Abstract
The E3 ubiquitin ligase UBE3A, also known as E6-AP, has a multitude of ascribed functions and targets relevant to human health and disease. Epigenetic regulation of the UBE3A gene by parentally imprinted noncoding transcription within human chromosome 15q11.2–q13.3 is responsible for the maternal-specific effects of 15q11.2–q13.3 deletion or duplication disorders. Here, we review the evidence for diverse and emerging roles for UBE3A in the proteasome, synapse and nucleus in regulating protein stability and transcription as well as the current mechanistic understanding of UBE3A imprinting in neurons. Angelman and Dup15q syndromes as well as experimental models of these neurodevelopmental disorders are highlighted as improving understanding of UBE3A and its complex regulation for improving therapeutic strategies.
Keywords: : Angelman syndrome, Dup15q syndrome, imprinting, neurodevelopment, proteosome, ubiquitin
The gene encoding E3 ubiquitin ligase protein 3a (UBE3A) is of interest to human geneticists because of the effect of mutations, deletions and duplications on the human neurodevelopmental disorders Angelman syndrome (AS) and chromosome 15q11.2–q13.3 duplication syndrome (Dup15q syndrome). UBE3A is also of interest to the field of epigenetics, as its regulation involves parental imprinting and noncoding RNAs. Figure 1 diagrams the cytogenetic alterations leading to the human disorders affecting UBE3A expression and copy number and the influence of imprinting on its expression. Here, we will first review what is known about UBE3A function and regulation, discuss the complex epigenetic mechanisms regulating UBE3A in neurons, and discuss the phenotypes and experimental models for both AS (UBE3A deficiency) and Dup15q syndrome (UBE3A excess).
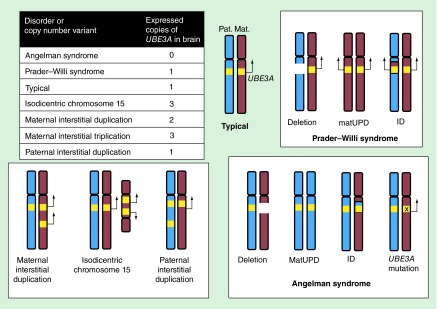
Figure 1. . Parent of origin expression patterns of the 15q11.2–q13.3 disorders.
The chromosomal abnormalities leading to copy number changes of UBE3A are diagrammed, with maternal chromosomes in red and paternal chromosomes in blue and the UBE3A copies represented as yellow bands. Specific genetic subtypes of Prader–Willi syndrome and Angelman syndrome are indicated. For Prader–Willi syndrome, these include deletion (˜75%), maternal uniparental disomy (˜25%) and imprinting defect (˜1%). For Angelman syndrome, these include deletion (˜75%), paternal uniparental disomy (˜2%), imprinting defect (˜2%) and UBE3A mutation (˜20%). Due to parental imprinting, the paternal copy of UBE3A is repressed in postnatal neurons, so the number of expressed copies of UBE3A is shown for each genetic disorder in the left table.
ID: Imprinting defects; Mat: Maternal; MatUPD: Maternal uniparental disomy; Pat: Paternal.
UBE3A function & expression
Ubiquitin ligase functions
The ubiquitin proteasome system (UPS) recycles proteins by transferring the 76 amino acid Ub to proteins marked for degradation, activation or relocalization in the cell. Three types of UPS proteins are involved in the process of marking substrate proteins: the E1 ubiquitin activating enzymes, which bring Ub to the E2 ubiquitin conjugating enzymes, which in concert with the E3 ubiquitin ligase transfer Ub to the substrate protein. Ub molecules are then typically elongated producing poly-Ub chains that are recognized by the UPS for degradation [1].
The UBE3A gene encodes a member of the large family of E3 ubiquitin ligase proteins. The UBE3A protein was first described as ‘E6-associated protein’ (E6-AP) that helps degrade the cell cycle regulatory protein p53 [2]. However, the UBE3A-directed degradation of p53 only occurs during papilloma virus infection and requires the E6 viral protein [3]. The family of E3 ligase proteins shares a C-terminal domain containing the active site of the ubiquitin ligase, where ubiquitin from the E2 conjugation enzyme is transferred to the substrate (reviewed in [4,5]). This domain is known as the HECT domain, ‘homologous to E6AP C-terminus’. The crystal structure of UBE3A bound to UbcH7, the associating E2 conjugation enzyme, shows that both UbcH7 and substrate proteins simultaneously bind to UBE3A during the process of the transfer of ubiquitin from E2 to the substrate [6]. UBE3A preferentially associates with UbcH7 E2 conjugation enzyme [7], but can associate with other E2 conjugation enzymes like UbcH8, which may alter specificity of substrate selection [8]. Although UBE3A helps identify substrate proteins, it only transiently associates with these substrates during ubiquitin transfer from the E2 conjugation enzyme. This makes it difficult to identify all UBE3A binding proteins that lead to UBE3A disease phenotypes.
Nuclear hormone receptor cofactor functions
Another function of UBE3A is as a transcriptional coactivator. UBE3A coactivates steroid hormone receptors such as progesterone, estrogen and other hormone receptors [9]. This coactivating function involves direct binding of the UBE3A protein to the transcription complex and is independent of the ubiquitin ligase function of the protein [10,11]. However, the role in regulating steroid hormone receptors may involve ubiquitination during degradation of the functional initiation complex by the UPS after transcriptional elongation [9]. Target genes coactivated by UBE3A include estrogen receptor responsive genes TFF1 and GREB1 [12], the androgen receptor responsive KLK3 gene [13], and the E6-dependent transactivation of human telomerase reverse transcriptase during viral infection in human cells [14].
In Drosophila melanogaster, several genes were upregulated at both the transcript and protein levels by the overexpression of a ubiquitin ligase defective version of Drosophila UBE3A (Dube3a) [15]. Dube3a transcriptionally upregulates GTP cyclohydrolase I (GCH1), encoding a rate limiting dopamine synthesis enzyme, in a protease-independent manner. The nonubiquitin ligase-dependent upregulation of Dube3a leads to elevated dopamine and dopamine precursors as well as hyperactivity in flies [16]. These effects were decreased, both neurotransmitter synthesis and activity, in Dube3a loss of function animals [16]. How transcriptional regulation involving UBE3A relates to the pathogenesis of UBE3A-related disorders remains unclear [17], but whole genome expression studies on both AS and 15q duplication syndrome derived cell lines [18] may shed some light on the transcriptional regulatory roles of UBE3A in neurons and other tissues.
Isoforms of UBE3A & subcellular locations
There are at least three different isoforms of UBE3A with distinct amino termini that result from alternative splicing of the first eight exons of UBE3A (grey exons in Figure 1) in humans. The isoform nomenclature is confusing because three UBE3A isoforms have also been described in mouse, but they have been numbered differently and at least one isoform is unique to either mouse or human (Table 1). In humans, there are three different protein isoforms with distinct N-termini, although alternative splicing of the 5′ untranslated exons are predicted to result in at least 12 different cDNA subtypes, with unknown functional roles ([9] and RefSeq accessions in Table 1). In mice, UBE3A isoform 1 utilizes an alternative polyadenylation site, resulting in a transcript truncated prior to the E3 ligase encoding exons and is localized to cytoplasm. The N-terminus of the predicted protein of mouse UBE3A isoform 1 is identical to isoform 3 (equivalent to isoform 1 in human), which localizes to the nucleus [19]. Mouse isoform 2 (equivalent to human isoform 3) has an additional 21 aa at the N-terminus and corrected the dendritic phenotype in pyramidal neurons [19]. The two other mouse isoforms (1 and 3) are regulated by neuronal activity and the MEF2 activity-dependent transcription factors [20]. Less is known about functional differences between the isoforms in other species, but transcripts corresponding to human isoform 3 are found in mouse, pig, rat and rhesus (Table 1).
