Abstract
Heparanase has generated substantial interest as therapeutic target for antitumor therapy, because its activity is implicated in malignant behavior of cancer cells and in tumor progression. Increased heparanase expression was found in numerous tumor types and correlates with poor prognosis. Heparanase, an endoglucuronidase responsible for heparan sulfate cleavage, regulates the structure and function of heparan sulfate proteoglycans, leading to disassembly of the extracellular matrix. The action of heparanase is involved in multiple regulatory events related, among other effects, to augmented bioavailability of growth factors and cytokines. Inhibitors of heparanase suppress tumor growth, angiogenesis and metastasis by modulating growth factor-mediated signaling, ECM barrier function and cell interactions in the tumor microenvironment. Therefore, targeting heparanase has potential implications for anti-tumor, anti-angiogenic and anti-inflammatory therapies. Current approaches for heparanase inhibition include development of chemically-modified heparins, small molecule inhibitors and neutralizing antibodies. The available evidence supports the emerging utility of heparanase inhibition as a promising antitumor strategy, specifically in rational combination with other agents. The recent studies with compounds designed to block heparanase (e.g., modified heparins) provide a rational basis for their therapeutic application and optimization.
Keywords: Heparanase, heparanase inhibitors, drug target, antitumor therapy
Graphical Abstract
1. Introduction
Heparanase, an endoglucuronidase responsible for heparan sulfate (HS) cleavage, regulates the structure and function of heparan sulfate proteoglycans, thus resulting in structural alterations of the extracellular matrix (ECM) and release of bioactive saccharide fragments and HS-bound growth factors and cytokines (Figure 1). Heparanase is a multifaceted protein endowed with enzymatic and non-enzymatic functions that appear to participate in major human pathological processes (1 and quoted references). Since the cloning of the human heparanase gene in 1999, heparanase was advanced from being an obscure enzyme with a poorly understood function to a promising drug target. While most attention was addressed to heparanase function in tumor biology, emerging evidence indicate that heparanase is also engaged in other disease conditions often associated with degradation of HS, release of bioactive molecules anchored within the ECM network, disregulated signalling cascades, gene transcription, and activation of innate immune cells. Among these diseases are chronic inflammation (i.e., inflammatory bowel disease, rheumatoid arthritis), autoimmunity (i.e., type 1 diabetes, psoriasis), diabetic nephropathy, bone osteolysis, thrombosis and atherosclerosis [2–9]. There is growing evidence that heparanase upregulates expression of genes that participate in cancer metastasis and angiogenesis, glucose metabolism, immune response, inflammation and atherosclerosis, suggesting that heparanase belongs to an emerging class of proteins that play a significant role in regulating transcription in addition to their well-recognized extra-nuclear functions [1,5,6,8,9].
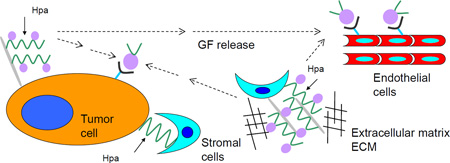
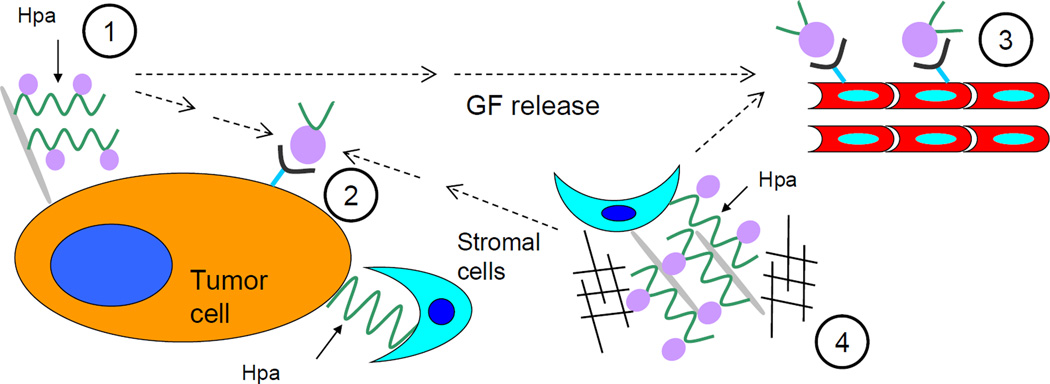
Figure 1. Scheme showing the effects of heparanase (Hpa) involving tumor/microenviroment interactions.
1) cleavage of heparan sulfate (HS) by heparanase (as indicated by the solid arrows) and release of heparan sulfate-bound growth factors (GF) from proteoglycans; 2) growth factor-mediated signaling; 3) proangiogenic signaling; 4) disassembly of extracellular matrix and release of HS-bound growth factors.
Several up-to-date reviews summarize basic aspects related to the involvement of heparanase in cancer progression and inflammation [10–14]. The present commentary provides information on the biology of the heparanase protein in cancer and inflammation, with emphasis on translational aspects of heparanase-inhibiting strategies.
2. Heparanase and heparan sulfate proteoglycans
Heparan sulfate proteoglycans (HSPGs)
HSPGs exert their multiple functional repertoires via several distinct mechanisms that combine structural, biochemical and regulatory aspects. By interacting with other macromolecules such as laminin, fibronectin, and collagens I and IV, HSPGs contribute to the structural integrity, self-assembly and insolubility of the extracellular matrix (ECM) and basement membrane (BM), thus intimately modulating cell-ECM interactions [15,16]. HSPGs also directly transfer information from the extracellular space to intracellular kinases and cytoskeletal elements, thus affecting cell signaling, adhesion and motility [16]. The sulfated saccharide domains of HS provide numerous docking sites for a multitude of protein ligands, ensuring that a wide variety of bioactive molecules (i.e., cytokines, chemokines, growth factors, enzymes, protease inhibitors, ECM proteins) bind to the cell surface and ECM [15,18] and thereby function in the control of normal and pathological processes, among which are morphogenesis, tissue repair, vascularization, cancer metastasis, inflammation, atherosclerosis, thrombosis and diabetes [18,19]. Heparanase-mediated cleavage of HSPGs would ultimately release these proteins and convert them into bioactive mediators, ensuring rapid tissue response to local or systemic cues. As a result, HS provides cells with a rapidly accessible reservoir, precluding the need for de novo synthesis when the requirement for a particular protein is increased (Figure 1) [6,7].
The biosynthesis of HS takes place in the Golgi system and has been studied in great detail. Briefly, the polysaccharide chains are modified at various positions by sulfation, epimerization and N-acetylation, yielding clusters of sulfated disaccharides separated by low or non-sulfated regions [18,19]. Unlike the well resolved biosynthetic pathway, the mode of HS breakdown is less characterized. While synthesis and modification of HS chains require the activity of an array of enzymes, degradation of mammalian HS is primarily carried out by one enzyme, heparanase (HPSE), which cleaves the HS side chains of HSPGs into fragments of 10–20 sugar units. Cleavage of HS by heparanase has multiple downstream effects due to the broad regulatory activity of HS. For example, HS promote growth factor signaling, mediate cell adhesion and sequester growth factors within the ECM, thereby facilitating storage of growth factors and the establishment of growth factor/chemokine gradients [19]. Additionally, heparanase upregulates expression of the HSPG syndecan-1 and also enhances its shedding from the cell surface. This is important because shed syndecan-1 is known to regulate tumor growth, metastasis and angiogenesis, largely by promoting growth factor signaling within the tumor microenvironment [6].
