Abstract
Mutations that inhibit differentiation in stem cell lineages are a common early step in cancer development, but precisely how a loss of differentiation initiates tumorigenesis is unclear. We investigated Drosophila intestinal stem cell (ISC) tumors generated by suppressing Notch (N) signaling, which blocks differentiation. Notch-defective ISCs require stress-induced divisions for tumor initiation and an autocrine EGFR ligand, Spitz, during early tumor growth. Upon achieving a critical mass these tumors displace surrounding enterocytes, competing with them for basement membrane space and causing their detachment, extrusion and apoptosis. This loss of epithelial integrity induces JNK and Yki/YAP activity in enterocytes and, consequently, their expression of stress-dependent cytokines (Upd2, Upd3). These paracrine signals, normally used within the stem cell niche to trigger regeneration, propel tumor growth without the need for secondary mutations in growth signaling pathways. The appropriation of niche signaling by differentiation-defective stem cells may be a common mechanism of early tumorigenesis.
Introduction
As in mammals, Drosophila ISCs maintain gut homeostasis by dividing to replace lost cells. ISCs generate transient progeny, enteroblasts (EBs), that can differentiate into absorptive enterocytes (ECs) or secretory enteroendocrine cells (EEs)1, 2. These epithelial cells grow on a basement membrane (BM) overlaying the visceral muscle (VM), which together with ECs, EEs, and EBs comprises the stem cell niche. Fly midgut homeostasis is regulated by Notch, cytokine/JAK-STAT, EGFR/Ras/MAPK, JNK, Hippo, insulin, Wnt, PDGF/VEGF, Hh and BMP/TGFβ signaling3. The various niche cells (EBs, ECs and VM) collectively provide these signals to regulate ISC growth, self-renewal and differentiation. Delta-Notch signaling is the primary trigger of EB to EC differentiation1, 2, 4. ISCs express a Notch (N) ligand, Delta (Dl), which activates the Notch receptor in EBs and promotes their differentiation into ECs2, 4. Loss of N, Dl, or other Notch pathway components in progenitor cells (ISCs and EBs) results in the rapid expansion of differentiation-defective, escargot- positive (esg+), Delta- positive (Dl+) ISC-like cells that form multi-layered neoplastic tumors, typically admixed with excess Prospero-positive (Pros+) EE cells that are also produced1, 2, 4–6.
According to current models differentiation-defective stem cells should remain dependent on growth and survival factors in their niche7, and require secondary mutations to initiate the run-away growth characteristic of tumors8. While the importance of immune cells, fibroblasts, and vasculature recruited as a tumor builds its microenvironment is appreciated9, how tumor initiating cells interact with the stem cell niche prior to microenvironment formation is poorly understood. In flies, differentiation-defective larval neural or adult germline stem cell tumors can overgrow in their respective organs, but whether the stem cell niche is required to propel the growth of these cells has not been tested10. In the fly midgut, the growth of ISC-derived tumors (e.g Apc− or RasV12 dlg−) has been proposed to require niche signals10–13, however the mechanisms controlling the activation of these signals, and the functional importance of the niche cells that produce them, have not been directly tested. In the mouse intestine, stem-like tumor initiating cells (LGR5+ APC−) in early adenomas remain associated with Paneth cells - an essential part of the normal ISC niche14 - but whether these tumor initiating cells are dependent on growth signals from Paneth cells is unknown15. Here we examine how differentiation-defective Notch− ISC tumors grow in the adult fly ISC niche.
Notch− ISC tumor cells are proliferative endocrine progenitors
We first investigated the identity of the tumor cells that arise from depleting Notch with RNAi in progenitors, using the esgtsGAL4 system (esgts). We found N− tumors to be an admixture of Proshigh EE-like cells and neoplastic ISC-like cells that express high levels of ISC markers (Dlhigh esghigh) but also low levels of EE markers (Pros) (Supplementary Fig. 1a–a’, c–c’’’). ISC-like tumors cells did not show markers of active Notch signaling (SuH-lacZ) or EC differentiation (Pdm1; Supplementary Fig. 1b–b’, d–d’’’). mRNA-seq analysis showed that ISC-like N– tumor cells (Dlhigh esghigh) expressed many ISC-associated genes (i.e. Dl and spdo)2, 4, 6, 16 and EE-associated genes (i.e. pros and Allatostatin, AstA)1, 2, 17 (Supplementary Fig. 1g–j), and functional tests showed that these cells differentiated into EEs, rather than ECs, when NRNAi expression was extinguished (Supplementary Fig. 1e–f’). Together these data indicate that ISC-like tumor cells are actually committed EE precursors rather than multipotent progenitors. We did not find mitotic (phospho-Ser 10-Histone 3 positive) Pros+ EE-like cells within the tumors; indeed only esg+ ISC-like tumor cells were proliferative (Fig. 3a, 5c). To determine the role of the excess EEs in tumor growth, we suppressed Notch signaling by expressing RNAi towards Suppressor of Hairless (SuH), in which tumors without excess EEs are generated (Supplementary Fig. 5d)17. In this case large esg+ tumors formed similar to those seen with N− tumors containing EEs, indicating that the excess EEs present in most N− tumors have little if any role in tumor growth.
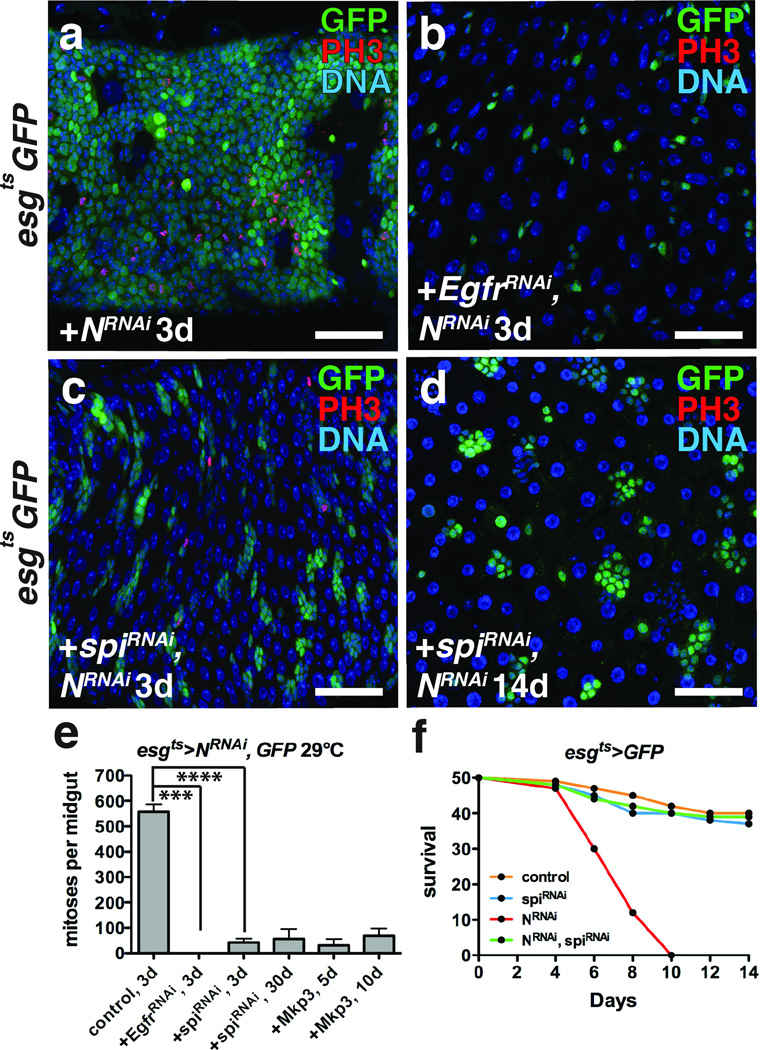
Figure 3. ISC tumor initiation and outgrowth requires autonomous Spi/EGFR signaling.
(a–c) Phosphorylated histone H3 Ser10 (red) and GFP (green) after expression of NRNAi (a), EGFRRNAi and NRNAi (b) and spiRNAi and NRNAi (c) with esgts for 3 days.
(d) Phosphorylated histone H3 Ser10 (red) and GFP (green) after expression of spiRNAi and NRNAi with esgts for 14 days.
(e) Mean number of phosphorylated histone H3 Ser10 positive cells per midgut with s.e.m. after expression with esgts of NRNAi and GFP (n=30 midguts), NRNAi and EgfrRNAi (n=30 midguts, Mann-Whitney: p=0.0001), NRNAi and spiRNAi for 3 (n=33 midguts, Mann-Whitney: p<0.0001) or 30 days, and NRNAi and Mkp3 for 5 or 10 days. Midguts were pooled from 3 independent experiments.
(f) Survival after expression of GFP alone (control), spiRNAi, NRNAi, and NRNAi and spiRNAi with esgts (n=100 flies per genotype pooled from 2 independent experiments).
DNA is a-d (blue). Scale bars in a–c, 40 µm; d, 50µm.