Table 1. . Protein isoforms of UBE3A.
|
Isoform characteristics |
Human |
Mouse |
Pig |
Rat (inferred) |
Rhesus macaque (inferred) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| UBE3A isoform 1 | UBE3A isoform 2 | UBE3A isoform 3 | Ube3a isoform 3 (human 1) | Ube3a isoform 1 | Ube3a isoform 2 (human 3) | Similar to human isoform 1 | Similar to human isoform 3 | Similar to human isoform 3 | Similar to human isoform 3 | |
| RefSeq mRNA number |
NM_130838.1 |
NM_000462.3 |
NM_130839.2 |
NM_001033962.1 |
NM_173010.3 |
NM_011668.2 |
NM_001258279 |
NM_001243181 |
NM_001191837 |
NM_001261040 |
| Protein size (aa) |
852 |
875 |
872 |
849 |
762 |
870 |
854 |
875 |
868 |
872 |
| Transcript size (bp) |
4491 |
5276 |
5211 |
4910 |
3888 |
5097 |
3560 |
3746 |
5795 |
2882 |
| Number of exons |
10 |
14 |
13 |
11 |
9 |
13 |
Unknown |
Unknown |
16 |
Unknown |
| ATG location |
First and second exon |
Fourth exon |
Third exon |
Second and third exon |
Second and third exon |
Third exon |
Unknown |
Unknown |
Third exon |
Unknown |
| HECT domain (aa) |
527–849 |
523–873 |
547–869 |
524–846 |
497–762 |
545–867 |
502–852 |
550–872 |
543–865 |
547–869 |
| Subcellular localization | Unknown | Unknown | Unknown | Nuclear | Cytoplasmic | Cytoplasmic | Unknown | Unknown | Unknown | Unknown |
UBE3A protein localizes in pre- and post- synaptic neuronal compartments and in both cytoplasmic and nuclear locations [23]. This synaptic localization may primarily regulate experience-dependent synaptic plasticity [24], although our understanding of the diverse roles of ubiquitination and protein recycling at the synapse is rapidly expanding [25]. The cytosolic and nuclear locations of UBE3A are consistent with its predicted roles in proteosome targeting and transcriptional regulation. Interestingly, the nuclear isoform 3 in mouse (isoform 1 in human) was dynamically regulated in early postnatal life and nuclear immunofluorescent staining patterns of UBE3A are predominate in mature neurons [19], suggesting that alternative splicing of UBE3A may be important in early life. Alternative polyadenylation of mouse/rat isoform 1 results in a noncoding isoform of Ube3a that promoted dendritic growth and spine maturation by acting as competing endogenous RNA to miR-134 [26].
Known targets of UBE3A
There are many proteins that are potential substrates of UBE3A based on reported interactions (BioGRID [27]). Table 2 summarizes a shorter list of UBE3A targets and interactors that do not depend on E6, were validated in more than one study and/or are ubiquitinated by UBE3A.
Table 2. . Protein targets and interactors of UBE3A.
| Category | Protein | Interaction | System | Subcellular localization | Predicted function | Ref. |
|---|---|---|---|---|---|---|
| Proteosome | RPN10/PSMD4 | Direct Ub | Flies/neuronal cells | Cytosol, nucleus | 26S proteasome regulation via deubiquitination | [28,29] |
| |
RPN11/PSMD14 |
Physical interaction |
In vitro/human cells |
Cytosol |
26S proteasome regulation via deubiquitination |
[28,29] |
| Ubiquitin processes | UBE2L3 | E3–E2 interaction | In vitro binding | Cytosol | Potential E2 binding partner for UBE3A | [6] |
| UBE2D1 | E3–E2 interaction | In vitro binding | Cytosol | Potential E2 binding partner for UBE3A | [28] | |
| UBE2D2 | E3–E2 interaction | In vitro binding | Cytosol | Potential E2 binding partner for UBE3A | [28] | |
| UBE2D3 | E3–E2 interaction | In vitro binding | Cytosol | Potential E2 binding partner for UBE3A | [28] | |
| UBC | Direct transfer from E2 to substrate | Flies/mice/cells | Cytosol | Ubiquitin C protein. Transferred by UBE3A to substrate | [30] | |
| UBE3A | Direct Ub | Flies/mice/cells | Nucleus, cytosol | E3 ubiquitin protein ligase, transcriptional coactivator | [30,31] | |
| RING1B/RNF2 | Direct Ub | Human/mouse | Nucleus | E3 ubiquitin protein ligase that mediates monoubiquitination of histone H2A (H2AK119Ub), member of PRC1 complex | [32] | |
| |
HERC2 |
Direct Ub |
Human cells |
Cytoskeleton, cytosol, nucleus, mitochondrion |
E3 ubiquitin protein ligase |
[33] |
| Transcription regulation | PRX1 | Direct Ub | Human cells | Nucleus | Homeobox transcription factor, coactivator of growth factor responses | [34] |
| ESR2 | interaction and degradation | Human cells | Nucleus, mitochondrion, extracellular | Estrogen receptor, nuclear receptor transcription factor | [35] | |
| |
BMAL1/CLOCK |
Direct Ub |
In vitro/mice |
Nucleus, cytosol |
Histone acetyltransferase and transcriptional regulator of circadian genes |
[36,37] |
| Other | Ephexin V | Direct Ub | Mice | Dendrite, cytosol, nucleus | Rho GTPase | [38] |
| ECT2 | Direct Ub/may not degrade | Flies/mice/cells | Cytoskeleton, cytosol, nucleus | Rho GTPase | [39] | |
| ATP-α | Indirect evidence of Ub | Flies/cells | Endoplasmic reticulum, golgi, plasma membrane, nucleus | Na+/K+ ATPase, neuronal homeostasis | [15] | |
| ARC | Likely indirect | Human cells/mice | Plasma membrane, endosome, cytoplasm | Activity-dependent synaptic plasticity | [20,33] | |
| HHR23A | Direct Ub | Human cells | Nucleus, cytosol, mitochonrion | Nucleotide excision repair through proteasomal ubiquitin-dependent protein degradation | [40] | |
| HSPA4 | Direct Ub | In vitro/human cells | Cytosol, aggrasomes, perinuclear compartment | Heatshock 70kDa chaparon stress response, misfolded proteins | [41] | |
| TSC2 | Indirect evidence of Ub | In vitro/human cells | Cytosol, Golgi, nucleus, plasma membrane | GTPase for RHEB, negative regulator of mTORC1 signaling | [42,43] |
ARC: AMPA subtype glutamate receptor regulatory protein; RHEB: Ras homologue enriched in the brain.
Most proteins in Table 2 are involved in some aspect of the ubiquitination or proteasome process, such as the E2 conjugating enzymes UBE2L3, UBE2D1, UBE2D2 and UBE2D3 [28], and the UBC protein involved in direct transfer of ubiquitin form E2 to substrate [30]. UBE3A also self-ubiquitinates in its active form as an oligomeric complex [31], so is frequently found as its own target. The Rpn10/PSMD4 26S proteasome non-ATPase regulatory subunit 4 and related Rpn11/PSMD14 were identified in several interaction studies [28,29] and in Drosophila neuronal cells as a direct ubiquitination substrate of UBE3A [44]. ATP-α, an indirect target of Dube3a in Drosophila neuronal cells is an sodium/potassium ATPase involved in neuronal homeostasis [15]. In humans, PSMD4 is a non-ATPase subunit of the 26S proteasome complex, which could profoundly affect many proteins when regulated by UBE3A in vivo. In vitro, UBE3A promotes the ubiquitination of the 26S proteasome [28]. Studies also suggest a physical interaction between HERC2 and UBE3A in vivo [28,45] and that HERC2, itself an E3 ubiquitin ligase, can regulate UBE3A function in vitro [33]. Thus regulation of the UPS by UBE3A may have a ripple effect on all proteins regulated by the 26S proteasome.
Several individual UBE3A substrates have been lauded as the key protein responsible for the neurological phenotypes associated with changes in UBE3A levels (Table 2). The AMPA subtype glutamate receptor regulatory protein (ARC), which is regulated by UBE3A in vivo [20], although not a valid ubiquitin substrate of UBE3A in vitro, is an apparent transcriptional target [33]. The heat shock protein HSPA4 involved in stress response was found to be a direct target of ubiquitination by UBE3A in human cells [41]. UBE3A substrates regulated in the mouse brain by UBE3A include the transcription factor BMAL1, which transcriptionally regulates circadian rhythms and diurnally expressed transcripts in mammals [36]. Regulation of BMAL turnover by UBE3A occurs in the mouse brain and circadian rhythm affected in Ube3a deficient mice [37]. Also of interest for diurnal metabolism is TSC2 [42], a negative modulator of mammalian target of rapamycin, which is encoded by a gene mutated in tuberous sclerosis. Based on the finding of UBE3A regulation of TSC2, rapamycin was successfully used in the Ube3a m-/p+ mouse model to rescue the motor deficits and Purkinje cell dendritic branching deficits in the cerebellum [43].
UBE3A also modulates Ring1B ubiquitin ligase, a component of the polycomb group repressive complex 1 in mice. This E3 ligase monoubiquitinates histone H2A and could substantially impact global gene expression [32]. Another transcriptional regulatory factor that interacts with and is degraded by UBE3A is the nuclear estrogen receptor, ESR2 [35]. Two other substrates that physically interact with UBE3A and appear to be released from repression in Ube3a deficient mice are the RhoA guanine nucleotide exchange factors ECT2 [39] and Ephexin V [38]. These two substrates support neuronal plasticity in the brain and could contribute to experience-dependent synaptic defects found in AS mouse models. In humans, dysregulation of ECT2 and E5 through loss of UBE3A expression in neurons could impair memory and learning. Finally, two other nuclear substrates are the DNA repair protein HHR23A/RAD23A [40] and the antioxidant protein peroxiredoxin 1 [34].
