Abstract
Thermal denaturation of proteins is critical to cell injury, food science and other biomaterial processing. For example protein denaturation correlates strongly with cell death by heating, and is increasingly of interest in focal thermal therapies of cancer and other diseases at temperatures which often exceed 50 °C. The Arrhenius model is a simple yet widely used model for both protein denaturation and cell injury. To establish the utility of the Arrhenius model for protein denaturation at 50 °C and above its sensitivities to the kinetic parameters (activation energy Ea and frequency factor A) were carefully examined. We propose a simplified correlated parameter fit to the Arrhenius model by treating Ea, as an independent fitting parameter and allowing A to follow dependently. The utility of the correlated parameter fit is demonstrated on thermal denaturation of proteins and cells from the literature as a validation, and new experimental measurements in our lab using FTIR spectroscopy to demonstrate broad applicability of this method. Finally, we demonstrate that the end-temperature within which the denaturation is measured is important and changes the kinetics. Specifically, higher Ea and A parameters were found at low end-temperature (50°C) and reduce as end-temperatures increase to 70 °C. This trend is consistent with Arrhenius parameters for cell injury in the literature that are significantly higher for clonogenics (45 – 50 °C) vs. membrane dye assays (60 –70 °C). Future opportunities to monitor cell injury by spectroscopic measurement of protein denaturation are discussed.
Keywords: Thermal therapy, Kinetics, FTIR, Differential scanning calorimetry, Enthalpy-entropy compensation, Protein Denaturation
1. INTRODUCTION
Proteins are large biological molecules that consist of one or more chains of amino acids, and possess complicated three-dimensional folded structures and unique functionality.[1] Protein denaturation refers to a conformational change resulting in partial or total unfolding of a protein from its native state, and/or structural damage to the molecule.[2] Specifically, the unfolding includes changes in secondary structures such as α-helices and β-sheets, and leads to change or inactivation of protein function. This process is important for a number of biotechnologies[2, 3], disease conditions and treatments.[4, 5] Thermally induced protein denaturation, in particular, has found important applications in food processing,[3] burn injury, and thermal ablation of diseased or cancerous tissue,[6, 7] and has been proposed to correlate with cell injury.[4, 8]
The dynamic measurement of protein denaturation is most readily achieved by optical and calorimetric approaches. Typical measurement techniques for protein denaturation, especially protein thermal denaturation, are listed in Table 1. Among all the methods, X-ray crystallography and NMR spectroscopy are known to determine the three dimensional structure of proteins,[1] but X-ray crystallography is limited to static measurements since it requires proteins in a crystalline state. Optical approaches such as birefringence, vibrational spectroscopy (i.e. Fourier transform infrared spectroscopy (FTIR) and 2D correlation infrared spectroscopy (IR)) and circular dichroism (CD) are more suited to measure the dynamic change in the secondary structure of proteins during denaturation, in particular α-helix and β-sheet structures (IR band changes[8]) or loss of structure (order to disorder in birefringence[9]). On the other hand, differential scanning calorimetry (DSC) measures the enthalpic change during protein denaturation and unfolding. In this study, FTIR is used to monitor protein denaturation during thermal treatment.
Table 1.
Measurement techniques for protein denaturation
| Technique | Measurement | Sample requirement | Static/Dynamic | Ref. |
|---|---|---|---|---|
| X-ray crystallography | 3D structure | Protein in crystal state | Static | [1] |
| NMR+ spectroscopy | 3D structure | Small proteins or protein domains (≤20kD) | Dynamic | [1, 48] |
| FTIR | Secondary structure | Surface or thin sample (<100µm) | Dynamic | [8] |
| 2D IR | Secondary structure and hydration | Surface or thin sample (<100µm) | Dynamic | [49] |
| Circular dichroism (CD) | Secondary structure | Minimal preparation. Minimize absorption of other species in UV range. | Dynamic | [50] |
| DSC | Calorimetric effect associated with protein structure change. | Sample volume range from ng (Nano DSC) to mg (traditional DSC) depending on the instrument | Dynamic | [4, 8] |
| Birefringence | Anisotropy in tissue | Optically accessible | Static | [9] |
| Dilatometry | Density change | Liquid sample | Dynamic | [51] |
| Rheometry | Viscosity change | Liquid sample | Dynamic | [52] |
NMR stands for Nuclear magnetic resonsance. Heat induced protein aggregation can be challenging to study with NMR.[48]
Different models have been used to describe the kinetics of protein denaturation. For example, protein denaturation can be described as an absolute rate process between a native state, the final denatured state and one or several macrostates in between. One of the best-known models is the Lumry-Eyring model (or three state model) [10–12] which assumes that protein denaturation occurs in two steps: (1) reversible unfolding of native protein (N); and (2) irreversible change of unfolded protein (U) to the final denatured state (D).
| (1) |
Furthermore, the two-step Lumry-Eyring model can be simplified to a one-step, two-state irreversible denaturation model when the irreversible step is significantly faster than reversible unfolding step. [11, 13]
| (2) |
This first-order irreversible reaction kinetic model has been widely used for the analysis of protein denaturation and thermal injury of cells and tissues.[7, 14, 15] The reaction rate of this irreversible process is described by a temperature dependent rate constant (Arrhenius equation)
| (3) |
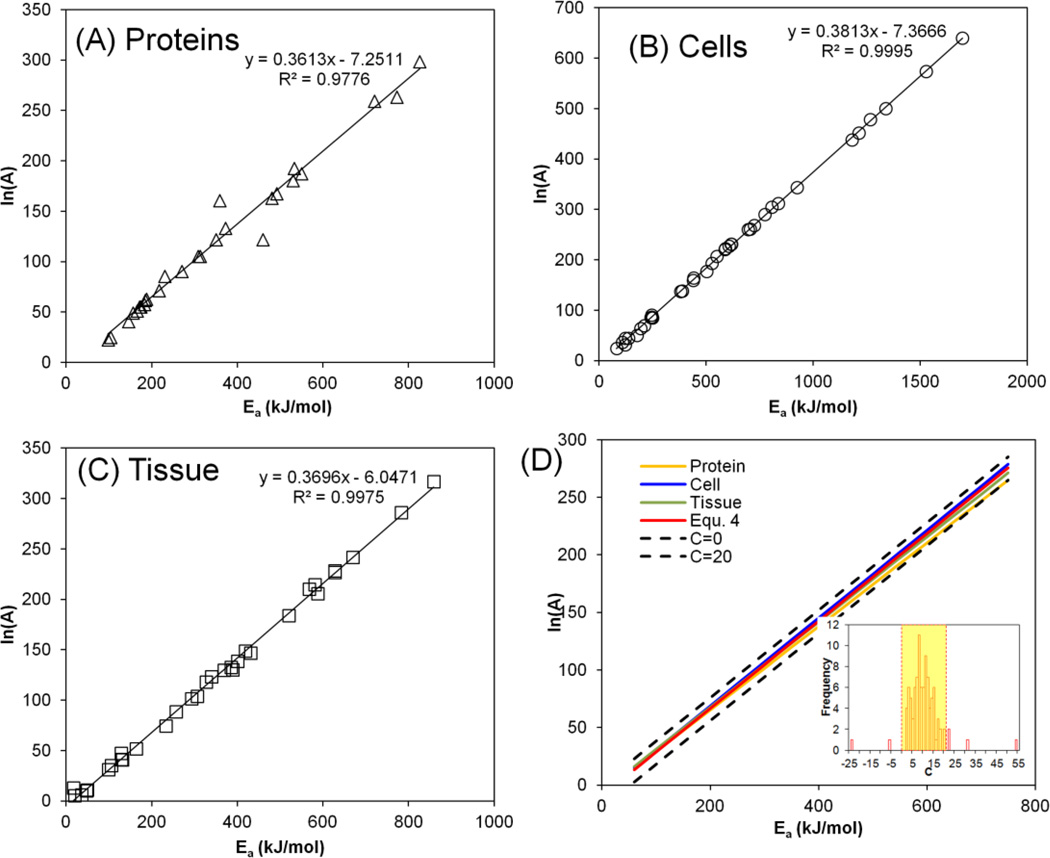
where A is the frequency factor (s−1), and Ea is the activation energy (kJ mol−1). Two parameters (Ea and A) are used to describe the kinetics of thermally induced protein denaturation where Ea is related to the reaction activation enthalpy and A depends on the reaction entropy according to the Eyring-Polyani equation.[16]. The activation energy in various protein systems varies between 100 and 800 kJ mol−1 and the frequency factor varies between 109 to 10129 s−1 (i.e. 20~300 for ln{A}) as shown in Figure 1.
