Abstract
The Influenza A virus is a great threat for human health, while various subtypes of the virus made it difficult to develop drugs. With the development of state-of-art computational chemistry, computational molecular docking could serve as a virtual screen of potential leading compound. In this study, we performed molecular docking for influenza A H1N1 (A/PR/8/34) with small molecules such as quercetin and chlorogenic acid, which were derived from traditional Chinese medicine. The results showed that these small molecules have strong binding abilities with neuraminidase from H1N1 (A/PR/8/34). Further details showed that the structural features of the molecules might be helpful for further drug design and development. The experiments in vitro, in vivo have validated the anti-influenza effect of quercetin and chlorogenic acid, which indicating comparable protection effects as zanamivir. Taken together, it was proposed that chlorogenic acid and quercetin could be employed as the effective lead compounds for anti-influenza A H1N1.
Influenza A virus is a type of orthomyxoviridae virus which is highly implicated in human health1. Through infecting the mucosa of upper respiratory tract, the influenza A virus could induce the acute respiratory disease1.Previously, a large number of biological studies have characterized the molecular basis of influenza A virus. Although there were a number of distinct types, in all influenza A viruses, 8 genes were conservatively encoded by RNA segments and could be translated into 11 different proteins through distinct open reading frames (ORFs)2.The classification of different influenza A virus subtypes were based on the two surface glycoproteins including hemagglutinin (HA) and neuraminidase (NA)3. Previously, it was identified that NA was critical for the replication and spread of influenza A virus, and NA inhibitor could serve as the anti-influenza A drugs4,5. These understandings provide great help to further biological and medical studies of influenza A.
Since the epidemic influenza A is threat to public health, it is critical to develop anti-influenza A drug. Currently, there are three anti-influenza A drugs including amantadine, oseltamivir and zanamivir6. Amantadine is the inhibitor of the matrix protein M2, while oseltamivir and zanamivir are inhibitors of neuraminidase (NA)6. Although these drugs are effective, the drug-resistant strains of influenza A virus also emerged with the wide usage of these drugs. So, it is critical to develop new anti-influenza A drug. Previously, abundant traditional Chinese medicines (TCMs) from herbaceous plants such as Forsythia suspense, Mangifera indica, Hypericum perforatum and Chaenomeles speciosa, were proved to be effective in clinical practice while the complication of their composition remains to be discovered7,8,9,10. With the advancement of the bioanalysis technology, a number of small molecules such as quercetin, chlorogenic acid, hypericin, rosmarinci acid and chrysophanol derived from the TCMs were found to be bioactive11,12,13,14. For example, as the most frequently studied flavonoid, quercetin was found to be good for heath through protecting low-density lipoprotein from oxidation15, antithrombic effects and antiviral properties, while recent studies also showed quercetin was anti-inflammatory and helpful for reduce the risk of pancreatic cancer for smokers16,17. Chlorogenic acid, a natural polyphenol product from various plants, has antiviral and antihypertensive activity, and could protect dopaminergic neuron from neuroinflammation18,19,20.
In this study, we employed the virtual screen method to explore the potential anti-influenza A activity of several TCM small molecules including quercetin and chlorogenic acid. The H1N1 (A/PR/8/34) subtype was employed for computational and experimental studies. Autodock was used to perform the molecular docking, and the structure of NA protein was modeled from previously reported N1 structure. The results showed that, it was found that the computed binding energy between NA and the two molecules were comparable with the results that from zanamivir. To test the antiviral ability of quercetin and chlorogenic acid, the experiments in vitro and in vivo were performed. The results suggested quercetin and chlorogenic acid have the antiviral ability. Further detailed analyses showed that these two molecules could serve as a good start for NA inhibitor-like anti-influenza A drug development.
