Abstract
Circadian rhythms are fundamental properties of most eukaryotes, but evidence of biological clocks that drive these rhythms in prokaryotes has been restricted to Cyanobacteria. In vertebrates, the gastrointestinal system expresses circadian patterns of gene expression, motility and secretion in vivo and in vitro, and recent studies suggest that the enteric microbiome is regulated by the host’s circadian clock. However, it is not clear how the host’s clock regulates the microbiome. Here, we demonstrate at least one species of commensal bacterium from the human gastrointestinal system, Enterobacter aerogenes, is sensitive to the neurohormone melatonin, which is secreted into the gastrointestinal lumen, and expresses circadian patterns of swarming and motility. Melatonin specifically increases the magnitude of swarming in cultures of E. aerogenes, but not in Escherichia coli or Klebsiella pneumoniae. The swarming appears to occur daily, and transformation of E. aerogenes with a flagellar motor-protein driven lux plasmid confirms a temperature-compensated circadian rhythm of luciferase activity, which is synchronized in the presence of melatonin. Altogether, these data demonstrate a circadian clock in a non-cyanobacterial prokaryote and suggest the human circadian system may regulate its microbiome through the entrainment of bacterial clocks.
Introduction
In contrast to the situation in the animal clock, which involves a transcriptional, translational feedback of “clock genes” such as Per, Cry, Bmal1 and Clock, prokaryotic circadian clocks, demonstrated only in the cyanobacterium Synechococcus elongatus, are post-transcriptional in nature. These bacteria express circadian patterns of gene expression, photosynthesis and nitrogen fixation [1–3], but the molecular mechanism for this cyanobacterial clock is the result of rhythmic autokinase activity of the hexamer-forming ATPase KaiC that is enhanced by KaiA binding and subsequent autophosphatase activity of KaiC that is modulated by KaiB binding to the KaiA-KaiC complex [4]. Remarkably, the three purified proteins, when provided free ATP, exhibit rhythmic phosphorylation of KaiC in vitro for many cycles [5]. Although S. elongatus is the only cyanobacterium studied in much detail, the Kai proteins are found extensively within the Phylum Cyanobacteria [6]
As stated above, vertebrate circadian organization results from the rhythmic expression of “clock genes” whose products interact in a dynamic transcription/translation feedback loop [1,7] “Positive elements” Clock and Bmal1 are transcribed and translated in the cytoplasm, where they dimerize, reenter the nucleus and activate expression of genes containing an E-box element in their promoter regions. Among these, “negative elements” Period (per1, 2 and 3) and Cryptochrome (cry1 and 2) are transcribed, translated and then feedback within the nucleus by interfering with Clock/Bmal1 transcriptional activation [1,7,8] The molecular feedback loop is expressed in multiple tissues in the body, where they regulate rhythmic processes locally, but they are coordinated by pacemakers such as the hypothalamic suprachiasmatic nucleus (SCN) in mammals [9].
Among the vertebrate peripheral tissues that express circadian rhythms is the gastrointestinal system, which exhibit circadian rhythms in gene expression (including clock genes), motility and secretion in vivo and in vitro [10–12]. These rhythms depend upon a patent molecular clock, since they are abolished in per1/per2 double knockout mice [12]. They are also coordinated by SCN input via the sympathetic nervous system [13].
The emerging role of the gut microbiome as an important modulator of gastrointestinal function has recently included the role of circadian rhythms. Recent studies have suggested that microbial signaling plays a critical role in homeostatic maintenance of intestinal function along with the host circadian mechanism [14,15]. Further studies have expanded this view and have shown that disruption of the circadian clock, either via dietary restriction or phase shifting (e.g. jet-lag) affects temporal distribution of the gut microbiome constituents [16–19]. While it is clear from these studies that commensal bacteria and gut tissues do communicate, it is not clear which signal or signals the microbiome exploits to sustain its own homeostasis.
Here we present evidence for one possible signal, the indole hormone melatonin, which is present at high levels in the gut and which induces swarming activity in a clinical isolate of Enterobacter aerogenes. Further investigation of the motility patterns in this bacterium evinced an endogenous circadian rhythm within cultures, which is enhanced and synchronized by melatonin.
Materials and Methods
Strains and culture conditions: E. aerogenes and E. coli clinical isolates (gift from Dr. John Seabolt, U. of Kentucky), K. pneumoniae Isolate-1 (NR-15410, BEI resources, NIAID, NIH), and DH5α with luxcdabe driven by the promoter region of MotA [20] (gift from Brian Ahmer, Ohio State University), were initially cultured in LB Broth at 37°C in a shaking incubator. Motility assays were conducted on Eosin-Methylene Blue Agar (EMB) plates [21] with a 50% reduction in agar to facilitate motility. All chemicals used in motility assays were purchased from Sigma (St. Louis, MO) and diluted in water.
Motility Assays: 100mm petri dishes were visually divided into quadrants, filled with 30mls of EMB agar with or without specified concentration of chemicals, and allowed to dry for ~4 hours in a sterile hood. 2μl of overnight culture were stabbed and released into the center of each quadrant and allowed to grow for 48 hours at 37°C. Each plate was imaged on a light box by digital camera using qCapture Pro software (Media Cybernetics, MD) and areas measured by ImageJ [22].
Transformation of MotA::luxcdabe into E. aerogenes: E. aerogenes were made competent by CaCl2 method and MotA::luxcdabe plasmid extracted from the host strain was transformed into E. aerogenes by heat shock. Transformants were selected for on tetracycline-supplemented medium and stored as glycerol stocks for future studies.