UBE3A deficiency in AS
Clinical features of AS
AS is characterized by severe developmental delay and motor abnormalities including ataxia and jerky motions [46]. AS is distinguished from other neurodevelopmental disorders by a classic constellation of: happy demeanor, excitability, frequent smiling and laughter and absent or minimal use of words [47]. Seizures with a characteristic EEG of notched delta and rhythmic theta activity and epileptiform discharges are observed in up to 80% of AS patients, with an onset of seizures usually beginning before 3 years of age and lasting through adulthood [48]. Other common features of AS include hypotonia, feeding problems in infancy, protruding tongue, abnormal sleep-wake cycles and the diminished need for sleep, as well as excessive mouthing/chewing behaviors.
Genetic subtypes of AS & genotype–phenotype differences
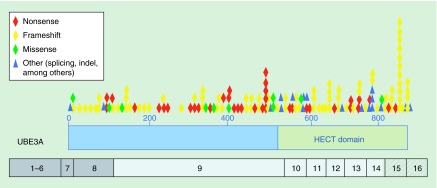
The genetic defects leading to AS share a deficiency in the maternally expressed copy of UBE3A including: large deletions, small deletions, mutations or epimutations. Large maternal deletions including UBE3A are the most frequent (˜75%), followed by point mutations in maternal UBE3A (˜20%), paternal uniparental disomy (2%) or imprinting defects (3%) [49]. The large deletions typically result from nonallelic homologous recombination due to repetitive blocks of low-copy repeats concentrated in the 15q11–q13.3 region. Point mutations in the maternal copy occur throughout the UBE3A gene (Figure 2). Frameshift mutations are the most abundant and concentrated in the HECT domain, but nonsense, missense and other types of mutations are dispersed throughout the gene [50,51].
Figure 2. . UBE3A mutations observed in Angelman syndrome.
The genetic locations of Angelman syndrome mutations are mapped relative to the HECT domain encoding E3 ligase function (green bar) and the encoding exons of UBE3A. Alternatively spliced exons 1–8 are shown in dark grey, consistent exons are in light grey and alternative exons due to polyadenylation differences are hatched grey. Current numbering of exons was according to [50].
HECT: Homologous to E6AP C-terminus.
Because of the diversity and low frequency of UBE3A mutations in AS, genotype–phenotype correlations are only useful in comparing the general genetic categories of large deletions, UBE3A mutations and uniparental disomy or imprinting defects (UPD/ID). AS patients with large deletions show the most severe and classical features of AS compared with other genotypes. Compared with the UPD or mutation genotypes, AS patients with class I (BP1-BP3) or class II (BP2-BP3) deletions exhibited significantly reduced receptive and expressive language as well as visual perception [52]. Autism Diagnostic Observation Schedule scores meeting the criteria for autism spectrum disorders (ASD) were also highest in deletion forms of AS (92% class I, 83% class II) compared with the combined mutation/UPD group (56%) [52]. Smaller studies with mainly deletion patients estimated ASD rates of 50–80% [53–55]. Epilepsy is also more common in AS deletion compared with nondeletion causes [56]. Among patients with large deletions, class I deletions are associated with more frequent and severe (e.g., status epilepticus) seizures than class II deletions [57]. Thus, while UBE3A is the predominant gene causing AS, additional genes within the 15q11–q13.3 locus can modify disease severity.
Among AS patient, categories without large deletions, obesity and hyperphagia are common [58,59]. These classes differ from large deletion AS cases in having two copies of paternally expressed transcripts that are normally expressed from only one allele (see genomic imprinting section below). Obesity and hyperphagia are more commonly associated with Prader–Willi syndrome (PWS, paternal deletions of 15q11–q13.3), but occurred in a case of Dup15q syndrome with PWS-like features [60]. These findings suggest that an imbalance in the ratio of paternal to maternal transcripts within 15q11–q13.3 can disrupt metabolic homeostasis.
Imprinting of UBE3A by noncoding RNA
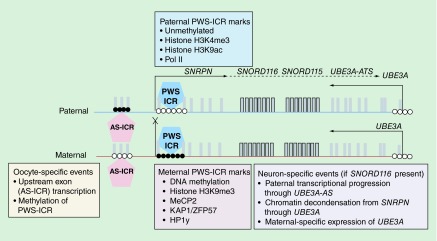
UBE3A is a parentally imprinted gene in which the paternal allele is selectively silenced in mature neurons. Therefore, it is the loss of maternal expression of UBE3A in neurons that adversely effects the postnatal brain, leading to AS. Figure 3 diagrams the parent-specific opposing coding and noncoding transcripts, imprinting marks and neuron-specific events within the imprinted region of 15q11.2–q13.3. The promoter of UBE3A is completely unmethylated and shows an active state on both parental copies in all tissues (Epigenome Roadmap [61]), unlike most imprinted genes in which the silent allele is marked by parental-specific DNA methylation and repressive chromatin marks. Instead, it is the epigenetic marks on the paternal allele that determine maternal silencing of UBE3A in postnatal neurons. The maternal allele of UBE3A is silenced by a paternally expressed noncoding antisense transcript (UBE3A-ATS) [62–65].
Figure 3. . Imprinting and neuron-specific epigenetic regulation of UBE3A.
The CpG sites of both maternal and paternal alleles of UBE3A are unmethylated (open circles), resulting in biallelic transcription of the 5′ end of UBE3A (arrows pointed left, exons represented as vertical bars). However, the Prader–Willi imprinting control region (PWS-ICR, in blue) is characterized by DNA methylation (closed circles) and repressive chromatin marks on the maternal allele (pink box) and active chromatin marks on the paternal allele (blue box). The Angelman imprinting control region (AS-ICR, pink) is characterized by paternal-specific DNA methylation because the maternal allele is protected from methylation at this region by the oocyte-specific transcription of noncoding upstream exons that serve to methylate the PWS-ICR on the maternal allele. While paternal expression of the protein coding gene SNRPN is observed in all tissues, transcriptional progression continues in postnatal neurons through the repetitive small nucleolar RNA clusters through to the antisense transcript for UBE3A (UBE3A-ATS). Specifically in neurons, the paternal allele undergoes chromatin decondensation and the maternal allele of UBE3A is silenced from expression of the UBE3A-ATS. AS-ICR: Angelman syndrome imprinting control region; PWS-ICR: Prader–Willi syndrome imprinting control region.
The mechanisms involved in the neuronal imprinting of UBE3A are illuminated by the molecular genetics of the oppositely imprinted disorder, PWS. Like AS, most PWS cases are caused by similar sized deletions of paternally derived 15q11.2–q13.3. Other causes of PWS involve maternal UPD or small deletions encompassing either the PWS imprinting control region (PWS-ICR) or the SNORD116 cluster of small nucleolar RNAs (snoRNAs) [66,67]. The PWS-ICR is where the major epigenetic differences lie between the maternal and paternal alleles. On the maternal allele, PWS-ICR is heavily methylated and marked by the repressive chromatin mark H3K9me3, but the paternal PWS-ICR is unmethylated and contains the mark of active promoters, H3K4me3. Therefore, transcription initiates exclusively on the paternal allele for the protein coding gene SNRPN.
However, SNRPN is not the only transcript expressed from the imprinted PWS-ICR promoter. In mature postnatal neurons, this paternally initiated transcript progresses through clusters of repetitive snoRNA subunits (SNORD116 and SNORD115) and their intervening spliced host genes (116HG and 115HG), ending as an opposite stranded transcript to UBE3A (UBE3A-ATS) [68–70]. In nonneuronal cell types, paternal transcription does not progress beyond SNORD116 in human or Snprn in mouse, so the UBE3A-ATS is not made and UBE3A/Ube3a remains biallelically expressed (Figure 3). In nonneuronal cells from a small deletion (SNORD116del) PWS patient, transcription progressed through to the UBE3A-ATS, resulting in UBE3A imprinting [71]. In mice and humans, the SNORD116/Snord116 locus was demonstrated to encode repetitive R-loop forming repeats that displace histones, induce chromatin decondensation and slow the transcriptional progression and monoallelic expression of Ube3a [68]. Transcriptional interference arising from the paternal transcriptional progression antisense through the UBE3A gene body is the current model for how UBE3A becomes imprinted specifically in neurons.