Results
Preparation the structures of A/PR/8/34 H1N1 NA and small molecules
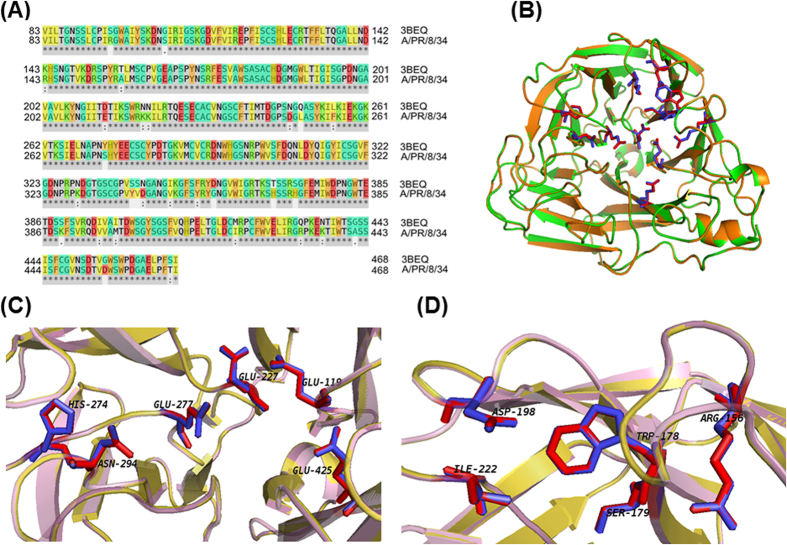
In this study, we selected the structure of NA from A/Brevig Mission/1/1918 H1N1 (PDB ID: 3BEQ) as the template21. Through sequence alignment with Clustal Omega22, the identity of the two sequences is computed as 93.25%, and the detailed alignment result was presented in Fig. 1A. Previously, it was found that a number of residues including Glu119, Arg156,Trp178, Ser179, Asp/Asn198, Ile222, Glu227, His274, Glu277, Asn294, and Glu425 were critical for the activity of NA21. It was observed that these key residues were conserved between the two sequences (Fig. 1A).
Figure 1. The sequence and structural alignment for the NA from A/PR/8/34 H1N1 and A/Brevig Mission/1/1918 H1N1.
(A) The sequence alignment result. (B) The structural alignment result. The details for structural alignment were presented in (C,D).
To further evaluate the modeling, the modeled structure was compared with the template, and the result was presented by PyMOL23. It was obvious that the two structures were nearly identical (Fig. 1B). The detailed results for the local structure of the key residues were presented in Fig. 1C,D, which indicated that the pocket for NA activity is structurally conserved. Taken together, it was observed that the modeled structure is reliable for further computational studies.
Molecular docking between NA and small molecules
To further investigate the potential binding between NA and the compounds, the molecular docking was performed. The molecular docking between zanamvir and NA was employed as the control to evaluate the binding ability of other molecules. The binding energies for the fifteen small molecules from docking results were summarized in Fig. 2, which showed that quercetin and chlorogenic acid had highest binding energies comparable with zanamvir. Thus, further investigations in this study were focused on quercetin and chlorogenic acid. The detailed docking results such as binding energies and inhibition constants were presented in Table 1. It was observed that the binding energies between NA and the two molecules are comparable with zanamvir. The docking energy for zanamvir, quercetin and chlorogenic acid were −11.24 kcal/mol, −10.23 kcal/mol and −11.05 kcal/mol, respectively. These results indicated that the NA binding ability of the small molecules were useful for further studies.
Figure 2. The top15 small molecules with high binding free energies with NA, while zanamvir served as the control.
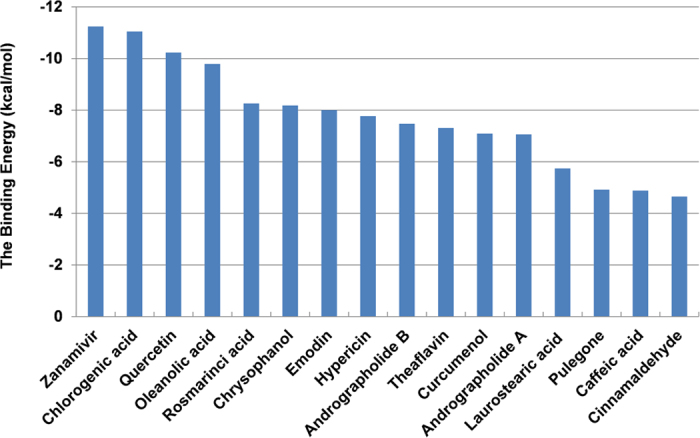
Table 1. The energies for the binding between the small molecules and NA.
| Molecules | (vdW + Hbond + desolv) energy(kcal/mol) | Electrostatic energy (kcal/mol) | Total Internal energy (kcal/mol) | The best docking energy (kcal/mol) | Inhibition constant (nM) |
|---|---|---|---|---|---|
| Zanamvir | −11.1 | −3.1 | −17.92 | −11.24 | 5.73 |
| Quercetin | −8.09 | −0.59 | −19.65 | −10.23 | 31.97 |
| Chlorogenic acid | −11.1 | −2.49 | −22.43 | −11.05 | 7.98 |
To further dissect the docking energies, the contributions of different energies were analyzed. For the interaction between NA and small molecules, the van der Waals’ interaction, hydrogen bond (H-bond), desolvation energy and electrostatic energy contributed as the major part (vdW + Hbond + desolv). In the docking results, it was observed that the vdW + Hbond + desolv energy for chlorogenic acid is −11.1 kcal/mol, which is equal with zanamvir. For quercetin, the result was smaller as −8.09 kcal/mol, respectively. Zanamvir aslo has the highest electrostatic energy as −3.1 kcal/mol, while the electrostatic energies for quercetin and chlorogenic acid were −0.59 kcal/mol and −2.49 kcal/mol, respectively. The internal energy and inhibition constant were also calculated and presented in Table 1, while all the results indicated these small molecules have the potential to binding NA. Furthermore, the inhibition constants calculated for zanamivir, quercetin and chlorogenic acid were 5.73, 31.97 nd 7.98nM, respectively (Table 1), which indicated that quercetin and chlorogenic acid might have considerable NA inhibition ability.
