Abstract
As glyphosate is a broad spectrum herbicide extensively used in agriculture worldwide, identification of new aroA genes with high level of glyphosate tolerance is essential for the development and breeding of transgenic glyphosate-tolerant crops. In this study, an aroA gene was cloned from a Janibacter sp. strain isolated from marine sediment (designated as aroAJ. sp). The purified aroAJ. sp enzyme has a Km value of 30 μM for PEP and 83 μM for S3P, and a significantly higher Ki value for glyphosate (373 μM) than aroAE. coli. AroAJ. sp is characterized as a novel and naturally occurring class I aroA enzyme with glyphosate tolerance. Furthermore, we show that aroAJ. sp can be used as an effective selectable marker in both japonica and indica rice cultivar. Transgenic rice lines were tested by herbicide bioassay and it was confirmed that they could tolerate up to 3360 g/ha glyphosate, a dosage four-fold that of the recommended agricultural application level. To our knowledge, it is the first report of a naturally occurring novel class I aroA gene which can be efficiently utilized to study and develop transgenic glyphosate-tolerant crops, and can facilitate a more economical and simplified weed control system.
The shikimate pathway is essential for the biosynthesis of aromatic amino acids in plants, fungi and microorganisms1. The enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS, EC2.5.1.19), also called aroA enzyme, plays a crucial role in the penultimate step of the shikimate pathway by catalyzing the transfer of the enolpyruvyl moiety of phosphoenolpyruvate (PEP) to the 5-hydroxyl group of shikimate-3-phosphate (S3P) to form 5-enolpyruvylshikimate-3-phosphate (EPSP). Glyphosate is the most widely used broad spectrum herbicide that mimics the carbocation state of PEP and binds EPSPS competitively2,3,4. The inhibition of EPSPS is a reversible reaction in which glyphosate binds to the binding site of PEP and forms a stable but non-covalent ternary complex with the enzyme and S3P (EPSPS-S3P-glyphosate)5. The ternary complex blocks the formation of EPSP, affecting the growth of the organism. Glyphosate provides an effective, efficient and economical way to control weeds6. As glyphosate is an unselective herbicide, the development of glyphosate-tolerant crops has dramatically changed weed management practices and increased the yield of crops.
Since the mutant aroA gene cloned from Salmonella typhimurium was first reported in 19837, the aroA enzyme (EPSPS) identified as the target of glyphosate resistance has been studied extensively over the past three decades8. Different aroA enzymes from various organisms have been divided into two classes on the basis of their intrinsic glyphosate sensitivities and their substantial sequence variations9. In general, class I aroA enzymes are naturally sensitive to glyphosate and generally found in Escherichia coli, Aeromonas salmoncida, Petunia hybrida and Arabidopsis thaliana. Class II aroA enzymes share less than 30% sequence similarity with class I enzymes, and can retain their activity at a high concentration of glyphosate, which are isolated from Agrobacterium tumefaciens CP4, Pseudomonas sp. strain PG2982, Bacillus subtilis, Ochrobactrum anthropi, Staphylococcus aureus and other bacteria species10,11,12,13,14,15,16. In class II enzymes, two conserved regions have been proved to be key regions involved in glyphosate tolerance: RXHXE and NXTR (X represents non-conserved amino acids), in both of which the positive charge of the side chain of Arg hinders the binding of glyphosate17.
Several class II enzymes have been used to generate transgenetic plants18,19,20,21,22. Among them, only the one from Agrobacterium tumefaciens CP4 has been used for the production of transgenic glyphosate-tolerant crops.
Glyphosate insensitivity can be also achieved in class I aroA enzymes through site-directed mutagenesis23,24,25 or natural selection26,27,28. However, most of the mutants show lower affinity for their substrate PEP. Among them, the double mutated aroA (TIPS) has been successfully used to produce the first commercial transgenic glyphosate-tolerant maize (GA21). In recent years, considerable attention has been paid to the exploration of new types of aroA enzymes with commercial feasibility.
In this study, a novel aroA enzyme from a glyphosate-tolerant strain Janibacter sp (designated as aroAJ. sp) was isolated from marine sediment and further functionally characterized. Through the sequence analyses and phylogenetic analyses, aroAJ. sp is characterized as a new class I aroA enzyme. The Km values for PEP and S3P of this enzyme were found to be similar to those of the analog from aroAE. coli, but its Ki value for glyphosate was significantly higher. Finally, the function of aroAJ. sp was evaluated in both Japonica and Indica rice cultivar, zhonghua11 (ZH11) and minghui86 (MH86), respectively. The results show that it can be used as an effective marker for direct selection and the generated transgenic rice lines were conferred high glyphosate tolerance (up to 3360 g/ha glyphosate). Taken together, our study indicates that the naturally occurring novel class I aroAJ. sp gene has promising potential for the development and breeding of transgenic glyphosate-tolerant crops.
Results
Isolation of gene conferring glyphosate tolerance
With the aim of finding novel aroA genes, the marine sediment sample was enriched with glyphosate and plated on M9 agar plates containing different concentrations of glyphosate. After plate-screening, one strain (named as L42) grew very well at a concentration of 150 mM glyphosate. The L42 strain was gram-positive and coccoid, whose 16S rRNA sequence (1487 bp) displayed the highest similarity (99%) with that of Janibacter.sp N2M. Thus, this strain was identified as a Janibacter species. To isolate the gene encoding aroA enzyme from L42 strain, a genomic library was constructed. After screening about 5,000 colonies from the library, one positive colony was found to possess the ability to grow on M9 agar containing 100 mM glyphosate, indicating that the recombinant plasmid in the colony might contain a gene involved in glyphosate tolerance. From this colony the plasmid (named as pZY3) was recovered.
Sequence analysis of aroA J. sp gene
DNA sequencing analysis revealed that the pZY3 plasmid contained a 1,299 bp DNA fragment that consisted of a complete open reading frame (ORF1) and encoded 432 amino acids with an estimated molecular weight of 45.1 kDa. The overall GC content of the ORF1 was 73.6%. The deduced amino acid sequence was then used to search for homologous sequences through the BLAST program, and the protein was found to share the highest homology (67% amino acid identity) with aroA enzyme from Janibacter sp HTCC2649 (GenBank: EAP99947.1), indicating that the ORF1 from L42 strain encoded an aroA enzyme. Thus, the aroA gene from the Janibacter.sp was designated as aroAJ. sp.
