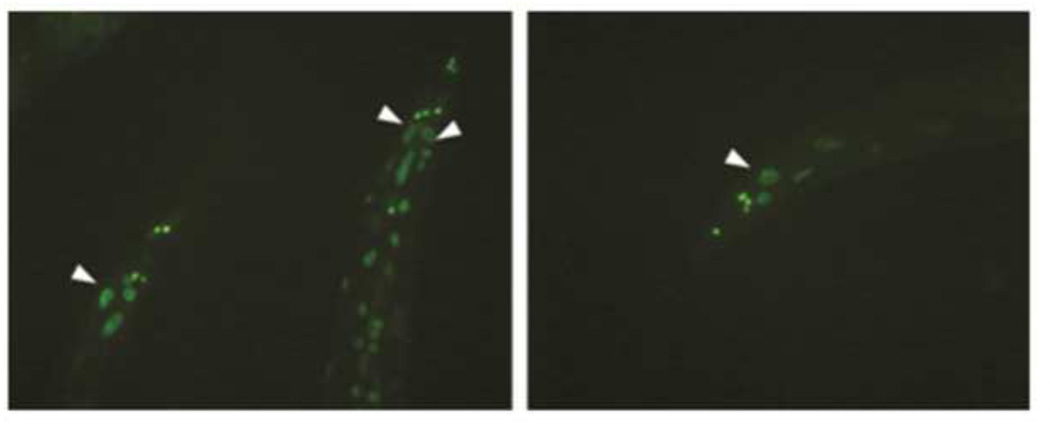
Tonsaker et al. state that there are two issues with the results previously published by us in Budovskaya et al. (2008). The first is whether elt-3 is expressed in the intestine in C. elegans. Tonsaker et al. claim that there is no elt-3 expression and we claimed that there is low level expression in the intestine. Figure 1 shows elt-3 expression from an elt-3::GFP transcriptional reporter used in Budovskaya et al., 2008 that includes 1999 bp from the upstream region of elt-3 driving expression of H2B::GFP. We consistently see elt-3 expression from this transcriptional reporter in the posterior part of the intestine.
Figure 1.
Expression of elt-3::H2B::GFP (SD1276 (pha-1(e2123)III, Ex[elt-3pro::GFP::H2B; pha-1(+)])). The elt-3 promoter includes the 2kb upstream of K02B9.4 transcript (genomic location X:13927330..13929329bp). The figure shows worms at day 2 of adulthood, and the arrowheads show GFP expression in the posterior two intestinal cells. Figures are from data used in Budovskaya et al., 2008.
We feel that the issue regarding intestinal expression of elt-3 in the intestine is not relevant to the main points of Budovskaya et al. 2008. We did not state that elt-3 functions in the intestine to modulate aging, and we feel that Tonsaker et al., have misinterpreted our statements. elt-3 may be expressed at a low level (our view) or not at all (their view) in the intestine, but in either case this does not address whether elt-3 functions in the intestine. Genetic mosaic analysis is required to show in which tissue elt-3 functions, but this analysis has not yet been performed to our knowledge. In principle, expression of elt-3 anywhere in the worm could affect expression of genes in the intestine via an indirect, cell non-autonomous fashion.
The second issue is whether elt-3 mutations can suppress the longevity conferred by a daf-2 mutation. This is one of the points used in Budovskaya et al., 2008 to argue that elt-3 has a role in worm aging. In Supplemental Table 1 of Budovskaya et al., 2008, we showed four examples of elt-3 suppression: three times using elt-3(RNAi) and one time using the elt-3(vp1) null allele performed by K.W. in the lab.