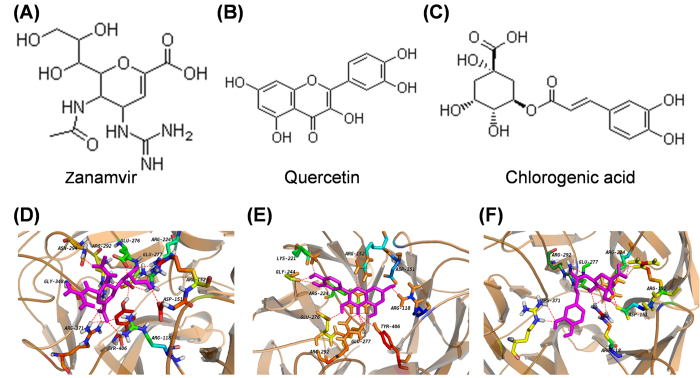
To further evaluate the druggabilities, toxicities and promiscuities of quercetin and chlorogenic acid, FAF-Drugs324 was employed to perform chemical informatic analyses for zanamvir, quercetin and chlorogenic acid. The structures of the molecules were presented in Fig. 3, while the results were showed in Table 2. For quercetin and chlorogenic acid, it was observed that: the molecular weights were similar as ~500; the logarithm of the partition coefficient between water and 1-octanol (known as logP) was less than five; the number of rotatable bonds is less than 10; the number of hydrogen atoms acceptors is less than 10; the number of hydrogen atoms donors is less than 10. Thus, quercetin and chlorogenic acid matched Lipinski’s rule of five25,26, which indicated that these two compounds had low toxicities and promiscuities, and high druglikeness.
Figure 3.
The structures of zanamvir (A), quercetin (B), chlorogenic acid (C), and the details of the binding between NA and zanamvir (D), quercetin (E), chlorogenic acid (F).
Table 2. The chemical informatics analyses of zanamvir, quercetin and chlorogenic acid.
| Molecules | MW | LogP | tPSA | Rotatable bonds | HB donors | HB acceptors | Oral Bioavailability (VEBER) | Oral Bioavailability (EGAN) |
|---|---|---|---|---|---|---|---|---|
| Zanamvir | 333.32 | −2.98 | 202.79 | 7 | 9 | 11 | ||
| Quercetin | 302.24 | 2.17 | 134.2 | 1 | 5 | 7 | Good | Good |
| Chlorogenic acid | 354.31 | −0.42 | 167.6 | 5 | 5 | 9 | Good | Good |
Structural insights of the docking complex
To further understanding the details of NA binding for the molecules, the molecular structures for the NA in complex with the molecules were analyzed. The complex structures for zanamvir, quercetin and chlorogenic acid were presented in Fig. 3. The key residues were labeled while the interaction details were visualized. As the important molecular mechanism, the H-bond were analyzed and shown as dotted line, while the residues from NA for H-bond were summarized in Table 3.
Table 3. The residues for the H-bonds between the small molecules and NA.
| Molecules | H-bond residues |
|---|---|
| Zanamvir | ARG118, ASP151, ARG152, ARG224, GLU276, GLU277, ARG292, ASN294, GLY348, ARG371, ARG406 |
| Quercetin | ARG118, ASP151, ARG152, LYS211, ARG224, GLY244, GLU276, GLU277, ARG292, TYR406 |
| Chlorogenic acid | ARG118, ASP151, ARG152, ARG224, GLU277, ARG292, ARG371 |
As the result presented in Fig. 3D, it was observed that there were 21 H-bonds between zanamvir and 11 residues of NA. The binding area was critical for the activity of NA, while the abundant H-bonds indicated that the H-bonds were important for the binding between small molecules and NA. Figure 4E shows that the binding of quercetin could generate 17 H-bonds, which indicated that quercetin could serve as a leading molecule as NA inhibitor. For chlorogenic acid, 15 H-bonds could be formed with NA (Fig. 4F), while the average energy for H-bonds was higher than zanamvir.