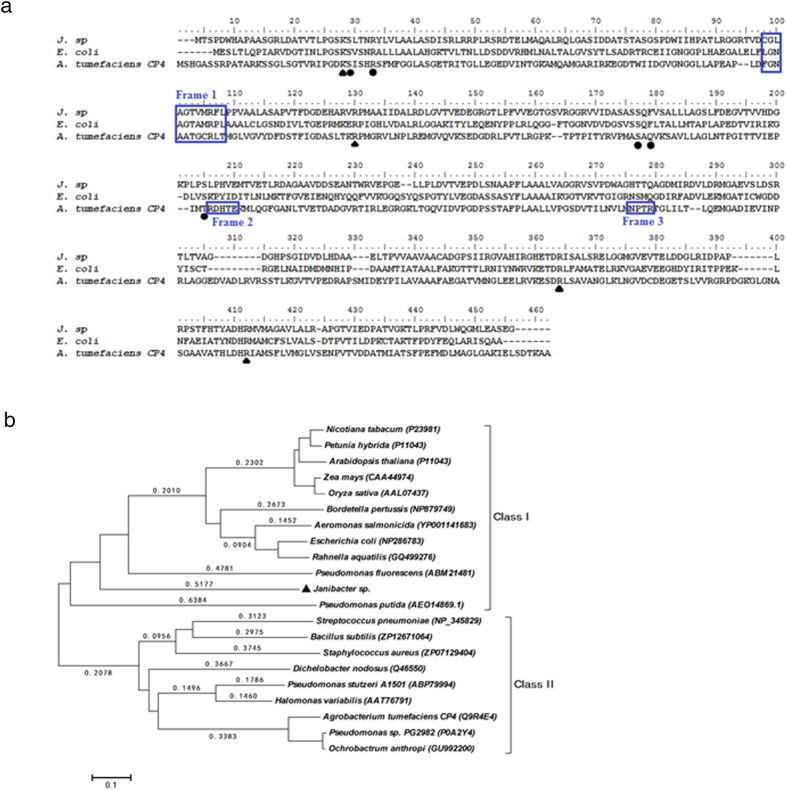
Multiple alignments of amino acids showed that aroAJ. sp shared only 26–35% amino acid identity with most class I and class II aroA enzymes. Several key residues involved in S3P and PEP binding were well conserved in aroAJ. sp, including S24, K25, S26, R127, S173, Q175, R348, R389, S202, R348 and R389, which are indicated in two classes of the aroA enzymes in Fig. 1. The highly conserved region containing residues XLGNAGTAXRXL (Fig. 1a, Frame 1) has been demonstrated to be critical for the substrate PEP binding in aroA enzymes26. It was indeed present in aroAJ. sp, and showed higher sequence identity with that in class I aroA enzymes than that in class II analogs. In addition, two other regions RXHTE (Fig. 1a, Frame 2) and NPTR (Fig. 1a, Frame 3), which are well conserved in the class II aroA enzymes, were completely absent in aroAJ. sp. Further phylogenetic analysis indicated that aroAJ. sp belonged to class I aroA but showed little sequence similarity with other class I aroA enzymes (Fig. 1b). Taken together, these results suggest that aroAJ. sp is a new member of class I aroA enzymes based on its amino acid sequence.
Figure 1.
(a) Multiple alignments of amino acid sequences from aroAJ. sp with representative class I and class II aroA enzymes using ClustalW program. Triangles, residues critical for PEP binding; and circles, residues critical for S3P binding. The two regions involved in glyphosate resistance in class II aroA enzymes are boxed in Frames 2 and 3. The motif important for interaction with PEP conserved in class I aroA enzymes is boxed in Frame 1. (b) The phylogenetic tree was based on homologous sequences of the aroA proteins and the neighbor-joining methods (MEGA4.0). The percentage of the tree from 1000 bootstrap resamples supporting the topology is indicated when above 50. Accession numbers or international patent publication numbers are shown in parentheses. The scale bar represents 0.1 substitutions per position.
Glyphosate insensitivity assay of aroA J. sp in E. coli
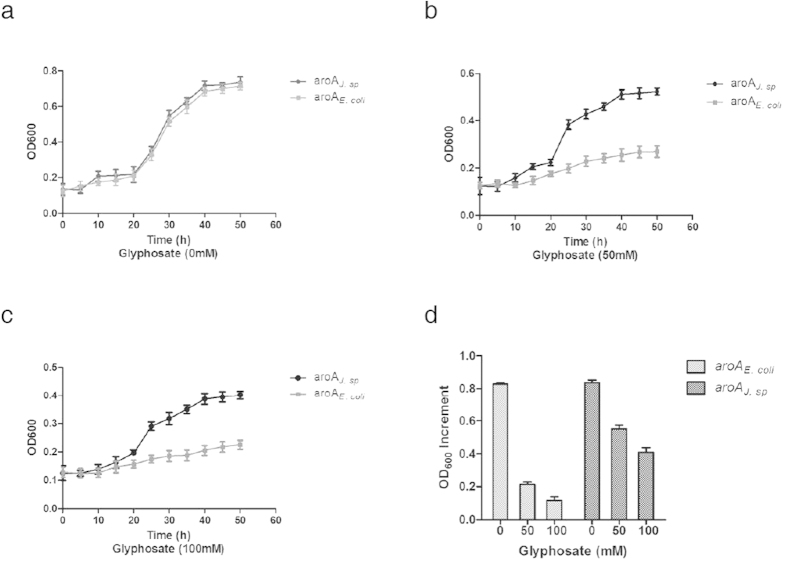
E.coli AB2829, which is deficient in aroA, can only grow in minimal medium when foreign aroA complements the deficiency. Plasmids pGEX-6p-1-aroAJ. sp and pGEX-6p-1-aroAE. coli were obtained by cloning aroAJ. sp and aroAE. coli into GST carrier vector pGEX-6p-1. Then, the function of the isolated aroAJ. sp gene in E. coli was investigated by comparing the growth characteristics of E. coli AB2829 harboring either plasmid pGEX-6p-1-aroAJ. sp or pGEX-6p-1-aroAE. coli at glyphosate concentrations of 0, 50 and 100 mM. All the strains grew well in M9 medium without glyphosate, and the growth of each strain was about 80%, suggesting that aroAJ. sp gene has no adverse effect on the bacterial growth under this condition (Fig. 2a). However, bacterial cells containing aroAE. coli were clearly inhibited in the presence of 50 mM glyphosate with the growth decreasing to 30% (Fig. 2b), and the cells were severely inhibited in the presence of 100 mM of glyphosate with the growth of only 20% (Fig. 2c). In contrast, cells carrying aroAJ. sp grew well under all the conditions with 50 or 100 mM of glyphosate. In medium containing 50 mM and 100 mM glyphosate, the growth of this strain was about 55% and 40%, which was 2.7 and 2-fold higher than that of the strain containing aroAE. coli (Fig. 2d). These results indicate that aroAJ. sp can functionally complement the deficiency of aroA in E. coli AB2829 and definitely carries a high glyphosate-tolerant capability, whereas the aroAE. coli gene derived from bacterium strain DH5α does not have such capability.
Figure 2.
Growth curve of E. coli AB2829 harboring either pGEX-6p-1-aroAJ. sp or pGEX-6p-1-aroAE. coli in liquid M9 minimal medium supplemented with glyphosate at concentrations of 0 mM (a) 50 mM (b) and 100 mM (c). (d) Bar charts of growth analysis of the strains. The results presented are the averages of two sets of experiments done in triplicate.
Expression and purification of aroA J. sp enzyme
After induction by IPTG, soluble expression of the aroAJ. sp-GST fusion was performed in bacterium strain BL21, and the induced fusion protein, which comprised an aroAJ. sp (47 kDa) and a GST tag (26 kDa), showed an expected molecular mass of 73 kDa on SDS-PAGE (Fig. S2a). Affinity-purified aroAJ. sp -GST fusion was cleaved by 3C protease, resulting in a 47 kDa aroAJ. sp protein with high purity as indicated by a single band on SDS-PAGE. Soluble protein was successfully produced in BL21 under the conditions used in this study, suggesting a proper folding and conformation for its functionality. GST carrier within the aroAJ. sp -GST fusion may play a role for this soluble expression. At the same time, the aroAE. coli-GST fusion was expressed in parallel and run on the same SDS-PAGE as a control, and aroAE. coli of a similar size was also obtained after cleavage of the fusion (data not shown). The concentration of the purified aroAJ. sp and aroAE. coli enzyme was 0.3 mg/ml and 1.5 mg/ml, respectively.
Kinetic properties of purified aroAJ. sp enzyme
As shown in Fig. S2b and S2c, aroAJ. sp enzyme had the highest activity at pH 8.0 and 40 °C, and maintained about 40% activity over a pH range from 6 to 10 and 30% activity over a temperature range from 10 to 60 °C, implying high stability and applicability of this enzyme under different conditions.