Mammalian heparanase
Heparanase cleaves HS side chains presumably at sites of low sulfation, releasing saccharide products with appreciable size (4–7 kDa) that can still associate with protein ligands and facilitate their biological potency. Elucidating the substrate specificity of heparanase has been complicated by the heterogeneity of heparan sulfate chains and the lack of highly pure homogeneous substrates. The enzyme cleaves the linkage between a GlcA unit and an N-sulfo glucosamine residue carrying either a 3-O-sulfo or a 6-O-sulfo group. In addition, heparanase cleaves such linkages with a 2-O-sulfated GlcA residue, but not a 2-O-sulfated IdoA residue, in proximity. This suggests that heparanase recognizes certain sulfation patterns rather than specific monosaccharide sequences and that cleavage occurs in the mixed domains between the sulfated and non-sulfated spacer domains. Use of structurally defined oligosaccharides indicates that heparanase displays different cleavage modes by recognizing structural features at the non-reducing ends of HS, thus suggesting a regulatory role in the release or preservation of specific HS structures [20].
Mammalian cells express a single dominant functional heparanase enzyme (heparanase-1) [21]. The heparanase mRNA encodes a 65 kDa pro-enzyme that is post-translationally cleaved into 8 and 50 kDa subunits that non-covalently associate to form the active heparanase. The heparanase structure delineates a TIM-barrel fold harboring the enzyme’ active site and substrate binding domains, and a C-terminus domain (C-domain) that is critical for heparanase secretion and signaling function [1]. Similar to other glycosyl hydrolases, heparanase has a common catalytic mechanism that involves two conserved acidic residues, a putative proton donor at Glu225 and a nucleophile at Glu343 [22]. Cellular processing of the secreted latent enzyme involves uptake and delivery into late endosomes and lysosomes followed by removal of a 6 kDa linker segment brought about by cathepsin L [23]. Importantly, heparanase functions beyond its enzymatic activity. Although enzymatic activity requires both the TIM-barrel and C-terminus domains, the C-domain can function independently of the TIM-barrel fold, promoting AKT signaling and leading to enhanced tumor growth in animal models [1]. Of increasing significance are observations that heparanase through both enzymatic and non-enzymatic activities promotes gene expression (i.e., VEGF, tissue factor, MMP-9, HGF, RANKL, TNFα) and signaling pathways (i.e., phosphorylation of Akt, Src, Erk, EGF-receptor, insulin receptor) of which some are mediated by its C-domain, devoid of heparanase enzymatic activity [1, 5–7, 21].
Heparanase in cancer progression
The clinical significance of heparanase in tumor progression emerged from a systematic evaluation of heparanase expression in primary human tumors. Immunohistochemistry, in situ hybridization, RT-PCR and real time-PCR analyses revealed that heparanase is up-regulated in essentially all major types of human cancer, namely carcinomas, sarcomas and hematological malignancies [7, 14, 21]. Notably, heparanase up-regulation in human tumors is associated with increased tumor size [7, 21]. Likewise, heparanase over-expression enhanced, while local delivery of anti-heparanase siRNA inhibited the progression of tumor xenografts [7]. A significant role of heparanase in tumor angiogenesis and lymphangiogenesis was demonstrated applying similar experimental approaches [21]. In fact, heparanase expression levels correlate with tumor vascularity in cancer patients, further indicating a significant role in tumor angiogenesis [7], altogether implying that heparanase function is not limited to tumor metastasis but is also engaged in accelerated growth of the primary lesion. Notably, cancer patients exhibiting high levels of heparanase had a significantly shorter postoperative survival time than patients whose tumors contained low levels of heparanase [7] further implicating heparanase as a master regulator of cancer progression and metastasis. The involvement of heparanase in tumor behaviour was reinforced by preclinical studies indicating a marked inhibition of tumor progression in mice treated with compounds that inhibit heparanase enzymatic activity [24–29]. Importantly, heparanase promotes cancer progression through its action on both the tumor cells and the tumor cell microenvironment [6].
3. Heparanase in inflammation
HS is known to control inflammatory responses at multiple levels, including sequestration of cytokines/chemokines in the extracellular space, modulation of leukocyte interactions with endothelium and ECM, and initiation of innate immune responses through interactions with toll-like receptor 4 (TLR4) [30–33]. Thus, HS enzymatic remodeling by heparanase may affect several aspects of inflammatory reactions, such as leukocyte recruitment, extravasation and migration towards inflammation sites; release of cytokines and chemokines anchored within the ECM or cell surfaces, as well as activation of innate immune cells. The link between inflammation and heparanase was first demonstrated when HS-degrading activity was discovered in immunocytes (neutrophils, activated T-lymphocytes) and found to contribute to their ability to extravasate and accumulate in target organs [34]. In subsequent studies, the notion that immunocytes represent the principal cellular source of the enzyme in inflammation was challenged by observations that heparanase expression occurs mainly in epithelial and/or endothelial compartment in numerous inflammatory settings, including delayed type hypersensitivity [35], vascular injury, chronic colitis [36], sepsis-associated lung injury [37], as well as in several auto-immune and auto-inflammatory human disorders, such as rheumatoid arthritis, atherosclerosis, psoriasis, ulcerative colitis and Crohn’s disease [9, 10, 11]. Collectively, a complex picture of the versatile role of heparanase in inflammation is evolving, whereby heparanase may act either in facilitating or limiting inflammatory responses, most likely depending on the cellular/extracellular framework.
Heparanase in acute inflammatory responses
Mounting evidence suggests that heparanase affects activities of several types of innate immunocytes, including neutrophils, macrophages, dendritic and mast cells [8, 10, 36–39]. Of those, neutrophils represent the important effectors in the acute inflammatory responses. The consequence of heparanase action on neutrophil behavior was highlighted in a recent report by Schmidt et al., focusing on enzymatic degradation of endothelial glycocalyx in a mouse model of sepsis-associated lung injury. In this model, rapid induction of heparanase activity (through TNFα-dependent mechanism) in pulmonary microvascular endothelial cells was shown to facilitate neutrophil recruitment through exposure of the endothelial surface and increased availability of cell adhesion molecules [37]. Moreover, sepsis associated loss of pulmonary glycocalyx and endothelial hyperpermeability were attenuated in heparanase-null mice and in mice treated with inhibitors of heparanase enzymatic activity [37]. On the other hand, constitutive over-expression of heparanase in heparanase transgenic (Hpa-tg) mice was shown to attenuate intraluminal crawling of neutrophils in the cremasteric muscle microvessels toward an extravascular chemokine source, reportedly due to reduction in endothelial surface HS chain length and altered ability of truncated HS to serve as a ligand for chemokines [38]. In addition, reports exploring acute inflammatory phenotypes of heparanase over-expressing Hpa-tg mice in models of inflammatory hyperalgesia and neuroinflammation [40] demonstrated that neutrophil recruitment and activation were attenuated in the presence of constitutively increased levels of heparanase in Hpa-tg mice. Thus, the overall effect of heparanase on neutrophil behavior may depend on the proportional contribution of glycocalyx removal (which is expected to facilitate neutrophil access to the blood vessel wall [37] vs. the disturbance of chemokine gradients at the endothelial cell surface (which attenuates neutrophil recruitment) [10, 38, 40].