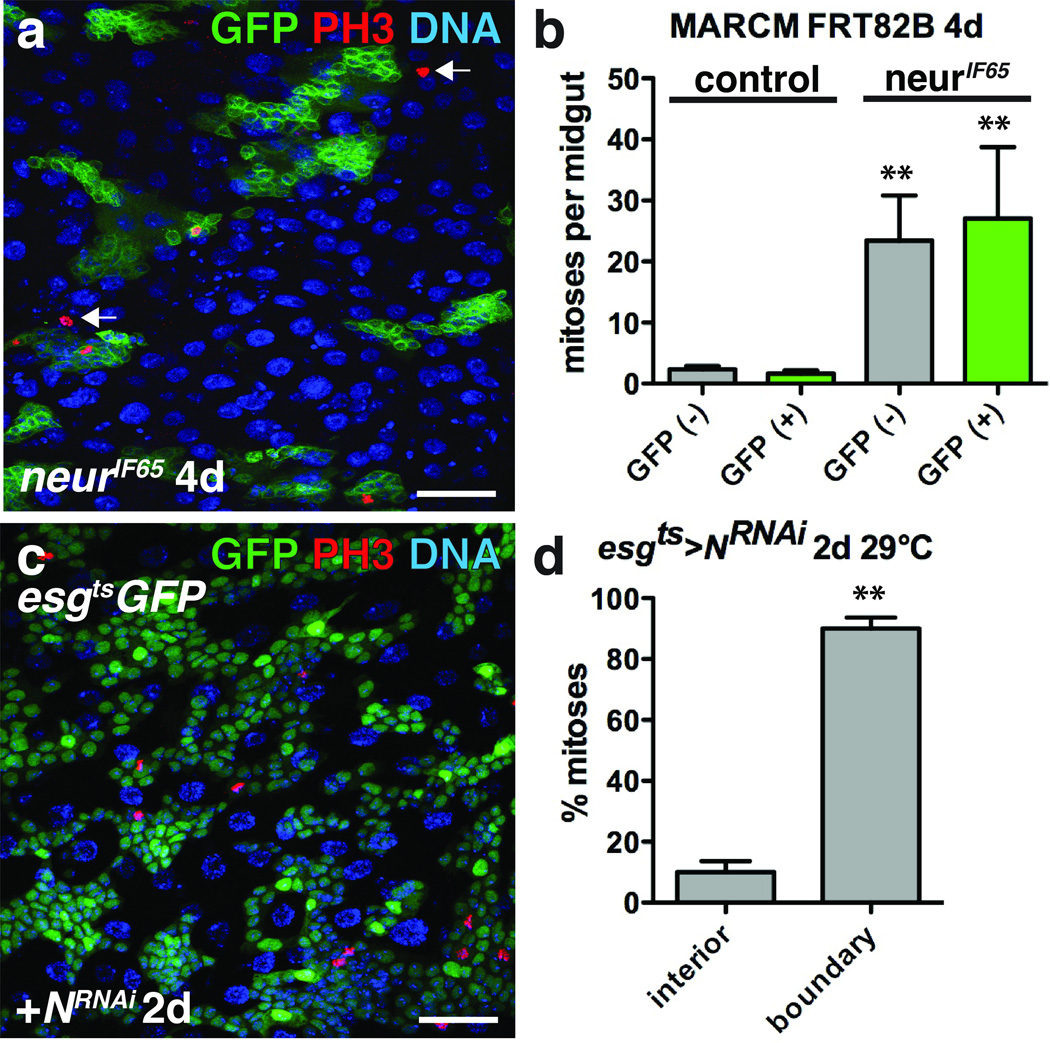
Figure 5. ISC tumor growth induces non-autonomous ISC proliferation.
(a) Phosphorylated histone H3 Ser10 (red) inside and outside (arrows) of 4 day neurIF65 clones (green).
(b) Mean number with s.e.m. of mitoses per midgut found inside (GFP+, green; for neur− clone, Mann-Whitney: p=0.0011) and outside (GFP–, grey; for neur− clone, Mann-Whitney: p=0.0012) of 4 day control (n=31 midguts) or neurIF65/− clones (n=24 midguts). Midguts were pooled from 3 independent experiments.
(c) Phosphorylated histone H3 Ser10 (red) in midguts expressing GFP (green) and NRNAi with esgts for 2 days.
(d) Mean percent of phosphorylated histone H3 Ser10 positive cells with s.e.m. found in the tumor interior or at the tumor boundary (n=3 fields; fields were selected from 3 posterior midguts from 2 independent experiments, paired t-test: p= 0.0080) in flies expressing GFP and NRNAi with esgts for 2 days.
DNA in a, c (blue). Scale bar in a, 40 µm; c, 35 µm.
Gut epithelial stress promotes ISC tumor initiation
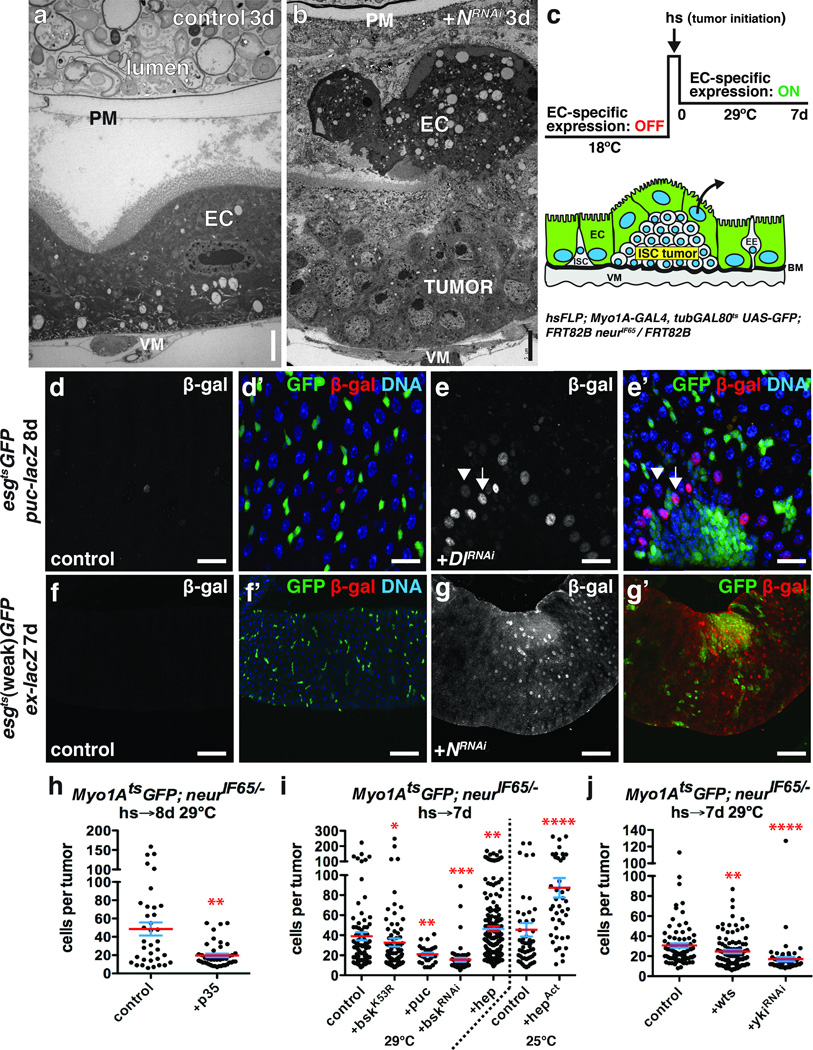
To understand how the niche might contribute to ISC-derived tumorigenesis, we investigated how N− tumors initiate. Despite a presumably uniform block to differentiation by the expression of NRNAi using esgts, tumors were not detected in all experimental animals. This indicated that simply blocking differentiation was not sufficient for tumor initiation. Hence we asked whether tumor initiation might be dependent on environmental factors. Previous reports showed that stress or damage to the midgut (e.g. by enteric infection) activates JNK and YAP/Yki signaling and that this stimulates the production of cytokines (Upd2,3) that are mitogenic for ISCs18–23. Previous work also showed that enteric infection could increase ISC tumor outgrowth and reduce host survival13, 18. For instance Apidianakis et al.13 found that the frequency of tumor initiation from N− ISCs could be enhanced by enteric infection. We confirmed these results by performing enteric infection with Pseudomonas entomophila (P.e.) prior to initiating ISC tumors by inducing NRNAi (Fig. 1f; Supplementary Fig. 6f). Since JNK signaling can be activated by enteric infection, we tested whether JNK signaling in ECs might influence the frequency of tumor initiation, as suggested by Apidianakis et al.13. We expressed a brief pulse of activated Hemipterous (HepAct, Jun Kinase Kinase), with the EC-specific Myo1AtsGAL4- UAS system (Myo1Ats), and then afterwards used Flp/FRT-mediated recombination to induce ISC clones mutant for the Notch signaling component, neuralized (neur), to inhibit stem cell differentiation (Fig. 1a, Supplementary Fig. 3a–c). Neur is an E3 ubiquitin ligase required for Notch signaling5 and its loss results in ISC tumors consisting of Dl+ ISC- like cells and Pros+ EE cells4, similar to NRNAi. Similarly to infection, delivering a pulse of JNK activity to ECs prior to Notch suppression resulted in more tumors per midgut (Fig. 1b) and more flies bearing tumors (Fig. 1c). However the JNK pulse did not affect tumor outgrowth after initiation (Fig. 1d), indicating that this transient stress specifically promoted tumor initiation. As JNK is known to induce the Upd cytokines18 and thereby activate ISCs for division, we asked whether mutant ISC division might be a prerequisite for tumor initiation. To test this we co-expressed Cyclin E and string (Cdc25), a gene combination that promotes ISC division24, together with NRNAi in progenitor cells. This also greatly increased tumor incidence, confirming that ISC division is sufficient to promote tumor initiation (Fig. 1e; Supplementary Fig. 6g). These results suggest that, in addition to loss of differentiation capacity, the formation of small clusters of ISC-like cells by stress-induced stem cell divisions may be a prerequisite for tumor formation.
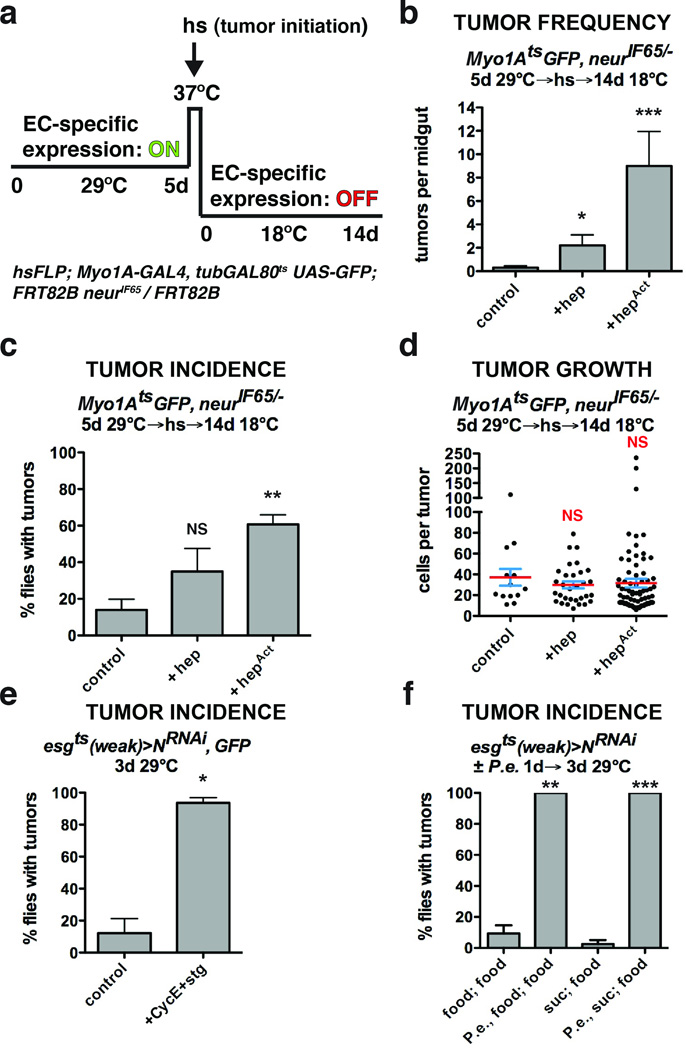
Figure 1. Tissue stress promotes ISC tumor initiation.