Bioluminescence monitoring: 2μl of overnight cultures were stabbed and released into the center of 35mm culture dishes containing 5mls of EMB agar with or without 1nM melatonin. Plates were sealed with 40mm cover glass by sterile vacuum grease and placed into an automated photomultiplier-based bioluminescence recorder (Lumicycle, Actimetrics, Il). Each sample was counted for 70 seconds on a rotating platform. Raw bioluminescence baselines were subtracted using a 24-hour running average via Lumicycle Analysis software (Actimetrics, IL). Cultures were photographed as above and used for illustrative purposes here.
Bioinformatics: Initial protein searches were performed by BLASTp (NCBI) using human MEL1A and MEL1B protein sequences against a protein database from the curated human microbiome project (HMP) repository (NCBI). Clock protein comparisons were performed by PsiBLAST program (NCBI) using Uniref_50 clusters against the available proteomes of E. aerogenes strains KCTC2190 and EA1509E (Uniprot.org taxonomy IDs 1028307 and 935296, respectively). Unique microbial proteins were aligned to the original clock gene clusters using MUSCLE and trees generated by PhyML software with 100 bootstraps.
Motif analysis: KAI complex protein sequences, including positive PsiBLAST hits above, were entered into the online MEME suite ([23] http://meme.nbcr.net/meme) under Multiple Em for Motif Elicitation (MEME) and subsequent Motif Alignment and Search Tool (MAST, [24]) The output of MAST for each protein is included here in supplemental data.
Statistics: Circadian rhythmicity was determined by Circwave, Circwave Batch software v3.3 [25] and periodogram analysis [26]. Each day of bioluminescence recording was separated and analyzed for periods of 24 hours (Circwave and periods between 19 and 28 hours (Circwave Batch, p<0.02). Periodogram analysis was performed using R statistical program and the GeneCycle package [26] followed by Fisher's exact g Test to obtain p-values of each culture. Spread/motility measures, periods, amplitudes, and damping coefficients were compared by 1- or 2-way ANOVA, where appropriate. All analyses were performed using SigmaStat software (Systat, CA).
Results and Discussion
We hypothesized that one potential human signal that may affect gastrointestinal microbiota is the secretion of melatonin into the lumen of the gut. Although melatonin is widely regarded as a pineal and retinal neuromodulator of circadian and photoperiodic function [27, 28], it is present throughout the gastrointestinal system [27–29], in part from pineal melatonin secretion [30, 31], but there is evidence for melatonin biosynthetic enzymes in biliary cholangiocytes, enterochromaffin cells and intestinal mucosa [31, 32]. In addition, many foods contain melatonin [27, 31]. In all, melatonin content has been reported to be 10-400x levels found in the serum [31, 32]. We identified from metagenomics data in GenBank several enteric bacteria that expressed sequences with 24–42% identity to known melatonin binding sites in the human genome (S1 Fig). These included receptors in Enterobacter aerogenes, but not in Escherichia coli or Klebsiella pneumoniae.
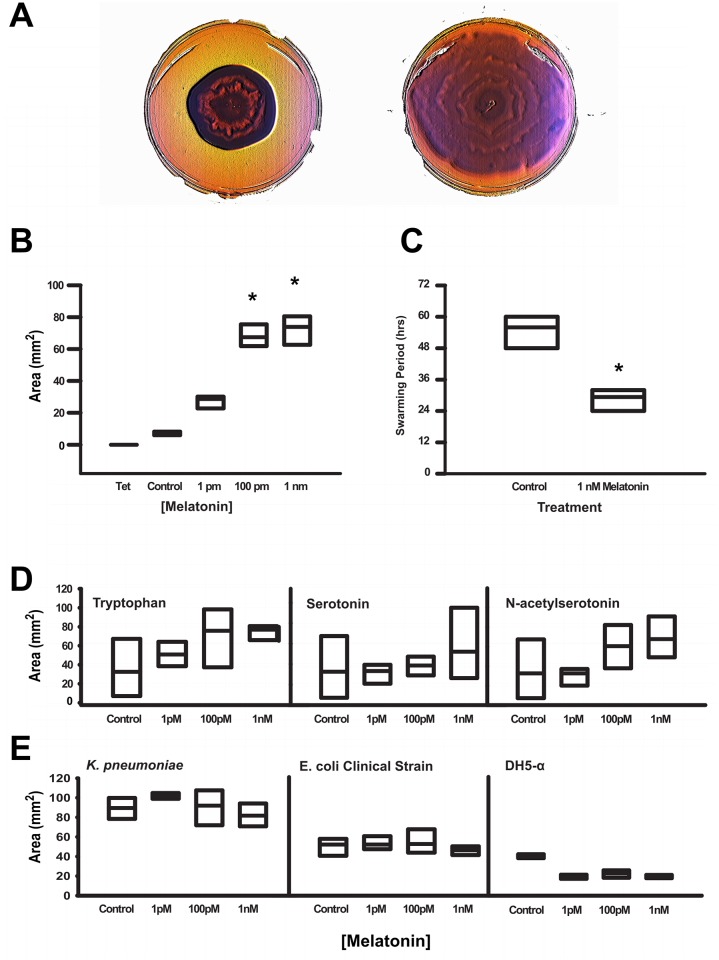
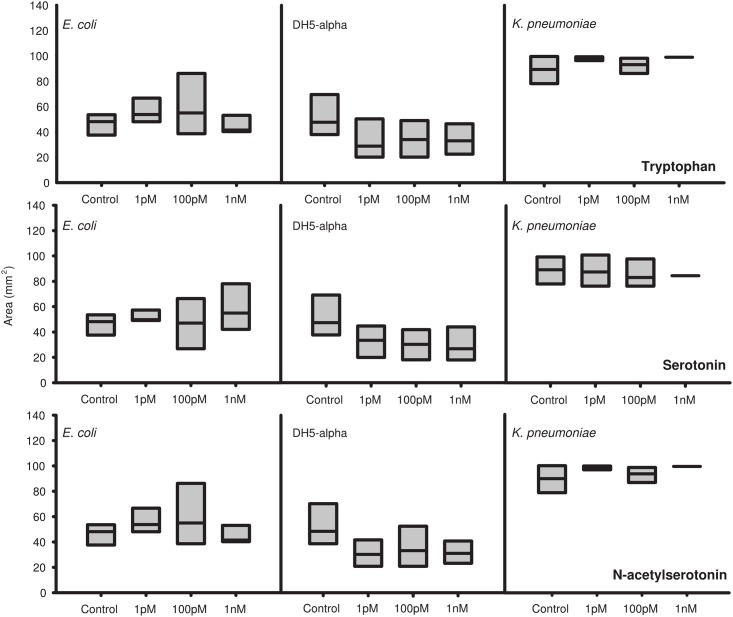
Colonies formed by clinical isolates of Enterobacter aerogenes, a Gram negative, indole-negative motile bacterium, proliferated on semi-solid Agar significantly more rapidly in the presence of melatonin in a specific, dose-dependent fashion, with maximal response coinciding within the physiological range of gut melatonin levels (Fig 1A and 1B and S2 Fig). This effect was specific for melatonin, as E. aerogenes spread further in the presence of melatonin than in the presence of equimolar concentrations of tryptophan, serotonin or N-acetylserotonin (Fig 1D). In contrast, Klebsiella pneumoniae, a closely related but non-motile, indole-negative member, and Escherichia coli, an indole-positive but motile member of the Enterobacteriacea Family, do not respond to melatonin or the other indoles tested (Figs 1E and 2).