Heparanase in chronic inflammation
When acute inflammation is not properly resolved, the composition of the infiltrating leukocytes changes from neutrophils to macrophages, dominant cellular players in chronic inflammation. Heparanase ability to modulate macrophages responses was highlighted in studies focusing on inflammatory bowel disease (IBD). In patients with IBD, an equilibrium between the immune response to pathogens and tolerance to the normal flora becomes unbalanced, leading to the uncontrolled uptake of proinflammatory substances (i.e., bacteria, bacterial products) from the gut lumen and triggering immune activation, cytokine release, and dysfunction of the epithelial barrier [41]. Given the important role of HS in maintaining the integrity of the gut wall [30], enzymatic degradation of HS is thought to significantly affect colon permeability and inflammatory reactions. Analysis of glycosaminoglycan content in normal colonic tissue and colons of IBD patients revealed loss of HS from the subepithelial BM and from the vascular endothelium in the submucosa [10]. In agreement, preferential expression of heparanase was reported in colonic epithelium, but not immunocytes, of IBD patients during both acute and chronic phases of the disease, suggesting that heparanase of epithelial origin modulates the inflammatory phenotype of macrophages towards a chronic inflammation pattern [36]. In support of this notion, exacerbated chronic inflammatory phenotype and augmented recruitment and activation of macrophages were detected in colonic mucosa of Hpa-tg mice following induction of DSS colitis [36]. Moreover, heparanase strongly augmented in vitro activation of macrophages by LPS, resulting in marked increase in production of TNFα, IL-6 and IL12 [36]. Activated macrophages can in turn induce epithelial heparanase expression (via a TNFα-dependent mechanism) and post-translational processing of the pro-enzyme via increased secretion of cathepsin L [36], fueling a self-sustaining inflammatory circuit.
Heparanase in inflammation-associated cancer
Chronic inflammatory conditions are present in the microenvironment of most tumors [42] and have been shown to contribute to cancer progression [43], among other mechanisms, through mobilization of tumor-supporting immunocyte populations (e.g., tumor associated macrophages, neutrophils) which supply bioactive molecules that foster survival, angiogenesis, invasion and metastasis [42, 44]. Progression of Barrett’s oesophagus to adenocarcinoma; chronic gastritis to intestinal-type gastric carcinoma, chronic hepatitis C to hepatocellular carcinoma pancreatitis to pancreatic adenocarcinoma and colitis to colorectal cancer are well-known examples of inflammation-driven tumorigenesis [10]. Remarkably, induction of heparanase prior to the appearance of malignancy was reported in essentially all of the above-mentioned inflammatory conditions, i.e., Barrett’s oesophagus [45], hepatitis C infection [46], chronic pancreatitis [47], Crohn disease and ulcerative colitis [36]. It is therefore conceivable that inflammation-induced heparanase may be involved in coupling inflammation and cancer. Findings obtained in a study utilizing a mouse model of colitis-associated colon carcinoma support this notion [36]. It appears that by sustaining continuous activation of macrophages that supply cancer-promoting cytokines (i.e., TNFα, IL-1, IL-6), heparanase participates in creating tumorigenic microenvironment characterized by enhanced NFκB and STAT3 signaling, augmented levels of cyclooxygenase 2 and increased vascularization (Figure 2) [36].
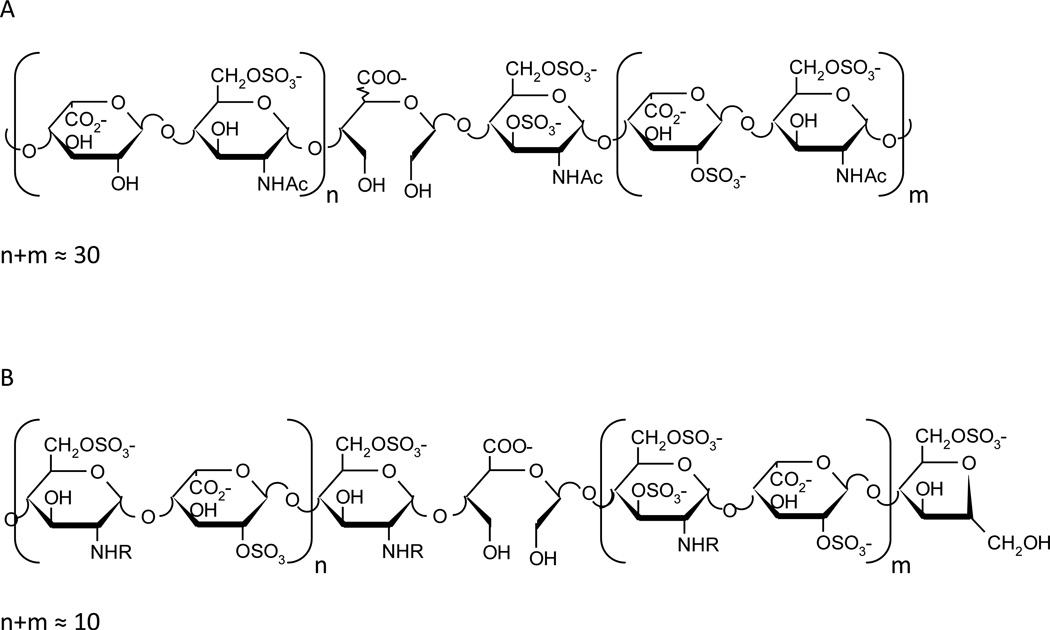
Figure 2. Structures of selected heparanase inhibitors.
A. Schematic representation of SST-0001 and chemical modification characterizing the heparin derivative. Given the polymeric nature of SST-0001 (average molecular weight, 20 kDa) a "statistical" representation of the structure is shown.
B. Schematic representation of M402 (average molecular weight, 6 kDa). As for SST-0001, a "statistical" representation of the structure is shown. R=SO3 or Ac.
4. Heparanase inhibitors
Heparanase promotes cancer progression by its actions within tumor, at the tumor cell surface and in the tumor cell microenvironment [7]. Since heparanase is secreted into the microenvironment, targeting heparanase has the therapeutic potential to concurrently block tumor growth and interfere with establishment and maintenance of the tumor microenvironment. Because there is only one single active heparanase enzyme, once the enzyme is blocked or inactivated, there are no backup molecules to perform its function. Also of therapeutic importance is the fact that heparanase expression in normal tissue is very low, allowing prediction of negligible side effects of blocking heparanase function.