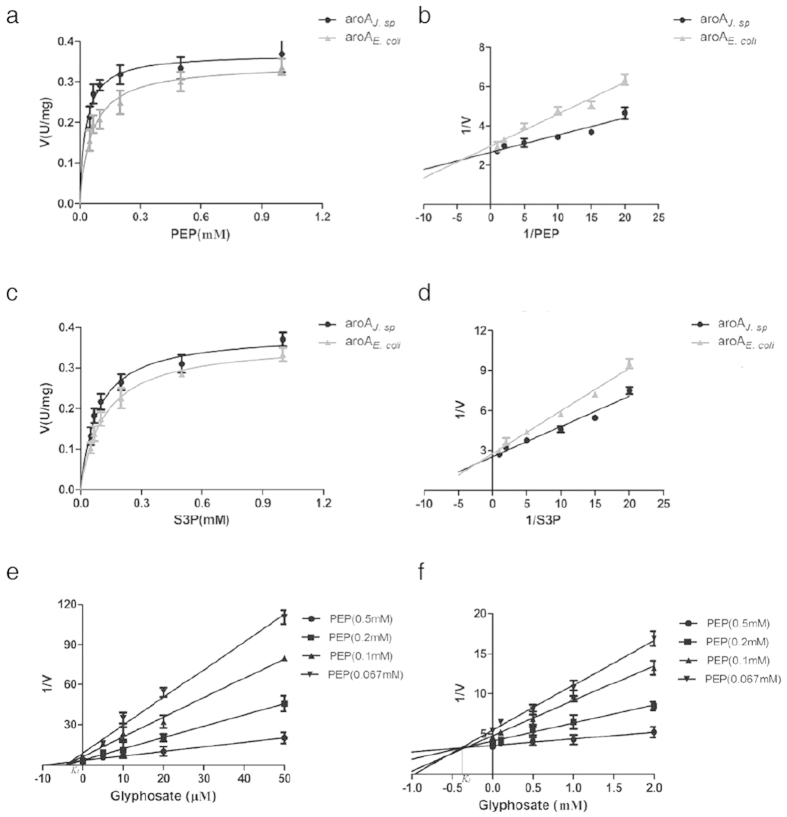
The proteins encoded by aroAJ. sp and aroAE. coli were purified separately with a GST system and used for enzymatic activity assay. Due to the structural difference from class II enzymes, class I aroA enzymes are known to be naturally glyphosate-sensitive. The Km value for PEP and the Ki value for glyphosate of the purified aroAE. coli enzyme were determined to be 60 and 0.9 μM in this experiment, respectively, and the corresponding Ki/Km ratio was 0.015. Kinetic constants of the aroAJ. sp enzyme were significantly different from those of the aroAE. coli enzyme (Table 1, Fig. 3). In the presence of KCl, The Kmvalues of the aroAJ. sp enzyme for PEP and S3P were 30 and 83 μM, respectively, which were lower than those of aroAE. coli, indicating a high substrate affinity of the aroAJ. sp enzyme. The IC50 and the Ki values for glyphosate of the aroAJ. sp enzyme were 3.6 mM and 373 μM, respectively, and the IC50 value was 90-fold that of the aroAE. coli enzyme. The Ki/Km value of aroAJ. sp enzyme was calculated to be 12.4, which is 800-fold that of aroAE. coli, suggesting that the aroAJ. sp enzyme has a higher level of glyphosate tolerance than the aroAE. coli counterpart.
Table 1. Kinetic properties of aroAJ. sp and aroAE. colia.
| Enzyme | Sp act (nKat/mg) | Km(PEP)b (μM) | Km(S3P)b (μM) | Ki c (μM) | IC50d (mM) | Ki/Km(PEP) |
|---|---|---|---|---|---|---|
| AroAJ. sp | 27.9 ± 3 | 30 ± 4 | 83 ± 11 | 373 ± 50 | 3.6 ± 0.2 | 12.4 |
| AroAE. coli | 15.6 ± 4 | 60 ± 7 | 115 ± 10 | 0.9 ± 0.09 | 0.04 ± 0.01 | 0.015 |
aThe results are the averages of two sets of experiments conducted in triplicate.
bThe PEP or S3P concentration was set at 0.05, 0.067, 0.1, 0.2, 0.5, and 1 mM, while the concentration of the other one was fixed at 1.0 mM.
cCompetitive inhibition by glyphosate with respect to PEP was demonstrated by lines converging on the x axes of Lineweaver-Burk plots. The PEP concentration was set at 0.067, 0.1, 0.2, and 0.5 mM, respectively, while the glyphosate concentration was 0, 5, 10, 20, and 50 μM in determining the inhibition of aroAE. coli; and the glyphosate concentration was 0, 0.1, 0.5, 1, 2 and 5 mM in determining the inhibition of aroAJ. sp. S3P concentration was fixed at 1 mM.
dThe glyphosate concentration causing 50% inhibition of enzyme activity, which was determined by fitting the data to the equation: V = V min + (V max −V min)/(1 + ([I]/IC 50)n), and V was determined at 1 mM PEP and 1 mM S3P with the glyphosate concentration ranging from 0.0001 mM to 100 mM.
Figure 3.
(a) The V-S curve of aroAJ. sp and aroAE. coli assayed at fixed S3P and various PEP concentrations. (b) The Lineweaver-Burk plots aroAJ. sp and aroAE. coli assayed at fixed S3P and various PEP concentrations. (c) The V-S curve of aroAJ. sp and aroAE. coli assayed at fixed PEP and various S3P concentrations. (d) The Lineweaver-Burk plots aroAJ. sp and aroAE. coli assayed at fixed PEP and various S3P concentrations. (e) The Ki values of aroAE. coli determined by lines converging on the x-axis of Lineweaver-Burk plots. (f) The Ki values of aroAJ. sp determined by lines converging on the x-axis of Lineweaver-Burk plots.
Nuclear transformation directly using aroA J. sp. as selectable marker
Since the aroAJ. sp showed high glyphosate-tolerant capability in E.coli and had a high Ki/Km value, it was tested in a japonica rice variety zhonghui11 (ZH11) to explore the possibility of employing aroAJ. sp as a selectable marker in crops. To ensure efficient expression of aroAJ. sp, maize Ubi promoter was used, because it conferred a higher level of foreign gene expression than Act1 and CaMV 35S promoters in monocot study29. Since aroA was located in the chloroplast in plants, a chloroplast transit peptide from Arabidopis thaliana (CTP) was added to the N-terminus of aroAJ. sp to target the protein into the chloroplasts in rice. The resulted pU130-aroAJ. sp (Fig. 4a) was transformed into ZH11.
Figure 4.