Fig 1. Swarming behavior in E. aerogenes is induced by melatonin and occurs with a circadian frequency.
A Swarming behavior in control treated (left cultures vs. treatment with 1nM melatonin (right. Images were equally enhanced using “Bump Map” in GIMP software to highlight banding patterns. B The increase in swarming was only seen at 100pM and 1nM concentrations of melatonin, * = p value < 0.001 compared to vehicle treated cultures, n = 16 cultures per treatment. C Period of swarming as calculated by the number of rings observed per culture period of 4 days in n = 16 cultures per treatment, * = p value < 0.001. D Area of bacterial spread was unaffected by tryptophan (left, serotonin (middle and N-acetylserotonin (right, n = 16 cultures per treatment. E Melatonin did not affect growth in K. pneumoniae (left or clinical or lab strains of E. coli (middle and right, respectively, n = 16 cultures per strain per treatment.
Fig 2. Neither lab nor clinical strains of E. coli nor K. pneumoniae show swarming response to other indoles.
Cultures of clinical isolates of E. coli (left panels, DH5-α (middle panels, and K. pneumoniae (right panels were tested for swarming/growth in the presence of tryptophan (top row, serotonin (middle row, and N-acetylserotonin (bottom row, n = 16 cultures per strain per treatment.
The larger cultures of E. aerogenes in the presence of melatonin exhibited patterns of swarming within the cultures, evidenced by stereotypical, concentric rings of colonies (Fig 1A), similar to recently reported diurnal swarming in Listeria monocytogenes [33] and identical to the bulls-eye pattern of swarming commonly seen in Proteus mirabilis, another intestinal commensal that is also in the Enterobacter family [34]. These patterns were less apparent in the smaller, control cultures of E. aerogenes in melatonin’s absence (S2 Fig). Remarkably, the number of rings consistently coincided with the number of incubation days. Calculation of banding periodicity—the number of bands visually observed divided by the number of hours of incubation—revealed a period of much greater than 24 hours in control-treated cultures. In contrast, in 1nM melatonin’s presence, the period of swarming behavior was 25.1 ± 1.4 (S.D. hours (Fig 1C).
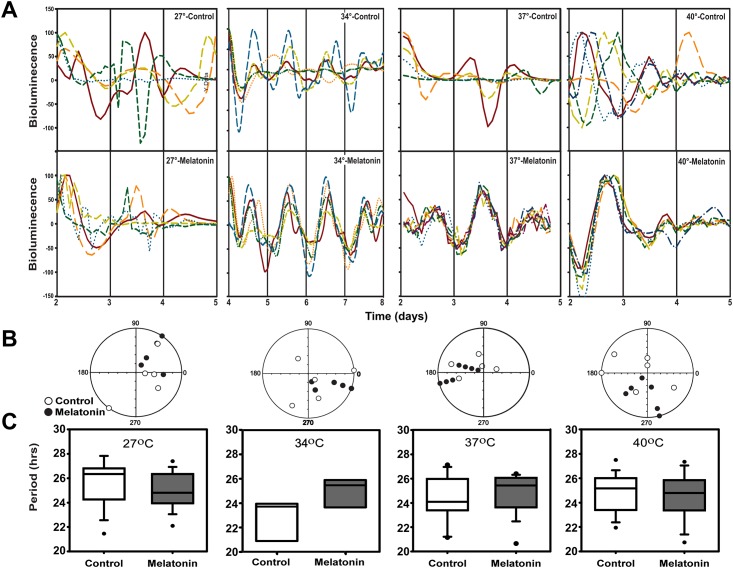
From the above banding period data, we hypothesized that the swarming rhythms might represent the output of a circadian clock. To test this, cultures of E. aerogenes were transformed to express luciferase using a luxcdabe construct driven by the MotA promoter [20] (S3A Fig). Bioluminescence from these cultures measured in a Lumicycle photomultiplier system indicated robust circadian patterns in 31–44% of cultures when maintained in temperatures ranging from ambient 27°C to those corresponding to human body temperatures (TB) of 34°C, 37°C and 40°C (Fig 3A–3C). The circadian periods of these bioluminescence rhythms were temperature compensated with a Q10 = 0.96 from 27°C to 40°C. While there was no effect of melatonin on circadian period (Fig 3C), there was a significant effect of melatonin on the phase of peak bioluminescence (Fig 3B). In the absence of melatonin, the circadian phases of multiple replicates were highly variable. However, in the presence of 1 nM melatonin the phases of these rhythms were synchronized, especially at temperatures closely corresponding to body temperature (34–37°C) (Fig 3A and 3B). In contrast, the plasmid donor strain of DH5-α E. coli failed to exhibit daily patterns of bioluminescence in the presence or absence of melatonin, despite having a 5-fold higher raw bioluminescence level (S3B and S3C Fig), which may be attributed to a higher plasmid copy number.