The understanding of the role of heparanase in malignant behavior of tumor cells has stimulated efforts to identify selective inhibitors as potentially useful agents in cancer therapy. The approaches proposed for targeting heparanase include the development of small molecule inhibitors, the use of monoclonal antibodies to block enzyme function on regulatory domains or modified heparins to inhibit enzyme activity as competitive substrate inhibitors [13]. The design of selective enzyme inhibitors is still limited by the absence of the three-dimensional structure of heparanase. In the absence of structural information, sequence homology models with proposed binding mode for heparan sulfate substrates have been developed to be employed in the rational design of novel inhibitors [48–50].
Heparin is one of the closest mimics of HS and is a natural choice as heparanase inhibitor. Heparin, however, is not clinically useful as an anti-cancer/anti-inflammatory drug because of its potent anticoagulant activity. Considerable efforts have thus been expended in the development of modified heparins and related polysulfated compounds with reduced anticoagulant activity. Modified heparins and heparin mimetics are the best studied heparanase inhibitors [25] and some compounds of this class are currently in various stages of preclinical/clinical development [6]. Chemically modified heparins have been developed in an attempt to abolish the anticoagulant activity and to retain or enhance their affinity for heparanase [25]. These features are critical requirements for their clinical use, because high doses and protracted treatment schedules should be used to fully exploit the therapeutic potential of heparin derivatives. Modified heparins or sulphated oligosaccharides, including SST0001, M402, PI-88 and PG545, have been developed as potent heparanase inhibitors (IC50 in the nM range).
SST0001 (Roneparstat, currently in Phase I/II clinical trial in myeloma patients) is a modified heparin with a mean molecular size of 20kDa, characterized by 100% N-acetylation, and 25% glycol splitting [25]. The N-acetylation is responsible for the loss of the anticoagulant activity and glycol splitting likely contributes to enhance affinity for heparanase [25, 51]. SST0001 was found to be effective as inhibitor of heparanase in vivo and as modulator of levels of growth factors, including VEGF and HGF [24, 27]. The antiangiogenic activity of SST0001 reflects the downregulation of angiogenic heparin-binding factors and the impact of heparanase inhibition on tumor-microenvironment interactions [6]. SST0001 also inhibit shedding of syndecan-1, a process regulated by heparanase and known to be implicated in signalling pathways in both tumor and endothelial cells [6]. Preclinical studies indicate that SST0001 is effective in the treatment of selected human tumor models including myeloma, pediatric sarcoma and pancreatic carcinoma [24, 27, 28, 52]. The good tolerability of SST0001 in protracted treatment schedules is consistent with the selective heparanase inhibition and lack of anticoagulant activity [24].
M402 is a related glycol-split heparin having a smaller molecular size (6 kDa) [29]. This compound is N-sulfated instead of N-acetylated, which may provide it with broader growth factor binding activity. M402 showed efficacy in metastasis models [29] and progressed in 2012 to a Phase I/II clinical trial in combination with gemcitabine in patients with metastatic pancreatic cancer [6].
Other sulfated polysaccharides which inhibit heparanase are potent inhibitors of inflammation, tumor growth and metastasis, but these often suffer from unacceptably high anticoagulant activity. Among many efforts in this area has been the identification of two potent heparanase inhibitors, PI-88 and maltohexaose sulfate, with attenuated anticoagulant activity. PI-88 displays antimetastatic and antiangiogenic activity [53] and has progressed to Phase III clinical trials in post-resection hepatocellular carcinoma [54]. Whilst developed primarily for cancer indications, PI-88 and maltohexaose sulfate display promising anti-inflammatory activity. Recently, polysulfated hexasaccharides (designated STMCs or SMTCs) in which two maltotriose units are joined head-to-head by a novel C1-C1’ C-glycosidic bond were shown to inhibit metastases in a mouse model via inhibition of heparanase and P-selectin [55]. Building on the clinical progress of PI-88, new series of HS mimetics have been developed with improved properties [53, 56, 57]. Unlike PI-88 and other sulfated oligosaccharides which are mixtures, the new compounds are single chemical entities and contain lipophilic modifications which endow them with significantly improved pharmacokinetic properties and in vivo activity in preclinical cancer models, and milder anticoagulant activity. Among these, of particular note is PG545 [56] a tetrasaccharide which has demonstrated promising in vivo antiangiogenic and antimetastatic activity and potent anti-tumour activity in multiple preclinical models that are resistant to PI-88 treatment [26, 58]. PG545 has been reported to inhibit angiogenesis by sequestering angiogenic growth factors and preventing the binding to receptor [26]. PG545 entered Phase I clinical trials in late 2010 administered as a subcutaneous injection, however, the trial was halted due to unexpected injection site reactions [59].
A number of heparin-like compounds and low-molecular-weight heparins (LMWH) are being considered as therapeutic agents [25]. Their biological effects may be not dependent on heparanase inhibitions. A low-molecular-weight heparin (tinzaparin) has been reported to prevent lung metastatis by the human breast cancer model MDA-MB-231 [60] and the formation of hepatic metastases by the human colon carcinoma HCT-116 cells [61]. The antimetastatic effect of tinzaparin has been ascribed to the inhibition of the interaction between the chemokine CXCL12 and CXCR4 receptor. Heparin derivatives compete for the binding of CXCL12, thus preventing chemokine-driven invasion and metastasis. In addition, LMWHs appear to display their biological effects by modulation of the tumor microenvironment and to inhibit angiogenesis by preventing the FGF-2 or VEGF-mediated signaling. Since continuous parental use of heparin derivatives is not a practical modality of protracted administration, chemical conjugates of LMWHs have been develop to allow oral absorption [63]. Although potential antineoplastic effects have been associated with the use of LMWHs [64], their mechanism of action appears somewhat complex. It is unclear whether their antimetastatic and antiangiogenic activity is related to heparanase inhibition or to competitive inhibition of binding of growth factors to their receptors [25]. Given the multiple in vivo interactions of heparin derivatives, it is conceivable that their biological effects are related to various features, including molecular weight and nature of chemical modifications (e.g., extent of sulfation or glycol splitting).
Another drug being explored clinically is defibrotide, a polydisperse oligonucleotide isolated from porcine mucosa that has multiple biological effects including inhibition of heparanase expression. Defibrotide is currently being tested as a component of combination chemotherapy in myeloma patients [62].
In addition to HS mimetics, a number of other approaches to heparanase inhibitors have been described and reviewed [13, 59, 65] and continue to be actively investigated. These include various small molecule approaches such as iminosugars and other putative transition state analogues, substrate analogues, inhibitors discovered via screening of compound libraries, and natural products and their derivatives.