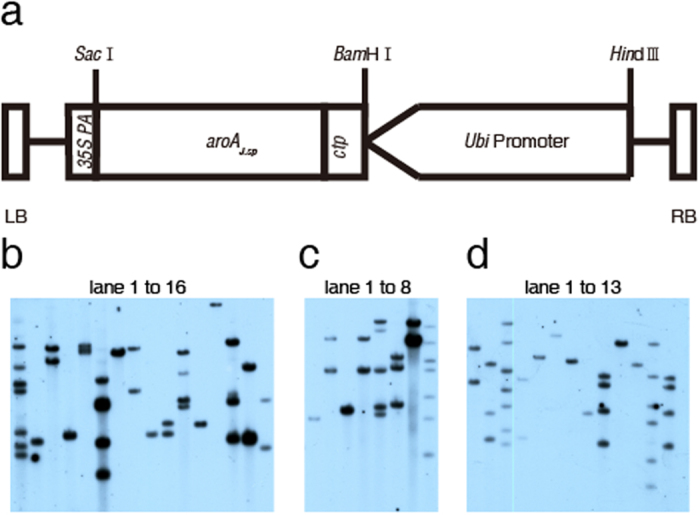
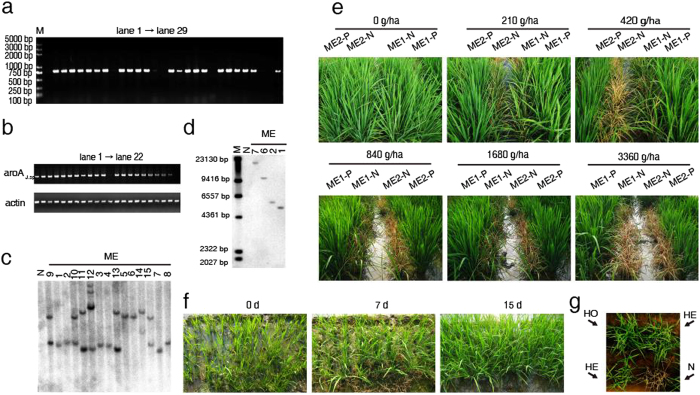
(a) the expression cassette of pU130-aroAJ. sp. The ctp-aroAJ. sp fusion gene was controlled by a maize ubiquitin promoter and 35S PolyA terminator. (b–d) Southern blot analysis of positive transgenic T0 Japonica plants of cv. ZH11 from 3 independent transformations. (b) lane 1–16, (c) lane 1–8 and (d) lane 1–13 represent different transgenic events.
From 3 independent transformation experiments, glyphosate-resistant calli were selected on medium containing 200 mg/L glyphosate. Resistant calli appeared after 5 weeks on average and could be transferred to the differentiation medium after one more week, resulting in a callus formation rate of around 50% (Table 2).
Table 2. Statistical analysis of transformation efficiency in japonica rice, ZH11.
| Replicates | Number of calli inoculated | Glyphosate-resistant calli | Resistant calli formation rate (%) | Resistant calli that regenerated plants | PCR positive independent lines | Single-copy independent lines |
|---|---|---|---|---|---|---|
| 1 | 148 | 84 | 56.8 | 16 | 16 | 6 |
| 2 | 124 | 67 | 54.0 | 8 | 8 | 2 |
| 3 | 165 | 75 | 45.5 | 13 | 13 | 6 |
16, 8 and 13 resistant lines were obtained out of 84, 67 and 75 resistant calli, respectively. All T0 lines were confirmed by PCR (data not shown) and Southern blot analysis (Fig. 4b–d). The results tentatively suggest that direct selection for glyphosate tolerance using aroAJ. sp as selectable marker can readily produce transgenic rice lines with a high transgenic positive rate.
Glyphosate tolerance of the transgenic indica rice
As aroAJ. sp could be used as direct selectable marker in japonica rice, we further applied it in a conventional indica rice variety, minghui86 (MH86). After 8 weeks of selection followed by the differentiation of calli, 28 putative transgenic plants were generated (named as P1-P28). The transformation efficiency for MH86 was about 5% based on the numbers of the total transformants and the calli used for transformation. PCR analysis showed that a specific fragment of 815 bp was amplified from 22 of the 28 T0 transgenic lines, indicating an integration of aroAJ. sp gene into the rice genome (Fig. 5a). Those PCR positive lines were then named as PP1-PP22, and chosen for RT-PCR analysis (Fig. 5b). The result demonstrated a proper transcription of the integrated aroAJ. sp gene in 20 rice transformants (marked as ME1-20). The copy number of T-DNA integration in these T0 transgenic lines (ME1-20) was determined by Southern blotting (Fig. 5c). T2 progenies from four lines (ME1, ME2, ME6 and ME7) were further tested by Southern blotting. The result confirmed that in these lines, the transgene was integrated into the genome as a single copy and was stably inherited (Fig. 5d). In order to further assay the tolerant capability for crop breeding, the ME1 line and ME2 line were chosen for further characterization, because their T-DNA insertions were mapped to the intergenic region (data not shown). The data collected from the field experiment for agronomic performance showed the homozygous T2 transgenic rice lines (ME1-P and ME2-P) had no significant differences from the their corresponding negative transgenic lines (ME1-N and ME2-N) (Table 3).
Figure 5.
(a) PCR analysis of transgenic T0 indica generation plants. M, Trans2K Plus DNA Marker; lane 1, untransformed MH86; lane 2 to 29, represents transgenic events P1-P28. (b) expression of aroAJ. sp cDNA in transgenic rice plants. Upper, Lanes 1 to 22, represents transgenic plants PP1-PP22, N, the non-transgenic rice control plant. Nether, the actin gene amplified from all the tested samples. (c) Southern blot analysis of transgenic T0 indica plants of cv. MH86 with Hind III digestion. N, untransformed MH86; lane 1–15, different transgenic lines, corresponding sample numbers were marked. (d) Southern blot analysis of transgenic T2 indica generation plants with SacI digestion. Lane 1, untrasformed MH86; lane 2–5, confirmed single copy, corresponding sample numbers were marked. (e) glyphosate tolernace of T2 transgenic rice plants under different glyphosate dosages. T2 rice seeds were germinated in field and grown to tillering stage. 0 g/ha, 210 g/ha, 420 g/ha, 840 g/ha, 1680 g/ha and 3360 g/ha glyphosate (the corresponding applied concentration of glyphosate to each dosage was 0 mg/L, 187.5 mg/L, 375 mg/L, 750 mg/L, 1500 mg/L and 3000 mg/L) were sprayed on ME1 and ME2 transgenic lines along with non-transformed control plant and photograph was taken 1 week after spray. ME1-P and ME2-P represent the homozygous transgenic lines of ME1 and ME2, while ME1-N and ME2-N are their corresponding negative transgenic lines. (f) glyphosate tolerance test of T4 generation seedlings of ME1 line. The 28-day-old ME1 T4 generation seedlings were transplanted in the field with weeds and sprayed with 840 g/ha glyphosate (the corresponding applied concentration of glyphosate was 840 mg/L). Photographs were taken by Y.C. at day 0, 7 and 15 after spray. (g) Selection of homozygous line by glyphosate. Homozyous (HO), heterzygous (HE) T2 transgenic rice plants along with untransformed control plants (N) were sprayed with 1% (vol/vol) solution of the herbicide Roundup containing 41% isopropylamine salt of glyphosate, and photograph was taken by Y.C. at 3 d after spray.
Table 3. Comparison of agronomic traits between transgenic homozygous lines and transgenic-negative lines under field conditionsa.
| Plant height (cm) | Panicle per plant | Panicle length (cm) | Seed-set rate | Weight per 1,000 grains (g) | Yield/plant (g) | |
|---|---|---|---|---|---|---|
| ME1-P | 110.50 ± 3.36 | 12.30 ± 2.00 | 25.75 ± 0.39 | 0.73 ± 0.02 | 23.36 ± 1.13 | 32.11 ± 2.70 |
| ME1-N | 112.00 ± 1.30 | 12.70 ± 0.44 | 27.33 ± 1.33 | 0.70 ± 0.03 | 22.76 ± 1.10 | 32.20 ± 1.74 |
| ME2-P | 107.05 ± 3.20 | 12.73 ± 1.76 | 24.80 ± 0.44* | 0.71 ± 0.06 | 23.86 ± 1.13 | 30.27 ± 9.22 |
| ME2-N | 112.87 ± 2.66 | 14.63 ± 1.14 | 25.94 ± 0.40 | 0.74 ± 0.01 | 24.29 ± 1.01 | 38.72 ± 2.54 |
ME1-P and ME2-P represent homozygous transgenic lines of ME1 and ME2, while ME1-N and ME2-N are their corresponding negative transgenic lines.
aThe parameters were given as means (±standard deviation) for data collected from 10 plants in triplicate for each plant type.