Fig 3. Bioluminescence recording of MotA::luxcdabe transformed E. aerogenes confirms a temperature compensated circadian rhythm.
A) Normalized bioluminescence rhythms from control-treated (top panels) and melatonin-treated (bottom panels) cultures show circadian rhythms at (from left to right) 27° (n = 5/treatment), 34° (n = 5/treatment), 37° (n = 5 control and 7 melatonin-treated) and 40° (n = 6 control and 6 melatonin-treated). Time scales represent days after plates were inoculated with bacteria, which varied in the amount of time needed to stabilize and begin outgrowth. B) Periodogram analysis-derived peak phases of rhythmic cultures from (A) reveal that control-treated cultures (white circles) show greater variation in peak phase than melatonin-treated cultures (black circles), which are more synchronized at all three temperatures. C) Periods of rhythms varied between 22 and 28 hours among temperature and melatonin treatments, but were not significantly affected by temperature or melatonin.
This is the first demonstration of a circadian clock in a prokaryote outside Phylum Cyanobacteria. The fact that this species exists primarily as a commensal bacterium raises the possibility that the circadian clockworks driving these rhythms in E. aerogenes may have arisen from horizontal gene transfer of human and/or ancestral vertebrate clock genes into these bacteria [35, 36]. However, comparison of the E. aerogenes proteome to known members of the vertebrate biological clock mechanism revealed no relationship between BMAL1, CLOCK, OR PER1 and any sequence within the E. aerogenes proteome (S4A, S4B and S4C Fig, respectively).
On the other hand, comparison of the E. aerogenes proteome data set to the cyanobacteria KAIABC complex revealed several sequences nested within trees for each of the Kai proteins (S4D, S4E and S4F Fig). Although position-specific iterated BLAST (PsiBLAST) provided significant alignments, motif-specific analysis using MEME and MAST software showed little similarity to conserved motifs within the KAI proteins (S5 Fig). Despite a lack of similarity at the sequence level, there may be an underlying similarity in cellular functions of related proteins. One KaiC ortholog found here, Dephospho-Coa Kinase, is also known to act with a phosphatase in bacteria and mammals, with the latter relationship in the form of a bi-functional single enzyme [37–39]. In S. elongatus, the Kai complex drives circadian rhythms of multiple processes through a post-translational molecular mechanism that persists even outside the bacterial cell; combination of the three Kai proteins and ATP reconstitutes a circadian pattern of phosphorylation and dephosphorylation for many cycles in vitro [5]. The major component of the complex, KaiC, expresses both kinase and phosphatase activities, the latter of which occurs in a manner similar to ATP synthase [40]. In vivo, this oscillator responds to light, temperature, and metabolic state thorugh the CikA, LdpA, and Pex pathways, each of which can entrain the Kai oscillator to environmental cues [2, 3, 41]. This relatively simple oscillator in turn regulates a wide array of processes through transcriptional regulation [42, 43]. Other factors must influence this oscillator, however, since in vivo, the periods of multiple circadian rhythms differ, depending on the promoter, the presence or absence of promoter recognition subunits of RNA polymerase, and environmental conditions, including light intensity and growth phase of the culture [44]. Our data cannot exclude this possibility in E. aerogenes, as we have only examined rhythmicity as it manifests in MotA motor protein expression, which—although an established proxy for motility—is likely to be an output of the mechanism. Alternatively, but not exclusively, circadian rhythms in E. aerogenes may derive from rhythmic peroxiredoxin activity, since this mechanism has been identified only recently in eukaryotes as well as prokaryotes [45]. Bioinformatics analysis reveals several sequences that share similarity to peroxiredoxin and thiol redoxins from various taxa (S5G Fig). Contrary to earlier studies investigating the structure and function of KaiB and SasA proteins [46, 47] our analysis showed no similarity between KaiB and thioredoxins or between KaiB orthologs and peroxiredoxin orthologs. However, the candidate proteins from our analysis are all linked to redox-sensitive pathways, including the manganese transporter MntH that initiated this investigation [48]. Recent reports of the anti-oxidant properties of melatonin in a neurodegenerative mouse model would support a mechanism involving melatonin and redox-state sensing [49]. We are currently exploring these candidate proteins to determine the mechanism or mechanisms behind the observed rhythms.