5. Tumor microenvironment and response to antitumor therapy
Several lines of evidence support the view that tumor microenvironment influences the efficacy of antitumor treatments [66–68]. The heterogeneous distribution of drugs in different tissues may limit the exposure of tumor cells to potentially lethal drug concentrations [68]. However, the role of the tumor microenvironment appears to be more complex, because the dynamic interplay among components of extracellular matrix and the interaction of tumor cells and host cells (fibroblasts, endothelial cells and immune cells) can be determinants of the malignant behavior of tumor cells, including proliferation, invasion and survival under stress conditions [67, 69]. Cellular signaling pathways may promote cell survival in response to chemotherapeutic agents. Interestingly, inhibition of stromal survival signals by CXCR4 antagonists (e.g., plerixafor) has been reported to enhance sensitivity of tumor cells to cytotoxic agents [70] and to therapeutic monoclonal antibodies [71]. Therefore, tumor microenvironment is now recognized as potential target of antitumor therapy in a context of combination treatment strategies [72].
On the basis of the putative role of heparanase, i.e. cleavage of heparan sulfate and structural remodeling of the extracellular matrix (Figure 1), inhibition of heparanase could be a promising approach to target critical components of the tumor microenvironment and may have potential to improve the efficacy of conventional antitumor agents. The local release of prosurvival growth factors, sequestered by heparan sulfate in the extracellular matrix, through heparanase action may contribute to tumor cell survival and protection against drug treatment. Thus inhibition of survival signals and disruption of the tumor/stroma interactions may influence sensitivity to chemotherapy.
In spite of the relevance of heparanase and heparan sulfate as regulators of signaling pathways, it is likely that the efficacy of heparanase inhibitors can be better exploited in rational combination therapy with other antitumor agents. Only few studies addressed this aspect of the therapeutic applications of heparanase inhibitors. The available preclinical studies support the therapeutic potential of heparanase inhibitors in combination with agents that display their effects on tumor microenvironment. The combination of SST0001 with dexamethasone exhibited improved efficacy in the treatment of myeloma models as compared to single-agent therapy [27]. The efficacy of the combination was also observed in a dexamethasone-resistant tumor subline. Since heparanase is involved in the inflammatory process [7], the efficacy of this combination is consistent with interpretation that tumor growth inhibition in vivo reflects dual targeting of the tumor and its microenvironment.
Based on role of heparanase in angiogenesis [6, 20], a process regulated by secretion of VEGF, a recent study was designed to investigate the efficacy of SST0001 in combination with antiangiogenic therapies, including the anti-VEGF antibody bevacizumab and the tyrosine kinase inhibitor, sunitinib, known to inhibit proangiogenic signaling [24]. In both approaches, the combination with SST0001 resulted in a synergistic interaction in the treatment of the TC71 Ewing’s sarcoma model, as documented by the high rate of complete tumor regressions, with no evidence of tumor regrowth in an appreciable number of animals. The synergistic effect of the combination of SST0001 with antiangiogenic agents may have therapeutic implications in clinical setting, because of the modest single-agent efficacy of antiangiogenic drugs.
In addition to the documented therapeutic interest of heparanase inhibitors combined with antiangiogenic therapies, other potentially useful combinations could be envisaged, because heparanase is involved in multiple regulatory pathways. Indeed like VEGF, the hepatocyte growth factor (HGF) is known to be regulated by the heparanase/syndecan-1 axis and aberrant HGF expression is implicated in several biological processes, including angiogenesis and cell migration/metastases [6]. A number of inhibitors of MET, the tyrosine kinase receptor of HGF, are now available [74] and could be explored for their antitumor/antimetastatic efficacy in combination with heparanase inhibitors.
6. Discussion
The role of heparanase in modulating critical biological or pathological processes mediated by release of bioactive molecules makes this enzyme as a promising target for novel approaches in antitumor treatment. The preclinical studies have focused on the effects on metastasis and angiogenesis. Indeed, emerging evidence support that heparanase inhibitors may have potential interest for antitumor/antimetastatic and antiangiogenic therapies [6]. Although the primary focus of heparanase inhibitors has been antitumor therapy, the available evidence provides support for their application in other pathological conditions, including inflammatory and vascular diseases [6, 7].
In spite of the critical functions of heparanase in tumor biology, the therapeutic potential of enzyme inhibition remains to be explored in various tumor types and the optimal approach to exploit the therapeutic potential of heparanase inhibitors remains to be identified.
Preclinical studies document the efficacy of approaches based on heparanase inhibition in specific tumor types, including multiple myeloma and pediatric sarcoma [24, 27, 28]. Based on the known functions of heparanase, one could speculate that tumors which are critically dependent on signals delivered by the microenvironment could be the most responsive to heparanase inhibitors. The efficacy of the glycol-split heparin derivative SST0001 against pediatric sarcoma models is consistent with this prediction and could reflect the ability of SST0001 to suppress the release of factors implicated in promoting tumor growth and angiogenesis [24]. Specifically, the function of VEGF is not restricted to angiogenesis, since VEGF/VEGFR signaling contributes to various aspects of tumor biology [75] as observed for Ewing’s sarcomas [76].
Although SST0001 and other available heparanase inhibitors may exhibit antitumor activity as single agents in some tumor models [24, 27, 28], it is likely that, as observed for other target-specific agents, single-agent therapy is not sufficient to control tumor growth. Also based on preclinical evidence, it is conceivable that rational drug combinations with heparanase inhibitors could achieve superior efficacy over single-agent therapy. This prediction is supported by the expected contribution of targeting tumor cells and disruption of tumor microenvironment. On the basis of the evidence of downregulation of proangiogenic factors induced by heparanase inhibitors, it is likely that one of their prominent effects is the inhibition of angiogenesis. This mechanism is supported by the observation that SST0001 exhibits synergistic interaction in combination with agents effective in modulation the angiogenesis process [24]. A number of agents effective in the clinical setting, including cytotoxic agents, may have also the potential to improve the control of tumor growth through inhibition of angiogenesis [77].
Although some small molecule heparanase inhibitors have been identified [13], their development has been hampered by the lack of crystal structure for heparanase. A deeper understanding of heparanase mechanism of action and resolution of heparanase crystal structure and substrate specificity will lead to identification of small molecule inhibitors and to improve the design of modified heparins to block heparanase activity. A number of heparin/heparan sulfate mimetics have exhibited significant activity in preclinical studies and some promising heparin-derived compounds are now under clinical evaluation [6]. The identification of the clinical setting which may benefit by the use of heparanase inhibitors will be a relevant challenge of their clinical development. The preclinical evidence supports the optimal efficacy of SST0001 when administered during the early phase of tumor growth following tumor cell inoculation. Indeed, the inhibition of release/activation of growth factors that provide a favorable microenvironment for tumor growth might be the critical event implicated in the control of primary tumor growth. This observation would suggest a therapeutic potential of heparanase inhibitors in the control of minimal residual disease, which requires the support of microenvironment, rather than in the treatment the bulky progressive disease.
A better understanding of molecular/biological features of tumor types, responsive to heparin mimetics could provide a rational basis to exploit their therapeutic potential and to optimize their combinations.