*Means significantly different from the control (P < 0.05).
Spray with glyphosate resulted in a strong inhibition of growth and the ultimate death of the non-transgenic rice plants (210 g/ha), but did not influence the normal growth of the homozygous T2 transgenic rice plants at 3360 g/ha glyphosate (Fig. 5e).
In addition, T4 generation seedlings of ME1 line were transplanted into a weedy field without weed control. Only 7 days after spray with 840 g/ha glyphosate, the weeds were dead, while the transgenic rice plants reached to tillering stage, and grew normally 15 days after spray (Fig. 5f), suggesting the applicable potential of aroAJ. sp in weed control.
Discussion
In modern agricultural system, herbicides have greatly contributed to the reliable global food production as they can easily remove weeds. Among these chemicals, glyphosate is the most widely used one due to its broad-spectrum herbicidal activity and minimal human and environment toxicity. Since the glyphosate use pattern commenced in 1996, the planting area of transgenic herbicide-tolerant crops accounts for more than 80% of the total planting area of transgenic crops in 2014 (ISAAA Annual Report: 2014 http://www.isaaa.org/resources/publications/annualreport/2014/pdf/ISAAA-Aannual_ Report-2014.pdf). In the last three decades, a number of promising enzymes were identified. However, among those enzymes, only two aroA variants have been utilized for developing commercial glyphosate-tolerant crops: CP4, a naturally occurring class II type, and TIPS, a mutant class I type. Therefore, identification of more novel glyphosate-insensitive aroA genes which can be used to generate glyphosate-tolerant crops with commercial feasibility will bring great positive impact on global food security by facilitating a more economical and simplified weed control system.
Mutagenesis is one method to improve the glyphosate tolerance of aroA enzymes, but when the tolerance for glyphosate is increased, the binding affinity for PEP and catalytic efficiency may be decreased at the same time, as glyphosate and PEP bind to the same site. To date, TIPS is the only class I enzyme which is essentially insensitive to glyphosate but maintains high affinity for PEP. Thus, the exploration of new types of aroA enzymes with both intrinsic glyphosate tolerance and high affinity with PEP is very necessary.
Microbial biodiversity provides opportunities to extract genes and proteins with unique properties for industrial and environmental applications. As a result, in this study, a Janibacter sp strain that could grow at a 150 mM concentration of glyphosate was isolated from marine sediment, and the aroA gene from this strain was cloned and named as aroAJ. sp. Phylogenetic analysis and conserved domain analysis revealed that aroAJ. sp is characterized as an novel and naturally occurring class I aroA enzyme but with glyphosate tolerance.
Ki/Km of aroA enzyme is often seen as an important indicator of enzymatic activity in the presence of glyphosate. The overexpression of class I type AM79 aroA enzyme, whose Ki/Km value is 10.6, resulted in high glyphosate tolerance in tobacco27. The double mutated maize aroA (TIPS), which also belongs to class I type aroA enzyme, has also relatively high Ki/Km ratio (5.8)30. According to ISSSA GM approval database, TIPS has been used for generating glyphosate-tolerant crops, resulting in 38 events (http://www.isaaa.org/gmapprovaldatabase/default.asp). The Ki/Km value of aroAJ. sp enzyme was calculated to be 12.4 (Table 1), suggesting that aroAJ. sp maintains a high affinity with PEP and intrinsic glyphosate tolerance.
The mechanism of resistance to glyphosate of class I type has been extensively investigated. Take aroAE.coli for example: G96A substitution was shown to increase the glyphosate tolerance to 100 folds, but decrease the binding affinity for PEP. Interestingly, the corresponding site in CP4 gene (class II type) is also an A. We attempted to conduct G96A substitution. Surprisingly, it turned out that the enzyme activity dramatically decreased to be almost inactive (data not shown). It was also reported that the P106L mutant of rice (Oryza sativa) aroA enzyme has a high glyphosate resistance while retaining relatively high catalytic efficiency31. Besides, it was claimed that substituting T97 with I or L and P101 with T, G, C, A or I could ensure both intrinsic glyphosate tolerance and high affinity with PEP30. P101 substitution confers relatively low glyphosate tolerance while maintains high catalytic efficiency, but only the mutation of both T97 and P101 can provide the conformational changes and lead to high catalytic efficiency and glyphosate tolerance. For our newly identified aroAJ. sp, the corresponding amino acids are T and F, which get around patent protection. Therefore, this newly identified aroAJ. sp might provide a new aspect to reveal the function mechanism of class I type aroA enzymes. And further mutagenesis work is needed to further improve the glyphosate tolerance of this enzyme.
Selectable marker gene is essential in the production of transgenic plants. Most of the marker genes are usually either antibiotic or herbicide resistant genes32, such as hpt (hygromycin phosphotransferase gene), nptII (neomycin phosphotransferase gene), and bar (phosphinothricin acetyltransferase gene)33. Although glyphosate tolerant gene aroA has been used directly as selectable marker gene in Arabidopsis and other species34,35,36, there are few reports of its application in rice. In most of the reported glyphosate-tolerance rice events, hpt was used as the selectable marker gene22,37. Zhao et al. used a glyphosate-tolerant aroA variant G6 gene as the selectable marker gene in a japonica rice XiuShui-11021. Although several transgenic lines were generated, the PCR positive rate was rather low, and the Southern blotting results indicated that most of the lines were multiple-copy integration lines. In our study, the transformed calli from both japonica and indica rice were selected on medium containing 200 mg/L glyphosate, and transgenic rice lines were readily obtained (Table 2 and data not shown). Therefore, it can be speculated that aroAJ. sp can be directly used as a selectable marker gene for its high glyphosate tolerance capability.
Rice can be significantly influenced by glyphosate, and the improper use of glyphosate can cause severe injury to rice and reduce grain yield38. The planting area of transgenic herbicide-tolerant crops accounts for more than 80% of the total planting area of transgenic crops in 2014 (ISAAA Annual Report: 2014 http://www.isaaa.org/resources/publications/annualreport/2014/pdf/ISAAA-Aannual_ Report-2014.pdf). However, when other glyphosate-tolerant crops have been adopted9,25, no glyphosate-tolerant rice varieties have been commercialized. In our study, the untransformed MH86 was sensitive to glyphosate, and were dead 7 days after 420 g/ha glyphosate treatment; while transgenic MH86 rice plants expressing aroAJ. sp exhibited robust tolerance to glyphosate at a dosage of 3360 g/ha, which is four-fold that of the recommended dosage (Fig. 5e) The tolerant level was comparable with that resulted from the expression of several class II aroA genes in other plants20,22,39.
At the early stage of transplanting, the rice growth is often affected by weeds. Here, our results show that the spray of the recommended amount of glyphosate (840 g/ha) can efficiently control the weeds and has no apparent effect on the growth of aroA transgenic lines (Fig. 5f). The field assay indicated that glyphosate-tolerant line ME1 can be powerful in fighting against weeds, and thus can be used in no-till or conventional till system. In addtion, we observed that the homozygous and heterzygous T2 lines can be easily distinguished from nontransformant lines with the spray of 1% (vol/vol) solution of the herbicide Roundup which contains 41% isopropylamine salt of glyphosate (Fig. 5g). MH86 has been used as an importnat conventional restored line in hybrid seed production. When the aroAJ. sp MH86 rice line is used as the paternal line, its next generation will keep the glyphosate tolerance ability. This characteristic facilitates the identification of false hybrids, which helps to improve the qutlity of hybrids and to ensure the sustained development of food production.