Figure 1.
Empirical correlation between the activation energy (Ea, kJ mol−1) and natural logarithm of frequency factor (ln{A}, s−1) values for (A) proteins, (B) cells, and (C) tissues. (D) shows that the range of C chosen encompasses the linear fits for proteins, cell and tissues shown in (A–C). The kinetic parameters are plotted from tables in He et al.[7] and updated with recently reported data.[25, 53, 54] Inset histogram shows the distribution of C values and the highlighted region represents the range of C for the fit (0<C<20).
Despite the wide variation of the activation energy and frequency factor, it has been suggested that there is a relationship between the parameters (enthalpy-entropy compensation or alternatively Meyer-Neldel rule).[17, 18] The compensation law states that the kinetic rate varies exponentially with temperature, and a linear relationship exists between the thermal activation energy and logarithm of the prefactor, or frequency factor. This has been demonstrated for a variety of thermally activated processes including semiconductor conductivity, annealing phenomena, aging of insulating polymers, chemical reactions and biological death rates.[18, 19] Rosenberg was among the first to report the compensation behavior for thermal inactivation of different protein systems (pure protein, virus, yeasts, and bacteria).[17] The linear relation between the activation energy and ln{A} has also recently been demonstrated in more complex protein systems such as cells and tissues. Here, He et al. reported a linear correlation between Ea and ln{A} which likely represents the compensation law:[7]
| (4) |
Around the same time, Wright et al. also reported similar correlation (ln{A} = 0.3832*Ea - 10.042).[20] The correlations reported by He et al. and Wright et al. are similar to the one reported by Rosenberg (ln{A} = (0.366~0.370)*Ea −14.5).[17] Figure 1 shows Ea versus ln{A} plots of thermally induced denaturation of pure proteins (A), cells (B) and tissues (C) demonstrating a linear correlation in each case. It has also been suggested that random and systematic errors can lead to apparent compensation behavior which necessitates more careful considerations.[21, 22] In addition, modification of the Arrhenius model has also been reported to reduce the difficulty to fit the correlated kinetics parameters by introducing an optimum reference temperature.[23]
In this report, we first performed a detailed analysis on the characteristics of the Arrhenius model by examining its sensitivities to the kinetic parameters (Ea and A). We propose a correlated parameter fit that leverages the empirical correlation between the two kinetic parameters for protein denaturation in biological systems, and demonstrated the utility of this correlated parameter fit with several protein and cell systems measured by FTIR spectroscopy. We envision that the correlated parameter fit will simplify the measurement and analysis of protein thermal denaturation, and project towards future opportunities to monitor cell injury by spectroscopic measurement of protein denaturation.
2. MATERIALS AND METHODS
Cell culture
Several different cell types were used in the study including human dermal fibroblasts (HDF) and LNCaP Pro 5 tumor cells. HDF cells were obtained from cryopreserved stock (Cambrex, East Rutherford, NJ) and cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (p/s) in saline (Invitrogen, Carlsbad, CA). LNCaP cells were obtained from cryopreserved stock (MD Anderson Cancer center, Houston, TX) and cultured in DMEM F-12 media (Gibco, Grand Island, NY) and supplemented with 10% FBS and 1% p/s in saline. All cells were trypsinized using 0.05% Trypsin and 0.53mM EDTA (Gibco, Gaithersburg, MD). The trypsin was inactivated using serum filled media. Cells were then centrifuged at 400g for 10min, the media removed, and 12µL of the cell pellet was sandwiched between CaF2 windows for FTIR analysis. Historical data on dunning AT-1 prostate cancer cell (ref #8) and HuH-7 liver cancer cell (ref #8) were performed with the same protocol.
Measurement of Protein Denaturation with FTIR
FTIR spectra were obtained using a Nicolet Magna 750 spectrometer (Thermo-Nicolet, Madison, WI) equipped with a TGS detector similar to that used by Wolkers et al.[24] Spectra were acquired at 4cm−1 resolution using 32 co-added interferograms between 4000-900 cm−1 wave number ranges. Strong vibration bands from water are observed at ~3200–3600 cm−1, ~2200 cm−1 and ~1650 cm−1 corresponding to the –OH stretching vibration, the libration and bending combination vibration, and the scissoring vibration, respectively. The symmetric and asymmetric –CH2 stretching bands of lipid acyl chains are visible between 3000-2800 cm−1. Characteristic protein bands are visible at ~1650 cm−1, ~ 1550 cm−1 and ~1250 cm−1 corresponding to the amide-I, amide-II and amide-III bands respectively.
The samples were mounted into a specialized temperature cell whose temperature was regulated using a temperature controller (Minco products Inc., Minneapolis, MN). Sample temperature, also monitored separately using a thermocouple, was increased from room temperature to 90°C at 1°C min−1. Spectral analysis was carried out using Omnic software (Thermo-Nicolet, Madison, WI). Protein denaturation was determined by monitoring area changes of the raw spectra in the α helical (~1315cm−1) and the β sheet (~1235cm−1) regimes in the amide III region as a function of temperature as previously described.[24] The fractional denaturation (FD) of protein was calculated by the change in the β sheet. Protein denaturation is then normalized within the temperature range of interest (onset to end–temperature 50, 60, 70 or 80 °C). The onset temperature of protein denaturation is defined as the temperature where the fractional denaturation displays an abrupt increase above 37°C (Supplemental 1).[8]
Kinetic model and correlated parameter fitting method
A rate constant can be derived from the one step irreversible Arrhenius denaturation model using Equation (3). The fractional denaturation (FD) is then calculated by integrating the rate (k) and considering constant rate heating from low to high temperatures
| (5) |
where B is the rate of temperature change (or heating rate, K s−1).
For the traditional fit, the entire Ea-ln{A} parameter space is scanned and every combination of Ea and ln{A} is evaluated. This procedure consists of two steps. First, a grid search was performed on a selected 2D matrix so that all the possible combinations of Ea and ln{A} can be tested. The goodness of the fit (R2) was evaluated as follows
| (6) |
where Yi and Fi are the ith experimental data point and model fit respectively, and Ȳ represent the average of all experimental data. This grid search procedure yields a Ea-ln{A} combination with the maximum R2. Next gradient descent procedure was performed to further find a local optimal R2. In brief, the gradient at current point is calculated and steps are taken to calculate the R2 at the new point. If the R2 is higher than the current point, then the optimization step is accepted and it proceeds to the next optimization. On the other hand, if the R2 is lower than the current point, this optimization step is rejected and then a smaller step is taken.
The characteristics of the Arrhenius model were studied by testing the sensitivities of the fit to the variations by changing Ea & ln{A} separately, and by changing them at the same time according to Equation 4.
For the correlated parameter fit, Ea is chosen and then only Ea and ln{A} values close to the Ea-ln{A} correlation (Equation 4) are evaluated (Figure 1D between the dashed lines), followed by the gradient descent procedure as described above. The fitting procedure was performed with programs written in MATLAB. It is likely that the best Ea-ln{A} fit will not always appear on the Ea-ln{A} line. As a result, the intercept (9.36, in Equation 4) is allowed to vary within a small range by introducing a floating constant (C) that varies within 0 to 20. The kinetic rate can be thus expressed as
| (7) |
Although a smaller range of C could be selected, this current selection is a conservative choice. The frequency factor - or more frequently the natural logarithm of frequency factor, if needed - can be calculated by
| (8) |
3. RESULTS
First, we performed detailed analysis of the characteristics of the Arrhenius model using protein denaturation data from the literature. Then we validated our correlated parameter fit approach with literature data including denaturation of a pure protein (collagen peptides)[25] and a cell line (AT-1)[8], and demonstrated the close agreement of our proposed protocol with the literature data. Finally, we demonstrated the utility of the correlated parameter fit on thermally induced denaturation of a pure protein (bovine serum albumin) and several cell lines (new data on HDF and LNCaP; historial data on AT-1 (ref# 8 and HuH-7 (ref# 5)) as measured by FTIR spectroscopy.