A subclass of AS patients has ID and lacks DNA methylation at the PWS-ICR [72]. While some of these individuals had small overlapping deletions that defined the AS imprinting control region (AS-ICR), others lacked the methylated PWS-ICR mark with no apparent genetic mutation. AS individuals arising from IVF-derived pregnancies are primarily in the nonmutation ID class [73,74], suggesting that environmental factors in the oocyte and early embryo may alter AS-ICR activity. Early in utero exposure to the plastic-derived pollutant Bisphenol A altered PWS-ICR methylation levels, imprinting of Snrpn in placenta and Ube3a levels in placenta and brain [75]. These findings suggest that both genetic and environmental factors can cause imprinting defects affecting UBE3A.
Animal models and reproductive tissues suggest that upstream transcription is the mechanism of AS-ICR action on the PWS-ICR locus. Multiple upstream exons of PWS-ICR/SNRPN are observed in human, mouse and cow, although the locations and exact sequences are not well conserved [76–78]. What is conserved is the transcriptional activity of these upstream exons in oocytes, but not sperm or other tissues [78]. Therefore, the PWS-ICR is no longer the promoter CpG island of the oocyte Snrpn transcript, it is instead within the gene body and becomes methylated. This post-transcriptional event in oocytes is what in turn silences the maternal SNRPN/SNORD116/UBE3A-ATS transcript in brain to ensure that one copy of UBE3A remains expressed, an example of long-lived epigenetic memory.
Mouse models & human-induced pluripotent stem cells models of AS
Several mouse models of AS recapitulate some of the core neurological deficits of AS. The most widely used AS mouse model is a gene knockout of exon 2 (now exon 8 in Figure 1), resulting in UBE3A protein deficiency with maternal, but not paternal inheritance [79]. Maternal Ube3a deficiency in this model results in deficits in contextual learning, dendritic spine development and experience-dependent contextual learning [23,79–80]. Mice engineered without a functional HECT domain (current exons 10–11) have similar neurobehavioral and electroencephalographic abnormalities [81]. This Ube3a m-/p+ model also showed some of the sleep alterations characteristic of AS, including increased waking and deterioration of paradoxical sleep [82].
Mice with a 1.6 Mb deletion from Ube3a to Gabrb3 exhibited impaired learning and abnormities in neonatal ultrasonic vocalizations [83]. Both Ube3a m-/p+ knockout and Ube3a-Gabrb3 deletion mice showed alterations in circadian rhythms and light/dark entrainment [37]. An even larger mouse deletion including the entire syntenic imprinted region occurred in a transgene insertion line, resulting in the loss of Ube3a in cerebellum when inherited maternally [84]. Furthermore, two engineered small mutations upstream of Snrpn exon 1 appear to model the imprinting mutation forms of AS, as maternal inheritance of these deletions resulted in unmethylated PWS-ICR, resulting in reduced brain levels of UBE3A [85].
Human induced pluripotent stem cells (iPSCs) were successfully generated from fibroblasts of two AS and one PWS large deletion patients and are useful models of human-specific neuronal phenotypes [86]. AS and PWS iPSC lines maintained the imprinted methylation marks of the remaining parental allele following reprogramming and after neuronal differentiation. Differentiation of AS iPSCs into neurons resulted an upregulation of the UBE3A-ATS that corresponded to a downregulation of UBE3A, and lower levels of UBE3A compared with control iPSC-derived neurons.
Therapeutic strategies to reverse paternal UBE3A silencing
Since individuals with AS have an intact copy of UBE3A on the paternal allele of 15q11.2–q13.3, the most attractive molecular strategy to treat AS is to reverse the paternal silencing of UBE3A in neurons. A high-throughput screen for drug compounds using a paternally inherited Ube3a-YFP reporter gene in mouse primary neurons uncovered several topoisomerase inhibitors that reversed the paternal silencing [87]. One of these inhibitors, topotecan, was shown to be effective at reversing the silencing of Ube3a by reducing the levels of Ube3a-ATS on the paternal allele [87]. Topotecan acts to inhibit topoisomerase I, which helps resolve transcriptional progression through the Ube3a-ATS by removal of R-loops [68] and facilitates expression of long genes [88]. Fibroblasts from a PWS patient lacking the SNORD116 locus express UBE3-ATS, unlike controls that lack transcription through the UBE3A-ATS in nonneuronal cells [71]. Together these results suggest that topetecan acts directly on the upstream PWS SNORD116 locus to reduce transcript levels of the downstream UBE3A-ATS. The effects of topotecan are not specific for only the PWS/AS locus, as topetecan treatment of mouse neurons reduces the level of multiple synaptic genes, including many that are implicated in ASD such as Cntnap2, Nrxn3 and Cntn5 [88,89]. Although topotecan may not be a suitable therapy for AS because of this lack of specificity, these studies do establish that a small molecule approach to the regulation of UBE3A in human neurons would be plausible.
A more target-specific therapeutic approach to degrade or truncate the long paternal transcript could increase paternal Ube3a expression. A mouse line engineered with a poly(A) cassette between Snord115 and the 3′ end of Ube3a restored paternal Ube3a expression and ameliorated phenotypic deficits in the AS mouse model [65]. More relevant for human AS therapy are antisense oligonucleotides (ASOs) targeted to the Ube3a-ATS [90]. ASOs that target the Ube3a-ATS to a region just proximal to the 3′ end of Ube3a were most effective and these only affected transcription downstream but not upstream of the ASO-target locations. So, while the ASOs do not appear to work as predicted by degrading the entire transcript, they were effective at reversing paternal Ube3a silencing by reducing transcription of the distal Ube3a-ATS and at reducing the fear response (measured by contextual freezing) [90]. In contrast, no differences were observed in other behavioral measures with ASO treatment, including marble burying, open field or rotarod tests. Thus, targeting the paternal transcriptional progression just before the Ube3a gene body through ASOs or other similar approaches may be an effective molecular strategy for AS treatments.
For any strategy of increasing Ube3a expression, the timing of reintroduction is expected to be critical. A recent Ube3a Stop/Cre reintroduction mouse model demonstrated a critical window of embryonic to 3-week postnatal Ube3a expression for rescuing phenotypic deficits in marble burying, open field, nest building and forced swim tests, although motor deficits could be improved prior to adulthood [91]. Targeting Arc, a downstream target of UBE3A, by a genetic cross was effective at reducing audiogenic-induced seizures in young Ube3a deficient mice, but no changes to the motor deficits or increased ultrasonic vocalizations was observed [92].
UBE3A duplication in Dup15q syndrome
Genetic subtypes of Dup15q syndrome
There are two major genetic subtypes of Dup15q syndrome: interstitial and isodicentric duplications involving the 15q locus (Figure 1) [93]. Both duplications arise almost exclusively from the maternal chromosome during oogenesis. While paternal duplications of 15q exist, most affected patients have interstitial duplications and their predominant problems are sleep disturbances and anxiety disorder [93,94]. Individuals with supernumerary isodicentric 15q duplication (idic[15]) are tetrasomic for the 15q11.2–q13.3 region, having three maternal copies and one paternal copy of the locus. Interstitial duplications consist of one extra copy of the 15q11.2–q13.3 region inserted in tandem within the q arm of maternal chromosome 15. Individuals with maternal interstitial triplications are tetraploid for the 15q11.2–q13.3 region. Individuals with more than two additional copies of 15q11–q13 are rare [95].
Spectrum of clinical features of Dup15q syndrome
Chromosome 15q11.2–q13 duplication (Dup15q) syndrome shares many characteristics with AS, including developmental delay, intellectual disability, speech and language impairment and epilepsy. Hypotonia occurs in almost all individuals with Dup15q syndrome, and it can be severe, resulting in difficultly with ambulation, feeding difficulties, joint hyperextensibility and excessive drooling [96]. The majority of patients with Dup15q syndrome walk independently, distinguishing them from AS individuals who often have severe ataxia. Seizures affect 60% of individuals with Dup15q syndrome, and many more have abnormal EEG activity [94]. Seizures often present as infantile spasms or other seizure types before age five [97].
Cognitive delays occur in all children with Dup15q; intellectual disability is frequently severe to profound in idic(15) children. Speech and language delays are common, with expressive language most severely impacted. Language is often echolalic with pronoun reversal [96] and social impairment includes inappropriate social interactions, including gaze and contact avoidance, lack of symbolic play and loss of interest in peers. Difficult behaviors such as tantrums, shouting and aggressiveness often occur, as do stereotypies. The impairments in speech and language, social interactions, as well as behavioral difficulties fulfill diagnostic criteria for ASD for most children with Dup15q [94].