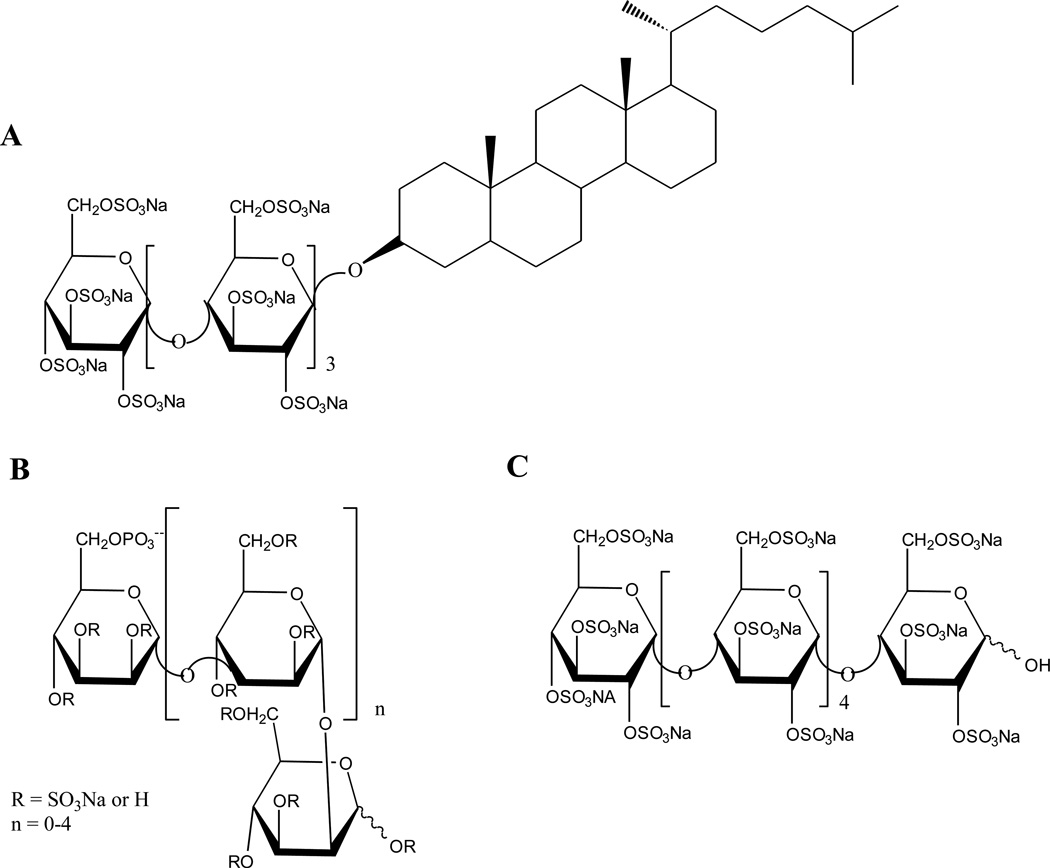
Figure 3. Structures of heparan sulfate mimetics.
A. Structure of the heparan sulfate mimetic, PG545, a fully sulfated oligosaccharide conjugated with a lipophilic moiety.
B. Structure of PI-88, a mixture of sulfated di- to hexasaccharides.
C. Structure of maltohexaose sulfate.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grant CA106456; the Israel Science Foundation (ISF grant 593/10); the MOST-DKFZ program for Cancer Research, and by a research contract from Sigma-Tau Research Switzerland S.A. I. Vlodavsky is a Research Professor of the Israel Cancer Research Fund (ICRF). We thank Dr. A. Naggi, Dr. S. Penco and Prof. S. Dallavalle for their help and collaboration.
The authors apologize for the inability, due to space limitations, to reference all studies relevant to this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sci. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gil N, Goldberg R, Neuman T, Garsen M, Zcharia E, Rubinstein A, van Kuppevelt T, et al. Heparanase is essential for the development of diabetic nephropathy in mice. Diabetes. 2012;61:208–216. doi: 10.2337/db11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li RW, Freeman C, Yu D, Hindmarsh EJ, Tymms KE, Parish CR, et al. Dramatic regulation of heparanase activity and angiogenesis gene expression in synovium from patients with rheumatoid arthritis. Arthritis and Rheumatism. 2008;58:1590–1600. doi: 10.1002/art.23489. [DOI] [PubMed] [Google Scholar]
- 4.Osterholm C, Folkersen L, Lengquist M, Ponten F, Renne T, Li J, et al. Increased expression of heparanase in symptomatic carotid atherosclerosis. Atherosclerosis. 2013;226:67–73. doi: 10.1016/j.atherosclerosis.2012.09.030. [DOI] [PubMed] [Google Scholar]
- 5.Parish CR, Freeman C, Ziolkowski AF, He YQ, Sutcliffe EL, Zafar A, et al. Unexpected new roles for heparanase in Type 1 diabetes and immune gene regulation. Matrix Biol. 2013;32:228–233. doi: 10.1016/j.matbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Ramani VC, Purushothaman A, Stewart MD, Thompson CA, Vlodavsky I, Au JL, et al. The heparanase/syndecan-1 axis in cancer: mechanisms and therapies. FEBS J. 2013;280:2294–2306. doi: 10.1111/febs.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vlodavsky I, Beckhove P, Lerner I, Pisano C, Meirovitz A, Ilan N, et al. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 2012;5:115–132. doi: 10.1007/s12307-011-0082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlodavsky I, Blich M, Li JP, Sanderson RD, Ilan N. Involvement of heparanase in atherosclerosis and other vessel wall pathologies. Matrix Biol. 2013;32:241–251. doi: 10.1016/j.matbio.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Ren Y, Ramani VC, Nan L, Suva LJ, Sanderson RD. Heparanase enhances local and systemic osteolysis in multiple myeloma by upregulating the expression and secretion of RANKL. Cancer Res. 2010;70:8329–8338. doi: 10.1158/0008-5472.CAN-10-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg R, Meirovitz A, Hirshoren N, Bulvik R, Binder A, Rubinstein AM, et al. Versatile role of heparanase in inflammation. Matrix Biol. 2013;32:234–240. doi: 10.1016/j.matbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermano E, Lerner I, Elkin M. Heparanase enzyme in chronic inflammatory bowel disease and colon cancer. Cell Mol Life Sci. 2012;69:2501–2513. doi: 10.1007/s00018-012-0930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JP, Vlodavsky I. Heparin, heparan sulfate and heparanase in inflammatory reactions. Thromb Haemost. 2009;102:823–828. doi: 10.1160/TH09-02-0091. [DOI] [PubMed] [Google Scholar]
- 13.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Bri J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 16.Hacker U, Nybakken K, Perrimon N. Heparan sulphate proteoglycans: the sweet side of development. Nature Rev. 2005;6:530–541. doi: 10.1038/nrm1681. [DOI] [PubMed] [Google Scholar]
- 17.Couchman JR. Transmembrane signaling proteoglycans. Annual Rev Cell Dev Biol. 2010;26:89–114. doi: 10.1146/annurev-cellbio-100109-104126. [DOI] [PubMed] [Google Scholar]
- 18.Lindahl U, Li JP. Interactions between heparan sulfate and proteins-design and functional implications. Int Rev Cell Mol Biol. 2009;276:105–159. doi: 10.1016/S1937-6448(09)76003-4. [DOI] [PubMed] [Google Scholar]
- 19.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–1031. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peterson SB, Liu J. Multi-faceted substrate specificity of heparanase. Matrix Biol. 2013;32:223–227. doi: 10.1016/j.matbio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem & Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Hulett MD, Hornby JR, Ohms SJ, Zuegg J, Freeman C, Gready JE, et al. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry. 2000;39:15659–15667. doi: 10.1021/bi002080p. [DOI] [PubMed] [Google Scholar]