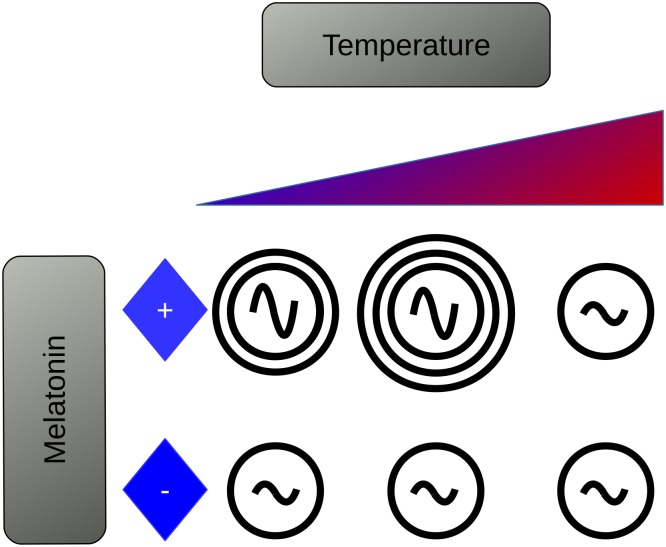
Importantly, the present observations indicate that at least one member of the human microbiome may synchronize to its host through synchronization of an endogenous, temperature-compensated circadian clock. The detailed mechanism for this synchronization is at this stage not completely known. However, it is remarkable that the presence of melatonin in the culture medium synchronizes the periodicity and phases of multiple clonal populations across different culture plates (Fig 4). The latter phenomenon suggests melatonin as a novel source of host-commensal communication within the gut, if not the internal Zeitgeber itself. The existence of a circadian rhythm within a commensal bacterium that responds to an endocrine signal that is regulated by the circadian mechanism of the host gives further credence to the concept of the microbiome as a “meta-organism”; one with an endogenous clock that is entrained by its host's clock-driven signals. If we regard our own circadian mechanism as an evolved adaptation to environmental phenomena governed by 24-hour periods, organs and organ systems could be perceived as the entraining environment for resident microflora. As such, perturbations to the environment (i.e. circadian disruptions) will affect rhythms within the microbiome as previously demonstrated [17,19]. However, it is not known whether or not, nor to what extent, the microbiome can recover from these challenges. Furthermore, the effect of host: commensal signaling is likely not relegated to one species, as we are limited to here, but to the community at large. If this phenomenon modulates quorum sensing, as is suggested by observations, there would be systemic alterations to the microbial community as a whole, as well as to the physiology of the host.
Fig 4. The data presented here show that a clinical isolate of E. aerogenes expresses a circadian rhythm in MotA expression and displays a swarming response to melatonin in a dose- and temperature-dependent manner.
Supporting Information
Alignments shown are a selection of positive BLAST hits (e-value < 0.001) aligned using MUSCLE that show several conserved residues and regions of high identity.
(TIF)
100mm EMB agar plates were inoculated with 2ul of overnight cultures (n = 4/plate, replicated with 4 different starter cultures) and incubated for 48 hours. Rosette patterns of swarming increased with increasing concentrations of melatonin on the plates.
(TIF)
A Representative map of plasmid pRG19 showing MotA upstream of luxcdabe complex and tetracycline resistance. B) DH5-α cultures (left) are not rhythmic regardless of presence of melatonin, however, raw trace of E. aerogenes cultures (right) transformed with MotA::luxcdabe plasmid show rhythmic expression with damping over time both in the presence and absence of melatonin. C) Melatonin increased the average amplitude of cultures exhibiting circadian rhythms at 27°C and 37°C, but not 40°C, * = p value < 0.05 as tested by one-way ANOVA. D) Neither temperature nor melatonin affected the damping rate of the cultures exhibiting circadian rhythms.
(TIF)
Bootstrapped trees (iterations shown between branches) show no homology among E. aerogenes proteins and vertebrate clock proteins BMAL1 (A), CLOCK (B), or PER1 (C). Similar analyses using Uniprot clusters of KAI A (D), KAI B (E), and KAI C (F) show potential homology with specific E. aerogenes proteins. G) E. aerogenes proteins share conservation with redox-related proteins across several taxa.
(TIF)
Proteins with sequence homology via PSI-BLAST share some motif-level sequences with A) KaiA and B) KaiB, but not C) KaiC.
(TIF)
Acknowledgments
The authors thank Drs. John Seabolt (UKY and Brian Ahmer (OSU for their generous gifts of bacterial strains and Ye Li, Clifford Harpole, Hannah Logsdon, Dr. Susan Golden, Dr. Sheldon Steiner, and Dr. Karla Lightfield for comments.
Data Availability
All relevant data are within the paper and/or supporting information files.
Funding Statement
The Cassone (VMC) laboratory is supported by National Institutes of Health R01 AG045833-01, “Circadian rhythms and melatonin signaling in the aging gastrointestinal system”; United States Department of Agriculture NIFA 2014-67012-21608, “The role of the premammillary nucleus in the regulation of broodiness in the domestic turkey” (Mentor; Dr. Ashli Moore post-doctoral fellow); and Howard Hughes Medical Institute Science Education Grant, “Student Retention and Success in STEM through collaborative and multi-layered STEMCats Freshmen Program.”