Figure 4.
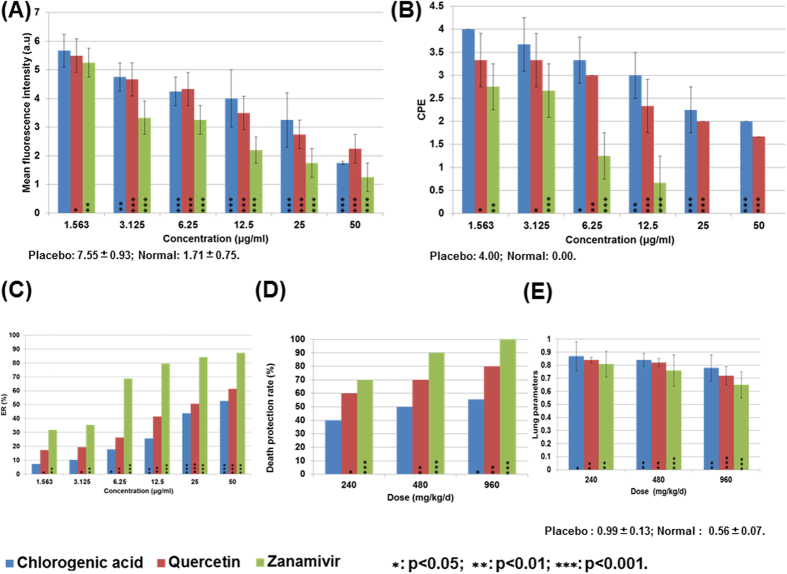
The summary of the experimental results for cytopathic effect (CPE) (A), mean fluorescence intensity (B), effect rate (C), death protection rate (D) and lung index (E).
Validation of NA inhibition ability for small molecules
To validate the NA inhibition ability for small molecules that showed high binding potential in the docking experiments, experiments including in vitro neuraminidase activity assay and cytopathic effect assay were carried out. Firstly, MTT cell proliferation assay was performed to obtain the maximum non-toxic dose (MNLD). It was observed that the MNLDs for zanamvir, quercetin and chlorogenic acid were 71.31, 61.54 and 73.72 μg/ml, respectively, which indicated that the toxicities of the three compounds might be similar. Then a series of concentrations including 1.56, 3.13, 6.25, 12.5, 25 and 50 μg/ml were employed, while the in vitro neuraminidase activity assay and cytopathic effect assay were parallel performed. From the results shown in Table 4 (Fig. 4A), it was observed that the three compounds could significantly inhibit the activity of neuraminidase at the concentrations of 1.56 μg/ml for zanamvir and quercetin and 3.13 μg/ml for chlorogenic acid (Fig. 4A). Furthermore, since the fluorescence intensity was reported as no significant difference in comparison with the control, it seems that the activity of neuraminidase was nearly fully inhibited by quercetin and chlorogenic acid under the concentration of 50 μg/ml. These results indicated that quercetin and chlorogenic acid showed considerably comparable neuraminidase inhibition abilities.
Table 4. Validation of NA inhibition ability for small molecules.
| Concentration (μg/ml) | Mean CPE ± S.D. | ER(%) | Mean fluorescence intensity (A.U ± S.D.) | |
|---|---|---|---|---|
| Chlorogenic acid | 50 | 2.00 ± 0.00*** | 52.7*** | 2.25 ± 0.39*** |
| 25 | 2.25 ± 0.50*** | 43.8*** | 3.31 ± 0.25*** | |
| 12.5 | 3.00 ± 0.50** | 25.6** | 4.11 ± 0.32*** | |
| 6.25 | 3.33 ± 0.50* | 17.8* | 4.43 ± 0.38*** | |
| 3.125 | 3.67 ± 0.58 | 10.2 | 4.75 ± 0.45** | |
| 1.563 | 4.00 ± 0.00 | 5.3 | 5.67 ± 0.57 | |
| Quercetin | 50 | 1.67 ± 0.00*** | 59.3*** | 1.74 ± 0.16*** |
| 25 | 2.00 ± 0.00*** | 50.6*** | 2.67 ± 0.20*** | |
| 12.5 | 2.33 ± 0.58*** | 41.5** | 3.50 ± 0.26*** | |
| 6.25 | 3.00 ± 0.00** | 26.3** | 4.34 ± 0.31*** | |
| 3.125 | 3.33 ± 0.58* | 19.4* | 4.63 ± 0.41*** | |
| 1.563 | 3.33 ± 0.58* | 17.7* | 5.48 ± 0.52* | |
| Zanamvir | 50 | 0.00 ± 0.00*** | 87.3*** | 1.24 ± 0.11*** |
| 25 | 0.00 ± 0.00*** | 84.1*** | 1.73 ± 0.15*** | |
| 12.5 | 0.67 ± 0.58*** | 79.6*** | 2.20 ± 0.25*** | |
| 6.25 | 1.25 ± 0.50*** | 68.8*** | 3.27 ± 0.29*** | |
| 3.125 | 2.67 ± 0.58*** | 35.3** | 3.34 ± 0.40*** | |
| 1.563 | 2.75 ± 0.50** | 31.7** | 5.35 ± 0.48** | |
| Placebo | – | 4.00 ± 0.00 | – | 7.54 ± 0.61 |
| Normal | – | 0.00 ± 0.00 | – | 1.71 ± 0.14 |
*p < 0.05, **p < 0.01, ***p < 0.001, the values were compared with placebo controls.