- 23.Abboud-Jarrous G, Atzmon R, Peretz T, Palermo C, Gadea BB, Joyce JA, et al. Cathepsin L is responsible for processing and activation of proheparanase through multiple cleavages of a linker segment. J Biol Chem. 2008;283:18167–18176. doi: 10.1074/jbc.M801327200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassinelli G, Lanzi C, Tortoreto M, Cominetti D, Petrangolini G, Favini E, et al. Antitumor efficacy of the heparanase inhibitor SST0001 alone and in combination with antiangiogenic agents in the treatment of human pediatric sarcoma models. Biochem Pharmacol. 2013;85:1424–1432. doi: 10.1016/j.bcp.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 25.Casu B, Vlodavsky I, Sanderson RD. Non-anticoagulant heparins and inhibition of cancer. Pathophysiol Haemost Thromb. 2008;36:195–203. doi: 10.1159/000175157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dredge K, Hammond E, Handley P, Gonda TJ, Smith MT, Vincent C, et al. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br J Cancer. 2011;104:635–642. doi: 10.1038/bjc.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, et al. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin Cancer Res. 2011;17:1382–1393. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shafat I, Ben-Arush MW, Issakov J, Meller I, Naroditsky I, Tortoreto M, et al. Pre-clinical and clinical significance of heparanase in Ewing’s sarcoma. J Cell Mol Med. 2011;15:1857–1864. doi: 10.1111/j.1582-4934.2010.01190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H, Roy S, Cochran E, Zouaoui R, Chu CL, Duffner J, et al. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PloS ONE. 2011;6:e21106. doi: 10.1371/journal.pone.0021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bode L, Salvestrini C, Park PW, Li JP, Esko JD, Yamaguchi Y, et al. Heparan sulfate and syndecan-1 are essential in maintaining murine and human intestinal epithelial barrier function. J Clin Investigation. 2008;118:229–238. doi: 10.1172/JCI32335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Conditional signaling by Toll-like receptor 4. FASEB J. 2005;19:872–874. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
- 32.Taylor KR, Gallo RL. Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 2006;20:9–22. doi: 10.1096/fj.05-4682rev. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Fuster M, Sriramarao P, Esko JD. Endothelial heparan sulfate deficiency impairs L-selectin- and chemokine-mediated neutrophil trafficking during inflammatory responses. Nat Immunol. 2005;6:902–910. doi: 10.1038/ni1233. [DOI] [PubMed] [Google Scholar]
- 34.Vlodavsky I, Eldor A, Haimovitz-Friedman A, Matzner Y, Ishai-Michaeli R, Lider O, et al. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis. 1992;12:112–127. [PubMed] [Google Scholar]
- 35.Edovitsky E, Lerner I, Zcharia E, Peretz T, Vlodavsky I, Elkin M. Role of endothelial heparanase in delayed-type hypersensitivity. Blood. 2006;107:3609–3616. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerner I, Hermano E, Zcharia E, Rodkin D, Bulvik R, Doviner V, et al. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J Clin Investigation. 2011;121:1709–1721. doi: 10.1172/JCI43792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt EP, Yang Y, Janssen WJ, Gandjeva A, Perez MJ, Barthel L, et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat Med. 2012;18:1217–1223. doi: 10.1038/nm.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Massena S, Christoffersson G, Hjertstrom E, Zcharia E, Vlodavsky I, Ausmees N, et al. A chemotactic gradient sequestered on endothelial heparan sulfate induces directional intraluminal crawling of neutrophils. Blood. 2011;116:1924–1931. doi: 10.1182/blood-2010-01-266072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Jia J, Zhang X, Zcharia E, Vlodavsky I, Pejler G, et al. Heparanase affects secretory granule homeostasis of murine mast cells through degrading heparin. J Allergy Clin Immunol. 2011;128:1310–1317. doi: 10.1016/j.jaci.2011.04.011. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Wang B, O’Callaghan P, Hjertstrom E, Jia J, Gong F, et al. Heparanase overexpression impairs inflammatory response and macrophage-mediated clearance of amyloid-beta in murine brain. Acta Neuropathol. 2012;124:465–478. doi: 10.1007/s00401-012-0997-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 42.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 43.Grivennikov SI, Karin M. Inflammation and oncogenesis: a vicious connection. Curr Opin Genet Dev. 2010;20:65–71. doi: 10.1016/j.gde.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brun R, Naroditsky I, Waterman M, Ben-Izhak O, Groisman G, Ilan N, et al. Heparanase expression by Barrett’s epithelium and during esophageal carcinoma progression. Mod Pathol. 2009;22:1548–1554. doi: 10.1038/modpathol.2009.115. [DOI] [PubMed] [Google Scholar]
- 46.El-Assal ON, Yamanoi A, Ono T, Kohno H, Nagasue N. The Clinicopathological Significance of Heparanase and Basic Fibroblast Growth Factor Expressions in Hepatocellular Carcinoma. Clin Cancer Res. 2001;7:1299–1305. [PubMed] [Google Scholar]
- 47.Koliopanos A, Friess H, Kleeff J, Shi X, Liao Q, Pecker I, et al. Heparanase expression in primary and metastatic pancreatic cancer. Cancer Res. 2001;61:4655–4659. [PubMed] [Google Scholar]
- 48.Gandhi NS, Freeman C, Parish CR, Mancera RL. Computational analyses of the catalytic and heparin-binding sites and their interactions with glycosaminoglycans in glycoside hydrolase family 79 endo-beta-D-glucuronidase (heparanase) Glycobiology. 2012;22:35–55. doi: 10.1093/glycob/cwr095. [DOI] [PubMed] [Google Scholar]
- 49.Gozalbes R, Mosulen S, Orti L, Rodriguez-Diaz J, Carbajo RJ, Melnyk P, et al. Hit identification of novel heparanase inhibitors by structure- and ligand-based approaches. Bioorg & Med Chem. 2013;21:1944–1951. doi: 10.1016/j.bmc.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 50.Sapay N, Cabannes E, Petitou M, Imberty A. Molecular model of human heparanase with proposed binding mode of a heparan sulfate oligosaccharide and catalytic amino acids. Biopolymers. 2012;97:21–34. doi: 10.1002/bip.21696. [DOI] [PubMed] [Google Scholar]
- 51.Naggi A, Casu B, Perez M, Torri G, Cassinelli G, Penco S, et al. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J Biol Chem. 2005;280:12103–12113. doi: 10.1074/jbc.M414217200. [DOI] [PubMed] [Google Scholar]