Characteristics of Arrhenius Model
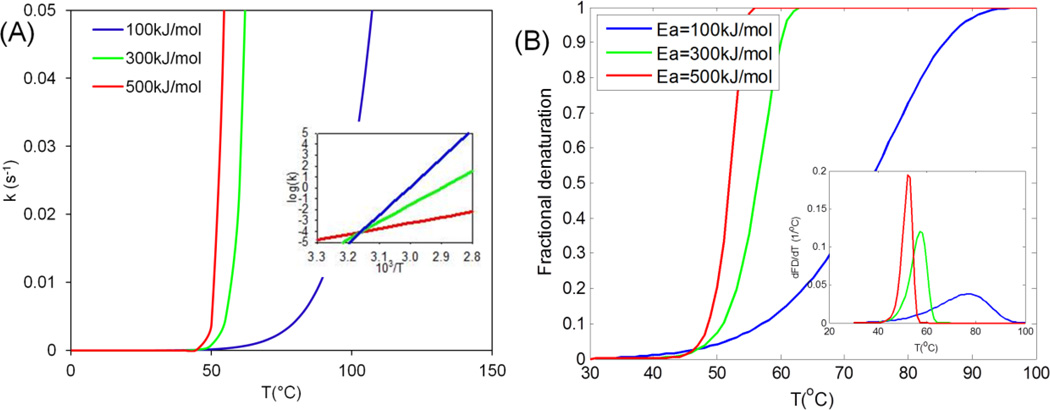
First we plotted the denaturation kinetics for low, medium and high activation energies (100, 300 and 500 kJ mol−1) as shown in Figure 2. The frequency factor is obtained using Equation (4), i.e. C=9.36. The reaction rate increases faster with temperature for higher activation energy (Figure 2A), and changes exponentially with the inverse of temperature, as would be expected experimentally during isothermal protocols (Figure 3A inset and Table 2). Figure 3B shows the expected denaturation rate (i.e. excess Cp for DSC) and fractional denaturation that can be measured by DSC or FTIR.
Figure 2.
Representative Arrhenius kinetic plots for low, medium and high activation energies. (A) Reaction rate vs. temperature. Inset shows the logarithm of k is linearly proportional to 103/T. (B) Fractional denaturation (FD) vs. temperature at 2°C min−1 heating rate. Inset shows rate of denaturation (dFD/dT) with temperature. Mathematically, it is the first derivative of fractional denaturation (FD) with temperature.
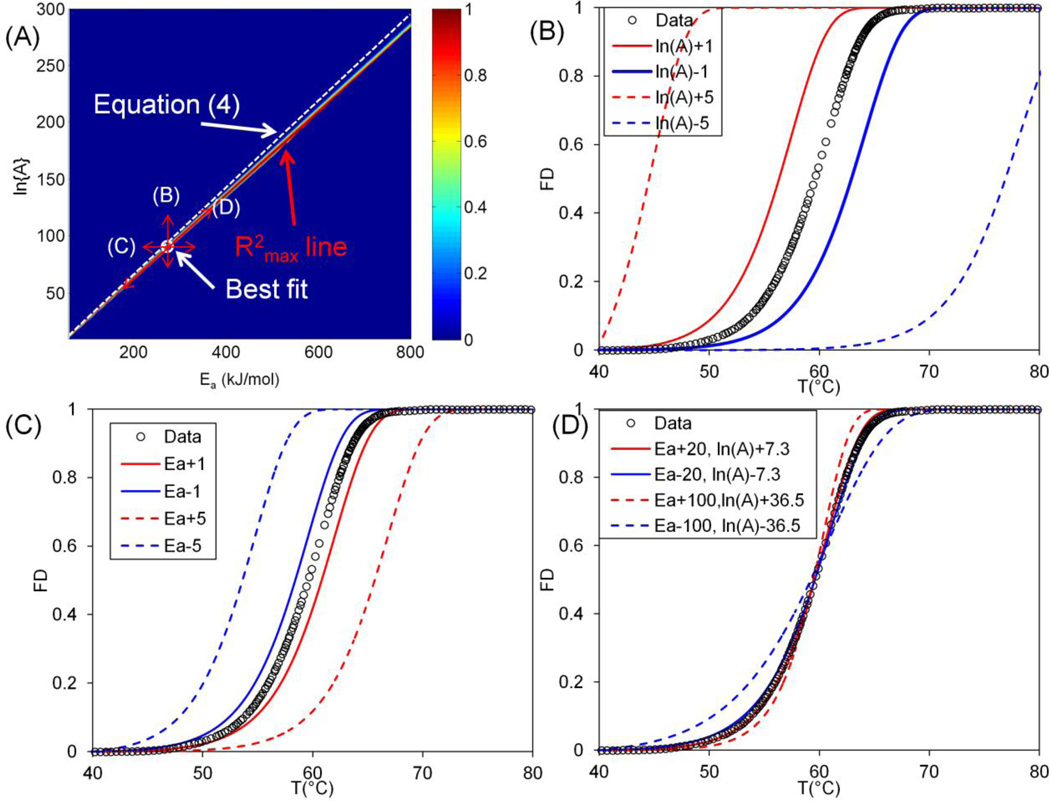
Figure 3.
Sensitivity analysis of the fitting results. (A) shows the distribution of R2 for Ea-ln{A} space for Miles et al. 0.1°C min−1. There exists a high R2 region and a line can be plotted along the highest R2 (R2max line). The R2max line is close to Equation 4. The white circle corresponds to the best fit. The arrows show the method of varying Ea and ln{A} with corresponding plots (B–D). (B) and (C) show the theoretical prediction when changing ln{A} and Ea by ±1s−1and ±1kJ mol−1 respectively. (D) shows the theoretical prediction when shifting the Ea value around the optimal Ea by ±20 and ±100kJ mol−1 along the R2max line. FD: Fractional Denaturation.
Table 2.
Protocols of measuring protein thermal denaturation and methods of interpreting protein denaturation data
| Heating method | Experiments | Result interpretation methods | Ref. |
|---|---|---|---|
| Isothermal (Temperature jump) | ≥ 5 temperatures, n=3 | (1) Plot ln{k} vs. 1/T (2) Linear fit to obtain Ea and ln{A} |
[27, 30, 32] |
| Controlled heating rate | N=3 | Scanning method | [25] |
| Integral transform | [25] | ||
| DSC thermogram shape analysis | [25, 26] | ||
| Flexible tolerance | [8] | ||
| Correlated parameter fit, Eqn. 7 | This work | ||
| Transient heating, T(t) with a single endpoint measurement. | N=3 | Paired Ea and ln{A} values that minimize the error in injury parameter yield an equivalent isothermal experiment. | [47] |
Next we examined the sensitivity and characteristics of the curve fit. The goodness of fit (R2) for the entire space was plotted in Figure 3A for the case of 0.1°C min−1 from Miles et al.[25] It shows a clear region where the R2 value is the highest, namely the R2max line. The linear correlation (Equation 4) was plotted as the white dashed line and is close to the R2max line. The best fit value (Ea=273.5 kJ mol−1, ln{A} = 90.9) is close to the line given by Equation (4) as well. Further, we analyzed the distribution of R2 along the R2max line (the line fitted from the value that gives the highest R2 at different Ea values) as shown in Supplemental 2. The peak value on the plot shows the optimal fit. Carefully examining the region close to the optimal fit shows that a range of Ea values give a good fit (Ea = 260 ~ 290 kJ mol−1, R2>0.9995). Note that this is based on the condition that corresponding ln{A} values were defined by the R2max line shown in Figure 3A. If plotting the R2 value by keeping either the ln{A} or Ea constant as shown in Figure S2–C and D respectively, the distribution is much sharper (ΔEa < 4kJ mol−1, and Δln{A} < 2 for R2>0.9). Further, we examined the result of theoretical prediction when shifting away from the best fit (Figure 3A). Specifically, changing only ln{A} by 1 ~ 5 while keeping Ea constant or changing only the Ea value by 1 ~ 5kJ mol−1 with ln{A} constant causes the theoretical prediction to shift away from the experimental data significantly (Figure 3B and C). However, changing the Ea value by 20 kJ mol−1 along the R2max line only slightly changes the slope of the fitted curve, as shown in Figure 3D.