In conclusion, aroAJ. sp is characterized as a new type of class I aroA enzyme with glyphosate tolerance, which provides new insights into the functions of class I aroA enzymes. We believe that aroAJ. sp gene has a promising potential for the development and breeding of transgenic glyphosate-tolerant crops, which will facilitate a more economical, complete and simplified weed control.
Methods
Materials and Mediums
Marine sediment was sampled at site IR-CTD5(16°59.9412′N 124°58.2958′E) on the south west Indian Ridge. The glyphosate tolerant bacteria Janibacter sp was isolated from the marine sediment. Bacterial strains, Escherichia coli DH5α, E. coli aroA mutant strain AB2829, E. coli BL21 (DE3) and the zhonghua11 japonica rice variety were stored in our laboratory. The pUC118 vector was purchased from Takara (Japan), and the vector pGEX-6p-1 was kept in our laboratory. Vector pCAMBIA1300 was a gift from the Center of the Application of Molecular Biology to International Agriculture, Australia. All enzymes used for restriction digestion and ligations were purchased from Takara (Japan). S3P was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). PEP and glyphosate were purchased from Sigma (St. Louis, MO, USA). All chemicals were of analytical grade. The medium used for isolating glyphosate tolerant strain was minimal medium M9 (6.8 g/L Na2HPO4, 3.0 g/L K2HPO4, 1.0 g/L NH4Cl, 0.5 g/L NaCl and 0.12 g/L MgSO4) prepared in artificial sea water (distilled water with 0.5 g/L MgCl2, 0.5 g/L MgSO4, 0.5 g/L CaCl2, 0.55 g/L KCl, 0.16 g/L NaHCO3, 0.08 g/L KBr, 34 mg/L SrCl, 22 mg/L H3BO3, 4 mg/L Na2SiO3, 2.4 mg/L NaF), and the minimal medium used for the glyphosate tolerance test in E. coli was M9 (6.8 g/L Na2HPO4, 3.0 g/L K2HPO4, 1.0 g/L NH4Cl, 0.5 g/L NaCl and 0.12 g/L MgSO4) supplemented with 0.4% (w/v) glucose as a carbon source plus the appropriate antibiotics.
Isolation of glyphosate-tolerant strain
The marine sediment was used for enrichment with glyphosate for 7 d, and then the enriched culture was diluted with distilled water and plated on M9 agar plates containing 60 mM glyphosate. After 48 h of incubation at 28 °C, the colonies were further screened at 100 and 150 mM glyphosate concentrations. One strain numbered L42 grew well in the presence of 150 mM glyphosate and was chosen for further study. The 16S rRNA sequence of L42 strain was used for species identification.
Isolation of gene associated with glyphosate tolerance
A genomic library of the L42 strain was constructed. The chromosomal DNA was extracted from the strain and partially digested with Sau3AI to produce 4–9 kb fragments. These DNA fragments were inserted into the pUC118 vector40. The ligation mixture was incubated at 4 °C overnight and transformed into E. coli DH5α. The transformants containing the recombinant pUC118 were transferred onto M9 agar plates containing 20 mM glyphosate and then the plates were incubated at 37 °C for 48 h. One colony exhibiting glyphosate tolerance was isolated from the plates containing a recombinant plasmid (pZY3) with an insert of approximately 3.5 kb.
Sequence analysis
The inserted fragment from the plasmid pZY3 was sequenced by the Genescript Company (Nanjing, China), and the nucleotide sequences were analyzed using the Softberry Gene Finding Tool (http://linux1.softberry.com/berry). Sequence alignments were performed using the BLAST program (http://blast.ncbi.nlm.gov/blast) combined with the ClustalW program software41. The phylogenetic tree of aroAJ. sp was constructed by MEGA542.
Construction of prokaryotic expression vectors pGEX-6p-1-aroA J. sp and pGEX-6p-1-aroA E. coli
The oligonucleotide primer sequences were as follows: primer 1 (5′-CGCGGATCC ATGACCAGTCCTGATTGGCATGC-3′) (the BamHI site is underlined); primer 2 (5′-CCGGAATTCTCAGCCCTCCGACGCCTCG-3′) (the EcoRI site is underlined), which was designed according to the sequence derived from the insert of plasmid pZY3 and supplied by the Genescript; primer 3 (5′-CGCGGATCCATGGAA TCCCTGACGTTACAACCCATCGCTCGTG-3′); and primer 4 (5′-CCGGAATTC TCAGGCTGCCTGGCTAATCCGCGCCAGCT-3′), which was specific for amplifying aroAE. coli gene and was designed based on the sequence available in GenBank (GenBank:X00557). The aroAJ. sp and aroAE. coli genes were amplified, using the recombinant plasmid pZY3 and the genomic DNA from E. coli as the template, respectively. The FastPfu DNA polymerase (Transgen, Beijing, China) was used for all the reactions under the following conditions: 30 cycles of 94 °Cfor 20 sec, 55 °C for 20 sec, and 72 °C for 1 min. After PCR, the amplified products were gel-purified, digested with BamHI and EcoRI, and ligated into the pGEX-6p-1 vector.
Cell growth in the presence of glyphosate
Strain E. coli AB282931 was transformed with pGEX-6p-aroAJ. sp plasmid and the transformants were cultured in 50 mL LB medium containing 100 μg/mL ampicillin at 37 °C until the OD600 values reached 0.1. The the culture was collected by centrifugation and washed twice using liquid M9 minimal medium. Bacteria in the culture were then grown with shaking at 37 °C in liquid M9 minimal medium supplemented with glyphosate at the concentrations of 0, 50 and 100 mM. The OD600 values of the cultures were determined at approximately 5-h intervals to record the growth rates of the strains till 50 h. OD600 increments were calculated as (ODa-ODb)/OD0, in which ODa and ODb represent the OD600 value before and after growing for 50 h, and OD0 is the final OD600 value without glyphosate. The results presented are the averages of two sets of experiments done in triplicate. The strain AB2829 harboring pGEX-6p-aroAE. coli was used as the control.
Expression and purification of aroA J. sp and aroA E. coli enzymes
E. coli BL21 (DE3) was transformed separately with pGEX-6p-1-aroAJ. sp. and pGEX-6p-1-aroAE. coli plasmids, and cultivated in LB medium containing 100 μg/mL ampicillin at 37 °C until the OD600 values reached 0.6–1.0. IPTG (0.5 mM) was then added, and the cultures were further incubated at 18 °C for 8 h. The cells were harvested and resuspended in 50 ml Hepes buffer (50 mM Hepes, 100 mM KCl, and 2 mM dithiothreitol, pH 7.0). After treatment by high pressure crushing, the cells were centrifuged at 10,000 × g for 40 min at 4 °C, and the supernatant was loaded onto a Glutathione-S-Transferase (GST) agarose at 4 °C. The glutathione-S-transferase (GST)-tagged aroA enzymes were purified using a GST Fusion System (GE Healthcare Sweden). The GST tag was removed by digestion with a 3C protease solution (10 U/μl, PreScission; Pharmacia). The molecular mass of the protein was determined by a 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and the enzyme concentration was measured by a Bradford Protein assay kit (Sangon, China).