The pathogenic role of increased UBE3A levels in Dup15q syndrome remains unproven, while the role of deficient UBE3A levels in AS is established. Interestingly, a recent family was described in which the proband had developmental delay and a maternally inherited small (129 kb) duplication encompassing only UBE3A that segregated with neuropsychiatric phenotypes across three prior generations [98]. Evidence implicating increased maternal copies of UBE3A is strongest for ASD phenotypes in patients with large interstitial duplications. Interstitial duplication of 15q11.2–q13 inherited from the mother resulted in an ASD a phenotype in the proband, while the mother who had an interstitial duplication inherited from her father, was unaffected [99]. Formal autism assessments show that maternal, but not paternal duplications result in ASD [94,100]. Consistent with this hypothesis, some studies have shown that individuals with PWS due to maternal UPD (i.e., two copies of the maternal chromosome) show an increase in autistic symptomatology [101]. However, the overlap between phenotypes in PWS deletion and PWS mUPD patients complicates the analysis of ASD [102].
Mouse models of Dup15q syndrome
Due to the dependence of the phenotypes of individuals on the parent-of-origin of the duplicated alleles in humans (Figure 1), it is hypothesized that UBE3A contributes significantly to the Dup15q syndrome; however, other genes could be involved. Transgenic mice engineered with a 2× increased UBE3A dosage showed impaired social behavior and communication, and increased repetitive behaviors, consistent with a role for Ube3a dosage in social behavior [103]. Mice with increased Ube3a dosage also showed reduced glutamatergic synaptic transmission, likely due to reduced presynaptic glutamatergic release. While implicating UBE3A dosage as the major cause of Dup15q-related phenotypes, a potential concern about this mouse model is that the C-terminal FLAG tag likely rendered the UBE3A protein ligase deficient [104,105]. Another hypothesis is that it is the transcriptional coactivator function of UBE3A that causes the phenotypes. Also, the mouse phenotype may be caused by a dominant-negative gain-of-function by the FLAG-tagged UBE3A, since the active form is an oligomer [31].
Taking advantage of the syntenic gene content between human 15q11–q13 and mouse chromosome 7C, Nakatanki et al. engineered mice with interstitial chromosomal duplication to model Dup15q syndrome [106]. Mice with a maternally inherited duplication showed increased levels of Ube3a, as well as the biallelically expressed genes, Gabrb3, Gabra5, Gabrg3 and Herc2. Inexplicably, mice with a maternally inherited duplication also showed increased mRNA level from the imprinted and paternally expressed gene Ndn. These mice showed no significant change in sociability, ultrasonic vocalizations, anxiety or behavioral inflexibility but did show significantly slower acoustic startle responses compared with wild-type mice. Conversely, mice with a paternally inherited duplication, showed no increase in Ube3a, but expected increases in the paternally-expressed genes, Ndn and Snrpn, as well as biallelically expressed genes. The paternal duplication mice showed decreased sociability, increased ultrasonic vocalizations, increased anxiety and behavioral inflexibility. In contrast to human cases (Figure 1), maternal interstitial duplication mice exhibit a subtle sensorimotor phenotype, while the paternal interstitial duplication mice present with a more pronounced phenotype consistent with ASD.
Human neuronal models of Dup15q syndrome
Human neuronal models provide another model to study mechanisms of disease in Dup15q syndrome. First, microcell mediated chromosome transfer copied an entire human maternal chromosome 15 human into SH-SY5Y neuroblastoma cells that maintained a maternalepigenotype [107]. SH(15M)-differentiated neurons exhibited deficits in homologous pairing of chromosome 15q11–q13 and transcript levels across 15q11–q13. Limitations of the SH(15M) model include that they carry an extra copy of the entire chromosome 15, not just 15q11–q13, and they are of neuroblastomal origin, so they do not differentiate entirely into mature neurons.
Human iPSCs were derived from fibroblasts or cord blood samples from individuals with maternal interstitial duplication, paternal interstitial duplication, and idic(15) [18]. iPSCs derived from Dup15q individuals maintained parental DNA methylation at the PWS-ICR and differentiated into fully functional, glutamatergic forebrain neurons over the course of 10 weeks. iPSC-derived neurons developed mature action potentials and spontaneous excitatory synaptic activity, suggesting that this cell culture model can be used to determine electrophysiological properties and synaptic plasticity of Dup15q neurons. However, iPSC-derived neurons are only equivalent in age to fetal neurons [108], and thus may not be reflective of gene expression in children and adults with Dup15q.
Finally, human postmortem brain from individuals with Dup15q syndrome provide another source for research. At least 16 postmortem brains from individuals with idic(15) have been studied for amyloid β deposition, gene and protein expression studies involving the 15q region, and genome wide gene expression studies in comparison to idiopathic autism brains [107,109–110]. Limitations of Dup15q brain tissue include their limited availability, variable quality due to postmortem interval, comorbid disorders, medications and cause of death, which is often due to epilepsy.
Transcriptional & epigenetic dysregulation in Dup15q compared with AS
Gene expression within the 15q region was investigated using the SH(15M) cell model system [107]. The paternally-expressed imprinted genes, NDN and SNRPN showed reduced expression in SH(15M) neuronal cells compared with the wild-type parent SH-SY5Y neuronal cells. The maternally-expressed imprinted gene, UBE3A, showed similar expression levels in SH(15M) and wild-type cells. ATP10A, which is imprinted in some individuals, was reduced in the SH(15M) neuronal cells, as were the biallelically-expressed genes, GABRB3 and CHRNA7. CYFIP1, another gene expressed from both alleles, was upregulated in the SH(15M) neuronal cells. Since most of the alterations reduced gene expression, an investigation of promoter methylation that found no differences between SH(15M) and wild-type cells, suggests that either higher-order chromatin interactions or interactions between homologues underlie the gene expression alterations in these cells. Homologous pairing of a region near GABRB3 was deficient in SH(15M) neuronal cells, and employed both MeCP2 and CTCF binding sites within the region. Subsequent studies also identified large chromatin loops within the 15q11–q13.3 region overlapping MeCP2 binding sites [111], further supporting a role for epigenetic regulation in gene expression in this region.
Gene expression in iPSCs and iPSC-derived neurons was also investigated [18]. In this model system, gene expression was largely, as expected, based on copy number for iPSCs. For instance, the biallelically-expressed genes, TUBGCP1, CYFIP1, NIPA1, GABRB3, HERC2 and CHRNA7 showed approximately twofold upregulation in idic(15) iPSCs compared with normal iPSCs. UBE3A showed slight upregulation in maternal and paternal interstitial duplications compared with normal iPSCs. Unexpectedly, NIPA1 showed decreased expression in the maternal interstitial duplication line. In iPSC-derived neurons, CYFIP1, HERC2 and CHRNA7 were increased in at least one of the maternal interstitial duplication lines more than expected by copy number. mRNA sequencing was used to quantify gene expression across the genome in iPSC-derived neurons from idic(15), AS and control samples [18]. Many genes differentially expressed in both idic(15) and AS iPSC-derived neurons were autism candidate genes, and genes involved in neuronal differentiation were downregulated in neurons from both disorders. Unexpectedly, genes differentially expressed in both AS and idic(15) were more often regulated in the same direction, rather than the opposite direction, suggesting that most of the differentially expressed genes are not transcriptional targets of UBE3A, and are likely secondary effects of the duplication or deletion of 15q11–q13. These data further support a role for higher order chromatin structure in regulating neuronal gene expression.
Gene expression across the chromosome 15q11–q13 region was quantified in postmortem tissue from idic(15) extrastriate visual cortices [60]. Quantitative reverse transcription-PCR showed that UBE3A expression was increased compared with control brain in most samples, but it was lower than expected based on copy number. GABRB3 expression was highly variable among the idic(15) brains, with some brains having much lower expression compared with controls. The paternally-expressed SNRPN gene showed reduced expression in idic(15) brains compared with control in most samples. Overall, the postmortem brain expression analyses showed that with the exception of UBE3A, most genes were not expressed as expected by parental origin or copy number at this late stage tissue of the disease progression.
Future perspective
While the genetics of AS is well understood, several important questions remain: which downstream targets of UBE3A are critical to the pathogenesis of AS?; can human AS therapies be developed that specifically block the UBE3A-ATS?; can circadian alterations in AS be targeted for therapies?
Four major questions underlie the genetics of Dup15q: which genes in the 15q11–q13 region contribute to the syndrome?; which specific genes contribute to specific features?; what is the effect of their increased dosage on the rest of the genome?; what are the epistatic effects of other genes on those in the 15q11–q13 region?
The observation of Dup15q syndrome in individuals with maternally but not paternally inherited duplications of chromosome 15q11–q13, suggest that UBE3A drives the ASD phenotype in this disorder [94]. The phenotypes associated with paternal 15q11–q13 duplications are therefore likely to be caused by genes other than UBE3A. Furthermore, a mouse model suggests that Ube3a plays a major role in the autism phenotype associated with Dup15q [103]. Secondly, a 15q12.2 duplication spanning only UBE3A was associated with a neuropsychiatric phenotype only when maternally inherited [98]. The influence of increased UBE3A on the expression of other synaptic genes throughout the genome remains an important area for future research.