References
- 1.Bell-Pedersen D, Cassone VM, Earnest DJ, Golden SS, Hardin PE, Thomas TL, et al. Circadian rhythms from multiple oscillators: lessons from diverse organisms. Nat Rev Genet [Internet]. 2005/06/14 ed. 2005. July [cited 2013 Jun 27];6(7):544–56. Available: http://www.ncbi.nlm.nih.gov/pubmed/15951747. Accessed 27 June 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson CH, Mori T, Xu Y. A cyanobacterial circadian clockwork. Curr Biol [Internet]. 2008. September 9 [cited 2015 Jan 29];18(17):R816–25. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2585598&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.1016/j.cub.2008.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackey SR, Golden SS, Ditty JL. The itty-bitty time machine genetics of the cyanobacterial circadian clock. Adv Genet [Internet]. 2011. January [cited 2015 Jan 29];74:13–53. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3319097&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.1016/B978-0-12-387690-4.00002-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J [Internet]. 2003. May 1 [cited 2015 Jan 8];22(9):2127–34. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=156084&tool=pmcentrez&rendertype=abstract. Accessed 8 January 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, et al. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science [Internet]. 2005. April 15 [cited 2015 Feb 25];308(5720):414–5. Available: http://www.ncbi.nlm.nih.gov/pubmed/15831759. Accessed 25 February 2015. [DOI] [PubMed] [Google Scholar]
- 6.Axmann IM, Hertel S, Wiegard A, Dörrich AK, Wilde A. Diversity of KaiC-based timing systems in marine Cyanobacteria. Mar Genomics [Internet]. 2014. April [cited 2015 Jan 29];14:3–16. Available: http://www.ncbi.nlm.nih.gov/pubmed/24388874. Accessed 29 January 2015. 10.1016/j.margen.2013.12.006 [DOI] [PubMed] [Google Scholar]
- 7.Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol [Internet]. 2013. July 31 [cited 2013 Aug 7]; Available: http://www.ncbi.nlm.nih.gov/pubmed/23916625. Accessed 7 August 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panda S, Hogenesch JB, Kay SA. Circadian rhythms from flies to human. Nature [Internet]. 2002. May 16 [cited 2015 Jan 13];417(6886):329–35. Available: http://www.ncbi.nlm.nih.gov/pubmed/12015613. Accessed 13 January 2015. [DOI] [PubMed] [Google Scholar]
- 9.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature [Internet]. 2002. August 29 [cited 2014 Oct 31];418(6901):935–41. Available: http://www.ncbi.nlm.nih.gov/pubmed/12198538. Accessed 31 October 2014. [DOI] [PubMed] [Google Scholar]
- 10.Hoogerwerf WA, Hellmich HL, Cornelissen G, Halberg F, Shahinian VB, Bostwick J, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology [Internet]. 2007/10/09 ed. 2007;133(4):1250–60. Available: http://www.ncbi.nlm.nih.gov/pubmed/17919497. [DOI] [PubMed] [Google Scholar]
- 11.Hoogerwerf WA, Sinha M, Conesa A, Luxon BA, Shahinian VB, Cornélissen G, et al. Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology [Internet]. 2008. December [cited 2015 Jan 29];135(6):2019–29. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2748881&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.1053/j.gastro.2008.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoogerwerf WA, Shahinian VB, Cornelissen G, Halberg F, Bostwick J, Timm J, et al. Rhythmic changes in colonic motility are regulated by period genes. Am J Physiol Gastrointest Liver Physiol [Internet]. 2009/11/21 ed. 2010;298(2):G143–50. Available: http://www.ncbi.nlm.nih.gov/pubmed/19926812. 10.1152/ajpgi.00402.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malloy JN, Paulose JK, Li Y, Cassone VM. Circadian rhythms of gastrointestinal function are regulated by both central and peripheral oscillators. Am J Physiol Gastrointest Liver Physiol [Internet]. 2012. August 15 [cited 2015 Jan 27];303(4):G461–73. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3423141&tool=pmcentrez&rendertype=abstract. Accessed 27 January 2015. 10.1152/ajpgi.00369.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell [Internet]. 2013. May 9 [cited 2014 Sep 3];153(4):812–27. Available: http://www.ncbi.nlm.nih.gov/pubmed/23663780. 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- 15.de Kivit S, Tobin MC, Forsyth CB, Keshavarzian A, Landay AL. Regulation of Intestinal Immune Responses through TLR Activation: Implications for Pro- and Prebiotics. Front Immunol [Internet]. 2014. January [cited 2015 Jan 29];5:60 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3927311&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.3389/fimmu.2014.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voigt RM, Forsyth CB, Green SJ, Mutlu E, Engen P, Vitaterna MH, et al. Circadian disorganization alters intestinal microbiota. PLoS One [Internet]. 2014. January [cited 2015 Apr 27];9(5):e97500 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4029760&tool=pmcentrez&rendertype=abstract. Accessed 27 April 2015. 10.1371/journal.pone.0097500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, et al. Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis. Cell [Internet]. 2014. October 23 [cited 2014 Oct 16];159(3):514–29. Available: http://www.ncbi.nlm.nih.gov/pubmed/25417104. Accessed 16 October 2014. 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- 18.Liang X, Bushman FD, FitzGerald GA. Time in Motion: The Molecular Clock Meets the Microbiome. Cell [Internet]. 2014. October 23 [cited 2014 Nov 20];159(3):469–70. Available: http://www.ncbi.nlm.nih.gov/pubmed/25417097. Accessed 20 November 2014. 10.1016/j.cell.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 19.Zarrinpar A, Chaix A, Yooseph S, Panda S. Diet and Feeding Pattern Affect the Diurnal Dynamics of the Gut Microbiome. Cell Metab [Internet]. 2014. December 2 [cited 2014 Dec 3];20(6):1006–17. Available: http://www.ncbi.nlm.nih.gov/pubmed/25470548. Accessed 3 December 2014. 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodier RI, Ahmer BM. SirA orthologs affect both motility and virulence. J Bacteriol [Internet]. 2001. April [cited 2015 Jan 29];183(7):2249–58. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=95131&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Difco Laboratories. Difco Manual Ninth. Laboratories D, editor. Detroit: Difco Laboratories, Inc; 1984. 679 p. [Google Scholar]