Historical Data Fits
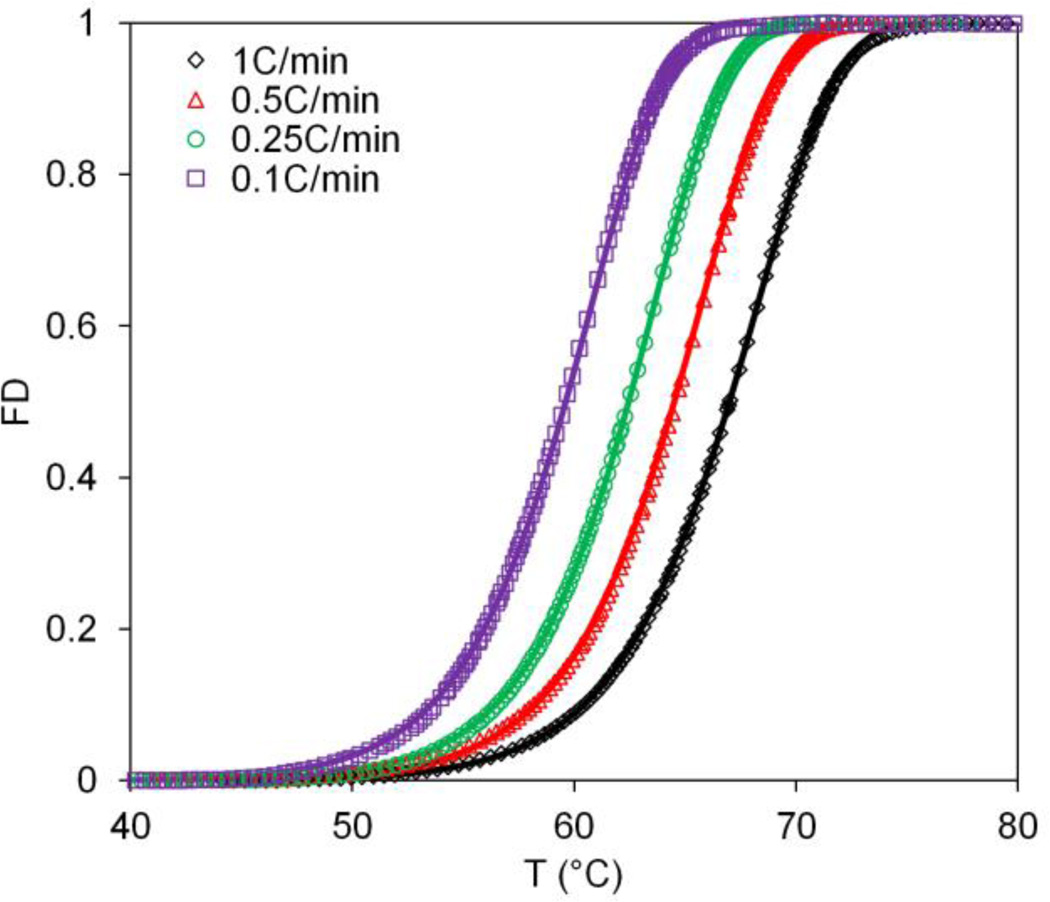
Miles et al. have performed seminal work on the thermal denaturation of collagen, an important ECM protein, using DSC.[25–27] Taking one of his recent publications on the thermal denaturation of a collagen-like peptide (Pro-Hyp-Gly)10[25] we show close agreement between our correlated parameter fit values with the previous fitting approach[25], as shown in Figure 4 and Table 3. Specifically, Miles reported the curve fit activation energy to be within 270 – 283kJ mol−1, and our approach matches reasonably for all of the cases (Table 3). Furthermore, previous data from our lab, where we had fit the thermal denaturation of proteins in AT-1 cells by both DSC and FTIR, were also re-analyzed. Here we are also able to show good agreement between the more complicated flexible-tolerance two-parameter method used by He et al.[8] and the more easily implemented correlated parameter fit shown here (Ea = 127.8 v.s. 141.5 kJ mol−1). It is worth noting here that the activation energy does not depend on heating rate (Table 3), indicating that activation energy is an intrinsic property of the protein. Higher heating rate, however, delays the denaturation events to higher temperatures (Figure 4) since protein experiences less time at each temperature.
Figure 4.
Correlated parameter fit for the thermal denaturation of collagen like peptide (Pro-Hyp-Gly)10. Four different heating rates (1, 0.5, 0.25, 0.1 °C min−1) were analyzed. Data from Miles et al. [25] 2007. FD: Fractional Denaturation.
Table 3.
Validation of correlated parameter fit (CP) with literature. Results and methods from Miles et al. [25] and He et al. [8] were compared and shown in bold.
| Range | Ea (kJ mol−1) | ln{A} | R2 |
|---|---|---|---|
| Pure protein | |||
| Miles et al.[25] | 270~283 | 89~95 | NA |
| CP fit of Miles et al. 1C min−1 | 264.4 | 87.8 | 1.0000 |
| CP fit of Miles et al. 0.5C min−1 | 273.4 | 91.0 | 1.0000 |
| CP fit of Miles et al. 0.25C min−1 | 282.6 | 94.3 | 1.0000 |
| CP fit of Miles et al. 0.1C min−1 | 273.5 | 90.9 | 0.9999 |
| Cell proteins | |||
| He et al.[8] AT-1 FTIR | 141.5 | 45.7 | 0.996 |
| CP fit of He et al. AT-1 FTIR | 127.8 | 40.7 | 0.9977 |
| He et al. AT-1 DSC | 159.7 | 52.2 | 0.997 |
| CP fit of He et al. AT-1 DSC | 147.1 | 47.6 | 0.999 |
Protein Denaturation Fit
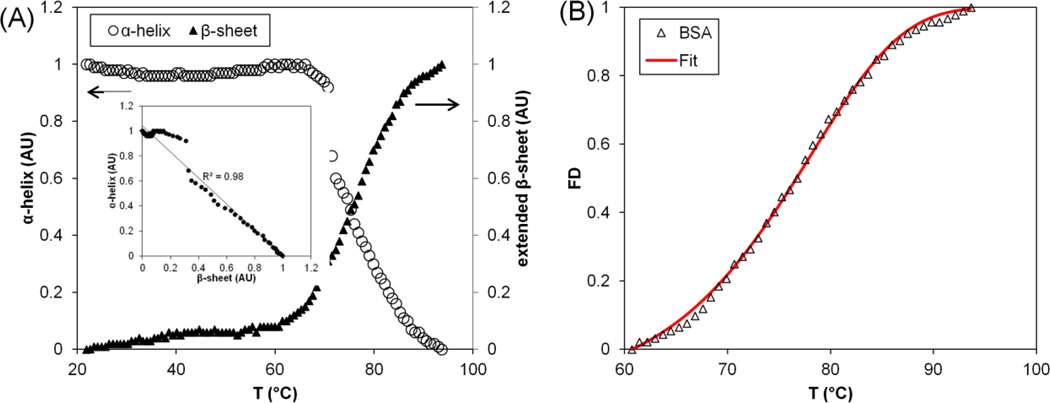
Thermal denaturation of a model pure protein, Bovine Serum Albumin (BSA), was studied by monitoring the infrared spectrum changes with FTIR. Figure 5 depicts the increase in extended β-sheet structures and the concomitant decrease in α-helical structures during thermal denaturation of BSA. An inverse linear correlation (R2=0.98) is obtained between the decrease in α-helical structure and increase in β-sheet structures (Figure 5A – inset). The onset temperature (Tonset) of BSA is 60.7°C, which is determined at zero first derivative of the fractional denaturation before fractional denaturation displays an abrupt change above 37 °C.[8] The correlated parameter fit was then applied to BSA (Figure 5B) and the activation energy determined to be 115.4 kJ mol−1.