Furthermore, the protection effect of the three compounds for cells were evaluated and compared. The cytopathic effect (CPE) and protective effect rate (ER) were calculated and presented in Table 4 and Fig. 4B,C. It was observed that the cytopathic effect (CPE) and protective effect rate (ER) for cells treated with quercetin and chlorogenic acid were significantly low and high, respectively, which indicated that the two compounds have protection effect for influenza-infected cells. Interestingly, the CPE results were consistent with neuraminidase activity assay that the quercetin and chlorogenic acid were effective at low concentrations of 1.56 μg/ml and 3.13 μg/ml, respectively. The MTT based protective effect rate (ER) assay for the three compounds showed a dose-dependent results (Table 4, Fig. 4C).
In vivo validation of anti-influenza abilities of quercetin and chlorogenic acid
Since the infection of influenza could cause the death, increase lung index and pneumonia, survival rate and lung index (Lung weight / Body weight) could be employed to evaluate the protection effect of the compounds27. In this study, influenza A H1N1 (A/PR/8/34) infected mice were treated with compounds of different doses including 240, 480 and 960 mg/kg/d through intragastric administration. As shown in Table 5 and Fig. 4D, treatments with chlorogenic acid and quercetin at the dose of 240 mg/kg/d could promote the survival rates from 40% and 60% to 56% and 80%, respectively. Furthermore, the inhibition rate of lung index were measured for the chlorogenic acid and quercetin treated mice as 20.2%, 14.4%, 12.2%, and 26.1%, 16.5%, 14.7% at the doses of 240, 480 and 960 mg/kg/d. It was observed that the inhibition rates were in a dose-dependent manner, while the survival rate and lung index for quercetin (80%, 0.72 ± 0.07) at 960 mg/kg/d were similar with zanamvir at 480 mg/kg/d (90%, 0.65 ± 0.10) (Table 5 and Fig. 4E).
Table 5. In vivo validation of anti-influenza abilities of quercetin and chlorogenic acid.
| Treatment | Dose (mg/kg/d) | Survivors/totals (%) | Mean lung parameters |
|
|---|---|---|---|---|
| Lung Index ± S.D. (%) | Lung indexInhibition % | |||
| Zanamivir | 960 | 10/10 (100)*** | 0.65 ± 0.10*** | 34.0 |
| 480 | 9/10 (90)*** | 0.74 ± 0.11*** | 25.3 | |
| 240 | 7/10 (70)*** | 0.81 ± 0.11** | 18.3 | |
| Quercetin | 960 | 8/10 (80)** | 0.73 ± 0.07*** | 26.3 |
| 480 | 7/10 (70)** | 0.82 ± 0.03** | 16.5 | |
| 240 | 6/10 (60)* | 0.84 ± 0.02** | 14.7 | |
| Chlorogenic acid | 960 | 5/9 (55.6)* | 0.79 ± 0.10** | 20.2 |
| 480 | 5/10 (50) | 0.84 ± 0.05** | 14.4 | |
| 240 | 4/10 (40) | 0.87 ± 0.11* | 12.2 | |
| Placebo | — | 1/10 (10) | 0.99 ± 0.13 | — |
| Normal | — | 10/10 (100) | 0.56 ± 0.07 | — |
*p < 0.05, ** p < 0.01, ***p < 0.001, the values were compared with placebo controls.