- 22.Rasband WS. ImageJ. U S Natl Institutes Heal Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997–2014. [Google Scholar]
- 23.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics [Internet]. 1998. January [cited 2015 Feb 10];14(1):48–54. Available: http://www.ncbi.nlm.nih.gov/pubmed/9520501. Accessed 10 February 2015. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol [Internet]. 1994. January [cited 2014 Dec 24];2:28–36. Available: http://www.ncbi.nlm.nih.gov/pubmed/7584402. Accessed 24 December 2014. [PubMed] [Google Scholar]
- 25.Oster H, Damerow S, Hut RA, Eichele G. Transcriptional profiling in the adrenal gland reveals circadian regulation of hormone biosynthesis genes and nucleosome assembly genes. J Biol Rhythms [Internet]. 2006. October [cited 2013 Oct 23];21(5):350–61. Available: http://www.ncbi.nlm.nih.gov/pubmed/16998155. Accessed 23 October 2013. [DOI] [PubMed] [Google Scholar]
- 26.Wichert S, Fokianos K, Strimmer K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics [Internet]. 2004. January 1 [cited 2015 Sep 15];20(1):5–20. Available: http://www.ncbi.nlm.nih.gov/pubmed/14693803. Accessed 15 September 2015. [DOI] [PubMed] [Google Scholar]
- 27.Acuña-Castroviejo D, Escames G, Venegas C, Díaz-Casado ME, Lima-Cabello E, López LC, et al. Extrapineal melatonin: sources, regulation, and potential functions. Cell Mol Life Sci [Internet]. 2014. August [cited 2014 Nov 10];71(16):2997–3025. Available: http://www.ncbi.nlm.nih.gov/pubmed/24554058. Accessed 10 November 2014. 10.1007/s00018-014-1579-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brzozowski T, Jaworek J. Basic and clinical aspects of melatonin in the gastrointestinal tract. New advancements and future perspectives. Curr Pharm Des [Internet]. 2014. January [cited 2015 Jan 29];20(30):4785–7. Available: http://www.ncbi.nlm.nih.gov/pubmed/24251669. Accessed 29 January 2015. [DOI] [PubMed] [Google Scholar]
- 29.Bubenik GA. Gastrointestinal melatonin: localization, function, and clinical relevance. Dig Dis Sci [Internet]. 2002. October [cited 2015 Jan 29];47(10):2336–48. Available: http://www.ncbi.nlm.nih.gov/pubmed/12395907. Accessed 29 January 2015. [DOI] [PubMed] [Google Scholar]
- 30.Bubenik GA, Brown GM. Pinealectomy reduces melatonin levels in the serum but not in the gastrointestinal tract of rats. Biol Signals [Internet]. 1997. January [cited 2015 Jan 29];6(1):40–4. Available: http://www.ncbi.nlm.nih.gov/pubmed/9098522. Accessed 29 January 2015. [DOI] [PubMed] [Google Scholar]
- 31.Reiter RJ, Rosales-Corral S, Coto-Montes A, Boga JA, Tan D-X, Davis JM, et al. The photoperiod, circadian regulation and chronodisruption: the requisite interplay between the suprachiasmatic nuclei and the pineal and gut melatonin. J Physiol Pharmacol [Internet]. 2011. June [cited 2015 Jan 29];62(3):269–74. Available: http://www.ncbi.nlm.nih.gov/pubmed/21893686. Accessed 29 January 2015. [PubMed] [Google Scholar]
- 32.Chen C-Q, Fichna J, Bashashati M, Li Y-Y, Storr M. Distribution, function and physiological role of melatonin in the lower gut. World J Gastroenterol [Internet]. 2011. September 14 [cited 2015 Jan 29];17(34):3888–98. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3198018&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.3748/wjg.v17.i34.3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaval KG, Hahn B, Tusamda N, Albrecht D, Halbedel S. The PadR-like transcriptional regulator LftR ensures efficient invasion of Listeria monocytogenes into human host cells. Front Microbiol [Internet]. 2015. January [cited 2015 Sep 8];6:772 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4517056&tool=pmcentrez&rendertype=abstract. Accessed 8 September 2015. 10.3389/fmicb.2015.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson MM, Rasko DA, Smith SN, Mobley HLT. Transcriptome of swarming Proteus mirabilis. Infect Immun [Internet]. 2010. June 1 [cited 2015 Oct 29];78(6):2834–45. Available: http://iai.asm.org/content/78/6/2834.full. Accessed 29 October 2015. 10.1128/IAI.01222-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aminov RI. Horizontal gene exchange in environmental microbiota. Front Microbiol [Internet]. 2011. January [cited 2014 Nov 17];2:158 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3145257&tool=pmcentrez&rendertype=abstract. Accessed 17 November 2014. 10.3389/fmicb.2011.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones B V. The human gut mobile metagenome: a metazoan perspective. Gut Microbes [Internet]. 2010. January [cited 2015 Jan 12];1(6):415–31. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3056110&tool=pmcentrez&rendertype=abstract. Accessed 12 January 2015. 10.4161/gmic.1.6.14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mishra P, Park PK, Drueckhammer DG. Identification of yacE (coaE) as the structural gene for dephosphocoenzyme A kinase in Escherichia coli K-12. J Bacteriol [Internet]. 2001. May [cited 2015 Apr 23];183(9):2774–8. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=99492&tool=pmcentrez&rendertype=abstract. Accessed 23 April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Worrall DM, Tubbs PK. A bifunctional enzyme complex in coenzyme A biosynthesis: purification of pantetheine phosphate adenylyltransferase and dephospho-CoA kinase. Biochem J [Internet]. 1983. October 1 [cited 2015 Apr 23];215(1):153–7. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1152375&tool=pmcentrez&rendertype=abstract. Accessed 23 April 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin DP, Drueckhammer DG. Separate enzymes catalyze the final two steps of coenzyme A biosynthesis in Brevibacterium ammoniagenes: purification of pantetheine phosphate adenylyltransferase. Biochem Biophys Res Commun [Internet]. 1993. May 14 [cited 2015 Apr 23];192(3):1155–61. Available: http://www.ncbi.nlm.nih.gov/pubmed/8389542. Accessed 23 April 2015. [DOI] [PubMed] [Google Scholar]