Figure 5.
Protein denaturation of Bovine Serum Albumin (BSA). (A) BSA thermal denaturation characterized by monitoring changes to the α-helical and extended β-sheet structures as a function of temperature increases from 20°C to 95°C. The inset depicts the correlation between the decrease in α-helical and the increase in extended β-sheet content during thermal denaturation. (B) Fitting of BSA thermal denaturation by the correlated parameter model. FD: Fractional Denaturation.
Cell Denaturation Fit
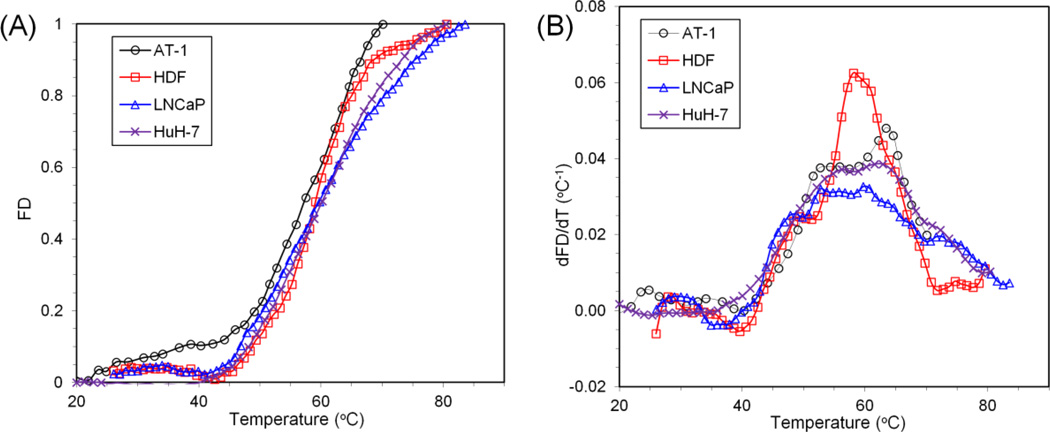
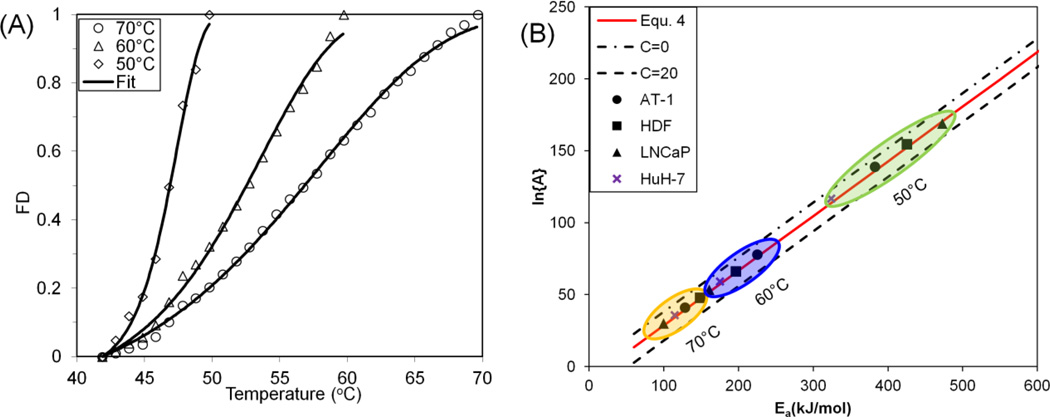
The fractional denaturation of two different cell types (HDF & LNCaP) was quantified during heating from room temperature to 90°C at a rate of 1°C min−1. In addition, AT-1 and HuH-7 cell denaturation data were obtained from He et al. (heating rate: 2°C min−1)[8] and Avaralli et al. (heating rate: 2°C min−1).[5] Figure 6A shows the increase in the β sheet structures for these cell types as a function of temperature. First derivative plots (Figure 6B) are included to identify peaks and patterns in the denaturation profiles. Specifically, it has been suggested that 5 to 8 specific protein groups can be identified to reside within different thermal regimes from higher order derivatives of protein denaturation plots, and that one or more group may be rate limiting to cell injury.[4, 8] This further highlights the cell-specific nature of protein denaturation (or “thermal fingerprinting”[28]) with profiles differing in the onset temperature of protein denaturation (Tonset), the slope of the curve, and the distribution of transition regions. The onset denaturation temperatures (Tonset) for AT-1, HDF, LNCaP and HuH-7 are 40.27°C, 42.53°C, 40.88°C and 40.03 °C respectively (Table 4). The activation energy of protein denaturation for the cells was assessed using the correlated parameter fit, and the results are listed in Table 4. The protein denaturation data is further analyzed at different end temperatures after renormalization. At lower end temperatures (50 °C) corresponding to the first transition regions, the activation energy is highest, as shown in Table 4 and Figure 7. Several other end-temperatures (60 and 70 °C) were used to encompass more of these transition regions. The effect of including more or fewer protein transitions is shown by varying the end temperature in Figure 7.
Figure 6.
Protein denaturation signature of four different cell types (AT-1, HDF, LNCaP, and HuH-7). (A) Comparison of the protein denaturation of four different cell types – Dunning AT-1 prostate cancer cells, human dermal fibroblasts (HDF), LNCaP prostate tumor cell, and HuH-7 liver cancer cell – characterized by monitoring changes to the extended β-sheet structures as a function of temperature increase from 20°C to 90°C. The error bars were omitted for easier data visualization. (B) First derivative of the average denaturation curves are plotted as a function of temperature. Note that AT-1 data is from He et al.[8] and HuH-7 data is from Avaralli et al.[5] FD: Fractional Denaturation.
Table 4.
Kinetic parameter fittings for four cell lines (AT-1, HDF, LNCaP, and HuH-7) with different end temperatures.
| Cell line | Tonset | Tend | Ea (kJ mol−1) | ln{A} | R2 |
|---|---|---|---|---|---|
| AT-1 | 40.27 | 70 | 128.6 | 41.0 | 0.9979 |
| 60 | 225.3 | 78.0 | 0.9976 | ||
| 50 | 382.4 | 139.0 | 0.9889 | ||
| HDF | 42.53 | 80.54 | 119.8 | 37.0 | 0.9950 |
| 70 | 148.7 | 47.8 | 0.9990 | ||
| 60 | 196.4 | 66.1 | 0.9851 | ||
| 50 | 425.1 | 154.5 | 0.9908 | ||
| LNCaP | 40.88 | 83.52 | 61.4 | 15.3 | 0.9983 |
| 70 | 99.7 | 29.9 | 0.9984 | ||
| 60 | 160.9 | 53.5 | 0.9960 | ||
| 50 | 472.7 | 169.0 | 0.9956 | ||
| HuH-7 | 40.03 | 80.4 | 84.1 | 24.3 | 0.9985 |
| 70 | 114.5 | 35.9 | 0.9985 | ||
| 60 | 175.1 | 59.3 | 0.9975 | ||
| 50 | 323.8 | 116.8 | 0.9900 |
Figure 7.
Summary of correlated parameter fit. (A) Fitting of protein denaturation for LNCaP cells at different end temperatures (70, 60, and 50°C); (B) Summary of correlated parameter fit at different end temperatures. FD: Fractional Denaturation.
4. DISCUSSION
Characteristics of Arrhenius Model and Debate on Compensation Behavior
While the correlation of the activation energy and frequency factor in Arrhenius kinetics has been recognized for years, the possible existence of the compensation law has stirred intensive debate.[17, 29] One possible explanation is that the parameters are correlated, but that complications from experimental errors and the form of the Arrhenius equation itself lead to the proposed compensation law behavior in multiple systems.[21, 22, 30] There are several ways that the linear relationship (i.e. compensation law behavior) between activation energy and logarithm of frequency factor can be derived, including from the transition state theory[7] and statistical mechanics. Yelon et al. [31] suggested that the compensation law holds as long as the activation energy is large compared with the typical excitations (i.e. infrared vibrations or phonons[19]) and with the thermal energy (k’T, k' = Boltzmann constant). Recently it has been suggested that random errors in experimental measurement can lead to an apparent compensation effect, and that a confidence ellipse should be calculated in order to eliminate this effect.[30] We have performed this calculation and demonstrate that there is correlation between the parameters even with random experimental errors (Figure S3).