Material and methods
Molecular structure preparing
The NA structure of A/PR/8/34 H1N1 was modeled from the structure of A/Brevig Mission/1/1918 H1N1 (PDB ID: 3BEQ)21 by SWISS-MODEL Workspace28,29,30. The identity of two NA sequences between A/Brevig Mission/1/1918 and A/PR/8/34 was 93.25%, which indicated that the modeled structure was reliable. All the visualizations of molecular structures were performed with PyMOL (Version 1.7.2, available at http://pymol.org)23. To find potential anti-influenza small molecules, we curated the plants from traditional Chinese medicine which were identified to have anti-influenza effects from published literature in Chinese. Then the components which were bioactive and identified in these plants were collected. In total, fifteen small molecules were collected for molecular docking inlcuding hypericin from Forsythia suspense, chlorogenic acid, laurostearic acid, oleanolic acid, caffeic acid, quercetin, theaflavin from Honeysuckle, andrographolide A, andrographolide B, curcumenol from Andrographis paniculata, chrysophanol, emodin from Rheum, pulegone, rosmarinci acid from Rosmarinus officinalis, and cinnamaldehyde from the root of Chinese thorowa. The structures for these small molecules were retrieve from the ZINC database, which is a curated collection of commercially available chemical compounds31.
Molecular docking
In this study, AutoDock software version 4.2 (http://autodock.scripps.edu) were employed to perform molecular docking as previously described32. AutoDock incorporates limited flexibility in the receptor, it combines an empirical free energy force field with a Lamarckian Genetic Algorithm, providing fast prediction of bound conformations with predicted free energies of association33,34. The binding energy is calcualted as the sum of the intermolecular energy and the torsional free-energy34,35,36. The modeled NA structure was employed as the receptor to docking with these small molecules. The AutoDockTools1.5.6 software was employed to prepare the docking procedures33. The default 0.375 Å spacing was adopted for the grid box, and the volume was set as 72 × 72 × 72. The key residues for NA were selected as the docking pocket, which contains eight residues including Arg118, Asp151,Arg152, Arg224, Glu276, Arg292, Arg371, Tyr4021,37,38,39.The number of runs was set as 100. Furthermore, FAF-Drugs3 software (http://fafdrugs3.mti.univ-paris-diderot.fr) was used to carried out chemical informatics analyses for the small molecules24.
Virus strain, cell culture and animals
Animals (BALB/c mice) were used according to the guidelines of the National Institutes of Health for care and use of laboratory Animals. All experiments requiring animals received prior approval from the Animal Care and Use Committee of the College of Medicine, Hunan Normal University. A/Puerto Rico/8/34(H1N1) virus was prepared as described in our preview research40. The influenza A/Puerto Rico/8/34(H1N1) virus and Madin-Darby canine kidney (MDCK) cells were preserved in Molecular Virology Lab, School of Life Sciences, Hunan Normal University. MDCK cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and incubated in an atmosphere of 5% CO2 at 37 °C which had been humidified. Female BALB/c mice (19–22g) were obtained from SJA Laboratory Animal Co., Ltd., Hunan, China.
Reagents and compounds
Chlorogenic acid (C3878), quercetin (Q4591), dimethyl sulfoxide (DMSO) (276855), sodium carboxymethyl cellulose (CMC) (C5678), zanamivir (102157A) and MTT (45956A) (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) were purchased from Sigma-Aldrich Trading Co., Ltd., Shanghai, China. Zanamivir was provided by Adamas Pharmaceuticals. Molecules were dissolved in DMSO and CMC for in vitro and in vivo experiments.
Cytotoxic Assay for Zanamivir, quercetin and chlorogenic acid
The noncytotoxic doses of three kinds of molecules on MDCK cells were determined by the methyl-thiazolyl-tetrazolium (MTT) method respectively and the maximum non-toxic concentrations (TC0) were calculated by R package ‘drc’41.
The cytopathic effect assay
MDCK cells grown in 24-well plates for 24 h were washed with serum-free medium and then infected with 400 μl of 100 TCID50 influenza virus A/PR/8/34 for 2 h at 37 °C. Unabsorbed virus was removed and the cells were treated with various doses of chlorogenic acid or quercetin solution (1.56, 3.13, 6.25, 12.5, 25.0, 50.0 μg/ml). The uninfected cell controls and placebo controls were included in each test. Zanamivir served as the positive control drug, which was used for estimating antiviral efficiency. Then, the plates were incubated at 37 °C. This experiment was repeated 3 times. The cell monolayers were examined by microscope every 12 h, and the cytopathic effect (CPE) induced by influenza virus was marked on a scale from 0 (no CPE) to 4 (100% CPE).