(a) System to independently express transgenes in ECs with Myo1Ats and subsequently initiate ISC- derived tumors by heat shock- induced Flp-FRT mediated recombination.
(b) Mean number of tumors per midgut with s.e.m. 14 days (18°C) after tumor induction following GFP (n=20 midguts), JNKK (hep) (n=12 midguts, Mann-Whitney: p=0.0203) or activated JNKK (hepAct) (n=10 midguts, Mann-Whitney: p=0.0002) expression in ECs with Myo1Ats for 5 days (29°C). Midguts pooled from 2 independent experiments.
(c) Mean percent of flies with tumors with s.e.m. from n=3 independent experiments 14 days (18°C) after tumor induction following GFP, JNKK (hep) or activated JNKK (hepAct) (paired t-test: p=0.0076) expression in ECs with Myo1Ats for 5 days (29°C).
(d) Cells per neurIF65/− tumor with mean (red line) and s.e.m. 14 days (18°C) after tumor induction following GFP (n=13 tumors pooled from 20 midguts), JNKK (hep) (n=31 tumors pooled from 12 midguts) or activated JNKK (hepAct) (n=77 tumors pooled from 10 midguts) expression in ECs with Myo1Ats for 5 days (29°C). Midguts pooled from 2 independent experiments.
(e) Mean percent of flies with tumors with s.e.m. from n=3 independent experiments in flies expressing GFP and NRNAi or NRNAi, Cyclin E (CycE) and cdc25/string (stg) (paired t-test: p= 0.0185) with esgts (weak) for 3 days.
(f) Mean percent of flies with tumors with s.e.m. from n=3 independent experiments after being fed food alone, P.e. containing food (paired t-test: p=0.0034), sucrose alone or P.e. containing sucrose (paired t- test: p=0.0007) for 1 day prior to tumor induction by expressing NRNAi with esgts (weak) for 3 days (food alone).
ISC tumor initiation and outgrowth requires autonomous Spi/EGFR signaling
We next sought to define the tumor autonomous factors that drive tumor growth after initiation. EGFR signaling is required for ISC proliferation25–28, so we checked its role. N− or neur− tumor growth increased the expression of the EGFR ligands spi and vn (Fig. 2g, Supplementary Fig. 2b) and mRNA-seq of FACS-isolated esg+ tumor cells revealed that the tumor cells themselves produced more spi but not vn (Fig. 2h). Notably, the highest level of spi expression in normal midguts was in Dl+ ISCs (Supplementary Fig. 2a). The expression of the EGFR pathway target argos was also increased in the tumor cells (Fig. 2h), which had higher levels of activated MAPK (ppMAPK) than normal ISCs (Fig. 2a–d). To determine whether EGFR/MAPK signaling was necessary for N− tumor growth we expressed the MAPK phosphatase 3 (Mkp3), or RNAi against EGFR or Spi in the tumor cells themselves. These treatments suppressed tumor growth. Thus N− tumor cells, like normal ISCs, require EGFR/MAPK activity to grow (Fig. 3a–d,e). Depleting spi for 2 weeks in wild-type animals did not deplete progenitor cells, suggesting that Spi, unlike the EGFR25, 26 is not an essential survival factor for normal ISCs. Remarkably, while flies bearing N− ISC tumors died within 10 days, depleting spi in the tumor cells resulted in viable flies with tumors that remained small even after 30 days (Fig. 3e–f). These results indicate that autocrine Spi/EGFR/MAPK signaling is required for ISC tumor growth. Since tumor initiation also requires ISC mitosis (Fig. 1), we speculate that Spi signaling may only be effective when multiple ISC-like Spi-expressing tumor cells are juxtaposed in a cluster, and that mitosis generates these clusters.
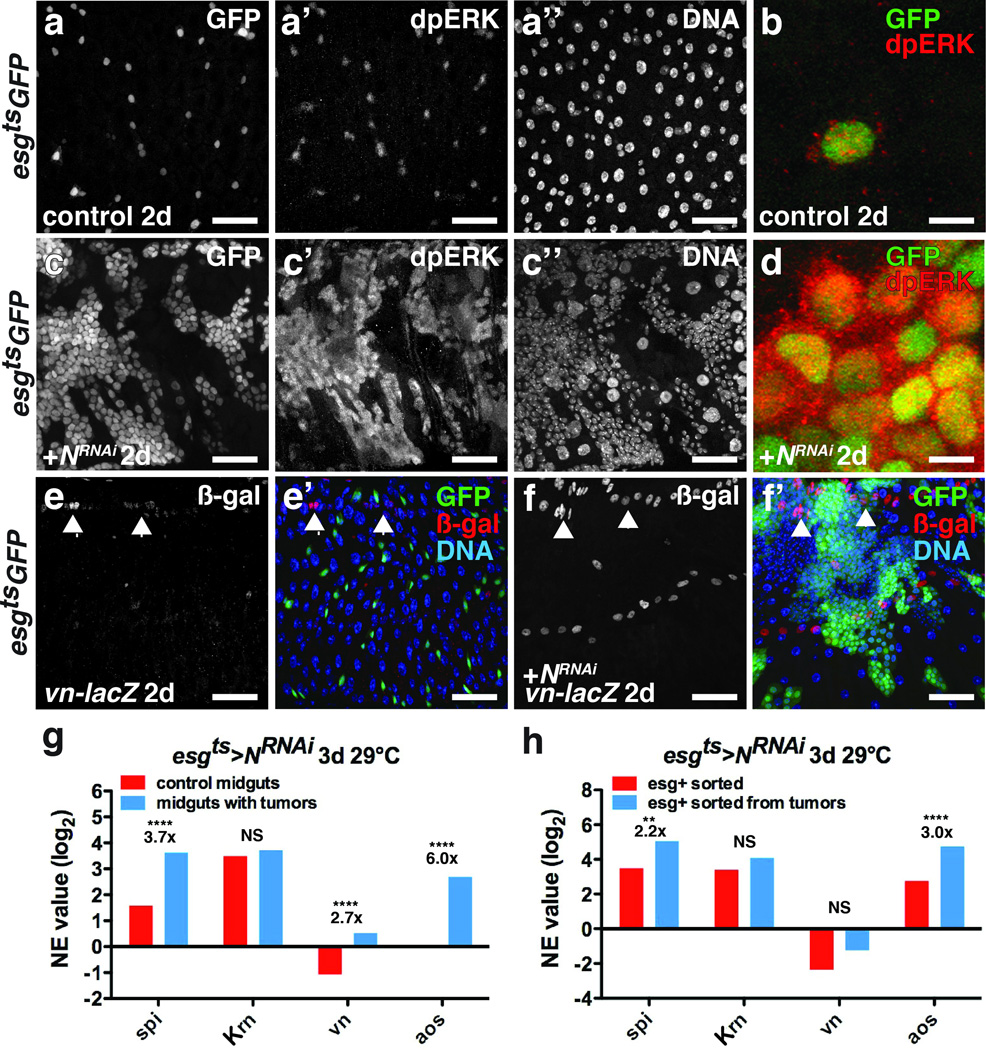
Figure 2. Spi/EGFR/MAPK signaling is induced in ISC tumors and the niche.
(a–d) Di-phosphorylated ERK (a’, c’; b, d, red) in midguts expressing GFP (a; b, green) or GFP and NRNAi (c; d, green) with esgts for 2 days.
(e–f) β-galactosidase (e,f; e’,f’, red) in midguts of flies bearing vn-lacZ (vnP1749) and expressing GFP (e’, green) or GFP and NRNAi (f’, green) with esgts for 2 days.
(g) Mean normalized expression (NE) value (RPKM, log2) from n=2 independent experiments of EGFR ligand (spi, Krn, vn) and signaling target (aos) mRNA determined by mRNA sequencing of midguts expressing GFP (control, red) or GFP and NRNAi (blue) with esgts for 3 days. The adjusted fold change in gene expression in tumorous midguts (blue), normalized to control midguts (red), is indicated. The Benjamini-Hochberg adjusted p-value for spi is 7.71e-322; vn, p= 3.25e-66; aos, p= 2.84e-264.
(h) Mean normalized expression (NE) value (RPKM, log2) from n=2 independent experiments of EGFR ligand (spi, Krn and vn) and signaling target (aos) mRNA determined by mRNA sequencing of esg+ cells sorted from control midguts expressing GFP (red) or from tumorous midguts expressing GFP and NRNAi (blue) with esgts for 3 days. The adjusted fold change in gene expression in sorted esg+ tumor cells (blue), normalized to esg+ cells from control midguts, is indicated. Benjamini-Hochberg corrected p-value for spi is 0.02924066; aos, 4.52e-05.
NS, not significant. DNA is in a’’ and c’’; e’ and f’ (blue). Scale bars in a–a’’, 35 µm; b, 10 µm; c–c’’, 40 µm; d, 10 µm; e–e’, 35 µm and f–f’, 45 µm.
ISC tumors promotes enterocyte detachment, extrusion, and death
Tumor cells might compete with normal epithelial cells for adhesion to the basement membrane (BM), which in the midgut consists of extracellular matrix (ECM) components including collagen IV, laminin and perlecan29, 30. Cell extrusion due to cell overcrowding has been observed in fish and mouse epithelia, and proposed as a mechanism for maintaining tissue homeostasis31. Accelerating epithelial replacement in the midgut results in the rapid loss of older ECs25, possibly also by cell extrusion. But little data is available on the extrusion of normal cells at tumor boundaries, or how this might impact tumor growth. In our case light and electron microscopy showed that ECs adjacent to and overlying the tumors were detached from the BM, and that a subset of these detached ECs were apoptotic (Fig. 4a–b, Supplementary Fig. 4a–a’, d). mRNA-seq analysis showed that N− tumor growth induced the pro-apoptotic genes rpr, skl and particularly grim in non-tumor cells (Supplementary Fig. 4b–c). Consistently, we observed pyknotic nuclei in non-tumor cells either adjacent or apical to 38.5% of neur− tumors (n=45) whereas pyknotic nuclei were rarely found in tumor-free regions. This suggested that ECs might initiate apoptosis during or after apical extrusion. Pyknotic nuclei were never associated with tumors smaller than 20 cells, suggesting that EC death depended upon the tumors achieving a minimum critical mass. To test whether apoptosis was required for EC detachment and extrusion we generated neur− tumors amongst ECs expressing the apoptosis inhibitor p35 (Fig. 4c). Non-apoptotic p35-expressing ECs could be found apical to tumors, suggesting that apoptosis was not required for EC detachment. However, tumors generated in p35-expressing epithelia were smaller than tumors growing in normal epithelia (Fig. 4h, Supplementary Fig. 3d–e), indicating that EC apoptosis enhanced tumor growth. Thus, N− tumors grow by promoting EC detachment from the BM, and their apoptosis- dependent death.