It will be critical to identify the protein targets of UBE3A essential for the molecular pathogenesis of both Dup15q and AS. The direct binding of HERC2 to UBE3A both in vitro and in vivo provides compelling evidence that this interaction is indeed regulatory, although perhaps not through the ubiquitin ligase function of HERC2 [33]. Notably, mutations in HERC2 can cause an Angelman-like syndrome in humans [112], suggesting that the regulation of UBE3A by HERC2 is involved in at least some aspects of the syndrome. Furthermore, elucidation of the role of nuclear and transcriptional targets of UBE3A will be needed to comprehend largescale transcriptional deficits in Dup15q syndrome.
A better understanding of the projected role for UBE3A in circadian rhythms may help improve treatments for the sleep and cognitive deficits in both AS and Dup15q syndrome. Interestingly, the paternal noncoding 116HG transcript also plays a role in dampening transcription of diurnally regulated genes that are activated by BMAL in the mouse cortex [69]. Together the circadian findings in both AS and PWS suggest that the maternally and paternally derived gene products may have counteracting feedback loops regulating circadian metabolic cycles that should be explored in future studies.
Executive summary.
UBE3A is an E3 ubiquitin ligase and many of its protein targets are part of the ubiquitin proteasome system with widespread effects on protein levels throughout the cell and at the neuronal synapse.
Another role for UBE3A in transcriptional regulation of nuclear steroid hormone receptors may contribute to the UBE3A-associated phenotypes in humans and mouse models.
UBE3A is an abundant protein in cytosolic and nuclear compartments and at both the pre- and post-synaptic locations, and thus may play a diverse role in multiple cellular and transcriptional events in neurodevelopment.
The epigenetic influences on UBE3A levels in neurons are complex, involving parental imprinting patterns inherited from oocytes and noncoding RNAs specific to neurons.
Angelman and Dup15q syndromes are disorders of maternal UBE3A deficiency or overexpression with distinct but partially overlapping clinical features.
Acknowledgements
The authors thank the Dup15q Alliance scientific advisory board members, particularly O Devinsky and S Jeste for their critical review of the manuscript and helpful comments.
Footnotes
Financial & competing interests disclosure
The authors thank the NIH NINDS (R01NS076263) and the Prader–Willi Research Foundation for ongoing support of research in this area. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Tu Y, Chen C, Pan J, Xu J, Zhou ZG, Wang CY. The ubiquitin proteasome pathway (upp) in the regulation of cell cycle control and DNA damage repair and its implication in tumorigenesis. Int. J. Clin. Exp. Pathol. 2012;5(8):726–738. [PMC free article] [PubMed] [Google Scholar]
- 2.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol. Cell. Biol. 1993;13(2):775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The first paper to identify the E3 ubiqutin ligase UBE3A as a protein associated with Papillomavirus E6 protein that mediates the degredation of p53.
- 3.Cooper B, Schneider S, Bohl J, Jiang Y, Beaudet A, Vande Pol S. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306(1):87–99. doi: 10.1016/s0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 4.Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 2007;354(2):329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scheffner M, Staub O. HECT E3s and human disease. BMC Biochem. 2007;8(Suppl. 1):S6. doi: 10.1186/1471-2091-8-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang L, Kinnucan E, Wang G, et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286(5443):1321–1326. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 7.Ronchi VP, Klein JM, Haas AL. E6AP/UBE3A ubiquitin ligase harbors two E2 ubiquitin binding sites. J. Biol. Chem. 2013;288(15):10349–10360. doi: 10.1074/jbc.M113.458059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eletr ZM, Kuhlman B. Sequence determinants of E2-E6AP binding affinity and specificity. J. Mol. Biol. 2007;369(2):419–428. doi: 10.1016/j.jmb.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramamoorthy S, Nawaz Z. E6-associated protein (E6-AP) is a dual function coactivator of steroid hormone receptors. Nucl. Recept. Sig. 2008;6:e006. doi: 10.1621/nrs.06006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nawaz Z, Lonard DM, Smith CL, et al. The angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol. Cell. Biol. 1999;19(2):1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This was the first time E6AP/UBE3A was assigned a transcriptional coactivation function separate from the ubiqutin ligase function of the protein.
- 11.Nawaz Z, Lonard DM, Dennis AP, Smith CL, O’Malley BW. Proteasome-dependent degradation of the human estrogen receptor. Proc. Natl Acad. Sci. USA. 1999;96(5):1858–1862. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Catoe HW, Nawaz Z. E6-AP facilitates efficient transcription at estrogen responsive promoters through recruitment of chromatin modifiers. Steroids. 2011;76(9):897–902. doi: 10.1016/j.steroids.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Khan OY, Fu G, Ismail A, et al. Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol. Endocrinol. 2006;20(3):544–559. doi: 10.1210/me.2005-0110. [DOI] [PubMed] [Google Scholar]
- 14.James MA, Lee JH, Klingelhutz AJ. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer. 2006;119(8):1878–1885. doi: 10.1002/ijc.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen LJ, Farook MF, Reiter LT. Proteomic profiling in Drosophila reveals potential Dube3a regulation of the actin cytoskeleton and neuronal homeostasis. PLoS ONE. 2013;8(4):e61952. doi: 10.1371/journal.pone.0061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferdousy F, Bodeen W, Summers K, et al. Drosophila UBE3A regulates monoamine synthesis by increasing GTP cyclohydrolase I activity via a non-ubiquitin ligase mechanism. Neurobiol. Dis. 2011;41(3):669–677. doi: 10.1016/j.nbd.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.El Hokayem J, Nawaz Z. E6AP in the brain: one protein, dual function, multiple diseases. Mol. Neurobiol. 2014;49(2):827–839. doi: 10.1007/s12035-013-8563-y. [DOI] [PubMed] [Google Scholar]
- 18.Germain ND, Chen PF, Plocik AM, et al. Gene expression analysis of human induced pluripotent stem cell-derived neurons carrying copy number variants of chromosome 15q11–q13.1. Mol. Autism. 2014;5:44. doi: 10.1186/2040-2392-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miao S, Chen R, Ye J, et al. The Angelman syndrome protein Ube3a is required for polarized dendrite morphogenesis in pyramidal neurons. J. Neurosci. 2013;33(1):327–333. doi: 10.1523/JNEUROSCI.2509-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greer PL, Hanayama R, Bloodgood BL, et al. The Angelman syndrome protein Ube3a regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.RefSeq: NCBI www.ncbi.nlm.nih.gov/refseq/
- 22.GeneCards: The Human Gene Database www.genecards.org
- 23.Dindot SV, Antalffy BA, Bhattacharjee MB, et al. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet. 2008;17(1):111–118. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]; • Provided a key reagent (Ube3a-GFP mice) to demonstrate paternal expression of Ube3a in specific brain regions, the observation of abnormal dendritic spine morphology, and the cellular localization of Ube3a to both the synapse and nucleus.