Evaluation of anti-H1N1 effect rate
MDCK cells were cultured in 96-well plates and cultured in minimum essential medium (MEM) for 2 days at 37 °C to >90% confluence. Cells were washed with serum-free Medium and infected with 100 μl of 100 TCID50 influenza virus A/PR/8/34 (H1N1) for 2 h at 37 °C. After removing the virus-containing medium, the cells were treated with various doses of chlorogenic acid, quercetin and zanamivir solution in MEM. Zanamivir served as positive control in all experiments, which was estimated for antiviral efficiency. Then, the 96-well microtiter plates were cultured at 37 °C and 5% CO2 for 48 h. The antiviral efficiency was determined with MTT method, the absorbance of the solution was measured spectrophotometrically at 570 nm with automated plate reader, while the effect rate (ER) was defined as follows:
 |
Enzymatic Assay of NA Activity by Fluorometric Substrate
When the monolayer MDCK cells cultured in 96-wells plates grew to 90% of plates, the maintenance medium was removed from replicates. The wells were washed with serum-free medium and inoculated with 100 μl of 100TCID50 influenza virus A/PR/8/34 (H1N1) for 2 h at 37 °C incubator which contained 5% CO2. The unabsorbed virus was removed, the cells were treated with molecules solution in a series of concentration from 1.563 ug/ml to 50 μg/ml. Zanamivir was used to evaluate the ability of molecules antiviral activity and served as the positive control. Then, the plates were sealed and incubated at 37 °C for 48 h. The cells were frozen and thawed three times. 8 min later, the supernatant of the cells was harvested for the assay of neuraminidase by Neuraminidase Inhibitors Identification Kit P0306-1.
In vivo anti-influenza virus experiments
A total number of 198 BALB/c female mice were randomly divided into 11 groups. All of mice were anesthetized by 1% pentobarbital sodium solution, and then infected with 24MLD50 mouse adapted influenza virus A/PR/8/34 suspension with 0.1% bovine serum albumin (BSA) in PBS through intranasal administration.
The oral administrations of chlorogenic acid and quercetin were suspended in 0.1% (w/v) sodium carboxymethylcellulose (CMC) solution, and 24 h after virus infection the suspension was delivered to the mouse with several doses of chlorogenic acid and quercetin solution (240, 480, 960 mg/kg/d) twice daily for 4d. Zanamvir and CMC were given as a positive controlled and placebo controlled, respectively.
Ten mice per group were used to observe for clinical signs of infection or death in 14d. On day 5 of infection with virus, lung infection parameters were assessed and per group had 8 animals. Mice were weighed and sacrificed, and then their lungs were removed and weighted. Body weight and lung weight were assigned to A and B, lung index of the drug-treated group and lung index of the infected controls were assigned to C and D, respectively. The lung index means B/A × 100%, the lung index inhibition means (D – C)/D × 100%.
Discussion
Recently, influenza A became a great threat to human health, while the effective drugs were limited because of the resistance to drugs generated from various mutation6,29,42,43,44,45. However, conventional drug development is time-consuming and labor-intensive, while computer-aid drug design could provide helpful information. In this study, computational screen was employed to discover small molecules with NA inhibition ablities and anti-influenza potential from TCMs. Since the protein structure is the basis for molecular docking based screening, the structure of NA (A/PR/8/34 H1N1) was computationally modeled from NA (A/Brevig Mission/1/1918 H1N1) through state-of-art homology-based computation. Through computational screen, we proposed that small molecules from TCMs including chlorogenic acid and quercetin might be highly potential anti-influenza candidate. The in vitro and in vivo experiments validated the protective effects of the two molecules. It was observed that chlorogenic acid and quercetin could protect influenza A H1N1 infected cells and mice at a comparable level of zanamvir, while quercetin acts at a lower dose. In this study, the small molecules were delivered through intragastric administration, which ensured the absorption. However, the rate of oral absorption of zanamvir is limited as 2%10, which indicated that the deliver efficiency of chlorogenic acid and quercetin should be carefully investigated. Furthermore, the in vivo absorption and metabolism of small molecules is complicated. Previous study showed that the metabolism product of oseltamivir carboxylate for oseltamivir is the active component with anti-influenza activity46. For quercetin, previous study showed that it was absorbed at the rate of 59.1% after hydrolyzing in intestinal tract47, while only 5.3% was absorbed directly48. The absorption of chlorogenic acid was also low and relied on methylation in intestinal tract49,50. Thus, further studies should be contributed to analyze the pharmacokinetics for these two small molecules. Taken together, the molecular docking results indicated that chlorogenic acid and quercetin could serve as leading molecules for the development of anti-influenza A drug, while this study provide an initial step for the further design and improvement.
Additional Information
How to cite this article: Liu, Z. et al. Computational screen and experimental validation of anti-influenza effects of quercetin and chlorogenic acid from traditional Chinese medicine. Sci. Rep. 6, 19095; doi: 10.1038/srep19095 (2016).