A further simple exercise demonstrates the correlation between the Arrhenius parameters by rewriting Equation (3), to obtain
Now considering the relatively small temperature range (1/RT ~ constant), ln{A} and Ea have to be inter-related to obtain a rate constant (k).1 For instance, as shown in Figure 3, changing Ea and ln{A} separately by a small magnitude (1 kJ mol−1and 1 s−1 respectively) leads to significant shift in the predicted curve, while changing Ea and ln{A} proportionally by a large magnitude (20 kJ mol−1and 7.3 s−1) along the R2max line changes the prediction curve only marginally.
Advantages of the Correlated Parameter Fit
Regardless of the existence of a compensation law, the proposed correlated parameter fit has several advantages over other approaches. One advantage is that it can significantly simplify the way kinetic parameters are obtained from experimental data. Traditionally, the kinetic parameters are fitted to isothermal temperature jump (T-jump) experiments as routinely performed for chemical kinetic experiments.[13] Specifically, the sample is held at various temperatures (usually ≥ 5 points) for extended amount of time. A linear fit (Arrhenius plot) can be correlated between ln{k} and 1/T. The activation energy and frequency factor can be determined from the slope and intercept of the linear fit, respectively.[7, 15, 32] This procedure involves tedious experiments under many different conditions over several orders of magnitude in heating time. With the controlled heating rate method proposed, it is possible to obtain the kinetic parameters from fewer experiments (3 runs, instead of 5 × 3 runs). For instance, it is possible to scan the samples with constant heating rates to certain temperatures. Then similar to the procedure described in this report, the kinetic parameters can be extracted with our proposed correlated parameter fit approach. This avoids the more difficult approach of fitting two parameters (2P, Ea and A) simultaneously, which necessitates time-consuming experimental repeats at different isothermal temperatures or complicated fitting routines in dynamically heated data (Table 2).
In both the calorimetric and FTIR spectroscopic measurement, the constant heating rate scan is a commonly-used and simpler method to measure protein denaturation, as discussed above. The heating rate or scanning rate is thus an important parameter and higher heating rate delays the denaturation events to higher temperatures (Figure 4) since protein experiences less time at each temperature. Since the scanning rate is incorporated in the fitting (Equation 6, parameter B), it does not affect the resulting kinetic parameters (Table 3).
Due to experimental errors, a tolerance range (C) has been used. The proposed tolerance range (0 < C < 20) can well encompass the experimental variation observed within the data tested in this report and beyond. The C values obtained from this report range maximally from 7 to 13, well within the proposed range (0–20). For the protein systems shown in Figure 1, the C values are within the proposed range, except for only 6 out of the 99 samples from the literature (Figure 1D inset). One of the 5 discordant samples has a very low activation energy (16.7 kJ mol−1), which may fall outside of the range where compensation behavior is typically observed. It may be possible to further refine this range in the future. However, a conservative choice was made for purposes of this study.
Relevance of Protein Denaturation to Cell Injury
The denaturation of pure proteins can be modeled by the Arrhenius model, as shown for the collagen like peptide and BSA in this study. The denaturation of protein mixtures and cells are more involved and may present multiple peaks from denaturation of different proteins that may partially overlap. For instance, the denaturation of egg white (60–65% ovalbumin, among other proteins[33]) displays two distinct peaks (at 65.7°C and 77.5°C as seen in Supplemental 4). In cells, the denaturation of proteins is believed to involve up to four to seven major protein transitions or groups[4, 8]. Each protein group can often be modeled as a separate first-order irreversible kinetic process (Arrhenius model). As a result, four to seven pairs of kinetic parameters can be calculated.[8] Further, deconvolution of protein denaturation in such complex systems may help uncover critical targets, and hence the rate limiting events for cell thermal injury. However, further work is needed to understand the function and location of these protein groups (i.e. membrane, cytoplasmic or cytoskeletal), and to directly observe them during denaturation. This may be possible in the near future by using advanced optical imaging techniques such as confocal Raman microspectroscopy, as has been demonstrated for chemical denaturation in cells.[5]
The proposed correlated parameter fit approach can be used to assess denaturation of additional protein groups by increasing the end-temperature limit. This has relevance to both the amount and type of protein denatured and the kinetics of cell death measured thereafter by various assays. As shown in this study, the activation energy increases with decreasing end temperature (Table 4 and Figures 7). This agrees with fits on deconvolved protein groups in previous work, where the initial protein group has the highest activation energy, and all protein groups together have lower activation energy.[8] Further, cell protein denaturation can be correlated with different cell injury assays, such as clonogenics and membrane integrity assays, after thermal treatment. For instance, the activation energies obtained from the clonogenic proliferation assay (Ea > 400 kJ mol−1) are usually much larger than the membrane integrity assay (e.g. Propidium Iodide – Ea < 300 kJ mol−1).[7, 34] A narrower temperature range for protein denaturation correlates well with clonogenic assays (close to 50°C) while a larger range of protein denaturation (up to roughly 60°C) correlates well with PI/Hoechst cell membrane integrity.[8]
Protein denaturation may be used as spectroscopic signature to assess cell injury in vivo. Currently, thermal therapies rely on predetermined protocols or advanced treatment planning to treat localized tumors.[35–37] Although there have been some efforts using imaging guidance,[38] employing real-time MR thermography and computation to adjust for patient-specific variations of the thermal responses,[39] very limited efforts have been focused on assessing cancer cell injury in vivo during thermal therapy. This work suggests new opportunities to use spectroscopic signatures from protein denaturation as a guide to assess cell injury in vivo. An earlier study suggested that only 5 – 10% of the total cellular protein denaturation, i.e. only the most thermally sensitive protein group(s), leads to 80 ~ 90% loss in viability by clonogenics.[4] This approach may become increasingly feasible as advanced imaging techniques become available.
Limitation of the Current Work
There are several limitations in this simplified correlated parameter model. Firstly, the complex process of protein folding and unfolding and its pathways are active areas of research themselves [40] and are not considered in the simple two-state irreversible kinetic model. For instance, typical protein unfolding involves several intermediate steps, each with different characteristics, as reviewed by Daggett et al.[41] Instead, here the protein denaturation is assumed to be a one-step first-order irreversible kinetic process in a lumped parameter sense. Many important protein interaction processes involve reversible steps,[42, 43] which were not included in this analysis. The impact of heat shock (and other chaperone) proteins in preventing thermal protein denaturation was also not considered (i.e. the development of thermo-tolerance), although it should be noted that heat shock proteins are expected to be most active in the range of 40 – 45 °C not above 50 °C. Secondly, only thermal denaturation has been considered. There are many other denaturants, such as chemical agents (e.g. urea), pressure, and pH, that can also affect the denaturation kinetics, and are not considered in this report. Thirdly, protein denaturation under higher temperatures and on short time scales appears to deviate from simple first order kinetics and compensation law behavior. It can be estimated that at 90 to 100°C or higher temperatures, proteins will denature on time scales of micro to nano seconds, [44, 45] and molecular dynamic (MD) simulations suggests very low activation energies (33 ~ 56 kJ mol−1, Supplemental 5).[44] Further work is needed to probe the protein denaturation kinetics in this temperature range, such as might occur for proteins attached or in close proximity to nanoparticles being heated by lasers[46] or at the edge of cavitation bubbles during high intensity focused ultrasound.