Figure 4. Growing ISC tumors induce changes in the niche.
(a–b) Transmission electron micrograph of anterior midgut expressing GFP (a) or NRNAi (b) with esgts for 3 days.
(c) System to independently initiate ISC- derived tumors by heat shock- induced Flp-FRT mediated recombination and subsequently express transgenes in ECs with Myo1Ats.
(d–e) β-galactosidase (d,e; d’,e’, red) in midguts of flies bearing puc-lacZ and expressing GFP (d’, green) or GFP and DlRNAi (e’, green) with esgts for 8 days. High β-galactosidase (e; e’, red; arrow) was observed in ECs adjacent to ISC tumors; lower β-galactosidase (arrowhead) in ECs further away.
(f–g) β-galactosidase (f,g; f’,g’, red) in midguts of flies bearing ex-lacZ and expressing GFP (f’, green) or GFP and NRNAi (g’, green) with esgts (weak) for 7 days.
(h–j) Cells per neurIF65/− tumor amongst ECs expressing GFP (control, n=35 tumors from 12 midguts, skewness= 1.117, kurtosis= 0.4573) or GFP and p35 (n=42 tumors from 28 midguts, p=0.0019, skewness= 1.586, kurtosis= 1.707) at 29 °C (h); GFP (control, n=83 tumors from 32 midguts, skewness= 2.650, kurtosis= 8.662), GFP and bskK53R (n=82 tumors from 45 midguts, p=0.0346, skewness= 3.805, kurtosis= 17.430), GFP and puc (n=29 tumors from 19 midguts, p=0.0046), GFP and bskRNAi (n=57 tumors from 44 midguts, p<0.0001, skewness= 3.857, kurtosis= 16.530) or GFP and hep (n=191 tumors from 68 midguts, p=0.0045, skewness= 1.508, kurtosis= 2.082) at 29 °C or GFP (control, n=51 tumors from 23 midguts, skewness= 2.414 and kurtosis= 5.719) or GFP and hepAct (n=42 tumors from 14 midguts, p<0.0001, skewness= 1.413 and kurtosis= 1.724) at 25°C (i); GFP (control, n=79 tumors from 41 midguts, skewness= 2.017, kurtosis= 4.577), GFP and wts (n=92 tumors from 33 midguts, p=0.0088) or GFP and ykiRNAi (n=55 tumors from 17 midguts, p<0.0001, skewness= 5.693, kurtosis= 36.85) at 29°C (j) with Myo1Ats, 7–8 days.
In h–j, midguts were pooled from 3 independent experiments; p-values from Mann-Whitney test; mean (red line) and s.e.m. (blue) are shown. DNA in d’, e’, f’ (blue). Scale bars in a–b, 5 µm; in d–d’, 25 µm; in e–e’, 20µm; in f–g’, 60 µm.
JNK and Yki activity in tumor-adjacent enterocytes promotes tumor growth
Epithelial stress activates both JNK and Hippo/Yki signaling in midgut ECs18, 20, 21, 23. Using reporters for JNK (puc-lacZ) and Yki (ex-lacZ) activity, we observed high JNK and Yki activity in detached ECs adjacent and apical to larger N− or Dl− ISC tumors, but not in the esg+ tumor cells themselves (Fig. 4e–e’, 4g–g’). Interestingly, both JNK and Yki activity spread several EC diameters away from tumors (Fig. 4e–e’, 4g–g’, Supplementary Video 1–3). To test whether JNK signaling in ECs affected tumor growth, we generated neur− tumors amongst ECs expressing a dominant-negative form of JNK (BskK53R) or bskRNAi, or the JNK phosphatase puckered (Puc). To avoid effects on tumor initiation these JNK suppressors were activated after tumor induction (Fig. 4c). Expression of BskK53R, bskRNAi, or Puc specifically in ECs suppressed neur− tumor growth (Fig. 4i, Supplementary Fig. 3f–g). Conversely, increasing JNK signaling in ECs by expressing Hep (JNK) or HepAct resulted in larger tumors (Fig. 4i) and increased pyknotic nuclei around tumors. This increased apoptosis was not due directly to increased JNK activity, since rapid EC detachment or pyknotic nuclei were not observed in regions without tumors. By determining the skewness and kurtosis for each tumor population we found that tumor populations were positively skewed, such that the majority of tumors were smaller than the mean size (see Fig. 4, 6 and Supplementary Fig. 7 legends). Decreasing tumor growth by JNK suppression increased the positive skew in the tumor size distribution, such that even more tumors were smaller than the mean and even fewer were larger. In contrast, enhancing tumor growth by overexpressing Hep or HepAct in ECs consistently shifted tumor sizes closer to a normal distribution. Using the same approach, we tested the non-tumor autonomous function of Hippo signaling in ISC tumor growth by expressing ykiRNAi or Warts (Wts), an inhibitor of Yki, in ECs. This also inhibited neur− tumor growth (Fig. 4j, Supplementary Fig. 3h–i). Thus the induction of JNK and Yki activity in ECs surrounding larger ISC tumors contributes indirectly to tumor growth.
Figure 6. ISC tumors co-opt niche derived signals for their growth.
(a–c) Flourescent in situ hybridization to upd3 mRNA (a–c’; a’,b’,c’, red) in control midguts expressing GFP (a’, green) or GFP and NRNAi (b’, c’, green) with esgts for 2 days. ECs near ISC tumors expressed upd3 (c–c’, arrows).
(d) D-GFP (d, green) and Delta (d’, red) in midguts of flies bearing 10X-STAT-D-GFP and expressing NRNAi with esgts (without UAS-GFP) for 5 days.
(e) Mean normalized expression (NE) value (RPKM, log2) from n=2 independent experiments of cytokine (upd1-3) and JAK-STAT target (Socs36) mRNA determined by mRNA sequencing of whole midguts expressing GFP (control, red) and GFP and NRNAi (blue) with esgts for 3 days. The adjusted fold change in gene expression in midguts bearing tumors (blue), normalized to control midguts (red), is indicated. The Benjamini-Hochberg adjusted p-value for upd2, upd3 and Soc36E is 7.71e-322. NS, not significant.
(f) Mean number of phosphorylated histone H3 Ser10 positive cells per midgut after expression of NRNAi (n=48 midguts), Upd1 and NRNAi (n=45 midguts, p=0.0011), and NRNAi and domeΔCYT (n=39 midguts, p<0.0001) with esgts for 2 days.
(g) Mean number of phosphorylated histone H3 Ser10 positive cells per midgut after expression of NRNAi with esgts (weak) for 14 days in control flies (n=27 midguts) and flies mutant for upd2 (upd2Δ, n=25 midguts, p=0.0176), upd3 (upd3Δ, n=34 midguts, p<0.0001) and upd2 and upd3 (upd2-3Δ, n=28 midguts, p<0.0001).
(h) Cells per neurIF65/− tumor amongst ECs expressing GFP (control, n=101 tumors from 38 midguts, skewness= 2.097, kurtosis= 4.754) or RNAis against upd2(1) (n = 41 tumors from 35 midguts, p=0.0181, skewness= 3.681, kurtosis= 15.980) and upd2(2) (n=27 tumors from 23 midguts, p=0.0033, skewness= 3.068, kurtosis= 10.720) and upd3(1) (n=55 tumors from 36 midguts, p=0.0233, skewness= 2.742, kurtosis= 8.061 and upd3(2) (n=37 tumors in 25 midguts, p=0.0010) with Myo1Ats for 7 days.
In f–h, midguts were pooled from 3 independent experiments; p-values from Mann-Whitney test; mean with s.e.m. are shown. Scale bars in a–b’, 40 µm; in c–c’, 30 µm; in d–d’, 25 µm. DNA in c’, d–d’ (blue).
Tumor growth induces mitogenic signals in the niche
In studying the tumorous neur− clones we noted increased mitoses both inside and outside of the tumors (Fig. 5a,b). In addition, most of the mitoses in tumor cells were at tumor boundaries rather than tumor interiors (Fig. 5c–d). Both observations suggested that the tumors might stimulate the production of diffusible mitogenic signals by surrounding niche cells. Previous studies showed that EC apoptosis or ectopic JNK or Yki activity in ECs can stimulate the expression of the EGFR ligands Vn and Krn, and the Upd2 and Upd3 cytokines, all of which are stress-inducible ISC mitogens18, 20–22, 25. Thus we suspected that stress from the N− tumors might also activate these niche-derived mitogens. Indeed, mRNA-seq and qPCR experiments on tumor-bearing midguts revealed increases in vn, upd2 and upd3 (Fig. 2g, 6e, Supplementary Fig. 2b, 5a,d). mRNA-seq experiments on sorted tumor cells showed that both vn and upd3 expression increased in the surrounding niche rather than in tumor cells (Fig. 2g–h, 6e, Supplementary Fig. 5b) whereas upd2 increased in tumor cells and also the niche (EC, VM) (Fig. 6e, Supplementary Fig. 5b). In contrast to a previous report32 Upd1 induction was detected neither inside nor outside of N− tumors (Fig. 6e, Supplementary Fig. 5b–d). Using a reporter we observed high vn expression in visceral muscle adjacent to the tumors (Fig. 2f–f’), but not in regions devoid of tumors (Fig. 2e–e’). in situ hybridization showed that many ECs adjacent to or located apically to ISC tumors expressed high levels of upd3 mRNA (Fig. 6b–c’, Supplementary Figs. 6a–b’), which was undetectable in controls (Fig. 6a–a’). Cytokine induction was not due to EE cell expansion nor to altered enteric bacterial load (Supplementary Fig. 5d, 6c–e), and so we infer that it was a direct result of tumor-induced EC stress. mRNA-seq experiments also revealed that the PDGF/VEGF (PVR) ligand Pvf2, the insulin-like peptide Dilp3 and wg, a Wnt, were non-autonomously induced by N− tumors, presumably in ECs and/or VM (Supplementary Fig. 2c–d). These signals have also been implicated as ISC mitogens or survival factors29, 33–37 and might also promote tumor growth.