- 24.Yashiro K, Riday TT, Condon KH, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat. Neurosci. 2009;12(6):777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mabb AM, Ehlers MD. Ubiquitination in postsynaptic function and plasticity. Ann. Rev. Cell Dev. Biol. 2010;26:179–210. doi: 10.1146/annurev-cellbio-100109-104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valluy J, Bicker S, Aksoy-Aksel A, et al. A coding-independent function of an alternative Ube3a transcript during neuronal development. Nat. Neurosci. 2015;18(5):666–673. doi: 10.1038/nn.3996. [DOI] [PubMed] [Google Scholar]
- 27.BioGRID: results summary http://thebiogrid.org/113185/summary/homo-sapiens/ube3a.html
- 28.Jacobson AD, Macfadden A, Wu Z, Peng J, Liu CW. Autoregulation of the 26S proteasome by in situ ubiquitination. Mol. Biol. Cell. 2014;25(12):1824–1835. doi: 10.1091/mbc.E13-10-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Chen CF, Baker PR, Chen PL, Kaiser P, Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46(11):3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz SE, Rosa JL, Scheffner M. Characterization of human HECT domain family members and their interaction with UbcH5 and UbcH7. J. Biol. Chem. 1998;273(20):12148–12154. doi: 10.1074/jbc.273.20.12148. [DOI] [PubMed] [Google Scholar]
- 31.Ronchi VP, Klein JM, Edwards DJ, Haas AL. The active form of E6-associated protein (E6AP)/UBE3A ubiquitin ligase is an oligomer. J. Biol. Chem. 2014;289(2):1033–1048. doi: 10.1074/jbc.M113.517805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaaroor-Regev D, De Bie P, Scheffner M, et al. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of angelman syndrome. Proc. Natl Acad. Sci. USA. 2010;107(15):6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhnle S, Kogel U, Glockzin S, et al. Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J. Biol. Chem. 2011;286(22):19410–19416. doi: 10.1074/jbc.M110.205211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasu J, Murakami K, Miyagawa S, et al. E6AP ubiquitin ligase mediates ubiquitin-dependent degradation of peroxiredoxin 1. J. Cell. Biochem. 2010;111(3):676–685. doi: 10.1002/jcb.22752. [DOI] [PubMed] [Google Scholar]
- 35.Picard N, Charbonneau C, Sanchez M, et al. Phosphorylation of activation function-1 regulates proteasome-dependent nuclear mobility and E6-associated protein ubiquitin ligase recruitment to the estrogen receptor beta. Mol. Endocrin. 2008;22(2):317–330. doi: 10.1210/me.2007-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gossan NC, Zhang F, Guo B, et al. The E3 ubiquitin ligase UBE3A is an integral component of the molecular circadian clock through regulating the BMAL1 transcription factor. Nucleic Acids Res. 2014;42(9):5765–5775. doi: 10.1093/nar/gku225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi SQ, Bichell TJ, Ihrie RA, Johnson CH. Ube3a imprinting impairs circadian robustness in Angelman syndrome models. Curr. Biol. 2015;25(5):537–545. doi: 10.1016/j.cub.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolis SS, Salogiannis J, Lipton DM, et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143(3):442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reiter LT, Seagroves TN, Bowers M, Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum. Mol. Genet. 2006;15(18):2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J. Biol. Chem. 1999;274(26):18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- 41.Mishra A, Godavarthi SK, Maheshwari M, Goswami A, Jana NR. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of HSP70-bound misfolded proteins. J. Biol. Chem. 2009;284(16):10537–10545. doi: 10.1074/jbc.M806804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng L, Ding H, Lu Z, et al. E3 ubiquitin ligase E6AP-mediated TSC2 turnover in the presence and absence of HPV16 E6. Genes Cells. 2008;13(3):285–294. doi: 10.1111/j.1365-2443.2008.01162.x. [DOI] [PubMed] [Google Scholar]
- 43.Sun J, Liu Y, Moreno S, Baudry M, Bi X. Imbalanced mechanistic target of rapamycin C1 and C2 activity in the cerebellum of Angelman syndrome mice impairs motor function. J. Neurosci. 2015;35(11):4706–4718. doi: 10.1523/JNEUROSCI.4276-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SY, Ramirez J, Franco M, et al. Ube3a, the E3 ubiquitin ligase causing angelman syndrome and linked to autism, regulates protein homeostasis through the proteasomal shuttle RPN10. Cell. Mol. Life Sci. 2014;71(14):2747–2758. doi: 10.1007/s00018-013-1526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; • The UBE3A substrate RPN10 is a shuttle protein for the ubiquitin proteasome system, so the downstream regulation of multiple proteins could have profound effects on UBE3A-related disorders.
- 45.Galligan JT, Martinez-Noel G, Arndt V, et al. Proteomic analysis and identification of cellular interactors of the giant ubiquitin ligase HERC2. J. Proteome Res. 2015;14(2):953–966. doi: 10.1021/pr501005v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan WH, Bird LM, Thibert RL, Williams CA. If not Angelman, what is it? A review of Angelman-like syndromes. Am. J. Med Genet. 2014;164A(4):975–992. doi: 10.1002/ajmg.a.36416. [DOI] [PubMed] [Google Scholar]
- 47.Jedele KB. The overlapping spectrum of rett and angelman syndromes: a clinical review. Sem. Ped. Neurol. 2007;14(3):108–117. doi: 10.1016/j.spen.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 48.Thibert RL, Larson AM, Hsieh DT, Raby AR, Thiele EA. Neurologic manifestations of Angelman syndrome. Ped. Neurol. 2013;48(4):271–279. doi: 10.1016/j.pediatrneurol.2012.09.015. [DOI] [PubMed] [Google Scholar]; • An EEG pattern was identified that is unique to Angelman syndrome (AS).
- 49.Chamberlain SJ, Lalande M. Angelman syndrome, a genomic imprinting disorder of the brain. J. Neurosci. 2010;30(30):9958–9963. doi: 10.1523/JNEUROSCI.1728-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadikovic B, Fernandes P, Zhang VW, et al. Mutation update for UBE3A variants in Angelman syndrome. Hum. Mut. 2014;35(12):1407–1417. doi: 10.1002/humu.22687. [DOI] [PubMed] [Google Scholar]
- 51.Fang P, Lev-Lehman E, Tsai TF, et al. The spectrum of mutations in UBE3A causing Angelman syndrome. Hum. Mol. Genet. 1999;8(1):129–135. doi: 10.1093/hmg/8.1.129. [DOI] [PubMed] [Google Scholar]
- 52.Mertz LG, Thaulov P, Trillingsgaard A, et al. Neurodevelopmental outcome in Angelman syndrome: genotype-phenotype correlations. Res. Dev. Disabil. 2014;35(7):1742–1747. doi: 10.1016/j.ridd.2014.02.018. [DOI] [PubMed] [Google Scholar]
- 53.Sahoo T, Peters SU, Madduri NS, et al. Microarray based comparative genomic hybridization testing in deletion bearing patients with Angelman syndrome: genotype-phenotype correlations. J. Med. Genet. 2006;43(6):512–516. doi: 10.1136/jmg.2005.036913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonati MT, Russo S, Finelli P, et al. Evaluation of autism traits in angelman syndrome: a resource to unfold autism genes. Neurogenetics. 2007;8(3):169–178. doi: 10.1007/s10048-007-0086-0. [DOI] [PubMed] [Google Scholar]
- 55.Peters SU, Beaudet AL, Madduri N, Bacino CA. Autism in angelman syndrome: implications for autism research. Clin. Genet. 2004;66(6):530–536. doi: 10.1111/j.1399-0004.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- 56.Valente KD, Fridman C, Varela MC, et al. Angelman syndrome: uniparental paternal disomy 15 determines mild epilepsy, but has no influence on EEG patterns. Epilepsy Res. 2005;67(3):163–168. doi: 10.1016/j.eplepsyres.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 57.Valente KD, Varela MC, Koiffmann CP, et al. Angelman syndrome caused by deletion: a genotype-phenotype correlation determined by breakpoint. Epilepsy Res. 2013;105(1–2):234–239. doi: 10.1016/j.eplepsyres.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 58.Mertz LG, Christensen R, Vogel I, Hertz JM, Ostergaard JR. Eating behavior, prenatal and postnatal growth in angelman syndrome. Res. Dev. Disabil. 2014;35(11):2681–2690. doi: 10.1016/j.ridd.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 59.Brennan ML, Adam MP, Seaver LH, et al. Increased body mass in infancy and early toddlerhood in angelman syndrome patients with uniparental disomy and imprinting center defects. Am. J. Med. Genet. Part A. 2015;167A(1):142–146. doi: 10.1002/ajmg.a.36831. [DOI] [PubMed] [Google Scholar]
- 60.Hogart A, Leung KN, Wang NJ, et al. Chromosome 15q11–13 duplication syndrome brain reveals epigenetic alterations in gene expression not predicted from copy number. J. Med. Genet. 2009;46(2):86–93. doi: 10.1136/jmg.2008.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.UCSC Genome Browser http://genomebrowser.wustl.edu
- 62.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1998;19(1):15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 63.Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10(23):2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 64.Yamasaki K, Joh K, Ohta T, et al. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 2003;12(8):837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- 65.Meng L, Person RE, Beaud, et al. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 2012;21(13):3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sahoo T, Del Gaudio D, German JR, et al. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40(6):719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cassidy SB, Schwartz S, Miller JL, Driscoll DJ. Prader–Willi syndrome. Genet. Med. 2012;14(1):10–26. doi: 10.1038/gim.0b013e31822bead0. [DOI] [PubMed] [Google Scholar]
- 68.Powell WT, Coulson RL, Gonzales ML, et al. R-loop formation at Snord116 mediates topotecan inhibition of Ube3a-antisense and allele-specific chromatin decondensation. Proc. Natl Acad. Sci. USA. 2013;110(34):13938–13943. doi: 10.1073/pnas.1305426110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Powell WT, Coulson RL, Crary FK, et al. A Prader–Willi locus lncRNA cloud modulates diurnal genes and energy expenditure. Hum. Mol. Genet. 2013;22(21):4318–4328. doi: 10.1093/hmg/ddt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vitali P, Royo H, Marty V, Bortolin-Cavaille ML, Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snorna gene arrays. J. Cell. Sci. 2010;123(Pt 1):70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 71.Martins-Taylor K, Hsiao JS, Chen PF, et al. Imprinted expression of UBE3A in non-neuronal cells from a Prader–Willi syndrome patient with an atypical deletion. Hum. Mol. Genet. 2014;23(9):2364–2373. doi: 10.1093/hmg/ddt628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buiting K, Gross S, Lich C, Gillessen-Kaesbach G, El-Maarri O, Horsthemke B. Epimutations in Prader–Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 2003;72(3):571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hiura H, Okae H, Miyauchi N, et al. Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproduction technologies. Hum. Reprod. 2012;27(8):2541–2548. doi: 10.1093/humrep/des197. [DOI] [PubMed] [Google Scholar]
- 74.Arnaud P, Feil R. Epigenetic deregulation of genomic imprinting in human disorders and following assisted reproduction. Birth Defects Res. 2005;75(2):81–97. doi: 10.1002/bdrc.20039. [DOI] [PubMed] [Google Scholar]
- 75.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9(4):e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev. Biol. 2005;286(2):587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 77.Landers M, Bancescu DL, Le Meur E, et al. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32(11):3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis MW, Brant JO, Kramer JM, et al. Angelman syndrome imprinting center encodes a transcriptional promoter. Proc. Natl Acad. Sci. USA. 2014;112(22):6871–6875. doi: 10.1073/pnas.1411261111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang YH, Armstrong D, Albrecht U, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]; • The first mouse model of AS recapitulated both seizure and ataxia phenotypes and maternal-specific expression of UBE3A in the brain.