- 40.Egli M, Mori T, Pattanayek R, Xu Y, Qin X, Johnson CH. Dephosphorylation of the core clock protein KaiC in the cyanobacterial KaiABC circadian oscillator proceeds via an ATP synthase mechanism. Biochemistry [Internet]. 2012 Feb 28 [cited 2015 Apr 23];51(8):1547–58. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3293397&tool=pmcentrez&rendertype=abstract. Accessed 23 April 2015. 10.1021/bi201525n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kondo T. A cyanobacterial circadian clock based on the Kai oscillator. Cold Spring Harb Symp Quant Biol [Internet]. 2007. January [cited 2015 Jan 29];72:47–55. Available: http://www.ncbi.nlm.nih.gov/pubmed/18419262. Accessed 29 January 2015. 10.1101/sqb.2007.72.029 [DOI] [PubMed] [Google Scholar]
- 42.Hosokawa N, Hatakeyama TS, Kojima T, Kikuchi Y, Ito H, Iwasaki H. Circadian transcriptional regulation by the posttranslational oscillator without de novo clock gene expression in Synechococcus. Proc Natl Acad Sci U S A [Internet]. 2011. September 13 [cited 2015 Jan 29];108(37):15396–401. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3174641&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.1073/pnas.1019612108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito H, Mutsuda M, Murayama Y, Tomita J, Hosokawa N, Terauchi K, et al. Cyanobacterial daily life with Kai-based circadian and diurnal genome-wide transcriptional control in Synechococcus elongatus. Proc Natl Acad Sci U S A [Internet]. 2009. August 18 [cited 2015 Jan 24];106(33):14168–73. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2729038&tool=pmcentrez&rendertype=abstract. Accessed 24 January 2015. 10.1073/pnas.0902587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clerico EM, Cassone VM, Golden SS. Stability and lability of circadian period of gene expression in the cyanobacterium Synechococcus elongatus. Microbiology [Internet]. 2009. March [cited 2015 Jan 29];155(Pt 2):635–41. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2729554&tool=pmcentrez&rendertype=abstract. Accessed 29 January 2015. 10.1099/mic.0.022343-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature [Internet]. 2012. May 24 [cited 2013 Aug 8];485(7399):459–64. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3398137&tool=pmcentrez&rendertype=abstract. Accessed 8 August 2013. 10.1038/nature11088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hitomi K, Oyama T, Han S, Arvai AS, Getzoff ED. Tetrameric architecture of the circadian clock protein KaiB. A novel interface for intermolecular interactions and its impact on the circadian rhythm. J Biol Chem [Internet]. 2005. May 13 [cited 2015 Apr 29];280(19):19127–35. Available: http://www.jbc.org/content/280/19/19127.full. Accessed 29 April 2015. [DOI] [PubMed] [Google Scholar]
- 47.Klewer DA, Williams SB, Golden SS, LiWang AC. Letter to the Editor: Sequence-specific resonance assignments of the N-terminal, 105-residue KaiC-interacting domain of SasA, a protein necessary for a robust circadian rhythm in Synechococcus elongatus. J Biomol NMR [Internet]. Kluwer Academic Publishers; [cited 2015 Apr 29];24(1):77–8. Available: http://link.springer.com/article/10.1023/A%3A1020649703380. Accessed 29 April 2015. [DOI] [PubMed] [Google Scholar]
- 48.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem [Internet]. 2008. January [cited 2015 Apr 18];77:755–76. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3057177&tool=pmcentrez&rendertype=abstract. Accessed 18 April 2015. 10.1146/annurev.biochem.77.061606.161055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng Y, Jiao C, Mi C, Xu B, Li Y, Wang F, et al. Melatonin inhibits manganese-induced motor dysfunction and neuronal loss in mice: involvement of oxidative stress and dopaminergic neurodegeneration. Mol Neurobiol [Internet]. 2015. March [cited 2015 Apr 29];51(1):68–88. Available: http://www.ncbi.nlm.nih.gov/pubmed/24969583. Accessed 29 April 2015. 10.1007/s12035-014-8789-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alignments shown are a selection of positive BLAST hits (e-value < 0.001) aligned using MUSCLE that show several conserved residues and regions of high identity.
(TIF)
100mm EMB agar plates were inoculated with 2ul of overnight cultures (n = 4/plate, replicated with 4 different starter cultures) and incubated for 48 hours. Rosette patterns of swarming increased with increasing concentrations of melatonin on the plates.
(TIF)
A Representative map of plasmid pRG19 showing MotA upstream of luxcdabe complex and tetracycline resistance. B) DH5-α cultures (left) are not rhythmic regardless of presence of melatonin, however, raw trace of E. aerogenes cultures (right) transformed with MotA::luxcdabe plasmid show rhythmic expression with damping over time both in the presence and absence of melatonin. C) Melatonin increased the average amplitude of cultures exhibiting circadian rhythms at 27°C and 37°C, but not 40°C, * = p value < 0.05 as tested by one-way ANOVA. D) Neither temperature nor melatonin affected the damping rate of the cultures exhibiting circadian rhythms.
(TIF)
Bootstrapped trees (iterations shown between branches) show no homology among E. aerogenes proteins and vertebrate clock proteins BMAL1 (A), CLOCK (B), or PER1 (C). Similar analyses using Uniprot clusters of KAI A (D), KAI B (E), and KAI C (F) show potential homology with specific E. aerogenes proteins. G) E. aerogenes proteins share conservation with redox-related proteins across several taxa.
(TIF)
Proteins with sequence homology via PSI-BLAST share some motif-level sequences with A) KaiA and B) KaiB, but not C) KaiC.
(TIF)
Data Availability Statement
All relevant data are within the paper and/or supporting information files.