Upd cytokines produced by tumor-adjacent enterocytes drive tumor growth
The most potent known effectors of ISC proliferation are the Upd cytokines. We found that tumor cells had high STAT activity (Fig. 6d,d’; Supplementary Fig. 5h–i’), expressed the STAT target socs36E (Fig. 6e), and could be stimulated to grow faster by increasing Upd signaling (Fig 6f). Expressing oncogenic Ras (RasV12G) in ECs - a treatment that induces JNK activity (Supplementary Fig. 7) and Upd325 in ECs - also strongly accelerated N− tumor growth (Supplementary Fig. 7e, 3l–m). A previous report38, 39 showed that STAT is required in N− ISC tumors for them to produce EEs, but the role of JAK-STAT signaling in tumor growth was not addressed. Expressing a dominant negative form of the Upd receptor, Domeless (DomeΔCYT) in N− tumors strongly suppressed their growth without altering cell identity (Fig. 6f, Supplementary Fig. 5g–g’’, i–j’), indicating that Upd signaling was required for tumor growth.
A recent study of Apc− tumors in the fly midgut11 also reported the induction of an upd3 reporter gene (upd3-lacZ) in tumor-adjacent ECs, and showed that domeless and Stat92E were required in the tumor cells for optimal tumor growth11. However, the requirement for upd3 was not functionally tested, and the Apc− tumor cells themselves also expressed upd3-lacZ11, leaving it unclear whether niche-derived Upd3 was important for tumor growth. To resolve this question we directly tested the requirement for niche-derived Upd2 and Upd3. First, we measured N− tumor growth in mutant flies completely lacking upd2 (upd2Δ), upd3 (upd3Δ) or both genes (upd2-3Δ). Second, to determine whether Upd2 and Upd3 were specifically required from ECs, rather than systemically, we generated neur− ISC tumors in flies in which upd2 or upd3 were specifically suppressed in ECs using targeted RNAi. In both cases tumor growth was suppressed (Fig. 6g–h, Supplementary Fig. 3j–k, 5e–f), confirming that the production of Upd cytokines by ECs promotes ISC tumor growth.
JNK, Yki and Upd3 are induced by detachment of enterocytes from the visceral muscle
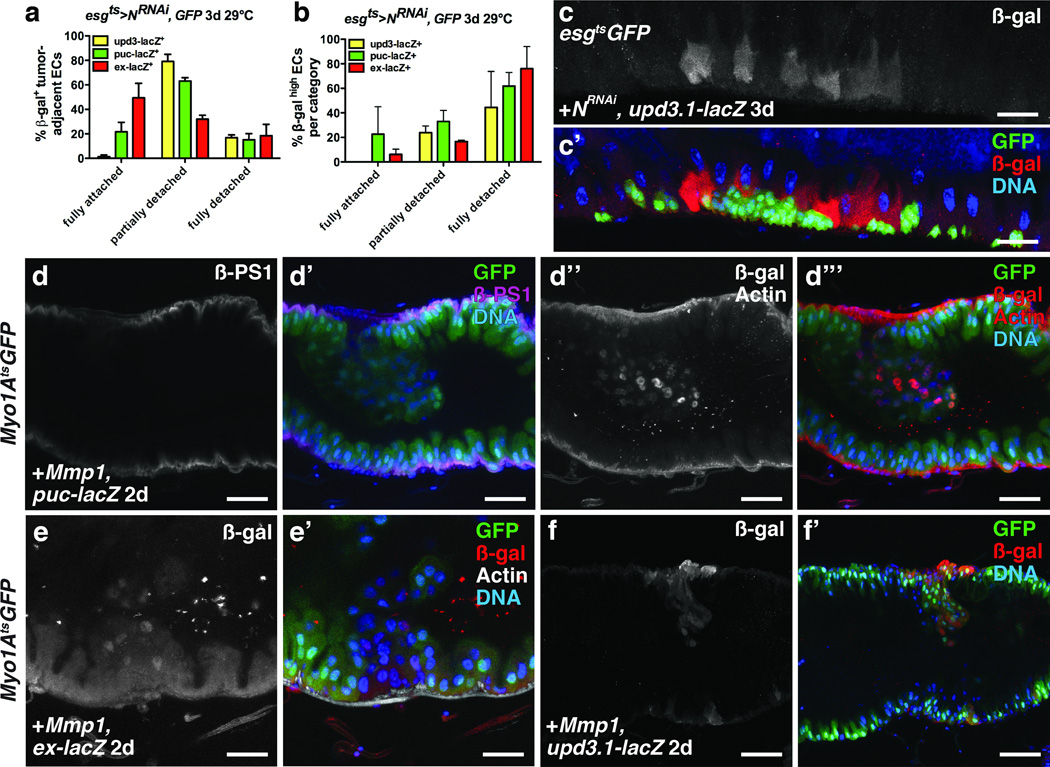
Cordero et al.11, proposed that hyperplastic Apc− tumors induce upd3 expression by stimulating EGFR signaling in surrounding ECs. However, we did not observe increased dpMAPK staining in ECs neighboring N− ISC tumors, even though these tumors produced high levels of Spi (Fig. 2a–d, h). Rather, we found high JNK and Yki activity and upd3 expression in ECs surrounding ISC tumors. To determine if EC detachment from the BM and VM was involved in the induction of JNK and Yki activity and Upd3 expression, we first scored how detachment correlated with these markers. Tumor-adjacent ECs positive for puc-lacZ, ex-lacZ or upd3.1-lacZ were scored in three categories: fully attached, partially detached or fully detached. upd3.1-lacZ was observed nearly exclusively in fully or partially detached ECs, whereas puc-lacZ and ex-lacZ were also observed in fully attached tumor-adjacent ECs (Fig 7a, c). When we scored only very strong signals for these markers, the trends were more obvious: high upd3.1-lacZ signal was predominantly confined to partially and fully detached ECs, whereas high Jnk and Yki signals were less tightly correlated with detachment and spread further from the tumors (Fig 7b, Fig 4e–e’, g–g’). These data indicate that JNK and Yki activity and upd3 expression are induced in detached ECs, and also that the induction of JNK and Yki activity can spread to neighboring fully attached ECs (Supplementary Fig. 8a).
Figure 7. Tumor- induced enterocyte detachment induces JNK and Yki activity and Upd3 expression.
(a) Mean percent with s.e.m. of β-galactosidase (β-gal)- positive N− tumor-adjacent enterocytes that were fully detached, partially detached or fully attached in midguts bearing upd3.1-lacZ (yellow, n=4 z-stacks), puc-lacZ (green, n=3 z-stacks) or ex-lacZ (red, n=3 z-stacks) and expressing GFP and NRNAi with esgts for 3 days.
(b) Mean percent with s.e.m. of N− tumor-adjacent enterocytes in each category (fully detached, partially detached and fully detached) that had high β-galactosidase positivity, in midguts bearing upd3.1-lacZ (yellow, n=4 z-stacks), puc-lacZ (green, n=3 z-stacks) or ex-lacZ (red, n=3 z-stacks) and expressing GFP and NRNAi with esgts for 3 days.
(c) β-galactosidase (red) in midguts bearing upd3.1-lacZ expressing GFP (green) and NRNAi with esgts for 3 days.
(d) β-PS1 integrin (d; d’, magenta) in midguts bearing puc-lacZ and expressing Mmp-1 and GFP (d’, green) with Myo1Ats for 2 days. β-galactosidase (d’’; d’’’, red (nuclear)) in ECs and phalloidin (Actin) (d’’; d’’’, red) in VM of midguts bearing puc-lacZ and expressing Mmp-1 and GFP (d’’’, green) with Myo1Ats for 2 days.
(e) β-galactosidase (e; e’, red) in ECs and phalloidin (e’, white) in VM of midguts bearing ex-lacZ and expressing Mmp1 and GFP (e’, green) with Myo1Ats for 2 days.
f) β-galactosidase (f; f’, red) in ECs of midguts bearing upd3.1-lacZ and expressing Mmp1 and GFP (f’, green) with Myo1Ats for 2 days.
In a–b, z-stacks acquired from 2 independent experiments. DNA in c’, d’, d’’’, e’ and f’ (blue). Scale bars in c–c’, 40µm; d–d’’’, 35µm; e–e’, 25 µm; f–f’, 60µm.
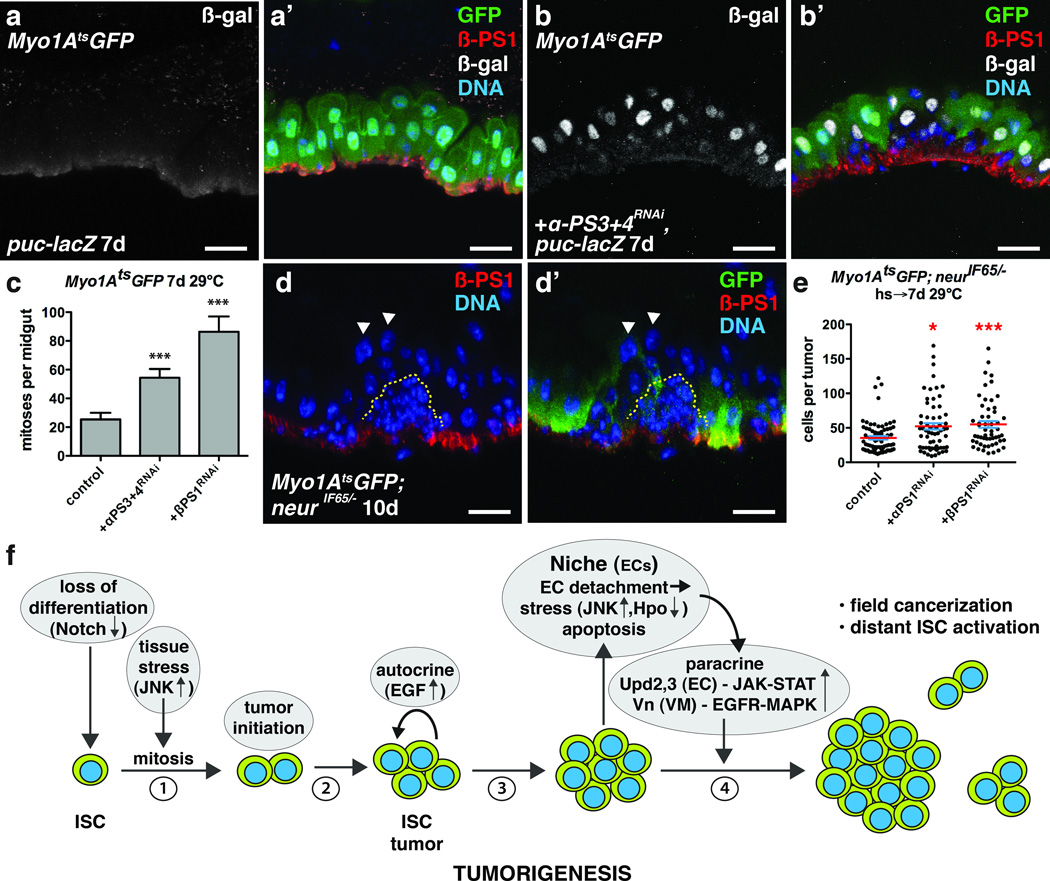
To determine if EC detachment is sufficient to induce JNK and Yki activity in ECs we expressed the matrix metalloproteinase Mmp-1, which is known to cleave ECM components. We also depleted integrins, which mediate cell adhesion to the ECM, from ECs. Loss of integrins from ISCs has been shown to affect their asymmetric division, proliferation and maintenance40–42, whereas loss of β integrin subunits from ECs can induce ISC proliferation42. MMP-1 expression in ECs resulted in their detachment from the VM (Fig. 7d–d’’’). Detaching ECs in this way caused them to lose βPS1 (Myospheroid, Mys) and to induce JNK and Yki activity (Fig. 7d–d’’’, e–e’) and upd3 expression (Fig. 7f–f’). Interestingly, we also found that ECs detached by tumors had reduced βPS1 expression (Fig. 8d–d’). Our mRNA-seq analysis of ECs revealed that in addition to βPS1 (mys), αPS1 (multiple edematous wing, mew) and αPS3 (scab, scb) were highly expressed in ECs (Supplementary Fig. 8b–c’’’’’), consistent with previous reports41. We found that depleting βPS1, αPS1 or both αPS3 and αPS4, but not αPS2 (inflated, if) in ECs, increased the number of detaching ECs and induced ISC proliferation (Fig. 8a–c). These ECs showed decreased basal surfaces and increased JNK activity (Fig. 8b–b’). JNK activity was often observed in partially detached ECs (Fig. 7a, 8b–b’) suggesting that decreased ECM adhesion, rather than decreased proximity to the VM, induces stress signaling. Notably, activating JNK directly by expressing HepAct in ECs did not result in their rapid detachment, indicating that JNK activation is not likely to be causal for EC detachment. Together these data demonstrate that EC detachment from the BM is sufficient to stimulate JNK and Yki activity, and upd3 expression (Supplementary Fig. 8a).
