. [Google Scholar]
- 93.Joo J, Lee YH, Song SI. Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA. Plant Biotechnology Reports 2014. November 01; 8(6): 431–41. [Google Scholar]
- 94.Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, et al. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol 2011. February 01; 52(2): 344–60. 10.1093/pcp/pcq196 [DOI] [PubMed] [Google Scholar]
- 95.Wu L, Chen X, Ren H, Zhang Z, Zhang H, Wang J, et al. ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 2007. September 01; 226(4): 815–25. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y, Chen C, Jin XF, Xiong AS, Peng RH, Hong YH, et al. Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 2009. August 31; 42(8): 486–92. [DOI] [PubMed] [Google Scholar]
- 97.Cheng MC, Liao PM, Kuo WW, Lin TP. The arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 2013. July 01; 162(3): 1566–82. 10.1104/pp.113.221911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Ying Y, Narsai R, Ye L, Zheng L, Tian J, et al. Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ 2013. January 01; 36(1): 224–36. 10.1111/j.1365-3040.2012.02569.x [DOI] [PubMed] [Google Scholar]
- 99.Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, et al. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol 2006. December 01; 142(4): 1718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, et al. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 2006. July 01; 34(Web Server issue): W293–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barragan V, Leidi EO, Andres Z, Rubio L, De Luca A, Fernandez JA, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell 2012. March 01; 24(3): 1127–42. 10.1105/tpc.111.095273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tarczynski MC, Jensen RG, Bohnert HJ. Stress protection of transgenic tobacco by production of the osmolyte mannitol. Science 1993. January 22; 259(5094): 508–10. [DOI] [PubMed] [Google Scholar]
- 103.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J 2006. February 01; 45(4): 523–39. [DOI] [PubMed] [Google Scholar]
- 104.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol 2011. June 01; 14(3): 267–74. 10.1016/j.pbi.2011.03.004 [DOI] [PubMed] [Google Scholar]
- 105.Wang W, Zhao X, Pan Y, Zhu L, Fu B, Li Z. DNA methylation changes detected by methylation-sensitive amplified polymorphism in two contrasting rice genotypes under salt stress. J Genet Genomics 2011. September 20; 38(9): 419–24. 10.1016/j.jgg.2011.07.006 [DOI] [PubMed] [Google Scholar]
- 106.Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, et al. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol 2008. October 01; 49(10): 1580–8. 10.1093/pcp/pcn133 [DOI] [PubMed] [Google Scholar]
- 107.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res 2008. January 01; 36(Database issue): D480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liang J, Zhou M, Zhou X, Jin Y, Xu M, Lin J. JcLEA, a novel LEA-like protein from Jatropha curcas, confers a high level of tolerance to dehydration and salinity in Arabidopsis thaliana. PLoS One 2013. January 20; 8(12): e83056 10.1371/journal.pone.0083056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tao Z, Kou Y, Liu H, Li X, Xiao J, Wang S. OsWRKY45 alleles play different roles in abscisic acid signalling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J Exp Bot 2011. October 01; 62(14): 4863–74. 10.1093/jxb/err144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xiao J, Cheng H, Li X, Xiao J, Xu C, Wang S. Rice WRKY13 Regulates Cross Talk between Abiotic and Biotic Stress Signaling Pathways by Selective Binding to Different cis-Elements. Plant Physiol 2013. December 01; 163(4): 1868–82. 10.1104/pp.113.226019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, et al. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J 2014. July 01; 79(1): 13–27. 10.1111/tpj.12538 [DOI] [PubMed] [Google Scholar]
- 112.Singh A, Giri J, Kapoor S, Tyagi AK, Pandey GK. Protein phosphatase complement in rice: genome-wide identification and transcriptional analysis under abiotic stress conditions and reproductive development. BMC Genomics 2010. January 20; 11: 435 10.1186/1471-2164-11-435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.You J, Zong W, Hu H, Li X, Xiao J, Xiong L. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant Physiol 2014. December 01; 166(4): 2100–14. 10.1104/pp.114.251116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh A, Jha SK, Bagri J, Pandey GK. ABA Inducible Rice Protein Phosphatase 2C Confers ABA Insensitivity and Abiotic Stress Tolerance in Arabidopsis. PLoS One 2015. January 20; 10(4): e125168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008. June 01; 228(1): 191–201. 10.1007/s00425-008-0729-x [DOI] [PubMed] [Google Scholar]
- 116.Li HW, Zang BS, Deng XW, Wang XP. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011. November 01; 234(5): 1007–18. 10.1007/s00425-011-1458-0 [DOI] [PubMed] [Google Scholar]
- 117.Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant 2008. September 01; 1(5): 732–50. 10.1093/mp/ssn047 [DOI] [PubMed] [Google Scholar]
- 118.Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, et al. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet 2006. February 01; 112(3): 570–80. [DOI] [PubMed] [Google Scholar]
- 119.Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, et al. Mapping of quantitative trait loci for salt tolerance at the seedling stage in rice. Mol Cells 2006. April 30; 21(2): 192–6. [PubMed] [Google Scholar]
- 120.Rai AN, Penna S. Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol Biol Rep 2013. November 01; 40(11): 6429–35. 10.1007/s11033-013-2757-2 [DOI] [PubMed] [Google Scholar]
- 121.Lin HX, Zhu MZ, Yano M, Gao JP, Liang ZW, Su WA, et al. QTLs for Na+ and K+ uptake of the shoots and roots controlling rice salt tolerance. Theor Appl Genet 2004. January 01; 108(2): 253–60. [DOI] [PubMed] [Google Scholar]
- 122.Kim DY, Kwon SI, Choi C, Lee H, Ahn I, Park SR, et al. Expression analysis of rice VQ genes in response to biotic and abiotic stresses. Gene 2013. October 25; 529(2): 208–14. 10.1016/j.gene.2013.08.023 [DOI] [PubMed] [Google Scholar]
- 123.Jami SK, Clark GB, Ayele BT, Roux SJ, Kirti PB. Identification and characterization of annexin gene family in rice. Plant Cell Rep 2012. May 01; 31(5): 813–25. 10.1007/s00299-011-1201-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(PDF)
(PDF)
(XLS)
(PDF)
Data Availability Statement
The sequence reads were submitted to GenBank GEO database under accession number GSE73181 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73181).