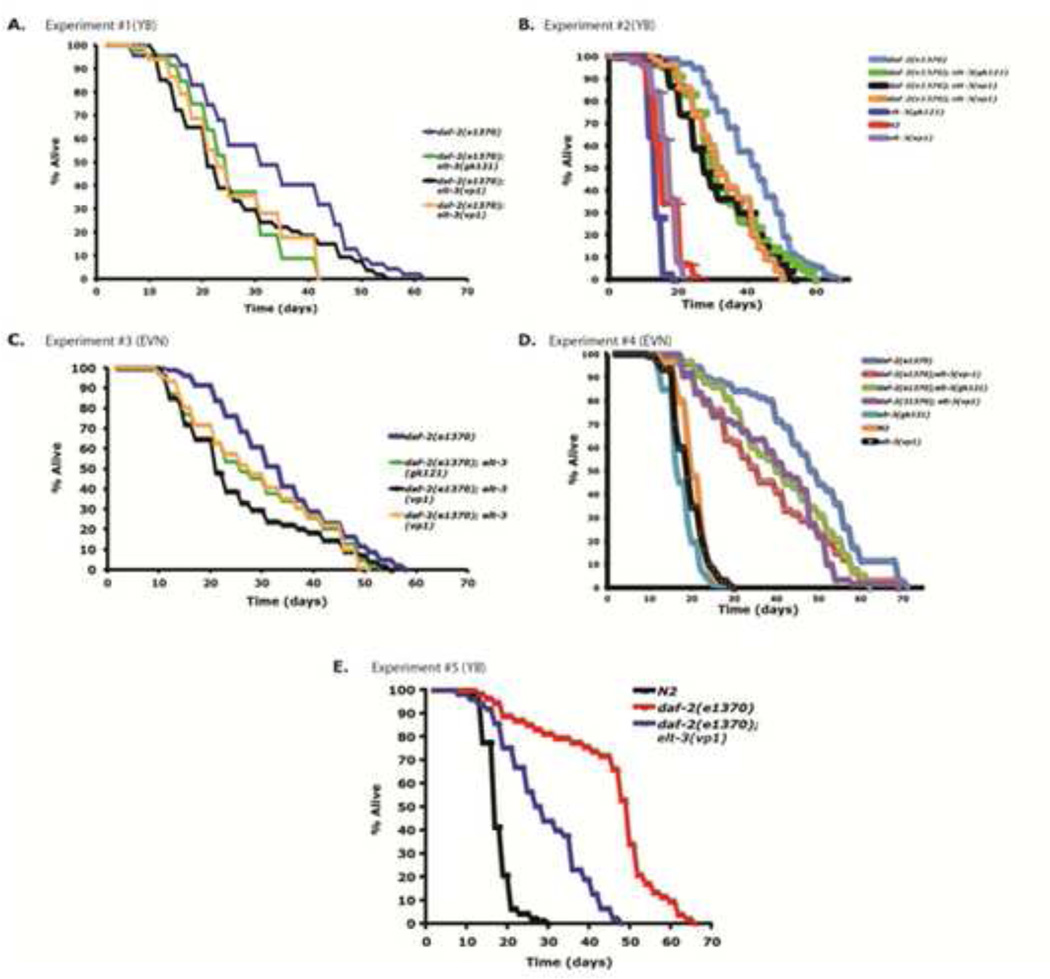
Since 2008, we have replicated our results five times. Figure 2E shows one replicate using the same daf-2(e1370); elt-3(vp1) strain (SD1294) used in Budovskaya et al., 2008 performed by YB in the lab. This experiment confirms that daf-2(e1370); elt-3(vp1) has a shorter lifespan than daf-2(e1370).
Figure 2.
elt-3(vp1) suppresses longevity conferred by daf-2(e1370). All experiments were performed at 20°C. We used on average 150-200 animals per experiment. L4 larvae stage was counted as day 0 of adulthood. At day 2, all animals were transferred to NGM plates containing FUDR to prevent their progeny from developing. During these experiments, we censored animals that displayed severe bagging, vulva protrusion, and gonad explosion.
Next, we repeated this experiment four times in a blinded experiment performed by two different people in the lab (YB and EVN)(Figure 2). These experiments included two new strains constructed using the elt-3(vp1) null mutation and one strain using the elt-3(gk121) null mutation. Table 1 shows the Mann-Whitney Rank statistics comparing daf-2 to daf-2; elt-3 from experiments 2, 4, and 5 in Figure 2. In all four experiments, we saw that mutations in elt-3 significantly suppress the longer lifespan conferred by daf-2(e1370), confirming results from Budovskaya et al., 2008.
Table 1.