Figure 8. Integrin loss from enterocytes induces JNK activity and promotes ISC tumor growth.
(a–b) β-galactosidase (nuclear) (a,b ; a’,b’, white) and β-PS1 (a’, b’, red) in ECs of midguts bearing puc-lacZ and expressing GFP (a’, green) or GFP and αPS3+4RNAi (b’, green) with Myo1Ats for 7 days.
(c) Mean number with s.e.m. of phosphorylated histone H3 Ser10 positive cells per midgut in flies expressing GFP (control, n=18 midguts), GFP and αPS3+4RNAi (n=19 midguts, Mann-Whitney test: p<0.0006) or βPS1RNAi (n=17 midguts, Mann-Whitney: p<0.0004) with Myo1Ats for 7 days.
(d) β-PS1 integrin (d, d’, red) in midguts bearing neurIF65/− tumors (yellow dashed line) and expressing GFP (d’, green) with Myo1Ats for 10 days. Arrowheads indicate detached ECs apical to tumors that have lost basal β-PS1 expression.
(e) Cells per neurIF65/− tumor with mean (red line) and s.e.m. amongst ECs expressing either GFP (control, n=74 tumors from 35 midguts, skewness= 1.876, kurtosis= 4.283) or GFP and αPS1RNAi (n=63 tumors from 10 midguts, Mann-Whitney: p= 0.0116, skewness= 1.221, kurtosis= 1.126) or GFP and βPS1RNAi (n=54 tumors from 23 midguts, Mann-Whitney: p= 0.0004, skewness= 1.262, kurtosis= 1.093) with Myo1Ats for 7 days.
(f) Model for Notch-dependent tumorigenesis in the adult Drosophila midgut.
In c and e, midguts pooled from 3 independent experiments. DNA in a’, b’, d–d’ (blue). The scale bars in a-b’ and d–d’, 20 µm.
ISC tumors and enterocytes compete for the substratum
A previous study in the fly midgut found that reducing integrins in N− or Apc− tumor cells inhibited their growth, and that these tumors were eliminated from the epithelium41. We confirmed this interesting observation, which suggests that tumor cells may compete with normal cells for BM attachment. To test the importance of tumor/EC competition in another way, we generated tumors amongst ECs in which integrins were depleted by targeted RNAi. We expected that this might reduce the ability of these ECs to compete with the tumor cells for BM attachment. Indeed, we found that these treatments enhanced tumor growth (Fig. 8e). Together these data support the idea that tumor cells compete with ECs for attachment to the BM, via integrin-mediated adhesion.
Discussion
Here we describe a step-wise series of events during the earliest stage of tumor development in a stem cell niche (Fig. 8f). First, the combination of environmentally triggered mitogenic signaling and a mutation that compromises differentiation generates small clusters of differentiation-defective stem-like cells. Autocrine (Spi/EGRF) signaling between these cells then promotes their expansion into clusters, which quickly reach a size capable of physically disrupting the surrounding epithelium and driving the detachment and apical extrusion of surrounding epithelial cells (ECs). This loss of normal cells appears to involve tumor cell/epithelial cell competition via integrin-mediated adhesion. Subsequently the loss of epithelial integrity (specifically, EC detachment) triggers stress signaling (JNK, Hpo/Yki) in the surrounding epithelium and underlying visceral muscle, and these stressed tissues respond by producing cytokines (Upd2,3) and growth factors (Vn, Pvf, Wg, dILP3). These signals are normally used within the niche to activate stem cells for epithelial repair, but in this context they further stimulate tumor growth in a positive feedback loop. It is noteworthy that in this example a single mutation that blocks differentiation is sufficient to drive early tumor development, even without secondary mutations in growth signaling pathways that might make the tumor initiating cells growth factor- and niche-independent (e.g. Ras, PTEN). Thus tumor cell-niche interactions can be sufficient to allow tumor-initiating cells to rapidly expand, increasing their chance to acquire secondary mutations that might enhance their growth or allow them to survive outside their normal niche8. Our study highlights the importance of investigating the factors that control paracrine stem cell mitogens and survival signals in the niche environment. Tumor-niche interactions may be important to acquire a sizable tumor mass prior to the recruitment of a tumor-specific microenvironment that supports further tumor progression. A careful analysis of similar interactions in other epithelia, such as in the lung, skin or intestine could yield insights relevant to the early detection, treatment, and prevention of cancers in such tissues.
Methods
Fly stocks
All experiments were performed using 5–10 day old, adult female Drosophila melanogaster. The following fly stocks were used: esgGAL4; tubGAL80ts UAS-GFP (esgts), myo1AGAL4; tubGAL80ts UAS-GFP (myo1Ats), tubGAL80ts UAS-GFP; how(24B)GAL4, Dl05151 (Dl-lacZ), Gbe-Su(H)-lacZ, pucE69 (puc-lacZ), 10XSTAT-DGFP, upd3.1-lacZ, vnP1749(vn-lacZ), UAS-CycE, UAS-hepAct, UAS-Mkp3, UAS-p35, UAS-RasV12G, UAS-stg, UAS-upd, FRT82B and the MARCM 82B stock: yw hsFLP UAS-GFP tubGAL4; +; FRT82B tubGAL80 and were previously described in Jiang et al., 2009, 2011. yw hsFLP; myo1AGAL4 tubGAL80ts UAS-GFP; FRT82B neurIF65 was generated for this study. FRT 82B neurIF65 was obtained from Hugo Bellen (Baylor College of Medicine, USA); Rab3-YFP from Suzanne Eaton (Max Plank Institute of Molecular Cell Biology and Genetics, Germany); ex697 (ex-lacZ) from Georg Halder (K.U. Leuven, Belgium); upd1-lacZ, Gbe-Su(H)GAL4; tubGAL80ts UAS-GFP and tubGAL80ts; Dl-GAL4, UAS-GFP from Steven Hou (National Cancer Institute, USA); UAS-hep, UAS-bskRNAi (GD34138) and UAS-bskK53R from Heinrich Jasper (Buck Institute, USA); upd2Δ, upd3Δ, upd2-3Δ, UAS-upd2RNAi (1) (NIG 5988R-3) and UAS-upd3RNAi (1) (Agaisse et al., 2003) from Bruno Lemaitre (EPFL, Switzerland); UAS-puc from Donald McEwen (UTHSC, San Antonio, USA); UAS-Mmp1 from Andrea Page-McCaw (Vanderbilt University, USA); esgGAL4 tubGAL80ts UAS-GFP from Norbert Perrimon (Harvard Medical School, USA) and UAS- domeΔCYT from Tian Xu (Yale University, USA). UAS-DlRNAi (3720GD), UAS-mysRNAi (29619GD), UAS-NRNAi (100002KK and 27228GD), UAS-αPS3-4RNAi (4891GD), UAS-SuHRNAi (103597KK), UAS-upd2RNAi (2) (14664GD), UAS-upd3RNAi (2) (106869KK) and UAS-ykiRNAi (104523KK) were obtained from the Vienna Drosophila RNAi Center (VDRC). UAS-sc, UAS-wts, UAS-EgfrRNAi (TRiP.JF01696) and UAS-spiRNAi (TRiP.JF03322) were obtained from the Bloomington Drosophila Stock Center. UAS-mewRNAi (1771R-3) was obtained from the National Institute of Genetics (NIG), Japan.
Drosophila genetics
Flies raised at 18°C were shifted to 29°C to induce UAS transgene expression in progenitor cells with esgGAL4; tubGAL80ts (esgts) or in ECs with the myo1AGAL4; tubGAL80ts (myo1Ats). To generate MARCM clones, flies were heat shocked at 37°C for 30 minutes and kept at 25°C until dissection. For myo1Ats; FLP-FRT experiments, flies were heat shocked as described above to generate neurIF65 FLP-FRT tumors and allowed to recover 1 day prior to a second heat shock. After recovery for 1 day, flies were then shifted for 7–8 days to 25°C or 29°C to allow UAS transgene expression in ECs with myo1Ats. Adult flies were orally infected with Pseudomonas entomophila as described in Jiang et al., 2009. No statistical method was used to predetermine sample size but typically between 5–20 flies were used per experiment. When selecting animals for an experiment, the parental genotype was not concealed since it was required to select pertinent progeny. Animals were first selected for their genotype and then randomly chosen to be used for an experiment.