While protein denaturation is linked to cell injury, the precise correlation between the amount and timing of protein denaturation to cell death remains a central question in this field.[4, 8, 47] For instance, some work suggests that immediate cell injury or death by protein denaturation can occur with as little as 5 to 10% of the cellular proteins being destroyed.[4] In this case, the Arrhenius approach suggested here may be useful in predicting the temperature and time necessary to achieve this. However, there are more subtle forms of delayed cell injury that can occur post heating, such as thermotolerance, receptor-induced and/or intrinsic apoptosis and necroptosis.[42, 43] These cell death processes are not immediate and can involve dynamic cascades and reversibility in systems of proteins that are outside the treatment of this paper and not amenable to the Arrehnius model. To model these complicated processes, more intricate mathematical constructs and parameters are required than Arrhenius kinetics provides, and are beyond the scope of this work.[42, 43]
5. CONCLUSIONS
This work brings together an improved framework for experimental and theoretical work on thermal protein denaturation. Specifically, we examined the characteristics of the Arrhenius model and further explore the correlation of activation energy to frequency factor, thereby allowing a more intuitive correlated parameter fit of the Arrhenius model. This simultaneously supports simpler experimentation (i.e. dynamic heating vs. isothermal heating) and easier fitting of protein denaturation data. Furthermore, we demonstrated that by selecting appropriate end temperatures the protein denaturation kinetics is different and can be potentially used to explain the cell injury kinetics measured by different cell viability assays after thermal treatment. Future opportunities to monitor cell injury by spectroscopic measurement of protein denaturation were discussed.
Supplementary Material
ACKNOWLEDGMENT
This project was financially supported by the National Institute of Health (NIH) R01-CA07528. W.F.W. performed work in Minnesota and Hannover for this project and was supported in part by funding from the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) for the Cluster of Excellence REBIRTH (From Regenerative Biology to Reconstructive Therapy). Z.Q was supported by an Interdisciplinary Doctoral Fellowship and Doctoral Dissertation Fellowship. J.C.B. was supported by McKnight Professorship and Carl and Janet Kuhrmeyer Chair in Mechanical Engineering. J.A.P. received partial support for his investigations from the T.L.L. Temple and O-Donnell Foundations, and from Transonic / Scisense Inc. We thank Dr. Neil Wright for his insightful comments.
Footnotes
Alternatively, the correlated parameter approach (equation 6) can also be re-written as , where 316K (or 43°C) can be considered as the reference temperature derived from past measurements (Figure 1).
REFERENCES
- 1.Alberts B. Essential cell biology: an introduction to the molecular biology of the cell. New York: Taylor & Francis; 1998. [Google Scholar]
- 2.Tanford C. Protein denaturation. Adv. Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 3.Relkin P, Mulvihill D. Thermal unfolding of β-lactoglobulin, α-lactalbumin, and bovine serum albumin. A thermodynamic approach. Crit. Rev. Food Sci. Nutr. 1996;36:565–601. doi: 10.1080/10408399609527740. [DOI] [PubMed] [Google Scholar]
- 4.Lepock JR, Frey HE, Ritchie KP. Protein Denaturation in Intact Hepatocytes and Isolated Cellular Organelles during Heat Shock. J. Cell Biol. 1993;122:1267–1276. doi: 10.1083/jcb.122.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aravalli RN, Choi J, Mori S, Mehra D, Bischof JC, Cressman EN. Spectroscopic and Calorimetric Evaluation of Chemically Induced Protein Denaturation in HuH-7 Liver cancer Cells and Impact on Cell Survival. Technol. Cancer Res. Treat. 2012;11:467–473. doi: 10.7785/tcrt.2012.500271. [DOI] [PubMed] [Google Scholar]
- 6.Sapareto SA, Hopwood LE, Dewey WC, Raju MR, Gray JW. Effects of Hyperthermia on Survival and Progression of Chinese Hamster Ovary Cells. Cancer Res. 1978;38:393–400. [PubMed] [Google Scholar]
- 7.He X, Bischof JC. Quantification of temperature and injury response in thermal therapy and cryosurgery. Crit. Rev. Biomed. Eng. 2003;31:355–421. doi: 10.1615/critrevbiomedeng.v31.i56.10. [DOI] [PubMed] [Google Scholar]
- 8.He X, Wolkers WF, Crowe JH, Swanlund DJ, Bischof JC. In situ thermal denaturation of proteins in dunning AT-1 prostate cancer cells: implication for hyperthermic cell injury. Ann. Biomed. Eng. 2004;32:1384–1398. doi: 10.1114/b:abme.0000042226.97347.de. [DOI] [PubMed] [Google Scholar]
- 9.Thomsen S, Pearce JA, Cheong W-F. Changes in birefringence as markers of thermal damage in tissues. IEEE Trans. Biomed. Eng. 1989;36:1174–1179. doi: 10.1109/10.42111. [DOI] [PubMed] [Google Scholar]
- 10.Lumry R, Eyring H. Conformation changes of proteins. J. Phys. Chem. 1954;58:110–120. [Google Scholar]
- 11.Sanchez-Ruiz JM. Theoretical analysis of Lumry-Eyring models in differential scanning calorimetry. Biophys. J. 1992;61:921–935. doi: 10.1016/S0006-3495(92)81899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lepock JR, Ritchie KP, Kolios MC, Rodahl AM, Heinz KA, Kruuv J. Influence of transition rates and scan rate on kinetic simulations of differential scanning calorimetry profiles of reversible and irreversible protein denaturation. Biochem. 1992;31:12706–12712. doi: 10.1021/bi00165a023. [DOI] [PubMed] [Google Scholar]
- 13.Maron SH, Lando JB, Prutton CF. Fundamentals of physical chemistry. New York: Macmillan; 1974. [Google Scholar]
- 14.Dewey W. Arrhenius relationships from the molecule and cell to the clinic. Int. J. Hyperthermia. 1994;10:457–483. doi: 10.3109/02656739409009351. [DOI] [PubMed] [Google Scholar]
- 15.Pearce JA. Relationship between Arrhenius models of thermal damage and the CEM 43 thermal dose. SPIE BiOS: Biomedical Optics. 2009 pp 718104-718104-15. [Google Scholar]
- 16.Eyring H. The activated complex in chemical reactions. J. Chem. Phys. 1935;3:107. [Google Scholar]
- 17.Rosenberg B, Kemeny G, Switzer RC, Hamilton TC. Quantitative evidence for protein denaturation as the cause of thermal death. Nature. 1971;232:471–473. doi: 10.1038/232471a0. [DOI] [PubMed] [Google Scholar]
- 18.Yelon A, Movaghar B, Branz H. Origin and consequences of the compensation (Meyer-Neldel) law. Phys. Rev. B. 1992;46:12244. doi: 10.1103/physrevb.46.12244. [DOI] [PubMed] [Google Scholar]
- 19.Yelon A, Movaghar B, Crandall R. Multi-excitation entropy: its role in thermodynamics and kinetics. Rep. Prog. Phys. 2006;69:1145–1194. [Google Scholar]
- 20.Wright NT. On a relationship between the Arrhenius parameters from thermal damage studies. J. Biomech. Eng. 2003;125:300–304. doi: 10.1115/1.1553974. [DOI] [PubMed] [Google Scholar]
- 21.Krug RR, Hunter WG, Grieger RA. Enthalpy-entropy compensation. 2. Separation of the chemical from the statistical effect. J. Phys. Chem. 1976;80:2341–2351. [Google Scholar]
- 22.Barrie PJ. The mathematical origins of the kinetic compensation effect: 2. the effect of systematic errors. Phys. Chem. Chem. Phys. 2012;14:327–336. doi: 10.1039/c1cp22667c. [DOI] [PubMed] [Google Scholar]