Suppression of the daf-2 longevity phenotype by elt-3
| Experment #2 | p-value |
|---|---|
| daf-2(e1370) vs. daf-2(e1370); elt-3(vp1)#1 | 1.22 × 10−05 |
| daf-2(e1370) vs. daf-2(e1370); elt-3(vp1)#2 | 4 × 10−15 |
| daf-2(e1370) vs. daf-2(e1370); elt-3(gk121) | 9.61 × 10−07 |
| Experment #4 | |
| daf-2(e1370) vs. daf-2(e1370); elt-3(vp1)#1 | 7.57 × 10−5 |
| daf-2(e1370) vs. daf-2(e1370); elt-3(gk121) | 5.41 × 10−5 |
| daf-2(e1370) vs. daf-2(e1370); elt-3(vp1)#2 | 3.1 × 10−4 |
| Experiment #5 | |
| daf-2(e1370) vs. daf-2(e1370);elt-3(vp1) | 5.5 × 10−5 |
We also analyzed the data to determine whether suppression of the daf-2 longevity phenotype by elt-3 was partial or complete. To do this, we compared the lifespan of the daf-2; elt-3 strains to either N2 or elt-3. If the suppression is only partial, then daf-2; elt-3 will live significantly longer than N2 or elt-3. Using Mann-Whitney Rank statistics, we find that suppression is partial rather than complete in every case, indicating that there are likely other genes, besides elt-3, acting downstream of daf-2 to confer longevity (Table 2).
Table 2.
Suppression by elt-3 of the daf-2 longevity phenotype is partial.
| Experment #2 | p-value |
|---|---|
| N2 vs. elt-3(gk121) | 0.000154 |
| N2 vs elt-3(vp1) | 0.162 |
| N2 vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| N2 vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| N2 vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| Experment #4 | |
| N2 vs. elt-3(gk121) | 1.3 × 10−9 |
| N2 vs elt-3(vp1) | 0.0244 |
| N2 vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| N2 vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| N2 vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| elt-3(gk121) vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(vp1)#1 | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(vp1)#2 | 1 × 10−27 |
| elt-3(vp1) vs. daf-2(e1370); elt-3(gk121) | 1 × 10−27 |
| Experiment #5 | |
| daf-2(e1370) vs. N2 | 1 × 10−27 |
| daf-2(e1370);elt-3(vp1) vs. N2 | 1.6 × 10−15 |
In summary, we have compared the lifespan of daf-2; elt-3 to daf-2 a total of nine times and observed partial suppression in every case. These results cannot be due to an artifact from one strain or allele of elt-3 as we have used two elt-3 null alleles as well as elt-3(RNAi). These results are not likely to involve differences in genetic background, as we see suppression using elt-3(RNAi) which uses the same daf-2 genetic background. These differences are not likely to be due to artifacts in scoring from one person, as we obtained similar results performed by three people in the lab.
We are not sure why the elt-3 suppression result from Figure 2 of Tonsaker et al. does not agree with ours, and we are deeply committed to finding out the true answer. We have requested the strains from Dr. McGhee and will perform the appropriate experiments to determine the source of the difference. One possibility is that the daf-2(e1370) strain used by Tonsaker et al. (CB1370) has a background mutation that shortens lifespan itself. This background mutation could mask the effects of an elt-3 mutation on suppressing longevity. Backcross experiments using strains from both labs should reveal the correct result, and we will report this result as soon as possible.
In summary, we disagree with the two issues raised by Tonsaker et al. The first issue on intestinal expression is not relevant to the main results of our paper on the role of elt-3 in worm aging. We again apologize if other readers were misled into thinking that that our model required that elt-3 function in the intestine to modulate aging. The second issue on suppression of daf-2 longevity by mutations in elt-3 is not resolved. Our lab can clearly replicate the published lifespan results and it will be necessary to exchange strains to get at the source of the difference. Besides daf-2 suppression by elt-3, there are many other results that support our model that GATA transcription factors have a role in worm aging, such as lifespan extension by RNAi treatment of elt-5 GATA and elt- 6 GATA, decreasing expression of elt-3 GATA with age, increasing expression of elt-5 GATA and elt-6 GATA with age, decreasing expression of the targets of elt- 3 GATA with age, etc. Thus, we do not think that the points raised by Tonsaker et al. should cause one to discard our model for modulation of lifespan by elt-3. Since 2008, we have continued to study the role of elt-3 in aging and our new results support the main points of Budovskaya et al., 2008 and allow us to modify and extend the original model.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsaker T, Pratt RM, McGhee JM. Re-evaluating the Role of ELT-3 in a GATA Transcription Factor Circuit Proposed to Guide Aging in C. elegans. doi: 10.1016/j.mad.2011.09.006. [DOI] [PubMed] [Google Scholar]