Histology
After dissection and 8% paraformaldehyde fixation, midguts were washed in PBS, 0.1% Triton X-100, blocked in PBS, 0.1% Trinton X-100, 1% bovine serum albumin (BSA), 2% normal goat serum (NGS) and stained in blocking solution with mouse monoclonal anti-β–PS1 integrin (clone #CF.6G11, 1:20, Lin et al., 2013), anti- Delta (clone #C594.9B, 1:100, Ohlstein and Spradling, 2007), anti-Discs Large (clone #4F3, 1:100) or anti-Prospero (clone #MR1A, 1:100, Micchelli and Perrimon, 2006) from the Developmental Studies Hybridoma Bank (DHSB), rabbit anti-Prospero (1:2000, from Yuh Nung Jan, UCSF, USA, Bardin et al., 2010), rabbit anti-Pdm-1 (1:1000, from Xiaohang Yang, Institute of Molecular and Cell Biology, Singapore, Jiang et al., 2011), rabbit polyclonal anti-phospho Ser10 histone 3 (1:1000, Upstate Biotechnology/Millipore, #06-570), rabbit polyclonal anti-β-galactosidase (1:1000, Cappel), chicken polyclonal anti-β-galactosidase (1:1000, Abcam, #ab9361), mouse monoclonal anti-diphospho-ERK (clone# MAPK-YT, 1:200, Sigma, #M8159) and rabbit or chicken polyclonal anti-GFP (1:1000, Life Technologies, #A11122, #A10262). Midguts were then washed in PBS 0.1% Triton X-100, stained in PBS, 0.3% Triton X-100, 0.1% BSA with Alexa Fluor- conjugated secondary antibodies (Life Technologies), Alexa Flour 647- phalloidin (Life Technologies) and Hoechst 33258 (Life Technologies), and mounted in Vectashield (Vector Laboratories).
Mitotic indices were determined by counting the number of pH3 positive cells from whole female midguts from 2–3 independent experiments. The mean number of mitoses per midgut and s.e.m. are presented for each genotype or treatment. Cells per tumor were determined by counting the total number of nuclei within Dl+ Pros+ tumors ≥ 8 cells. The mean cell number per tumor obtained from 3 independent experiments and s.e.m. are presented for each genotype. Before quantifying the number of mitoses per midgut or the number of cells per tumor, the genotype of each sample was concealed. Samples were then randomly analyzed and the genotype was revealed only after completing analysis. Tumor frequency was determined by counting the number of tumors per midgut in affected flies; the mean number of tumors per midgut obtained from 3 independent experiments and s.e.m. is presented for each genotype. Tumor incidence was determined by counting the number of flies (or midguts) with tumors in the population; the mean percent of affected flies obtained from 3 independent experiments and s.e.m. is presented for each genotype.
Statistical Analysis
Statistical analyses, including the determination of skewness and kurtosis for each tumor population, were performed using GraphPad Prism 5. For data describing mitoses per midgut, cells per tumor, tumor frequency, the Mann-Whitney test (two-sided) was applied to determine statistical significance. For data describing tumor incidence or mitoses at the tumor interior/boundary, the paired t-test (two-sided) was applied to determine statistical significance.
In situ hybridization
Fluorescent in situ hybridization for upd3 mRNA was performed as described in Jiang et al., 2011. Midguts were dissected, fixed in 8% paraformaldehyde/PBS overnight at 4°C, permeabilized 3 times for 10 minutes in PBST (PBS, 0.1% Triton X-100) and then stored in 70% ethanol. Midguts were then hybridized with a fluorescently labeled (CAL Flour Red 590) DNA 20-mer probes (pool of 48) (Stellaris) against upd3 transcript.
Light microscopy
Samples were analyzed using Nikon Eclipse Ti, Leica DM5000B, Zeiss LSM 510 and Leica SP5 microscopes. Images were processed with ImageJ (NIH) and Adobe Photoshop CS5. Confocal images are presented as maximal intensity projection of images obtained every 0.25–1.0µm. When comparing protein or transcript levels, each z- stack was acquired with the same laser intensity and gain, maximal intensity projections of identical dimensions were created, and were further similarly processed with Adobe Photoshop CS5, except in the case of Supplementary Fig. 5j which was significantly brightened. Representative images presented were obtained from ≥2 independent experiments.
Transmission electron microscopy
Midguts were dissected, fixed in 0.5× Karnovsky’s fixative and post-fixed in OsO4. Midguts were then dehydrated in ascending concentrations of ethanol, embedded in epon and sectioned to obtain 70–90nm sections. Samples were analyzed with a JEOL 1230 transmission electron microscope with an Orius SC1000 Gatan CCD.
mRNA sequencing of whole midguts and sorted midgut cell populations
Whole midgut RNA was isolated from 15 guts using RNeasy RNA isolation kit (Qiagen) following manufacturer’s protocol. Cell type specific profiling was performed as previously described43. Briefly, 100 guts were dissected in RNAse free PBS and treated with 7.5 mg/ml collagenase or 4 mg/ml elastase for 1 hour at 27°C. Dissociated cells were pelleted at 300×g for 15 mins, resuspended in 1× RNAse free PBS, filtered using 25-micron filters (BD Falcon) and sorted using a FACS Aria II sorter (BD Biosciences) with 70 micron nozzle size. To exclude auto fluorescence, gates were set using control midguts from w1118 flies. Cell type- specific GAL4 drivers (esg-GAL4 (ISCs and enteroblasts), Dl-GAL4 (ISCs), Su(H)-GAL4 (enteroblasts), myo1A-GAL4 (enterocytes), how-GAL4 (visceral muscle) and Rab3-YFP and pros-GAL4 (EEs) were used to express green or yellow fluorescent protein (GFP or YFP) to sort each cell population. GFP or YFP- positive cells were sorted based on fluorescence intensity and cell size. A total of 2000 cells were sorted for each sample. Total RNA from each sorted cell type was isolated using the PicoPure RNA isolation kit (Arcturus). 2ng of isolated total RNA was used for RNA amplification using Arcturus® RiboAmp® HS PLUS RNA Amplification Kit for whole midgut and cell type- specific RNA profiling. Total RNA from each sample was reverse transcribed using a T7 promoter sequence containing oligodT primer and SuperScript®III Reverse Transcriptase (200-U) enzyme. Random hexamers were used for the second strand synthesis. aRNA was then produced by in vitro transcription using T7 RNA polymerase at 42°C for 6hrs. The amplified RNA (aRNA) integrity was determined with an Agilent 2100 bioanalyzer, enriched for 200–400bp mRNA and used directly for RNA sequencing. Amplified mRNA was then shattered by magnesium-catalyzed hydrolysis and used for cDNA library preparation. Adaptors were ligated and mRNA sequencing was performed using an Illumina HiSeq2000 sequencer with 50 bp read length.
Data analysis
all reads were inspected using Fastqc version 01.0.1 with default settings. Since no trimming was needed, the raw reads were mapped to the reference genome version 70 (ENSEMBL) using tophat2 (version 2.0.9) with default parameters (2 mismatches allowed), boost library 1.54.0, bowtie2 2.1.0 and samtools version 0.1.19. The resulting bam files were converted to sam files using samtools, and subsequently counted using HTSeqcount (0.5.4p5). Ambiguous reads, reads with low quality, unaligned reads, reads with non-unique alignment and reads aligning to non-features were discarded. Differential expression analysis was conducted using edgeR (3.2.4) with filtering for low expressed genes showing a cpm value above 1 in two biological replicates. P-values were adjusted using Benjamini-Hochberg correction. Genes with a cpm >1, a 1.5-fold change and an adjusted p-value <0.05 were considered significantly deregulated. The mean normalized expression (RPKM, log2) value from 2 independent experiments, adjusted fold change and adjusted p-value for each gene is presented. A Principle Component Analysis (PCA) was conducted using the function prcomp in the stats package of R (3.1.0) with scaling the variables to have unit variance and zero centering based on the mean RPKM values per cell type. Only genes showing RPKM values greater than zero were used for calculation.
Quantitative RT-PCR
RNA was isolated from 15–20 midguts either using TRIzol or RNAeasy kit (Qiagen), 250–500ng of RNA was used for cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad) or the Quantitect cDNA synthesis kit (Qiagen). qPCR was performed using the iScript One Step RT-PCR kit with SYBR green (BioRad) or with LightCycler 480 SYBER Green I Master (Roche). SYBR green incorporation during PCR was detected using the iQ5 system (BioRad) or Roche 480 II Lightcycler. The following primers were used: upd1 F: 5’-CCACGTAAGTTTGCATGTTG-3’, upd1 R: 5’-CTAAACAGTAGCCAGGACTC-3’, upd2 F: 5’-CACAAGTGCGGTGAAGCTAA-3’, upd2 R: 5’-GGCTCTTCTGCTGATCCTTG-3’, upd3 F: 5’-GCCCTCTTCACCAAACTGAA-3’, upd3 R: 5’-TTTCTTCTGGATCGCCTTTG-3’, spi F: 5’-CCTTCTATTTGCGCTTCGAG-3’, spi R: 5’-CGCATGTGGTAGGGTAGCTT-3’, krn F: 5’-CGTGTTTGGCAACAACAAGT-3’, krn R: 5’-TGTGGCAATGCAGTTTAAGG-3’, vn F: 5’-AACGCAGAGGTCACGAAGAT-3’, vn R: 5’-GCGCACTATTAGCTCGGAAC-3’, Dl F: 5’-TCTGTTTTAGGCGAGGGTTC-3’, Dl R: 5’- AAGCTGCAGCCATTAGTTGC-3’, Ast F: 5’-CCTGCCGGTCTATAACTTCG-3’ and Ast R: 5’- GATCTCGTTGTCCTGGTCGT-3’. We found that crq gene expression did not change after ISC tumor growth and thus the expression of each target gene was normalized to crq expression (reference) to ensure equivalent RNA or cDNA input. The primers used to detect crq expression were: crq F: 5’-CAGAGCTCTCCTCCGAATTG-3’, crq R: 5’-ATGCCGGTGATGAGAAAGAC-3.’ Each assay was performed in triplicate or in quadruplet on ≥ 3 independent biological replicates. Since primer efficiency (E)= 1, the relative expression of each gene was determined using the ΔΔCt method, where ΔΔCt (or log2 fold change) is the difference in threshold cycles for the experimental and control samples normalized to the threshold cycles for the reference gene.
ΔΔCt = ΔCt (experiment) -ΔCt (control)
ΔCt (experiment) or ΔCt (control) = Ct target gene - Ct reference gene
All data are presented as mean fold change (log2) with s.e.m.