- 52.Meirovitz A, Hermano E, Lerner I, Zcharia E, Pisano C, Peretz T, et al. Role of heparanase in radiation-enhanced invasiveness of pancreatic carcinoma. Cancer Res. 2011;71:2772–2780. doi: 10.1158/0008-5472.CAN-10-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferro V, Dredge K, Liu L, Hammond E, Bytheway I, Li C, et al. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Seminars Thromb Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 54.Liu CJ, Lee PH, Lin DY, Wu CC, Jeng LB, Lin PW, et al. Heparanase inhibitor PI-88 as adjuvant therapy for hepatocellular carcinoma after curative resection: a randomized phase II trial for safety and optimal dosage. J Hepatol. 2009;50:958–968. doi: 10.1016/j.jhep.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 55.Borsig L, Vlodavsky I, Ishai-Michaeli R, Torri G, Vismara E. Sulfated Hexasaccharides Attenuate Metastasis by Inhibition of P-selectin and Heparanase. Neoplasia. 2011;13:445–452. doi: 10.1593/neo.101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferro V, Liu L, Johnstone KD, Wimmer N, Karoli T, Handley P, et al. Discovery of PG545: a highly potent and simultaneous inhibitor of angiogenesis, tumor growth, and metastasis. J Med Chem. 2012;55:3804–3813. doi: 10.1021/jm201708h. [DOI] [PubMed] [Google Scholar]
- 57.Johnstone KD, Karoli T, Liu L, Dredge K, Copeman E, Li CP, et al. Synthesis and biological evaluation of polysulfated oligosaccharide glycosides as inhibitors of angiogenesis and tumor growth. J Med Chem. 2010;53:1686–1699. doi: 10.1021/jm901449m. [DOI] [PubMed] [Google Scholar]
- 58.Hammond E, Brandt R, Dredge K. PG545, a heparan sulfate mimetic, reduces heparanase expression in vivo, blocks spontaneous metastases and enhances overall survival in the 4T1 breast carcinoma model. PloS ONE. 2012;7:e52175. doi: 10.1371/journal.pone.0052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ferro V. Heparan sulfate inhibitors and their therapeutic implications in inflammatory illnesses. Expert Opin Ther Targets. 2013;17:965–975. doi: 10.1517/14728222.2013.811491. [DOI] [PubMed] [Google Scholar]
- 60.Harvey JR, Mellor P, Eldaly H, Lennard TW, Kirby JA, Ali S. Inhibition of CXCR4-mediated breast cancer metastasis: a potential role for heparinoids? Clin Cancer Res. 2007;13:1562–1570. doi: 10.1158/1078-0432.CCR-06-1987. [DOI] [PubMed] [Google Scholar]
- 61.Ma L, Qiao H, He C, Yang Q, Cheung CH, Kanwar JR, et al. Modulating the interaction of CXCR4 and CXCL12 by low-molecular-weight heparin inhibits hepatic metastasis of colon cancer. Investigational new drugs. 2012;30:508–517. doi: 10.1007/s10637-010-9578-0. [DOI] [PubMed] [Google Scholar]
- 62.Palumbo A, Larocca A, Genuardi M, Kotwica K, Gay F, Rossi D, et al. Melphalan, prednisone, thalidomide and defibrotide in relapsed/refractory multiple myeloma: results of a multicenter phase I/II trial. Haematologica. 2010;95:1144–1149. doi: 10.3324/haematol.2009.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee DY, Park K, Kim SK, Park RW, Kwon IC, Kim SY, et al. Antimetastatic effect of an orally active heparin derivative on experimentally induced metastasis. Clin Cancer Res. 2008;14:2841–2849. doi: 10.1158/1078-0432.CCR-07-0641. [DOI] [PubMed] [Google Scholar]
- 64.Robert F. The potential benefits of low-molecular-weight heparins in cancer patients. J Hematol Oncol. 2010;3:3. doi: 10.1186/1756-8722-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- 66.Correia AL, Bissell MJ. The tumor microenvironment is a dominant force in multidrug resistance. Drug Resist Updat. 2013;15:39–49. doi: 10.1016/j.drup.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ostman A. The tumor microenvironment controls drug sensitivity. Nat Med. 2013;18:1332–1334. doi: 10.1038/nm.2938. [DOI] [PubMed] [Google Scholar]
- 68.Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Nat Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 69.Paraiso KH, Smalley KS. Fibroblast-mediated drug resistance in cancer. Biochem Pharmacol. 2013;85:1033–1041. doi: 10.1016/j.bcp.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 70.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–4351. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hu Y, Gale M, Shields J, Garron C, Swistak M, Nguyen TH, et al. Enhancement of the anti-tumor activity of therapeutic monoclonal antibodies by CXCR4 antagonists. Leukemia & lymphoma. 2012;53:130–138. doi: 10.3109/10428194.2011.601698. [DOI] [PubMed] [Google Scholar]
- 72.Hanna E, Quick J, Libutti SK. The tumour microenvironment: a novel target for cancer therapy. Oral Diseases. 2009;15:8–17. doi: 10.1111/j.1601-0825.2008.01471.x. [DOI] [PubMed] [Google Scholar]
- 73.Ramani VC, Yang Y, Ren Y, Nan L, Sanderson RD. Heparanase plays a dual role in driving hepatocyte growth factor (HGF) signaling by enhancing HGF expression and activity. J Biol Chem. 2011;286:6490–6499. doi: 10.1074/jbc.M110.183277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Underiner TL, Herbertz T, Miknyoczki SJ. Discovery of small molecule c-Met inhibitors: Evolution and profiles of clinical candidates. Anti-cancer agents Med Chem. 2010;10:7–27. doi: 10.2174/1871520611009010007. [DOI] [PubMed] [Google Scholar]
- 75.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nature Rev. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dalal S, Berry AM, Cullinane CJ, Mangham DC, Grimer R, Lewis IJ, et al. Vascular endothelial growth factor: a therapeutic target for tumors of the Ewing’s sarcoma family. Clin Cancer Res. 2005;11:2364–2378. doi: 10.1158/1078-0432.CCR-04-1201. [DOI] [PubMed] [Google Scholar]
- 77.Cassinelli G, Zuco V, Petrangolini G, De Cesare M, Tortoreto M, Lanzi C, et al. The curative efficacy of namitecan (ST1968) in preclinical models of pediatric sarcoma is associated with antiangiogenic effects. Biochem Pharmacol. 2012;84:163–171. doi: 10.1016/j.bcp.2012.04.005. [DOI] [PubMed] [Google Scholar]