Enzyme assay
AroA enzyme activity was determined by measuring the release of inorganic phosphate using the malachite green dye assay method43. The reaction (final volume, 50 μL) was performed at 28 °C in 50 mM Hepes buffer (pH 7.0), 1 mM S3P, 1 mM PEP, and 0.4 μg of the purified enzyme. After incubation for 4 min, 0.8 mL malachite green (0.045%) /ammonium molybdate (4.2%) colorimetric solution was added; 1 min later, the reaction was terminated by adding 0.1 mL 34% (w/v) sodium citrate solution. After 30 min of incubation at room temperature, the absorbance of the samples was measured at 660 nm; the same reaction solution without S3P was used as the blank control. The Km values for substrates, Ki value and IC50 value for glyphosate were calculated as described by Tian et al.12.
Generation of transgenic rice
To evaluate the potential application of the isolated aroAJ. sp gene in developing glyphosate tolerant crops, a japonica rice variety, zhonghua11 (ZH11) and a indica rice variety, minghui86 (MH86), were selected as the plant materials for genetic transforamtion. The chloroplast targeting signal peptide (CTP) coding sequence is from Arabidopsis thaliana and contains 228 nucleotides (Genebank:AAB72287.1). The nucleotide sequence of the aroAJ. sp gene and CTP were codon-optimized according to the codon bias in rice and was chemically synthesized to produce ctp-aroAJ. sp fusion gene (Takara, Japan). The hpt expression cassette in the binary vector pCAMBIA1300 (provided by the Center for the Application of Molecular Biology in International Agriculture, Australia) was removed by using XhoI and EcoRI restriction sites, resulting in pU130 transformation vector. Maize Ubi promoter and ctp-aroAJ. sp fusion gene were subcloned into pU130 using mutiple clone sites, and pU130-aroAJ. sp (Ubi-1: aroAJ. sp: 35S polyA) was obtained. The pU130-aroAJ. sp was introduced into A. tumefaciens EHA105 by electroporation, and by following the callus culture and tansformation procedures as described by Hiei et al.44 the resistant calli were obtained from the selection medium containing 200 mg/L glyphosate.
Molecular analysis of transgenic rice
To confirm the correct integration of the aroAJ. sp gene into the rice genome, the genome DNA from young leaves of rice plants was extracted by CTAB method45 and used as the template for PCR to amplify an 815 bp fragment of aroAJ. sp with Primer 5 (5′-TTCCTTAAAGCGAAAACCCC-3′) and Primer 6 (5′-AGGAGGGCGGACACGAACTG-3′). Expression of aroAJ. sp gene in T0 transgenic rice was analyzed by RT-PCR. Total RNA from rice plants was extracted using the Trizol reagent (Transgen, China) according to the manufacturer’s instructions. For the first strand cDNA synthesis, 1 μg of the total RNA was used with M-MLV reverse transcriptase (Invitrogen, USA) after DNase I digestion (Invitrogen, USA). The endogenous actin gene was amplified with the primers actin-F (5′-GCCACACTGTCCCCATCTAT-3′) and actin-R (5′-GCGACCACCTTGATCTTCAT-3′) as an internal control. The ctp-aroAJ. sp fragment was amplified with primer 7 (5′-GCATGCTCTCCCCGGATTGG-3′) and primer 8 (5′-GAGCTCCTATCAGCCCTCGGA-3′), resulting in 1.3 kp amplified DNA product. For Southern blot analysis, 10 μg of genomic DNA digested with HindIII or Sac I was electrophoresed on 0.8% agarose gel and transferred onto a Hybond+ nylon membrane (GE Healthcare UK Limited). The aroAJ. sp probe was DIG labeled using Primer 5 (5′-TTCCTTAAAGCGAAAACCCC-3′) and Primer 6 (5′-AGGAGGGCGGACACGAACTG-3′). Hybridization and detection steps were performed according to the manufacturer’s instruction (Roche, Mannheim, Germany).
Glyphosate tolerance assay of transgenic rice
Glyphosate-tolerance of transgenic rice plants was assayed by herbicide spraying. The T0 and T2 transgenic rice plants containing aroAJ. sp gene were planted in the weed controlled field and sprayed with either different amounts of glyphosate or 1% (vol/vol) solution of the herbicide Roundup which contains 41% isopropylamine salt of glyphosate (Monsanto, Malaysia). In addtion, T4 generation seedlings of ME1 line were also transplanted into a weedy field without weed control to investigate the potential to be used in no-till system. In the weedy field, there were mainly Gramineae weeds (Echinochloa Crusgalli (L.) Beauv), Cyperaceae weeds (Cyperus difformis L. & Cyperus fuscus L.), Pontederiaceae weeds (Monochoria vaginalis (Burm.f.) Presl ex Kunth) and Scrophulariaceae weeds (Lindernia procumbens (Krock.) Borbas) in this research.
Additional Information
How to cite this article: Yi, S.- et al. A Novel Naturally Occurring Class I 5-Enolpyruvylshikimate-3-Phosphate Synthase from Janibacter sp. Confers High Glyphosate Tolerance to Rice. Sci. Rep. 6, 19104; doi: 10.1038/srep19104 (2016).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 31270162), the National Program of Transgenic Variety Development of China (2011ZX08001-001), and 863 Program of China (No. 2012AA10A304).
Footnotes
Author Contributions Y.L., Z.L. and F.Z. conceived the experiments. S.Y., Y.C. and Y.Z. conducted the experiments. S.Y., Y.C. and F.Z. analyzed the results. S.Y., Y.C. and F.Z. wrote the paper. Y.L., Y.C. and F.Z. revised the paper. All authors reviewed the manuscript.