Quantitation of stress reporter- positive ECs surrounding tumors
Fixed midguts were stained with anti- Dlg to outline the lateral boundary of ECs and with anti- Pros to mark EE cells. For upd3.1-lacZ, cytoplasmic β-gal signal was used to determine the lateral EC boundary. z-stacks (0.5µm step size) of posterior midgut regions (387.5µm × 387.5µm) bearing N− tumors were obtained by confocal microscopy. 3–4 z- stacks were analyzed per genotype. β-gal- positive ECs were classified into the following categories: 1) fully detached ECs (fd): ECs for which tumor cells were found along the entire basal surface of the EC; 2) partially detached ECs (pd), ECs for which tumor cells were observed basally to the EC but not along the entire basal surface; and 3) fully attached ECs (fa), ECs for which no tumor cells were found along their basal surface. Only β-gal- positive ECs that bordered a cluster of esg+ ISC tumor cells were considered tumor- adjacent, and were scored. Rare non- tumor adjacent β-gal- positive ECs were observed but are not reported. upd3.1-lacZ and ex-lacZ were not observed in esg+ tumors cells; puc-lacZ was also observed in EE-like esglow Proshigh tumor cells and in Pros+ EE cells, but these were excluded from the analysis. β-gal intensity (high or low) in ECs was determined prior to its classification as fully attached, partially detached or fully detached.
Survival analysis
100 adult flies per genotype were transferred to food vials (10 flies/vial) and shifted to 29°C. The number of dead flies in each vial were scored every 2 days and surviving adults were transferred to fresh food vials. Flies expressing NRNAi with esgts were scored until 50 flies were found dead. For all other genotypes, the number of dead flies was scored for 14 days.
Antibiotic treatment and detection of enteric bacteria levels
50 adults were fed for 3–4 days at 22°C normal fly food or food containing antibiotics (10,000U/ml penicillin, 10mg/ml streptomycin (1:150). Midguts were dissected from 20–25 flies, homogenized and used for total genomic DNA isolation. gDNA was then used for PCR for bacterial 16S rDNA with the following primers: F: 5’-AGAGTTTGATCCTGGCTCAG-3’ and R: 5’- GGTTACCTTGTTACGACTT-3’. 20–25 flies from the same cohort were shifted to 29°C for 2–3 days to express either GFP or GFP and NRNAi with esgts to assess tumor growth in midguts with normal or low luminal bacteria levels.
Accession Code
The raw mRNAseq data for control midguts (esgts> GFP), midguts with tumors (esgts> NRNAi, GFP), esg+ cells sorted from control midguts (esgts> GFP) and esg+ cells sorted from tumorous midguts (esgts> NRNAi, GFP) has been deposited at the National Center for Biotechnology Information Sequence Read Archive (NCBI- SRA) under the primary accession number, SRP049937.
Supplementary Material
Acknowledgements
We thank the Bloomington Drosophila Stock Center, Vienna Drosophila RNAi Center (VDRC), National Institute of Genetics (Japan), Developmental Studies Hybridoma Bank, Hugo Bellen, Suzanne Eaton, Georg Halder, Steven Hou, Yuh Nung Jan, Heinrich Jasper, Andrea Page-McCaw, Bruno Lemaitre, Donald McEwen, Nobert Perrimon, Tian Xu and Xiaohang Yang for fly stocks and antibodies. We thank Bobbie Schneider and Judy Bousman (FHCRC Electron Microscopy Resource), Monika Langlotz (ZMBH Flow Cytometry), David Ibberson (Deep Sequencing Core Facility, U. Heidelberg), Jeff Delrow (FHCRC Sequencing Shared Resource) and Christine Gläser (ZMBH, Bioinformatics). Funded by American Cancer Society Postdoctoral Fellowship (PF-08-040-01-DDC) to P.H.P. and NIH R01 GM51186, DKFZ A220, DFG SFB 873, and ERC Advanced Grant 26515 to B.A.E.
Footnotes
Author Contributions
The project was conceived and developed by P.H.P. and B.A.E. P.H.P. contributed to Figures 1–8, Supplementary Figures 1–8. D.D. contributed to Figure 2g–h, 6e, Supplementary Figs. 1g–k, 2a, 2c–d, 4b–c, 5b, 8b. mRNA-seq data was analyzed by both P.H.P. and D.D.. P.H.P and B.A.E. prepared the manuscript.
Competing financial interests
Authors declare no competing financial interests.
References
- 1.Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature. 2006;439:470–474. doi: 10.1038/nature04333. [DOI] [PubMed] [Google Scholar]
- 2.Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature. 2006;439:475–479. doi: 10.1038/nature04371. [DOI] [PubMed] [Google Scholar]
- 3.Jiang H, Edgar BA. Intestinal stem cells in the adult Drosophila midgut. Exp Cell Res. 2011 doi: 10.1016/j.yexcr.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- 5.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perdigoto CN, Schweisguth F, Bardin AJ. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development. 2011;138:4585–4595. doi: 10.1242/dev.065292. [DOI] [PubMed] [Google Scholar]
- 7.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 10.Patel PH, Edgar BA. Tissue design: How Drosophila tumors remodel their neighborhood. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Cordero JB, Stefanatos RK, Myant K, Vidal M, Sansom OJ. Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development. 2012;139:4524–4535. doi: 10.1242/dev.078261. [DOI] [PubMed] [Google Scholar]
- 12.Zhou J, et al. Dpp/Gbb signaling is required for normal intestinal regeneration during infection. Dev Biol. 2015;399:189–203. doi: 10.1016/j.ydbio.2014.12.017. [DOI] [PubMed] [Google Scholar]
- 13.Apidianakis Y, Pitsouli C, Perrimon N, Rahme L. Synergy between bacterial infection and genetic predisposition in intestinal dysplasia. Proc Natl Acad Sci U S A. 2009;106:20883–20888. doi: 10.1073/pnas.0911797106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 15.Durand A, et al. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci U S A. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maeda K, Takemura M, Umemori M, Adachi-Yamada T. E-cadherin prolongs the moment for interaction between intestinal stem cell and its progenitor cell to ensure Notch signaling in adult Drosophila midgut. Genes Cells. 2008;13:1219–1227. doi: 10.1111/j.1365-2443.2008.01239.x. [DOI] [PubMed] [Google Scholar]
- 17.Bardin AJ, Perdigoto CN, Southall TD, Brand AH, Schweisguth F. Transcriptional control of stem cell maintenance in the Drosophila intestine. Development. 2010;137:705–714. doi: 10.1242/dev.039404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H, et al. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell. 2009;137:1343–1355. doi: 10.1016/j.cell.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 2009;23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staley BK, Irvine KD. Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol. 2010;20:1580–1587. doi: 10.1016/j.cub.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;137:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ren F, et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci U S A. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karpowicz P, Perez J, Perrimon N. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;137:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlmaier A, et al. Src kinase function controls progenitor cell pools during regeneration and tumor onset in the Drosophila intestine. Oncogene. 2014;0 doi: 10.1038/onc.2014.163. [DOI] [PubMed] [Google Scholar]
- 25.Jiang H, Grenley MO, Bravo MJ, Blumhagen RZ, Edgar BA. EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell. 2011;8:84–95. doi: 10.1016/j.stem.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu N, et al. EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev Biol. 2011;354:31–43. doi: 10.1016/j.ydbio.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 27.Buchon N, Broderick NA, Kuraishi T, Lemaitre B. Drosophila EGFR pathway coordinates stem cell proliferation and gut remodeling following infection. BMC Biol. 2010;8:152. doi: 10.1186/1741-7007-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biteau B, Jasper H. EGF signaling regulates the proliferation of intestinal stem cells in Drosophila. Development. 2011;138:1045–1055. doi: 10.1242/dev.056671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amcheslavsky A, Jiang J, Ip YT. Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell. 2009;4:49–61. doi: 10.1016/j.stem.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.You J, et al. Drosophila perlecan regulates intestinal stem cell activity via cell-matrix attachment. Stem Cell Reports. 2014;2:761–769. doi: 10.1016/j.stemcr.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenhoffer GT, et al. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 2012;484:546–549. doi: 10.1038/nature10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W, Singh SR, Hou SX. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. 2010;109:992–999. doi: 10.1002/jcb.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bond D, Foley E. Autocrine Platelet-derived Growth Factor-Vascular Endothelial Growth Factor Receptor-related (Pvr) Pathway Activity Controls Intestinal Stem Cell Proliferation in the Adult Drosophila Midgut. J Biol Chem. 2012;287:27359–27370. doi: 10.1074/jbc.M112.378018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi NH, Lucchetta E, Ohlstein B. Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc Natl Acad Sci U S A. 2011;108:18702–18707. doi: 10.1073/pnas.1109348108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Brien LE, Soliman SS, Li X, Bilder D. Altered modes of stem cell division drive adaptive intestinal growth. Cell. 2011;147:603–614. doi: 10.1016/j.cell.2011.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin G, Xu N, Xi R. Paracrine Wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature. 2008;455:1119–1123. doi: 10.1038/nature07329. [DOI] [PubMed] [Google Scholar]
- 37.Cordero JB, Stefanatos RK, Scopelliti A, Vidal M, Sansom OJ. Inducible progenitor-derived Wingless regulates adult midgut regeneration in Drosophila. EMBO J. 2012 doi: 10.1038/emboj.2012.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beebe K, Lee WC, Micchelli CA. JAK/STAT signaling coordinates stem cell proliferation and multilineage differentiation in the Drosophila intestinal stem cell lineage. Dev Biol. 2010;338:28–37. doi: 10.1016/j.ydbio.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 39.Lin G, Xu N, Xi R. Paracrine unpaired signaling through the JAK/STAT pathway controls self-renewal and lineage differentiation of drosophila intestinal stem cells. J Mol Cell Biol. 2010;2:37–49. doi: 10.1093/jmcb/mjp028. [DOI] [PubMed] [Google Scholar]
- 40.Goulas S, Conder R, Knoblich JA. The Par complex and integrins direct asymmetric cell division in adult intestinal stem cells. Cell Stem Cell. 2012;11:529–540. doi: 10.1016/j.stem.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin G, et al. Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev Biol. 2013;377:177–187. doi: 10.1016/j.ydbio.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 42.Okumura T, Takeda K, Taniguchi K, Adachi-Yamada T. betanu integrin inhibits chronic and high level activation of JNK to repress senescence phenotypes in Drosophila adult midgut. PLoS One. 2014;9:e89387. doi: 10.1371/journal.pone.0089387. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 43.Dutta D, Xiang J, Edgar BA. RNA expression profiling from FACS-isolated cells of the Drosophila intestine. Curr Protoc Stem Cell Biol. 2013;27(Unit 2F 2) doi: 10.1002/9780470151808.sc02f02s27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.