- 23.Schwaab M, Pinto JC. Optimum reference temperature for reparameterization of the Arrhenius equation. Part 1: Problems involving one kinetic constant. Chem. Eng. Sci. 2007;62:2750–2764. [Google Scholar]
- 24.Wolkers WF, Balasubramanian SK, Ongstad EL, Zec HC, Bischof JC. Effects of freezing on membranes and proteins in LNCaP prostate tumor cells. Biochim Biophys Acta. 2007;1768:728–736. doi: 10.1016/j.bbamem.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miles CA. Kinetics of the helix/coil transition of the collagen-like peptide (Pro-Hyp-Gly)10. Biopolymers. 2007;87:51–67. doi: 10.1002/bip.20787. [DOI] [PubMed] [Google Scholar]
- 26.Miles CA, Burjanadze TV, Bailey AJ. The kinetics of the thermal denaturation of collagen in unrestrained rat tail tendon determined by differential scanning calorimetry. J. Mol. Biol. 1995;245:437–446. doi: 10.1006/jmbi.1994.0035. [DOI] [PubMed] [Google Scholar]
- 27.Wright N, Humphrey J. Denaturation of collagen via heating: an irreversible rate process. Annu. Rev. Biomed. Eng. 2002;4:109–128. doi: 10.1146/annurev.bioeng.4.101001.131546. [DOI] [PubMed] [Google Scholar]
- 28.Balasubramanian SK, Wolkers WF, Bischof JC. Thermal ˜Fingerprinting" of Cells Using FTIR, ASME 2007 Summer Bioengineering Conference. American Society of Mechanical Engineers. 2007:87–88. [Google Scholar]
- 29.Banks B, Damjanovic V, Vernon C. The so-called thermodynamic compensation law and thermal death. Nature. 1972;240:147–148. [Google Scholar]
- 30.Barrie PJ. The mathematical origins of the kinetic compensation effect: 1. the effect of random experimental errors. Phys. Chem. Chem. Phys. 2012;14:318–326. doi: 10.1039/c1cp22666e. [DOI] [PubMed] [Google Scholar]
- 31.Yelon A, Movaghar B. Microscopic explanation of the compensation (Meyer-Neldel) rule. Phys. Rev. Lett. 1990;65:618–620. doi: 10.1103/PhysRevLett.65.618. [DOI] [PubMed] [Google Scholar]
- 32.He X, Bhowmick S, Bischof JC. Thermal Therapy In Urologic Systems: A Comparison Of Arrhenius And Thermal Isoeffective Dose Models In Predicting Hyperthermic Injury. J. Biomech. Eng. 2009;131:074507. doi: 10.1115/1.3128671. [DOI] [PubMed] [Google Scholar]
- 33.Huntington JA, Stein PE. Structure and Properties of Ovalbumin. J. Chromatogr. B. 2001;756:189–198. doi: 10.1016/s0378-4347(01)00108-6. [DOI] [PubMed] [Google Scholar]
- 34.Bhowmick S, Swanlund DJ, Bischof JC. Supraphysiological Thermal Injury in Dunning AT-1 Prostate Tumor Cells. J. Biomech. Eng. 2000;122:51–59. doi: 10.1115/1.429627. [DOI] [PubMed] [Google Scholar]
- 35.Lung DC, Stahovich TF, Rabin Y. Computerized planning for multiprobe cryosurgery using a force-field analogy. Computer Methods in Biomechanics and Biomedical Engineering. 2004;7:101–110. doi: 10.1080/10255840410001689376. [DOI] [PubMed] [Google Scholar]
- 36.Elliott AM, Stafford RJ, Schwartz J, Wang J, Shetty AM, Bourgoyne C, O'Neal P, Hazle JD. Laser-induced thermal response and characterization of nanoparticles for cancer treatment using magnetic resonance thermal imaging. Med. Phys. 2007;34:3102–3108. doi: 10.1118/1.2733801. [DOI] [PubMed] [Google Scholar]
- 37.Salloum M, Ma R, Zhu L. Enhancement in treatment planning for magnetic nanoparticle hyperthermia: Optimization of the heat absorption pattern. Int. J. Hyperther. 2009;25:309–321. doi: 10.1080/02656730902803118. [DOI] [PubMed] [Google Scholar]
- 38.Carpentier A, Itzcovitz J, Payen D, George B, McNichols RJ, Gowda A, Stafford RJ, Guichard J-P, Reizine D, Delaloge S. Real-Time Magnetic Resonance-Guided Laser Thermal Therapy for Focal Metastatic Brain Tumors. Neurosurgery. 2008;63:ONS21–ONS29. doi: 10.1227/01.neu.0000335007.07381.df. [DOI] [PubMed] [Google Scholar]
- 39.Feng Y, Fuentes D, Hawkins A, Bass J, Rylander MN, Elliott A, Shetty A, Stafford RJ, Oden JT. Nanoshell-mediated laser surgery simulation for prostate cancer treatment. Eng. Comput. 2009;25:3–13. doi: 10.1007/s00366-008-0109-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daggett V. Molecular dynamics simulations of the protein unfolding/folding reaction. Acc. Chem. Res. 2002;35:422–429. doi: 10.1021/ar0100834. [DOI] [PubMed] [Google Scholar]
- 41.Daggett V. Protein Folding-Simulation. Chem. Rev. 2006;106:1898–1916. doi: 10.1021/cr0404242. [DOI] [PubMed] [Google Scholar]
- 42.Pearce JA. Comparative Analysis of Mathematical Models of Cell Death and Thermal Damage Processes. Int. J. Hyperthermia. 2013;29:262–280. doi: 10.3109/02656736.2013.786140. [DOI] [PubMed] [Google Scholar]
- 43.Eissing T, Conzelmann H, Gilles ED, Allgower F, Bullinger E, Scheurich P. Bistability Analyses of a Caspase Activation Model for Receptor-Induced Apoptosis. J. Biol. Chem. 2004;279:36892–36897. doi: 10.1074/jbc.M404893200. [DOI] [PubMed] [Google Scholar]
- 44.Yan C, Pattani V, Tunnell JW, Ren P. Temperature-Induced Unfolding of Epidermal Growth Factor (EGF): Insight From Molecular Dynamics Simulation. J. Mol. Graphics Modell. 2010;29:2–12. doi: 10.1016/j.jmgm.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayor U, Guydosh NR, Johnson CM, Grossmann JGn, Sato S, Jas GS, Freund SM, Alonso DO, Daggett V, Fersht AR. The Complete Folding Pathway of a Protein From Nanoseconds to Microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 46.Qin Z, Bischof JC. Thermophysical and Biological Responses of Gold Nanoparticle Laser Heating. Chem. Soc. Rev. 2012;41:1191–1217. doi: 10.1039/c1cs15184c. [DOI] [PubMed] [Google Scholar]
- 47.Thomsen SL, Pearce JA. Thermal Damage and Rate Processes in Tissues. In: Welch A, Van Gemert MJC, editors. Optical-Thermal Response of Laser-Irradiated Tissue. Dordrecht, Netherlands: Springer; 2011. pp. 487–549. [Google Scholar]
- 48.Vajpai N, Nisius L, Wiktor M, Grzesiek S. High-Pressure NMR Reveals Close Similarity Between Cold and Alcohol Protein Denaturation in Ubiquitin. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E368–E376. doi: 10.1073/pnas.1212222110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Murayama K, Myojo Y, Tsenkova R, Hayashi N, Ozaki Y. Two-dimensional Fourier transform near-infrared spectroscopy study of heat denaturation of ovalbumin in aqueous solutions. J. Phys. Chem. B. 1998;102:6655–6662. [Google Scholar]
- 50.Shanmugam G, Polavarapu PL. Vibrational Circular Dichroism Spectra of Protein Films: Thermal Denaturation of Bovine Serum Albumin. Biophys. Chem. 2004;111:73–77. doi: 10.1016/j.bpc.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 51.Zamyatnin A. Amino Acid, Peptide, and Protein Volume in Solution. Ann. Rev. Biphys. Bioeng. 1984;13:145–165. doi: 10.1146/annurev.bb.13.060184.001045. [DOI] [PubMed] [Google Scholar]
- 52.Frensdorff H, Watson M, Kauzmann W. The Kinetics of Protein Denaturation. IV. The Viscosity and Gelation of Urea Solutions of Ovalbumin. J. Am. Chem. Soc. 1953;75:5157–5166. [Google Scholar]
- 53.Weijers M, Barneveld PA, Stuart C, Martien A, Visschers RW. Heat-Induced Denaturation and Aggregation of Ovalbumin at Neutral pH Described by Irreversible First-Order Kinetics. Protein Sci. 2003;12:2693–2703. doi: 10.1110/ps.03242803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miles CA, Bailey AJ. Studies of the Collagen-like Peptide (Pro-Pro-Gly)10 Confirm that the Shape and Position of the Type I Collagen Denaturation Endotherm is Governed by the Rate of Helix Unfolding. J. Mol. Biol. 2004;337:917–931. doi: 10.1016/j.jmb.2004.02.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.