References
- Bentley R. The shikimate pathway-a metabolic tree with many branches. Crit. Rev. Biochem. Mol. Biol. 25, 307–384 (1990). [DOI] [PubMed] [Google Scholar]
- Boocock M. R. & Coggins J. R. Kinetics of 5-enolpyruvylshikimate-3-phosphate synthase inhibition by glyphosate. FEBS Lett. 154, 127–133 (1983). [DOI] [PubMed] [Google Scholar]
- Steinrücken H. C. & Amrhein N. 5-Enolpyruvylshikimate-3-phosphate synthase of Klebsiella pneumoniae 2. Inhibition by glyphosate [N-(phosphonomethyl)glycine]. Eur. J. Biochem. 143, 351–357 (1984). [DOI] [PubMed] [Google Scholar]
- Priestman M. A. et al. Interaction of phosphonate analogues of the tetrahedral reaction intermediate with 5-enolpyruvylshikimate-3-phosphate synthase in atomic detail. Biochemistry. 44, 3241–3248 (2005). [DOI] [PubMed] [Google Scholar]
- Schönbrunn E. et al. Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. USA. 98, 1376–1380 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green J. M. Current state of herbicides in herbicide-resistant crops. Pest Manag. Sci. 70, 1351–1357 (2014). [DOI] [PubMed] [Google Scholar]
- Comai L. & Sen L. C. & Stalker, D. M. An altered aroA gene product confers resistance to the herbicide glyphosate. Science 221, 370–371 (1983). [DOI] [PubMed] [Google Scholar]
- Pollegioni L., Schonbrunn E. & Siehl D. Molecular basis of glyphosate resistance-different approaches through protein engineering. FEBS J. 278, 2753–2766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funke T., Han H., Healy-Fried M. L., Fischer M. & Schönbrunn E. Molecular basis for the herbicide resistance of Roundup Ready crops. Proc. Natl. Acad. Sci. USA. 103, 13010–13015 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priestman M. A., Funke T., Singh I. M., Crupper S. S. & Schonbrunn E. 5-Enolpyruvylshikimate-3-phosphate synthase from Staphylococcus aureus is insensitive to glyphosate. FEBS Lett. 579, 728–32 (2005). [DOI] [PubMed] [Google Scholar]
- Dill G. M., CaJacob C. A. & Padgette S. R. Glyphosate-resistant crops: adoption, use and future considerations. Pest Manag. Sci. 64, 326–331 (2008). [DOI] [PubMed] [Google Scholar]
- Tian Y. S. et al. Isolation from Ochrobactrum anthropi of a novel class II 5-enopyruvylshikimate-3-phosphate synthase with high tolerance to glyphosate. Appl. Environ.Microb. 76, 6001–6005 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. S. et al. Functional characterization of Class II 5-enopyruvylshikimate-3-phosphate synthase from Halothermothrix orenii H168 in Escherichia coli and transgenic Arabidopsis. Appl. Environ.Microb. 93, 241–250 (2012). [DOI] [PubMed] [Google Scholar]
- Tian Y. S. et al. Complementary screening, identification and application of a novel class II 5-enopyruvylshikimate-3-phosphate synthase from Bacillus cereus. World J. Microb. Biot. 29, 549–557 (2013). [DOI] [PubMed] [Google Scholar]
- Wang L. et al. Characterization of a class II 5-enopyruvylshikimate-3-phosphate synthase with high tolerance to glyphosate from Sinorhizobium fredii. World J. Microb. Biot. 30, 2967–2973 (2014). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Characterization and site-directed mutagenesis of a novel class II 5-enopyruvylshikimate-3-phosphate (EPSP) synthase from the deep-sea bacterium Alcanivorax sp. L27. Enzyme Microb. Tech. 63, 64–70 (2014). [DOI] [PubMed] [Google Scholar]
- Li L. et al. A novel RPMXR motif among class II 5-enolpyruvylshikimate-3-phosphate synthases is required for enzymatic activity and glyphosate resistance. J. Biotechnol. 144, 330–336 (2009). [DOI] [PubMed] [Google Scholar]
- Ye G. N. et al. Plastid-expressed 5-enolpyruvylshikimate-3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. Plant J. 25, 261–270 (2001). [DOI] [PubMed] [Google Scholar]
- Kahrizi D., Salmanian A. H., Afshari A., Moieni A. & Mousavi A. Simultaneous substitution of Gly96 to Ala and Ala183 to Thr in 5-enolpyruvylshikimate-3-phosphate synthase gene of E. coli (k12) and transformation of rapeseed (Brassica napus L.) in order to make tolerance to glyphosate. Plant Cell Rep. 26, 95–104 (2007). [DOI] [PubMed] [Google Scholar]
- Yan H. Q. et al. Novel AroA from Pseudomonas putida confers tobacco plant with high tolerance to glyphosate. PLoS One. 6, e19732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Lin C. Y. & Shen Z. C. Development of transgenic glyphosate-resistant rice with G6 gene encoding 5-enolpyruvylshikimate-3-phosphate synthase. Agr. Sci. China. 10, 1307–1312 (2011). [Google Scholar]
- Chhapekar S. et al. Transgenic rice expressing a codon-modified synthetic CP4-EPSPS confers tolerance to broad-spectrum herbicide, glyphosate. Plant Cell Rep. 34, 721–731 (2015). [DOI] [PubMed] [Google Scholar]
- He M., Yang Z. Y., Nie Y. F., Wang J. & Xu P. A new type of class I bacterial 5-enopyruvylshikimate-3-phosphate synthase mutants with enhanced tolerance to glyphosate. Biochi. Biophys. Acta. 1568, 1–6 (2001). [DOI] [PubMed] [Google Scholar]
- Cao M. et al. Engineering higher yield and herbicide resistance in rice by mediated multiple gene transformation. Crop Sci. 44, 2206–2213 (2004). [Google Scholar]
- Funke T. et al. Structural basis of glyphosate resistance resulting from the double mutation Thr97→Ile and Pro101→Ser in 5-enolpyruvylshikimate-3-phosphate synthase from Escherichia coli. J. Biol.Chem. 284, 9854–9860 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y. C. et al. Novel AroA with high tolerance to glyphosate, encoded by a gene of Pseudomonas putida 4G-1 isolated from an extremely polluted environment in China. Appl. Environ.Microb. 71, 4771–4776 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao G. et al. A novel 5-enolpyruvylshikimate-3-phosphate synthase shows high glyphosate tolerance in Escherichia coli and tobacco plants. PLoS One. 7, e38718 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R. H. et al. A novel 5-enolpyruvylshikimate-3-phosphate synthase from Rahnella aquatilis with significantly reduced glyphosate sensitivity. PLoS One. 7, e39579 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledzewski K. & Mendel R. R. Quantitative transient gene expression: comparison of the promoters for maize polyubiquitin1, rice actin1, maize-derived Emu and CaMV 35S in cells of barley, maize and tobacco. Transgenic Res. 3, 249–255 (1994). [Google Scholar]
- Alibhai M. F. et al. Glyphosate resistant class I 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS). United Patent US 7, 723, 575 B2 (2010).
- Zhou M. et al. Identification of a glyphosate-resistant mutant of rice 5-enolpyruvylshikimate 3-phosphate synthase using a directed evolution strategy. Plant physiol. 140, 184–195 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar I. K. & Sakthivel N. Advances in selectable marker genes for plant transformation. J. Plant Physiol. 165, 1698–1716 (2008). [DOI] [PubMed] [Google Scholar]
- Hiei Y. & Komari T. Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nature protoc. 3, 824–834 (2008). [DOI] [PubMed] [Google Scholar]
- Howe A. R. et al. Glyphosate as a selective agent for the production of fertile transgenic maize (Zea mays L.) plants. Mol. Breeding. 10, 153–164 (2002). [Google Scholar]
- Shrawat A. K. & Loerz H. Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol. J. 4, 575–603 (2006). [DOI] [PubMed] [Google Scholar]
- Wang J. X., Zhao E. Y. & Xu P. Use of aroA-M1 as a selectable marker for Brassica napus transformation. Crop Sci. 46, 706–711 (2006). [Google Scholar]
- Chandrasekhar K. et al. Development of transgenic rice harbouring mutated rice 5-enolpyruvylshikimate 3-phosphate synthase (Os-mEPSPS) and Allium sativum leaf agglutinin (ASAL) genes conferring tolerance to herbicides and sap-sucking insects. Plant Mol. Biol. Rep. 32, 1146–1157 (2014). [Google Scholar]
- Koger C. H. et al. Rice (Oryza sativa) response to drift rates of glyphosate. Pest Manag. Sci. 61, 1161–1167 (2005). [DOI] [PubMed] [Google Scholar]
- Tian Y. S. et al. Improvement of glyphosate resistance through concurrent mutations in three amino acids of the Ochrobactrum 5-enopyruvylshikimate-3-phosphate synthase. Appl. Environ.Microb. 77, 8409–8414 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T. & Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene 26, 101–106 (1983). [DOI] [PubMed] [Google Scholar]
- Chenna R. et al. Multiple sequence alignment with the clustal series of programs. Nucleic Acids Res. 31, 3497–3500 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzetta P. A., Alvarez L. J., Reinach P. S. & Candia O. A. An improved assay for nanomole amounts of inorganic phosphate. Anal.Biochem. 100, 95–97 (1979). [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T. & Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6, 271–282 (1994). [DOI] [PubMed] [Google Scholar]
- Murray M. G. & Thompson W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.