- 80.Yashiro K, Riday TT, Condon KH, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat. Neurosci. 2009;12(6):777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miura K, Kishino T, Li E, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol. Dis. 2002;9(2):149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 82.Colas D, Wagstaff J, Fort P, Salvert D, Sarda N. Sleep disturbances in Ube3a maternal-deficient mice modeling Angelman syndrome. Neurobiol. Dis. 2005;20(2):471–478. doi: 10.1016/j.nbd.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 83.Jiang YH, Pan Y, Zhu L, et al. Altered ultrasonic vocalization and impaired learning and memory in Angelman syndrome mouse model with a large maternal deletion from Ube3a to Gabrb3. PLoS ONE. 2010;5(8):e12278. doi: 10.1371/journal.pone.0012278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gabriel JM, Merchant M, Ohta T, et al. A transgene insertion creating a heritable chromosome deletion mouse model of Prader–Willi and Angelman syndromes. Proc. Natl Acad. Sci. USA. 1999;96(16):9258–9263. doi: 10.1073/pnas.96.16.9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu MY, Chen KS, Bressler J, et al. Mouse imprinting defect mutations that model Angelman syndrome. Genesis. 2006;44(1):12–22. doi: 10.1002/gene.20179. [DOI] [PubMed] [Google Scholar]
- 86.Chamberlain SJ, Chen PF, Ng KY, et al. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader–Willi syndromes. Proc. Natl Acad. Sci. USA. 2010;107(41):17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This was the first demonstration of induced pluripotent stem cells derived from human Prader–Willi syndrome and AS patients that showed maintenance of neuronal imprinting and expression of the UBE3A-antisense transcript in induced pluripotent stem cell-derived neurons.
- 87.Huang HS, Allen JA, Mabb AM, et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2012;481(7380):185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.King IF, Yandava CN, Mabb AM, et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501(7465):58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mabb AM, Kullmann PH, Twomey MA, Miriyala J, Philpot BD, Zylka MJ. Topoisomerase 1 inhibition reversibly impairs synaptic function. Proc. Natl Acad. Sci. USA. 2014;111(48):17290–17295. doi: 10.1073/pnas.1413204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518(7539):409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates that in the mouse model for AS, noncoding RNAs can be used in vivo to decrease UBE3A-antisense transcript and rescue some of the neurological phenotypes of the disorder.
- 91.Silva-Santos S, Van Woerden GM, Bruinsma CF, et al. Ube3a reinstatement identifies distinct developmental windows in a murine angelman syndrome model. J. Clin. Invest. 2015;125(5):2069–2076. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mandel-Brehm C, Salogiannis J, Dhamne SC, Rotenberg A, Greenberg ME. Seizure-like activity in a juvenile angelman syndrome mouse model is attenuated by reducing arc expression. Proc. Natl Acad. Sci. USA. 2015;112(16):5129–5134. doi: 10.1073/pnas.1504809112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hogart A, Wu D, Lasalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2–q13. Neurobiol. Dis. 2010;38(2):181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Urraca N, Cleary J, Brewer V, et al. The interstitial duplication 15q11.2–q13 syndrome includes autism, mild facial anomalies and a characteristic EEG signature. Autism Res. 2013;6(4):268–279. doi: 10.1002/aur.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang NJ, Parokonny AS, Thatcher KN, et al. Multiple forms of atypical rearrangements generating supernumerary derivative chromosome 15. BMC Genet. 2008;9:2. doi: 10.1186/1471-2156-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Battaglia A. The inv dup (15) or idic (15) syndrome (tetrasomy 15q) Orphanet. J. Rare Dis. 2008;3:30. doi: 10.1186/1750-1172-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Conant KD, Finucane B, Cleary N, et al. A survey of seizures and current treatments in 15q duplication syndrome. Epilepsia. 2014;55(3):396–402. doi: 10.1111/epi.12530. [DOI] [PubMed] [Google Scholar]
- 98.Noor A, Dupuis L, Mittal K, et al. 15q11.2 duplication encompassing only the Ube3a gene is associated with developmental delay and neuropsychiatric phenotypes. Hum. Mut. 2015;36(7):689–693. doi: 10.1002/humu.22800. [DOI] [PubMed] [Google Scholar]
- 99.Cook EH, Jr., Lindgren V, Leventhal BL, et al. Autism or atypical autism in maternally but not paternally derived proximal 15q duplication. Am. J. Hum. Genet. 1997;60(4):928–934. [PMC free article] [PubMed] [Google Scholar]
- 100.Al Ageeli E, Drunat S, Delanoe C, et al. Duplication of the 15q11–q13 region: clinical and genetic study of 30 new cases. Eur. J. Med. Genet. 2014;57(1):5–14. doi: 10.1016/j.ejmg.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 101.Ogata H, Ihara H, Murakami N, Gito M, Kido Y, Nagai T. Autism spectrum disorders and hyperactive/impulsive behaviors in japanese patients with Prader–Willi syndrome: a comparison between maternal uniparental disomy and deletion cases. Am. J. Med. Genet. A. 2014;164A(9):2180–2186. doi: 10.1002/ajmg.a.36615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dykens EM, Lee E, Roof E. Prader–Willi syndrome and autism spectrum disorders: an evolving story. J. Neurodev. Disord. 2011;3(3):225–237. doi: 10.1007/s11689-011-9092-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith SE, Zhou YD, Zhang G, Jin Z, Stoppel DC, Anderson MP. Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci. Transl. Med. 2011;3(103):103ra197. doi: 10.1126/scitranslmed.3002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salvat C, Wang G, Dastur A, Lyon N, Huibregtse JM. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J. Biol. Chem. 2004;279(18):18935–18943. doi: 10.1074/jbc.M312201200. [DOI] [PubMed] [Google Scholar]
- 105.Kuhnle S, Mothes B, Matentzoglu K, Scheffner M. Role of the ubiquitin ligase E6AP/Ube3a in controlling levels of the synaptic protein arc. Proc. Natl Acad. Sci. USA. 2013;110(22):8888–8893. doi: 10.1073/pnas.1302792110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nakatani J, Tamada K, Hatanaka F, et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137(7):1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meguro-Horike M, Yasui DH, Powell W, et al. Neuron-specific impairment of inter-chromosomal pairing and transcription in a novel model of human 15q-duplication syndrome. Hum. Mol. Genet. 2011;20(19):3798–3810. doi: 10.1093/hmg/ddr298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brennand K, Savas JN, Kim Y, et al. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psych. 2014;20(3):361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wegiel J, Frackowiak J, Mazur-Kolecka B, et al. Abnormal intracellular accumulation and extracellular abeta deposition in idiopathic and Dup15q11.2–q13 autism spectrum disorders. PLoS ONE. 2012;7(5):e35414. doi: 10.1371/journal.pone.0035414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Voineagu I, Wang X, Johnston P, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yasui DH, Scoles HA, Horike S, et al. 15q11.2–13.3 chromatin analysis reveals epigenetic regulation of CHRNA7 with deficiencies in Rett and autism brain. Hum. Mol. Genet. 2011;20(22):4311–4323. doi: 10.1093/hmg/ddr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harlalka GV, Baple EL, Cross H, et al. Mutation of HERC2 causes developmental delay with Angelman-like features. J. Med. Genet. 2013;50(2):65–73. doi: 10.1136/jmedgenet-2012-101367. [DOI] [PubMed] [Google Scholar]