Acknowledgments
The authors thank Mr. Zhicheng Pan (HKUST) for kind help on molecular docking. This study was supported by grants from National Natural Science Foundation of China (31501069, 81100054), the Innovative Training Center for College Student of Life Sciences (201301012), and the Construct Program of the Key Discipline of Basic Medicine in Hunan Province. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions Z.C., H.C. and R.Z. designed and supervised experiments. Z.L., J.Z. and W.L. performed experiments and data analysis. S.H., J.T., J.D., L.S. and F.F. contributed to experiments and data analysis. Z.L., J.Z., W.L., Z.C., H.C. and R.Z. wrote the manuscript with contributions of all authors.
References
- Taubenberger J. K. & Morens D. M. The pathology of influenza virus infections. Annu Rev Pathol 3, 499–522, 10.1146/annurev.pathmechdis.3.121806.154316 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q. et al. The influenza A virus PB2, PA, NP, and M segments play a pivotal role during genome packaging. J Virol 86, 7043–7051, 10.1128/JVI.00662-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J. et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125, 10.1038/nature08182 (2009). [DOI] [PubMed] [Google Scholar]
- Nayak D. P. & Jabbar M. A. Structural domains and organizational conformation involved in the sorting and transport of influenza virus transmembrane proteins. Annu Rev Microbiol 43, 465–501, 10.1146/annurev.mi.43.100189.002341 (1989). [DOI] [PubMed] [Google Scholar]
- Michiels B., Van Puyenbroeck K., Verhoeven V., Vermeire E. & Coenen S. The value of neuraminidase inhibitors for the prevention and treatment of seasonal influenza: a systematic review of systematic reviews. PLoS One 8, e60348, 10.1371/journal.pone.0060348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pica N. & Palese P. Toward a universal influenza virus vaccine: prospects and challenges. Annu Rev Med 64, 189–202, 10.1146/annurev-med-120611-145115 (2013). [DOI] [PubMed] [Google Scholar]
- Ge H. et al. Anti-influenza agents from traditional Chinese medicine. Nat Prod Rep 27, 1758–1780 (2010). [DOI] [PubMed] [Google Scholar]
- Wang X., Jia W., Zhao A. & Wang X. Anti‐influenza agents from plants and traditional Chinese medicine. Phytother Res 20, 335–341 (2006). [DOI] [PubMed] [Google Scholar]
- Ma S.-C. et al. Antiviral Chinese medicinal herbs against respiratory syncytial virus. J Ethnopharmacol 79, 205–211 (2002). [DOI] [PubMed] [Google Scholar]
- Sawai R. et al. Anti-influenza virus activity of Chaenomeles sinensis. J Ethnopharmacol 118, 108–112 (2008). [DOI] [PubMed] [Google Scholar]
- Nahrstedt A. & Butterweck V. Biologically active and other chemical constituents of the herb of Hypericum perforatum L. Pharmacopsychiatry, 129–134 (1997). [DOI] [PubMed] [Google Scholar]
- Luan L., Wang G. & Lin R. Studies on the chemical constituents of extract with water from Forsythia suspensa. Zhong Yao Cai 33, 220–221 (2010). [PubMed] [Google Scholar]
- Yang Y. et al. Studies on the chemical constituents of Chaenomeles speciosa. Zhong Yao Cai 32, 1388–1390 (2009). [PubMed] [Google Scholar]
- Xianfei X., Xiaoqiang C., Shunying Z. & Guolin Z. Chemical composition and antimicrobial activity of essential oils of Chaenomeles speciosa from China. Food Chem 100, 1312–1315 (2007). [Google Scholar]
- Formica J. V. & Regelson W. Review of the biology of Quercetin and related bioflavonoids. Food Chem Toxicol 33, 1061–1080 (1995). [DOI] [PubMed] [Google Scholar]
- Stewart L. K. et al. Quercetin transiently increases energy expenditure but persistently decreases circulating markers of inflammation in C57BL/6J mice fed a high-fat diet. Metabolism 57, S39–46, 10.1016/j.metabol.2008.03.003 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothlings U., Murphy S. P., Wilkens L. R., Henderson B. E. & Kolonel L. N. Flavonols and pancreatic cancer risk: the multiethnic cohort study. Am J Epidemiol 166, 924–931, 10.1093/aje/kwm172 (2007). [DOI] [PubMed] [Google Scholar]
- Zhao Y., Wang J., Ballevre O., Luo H. & Zhang W. Antihypertensive effects and mechanisms of chlorogenic acids. Hypertens Res 35, 370–374, 10.1038/hr.2011.195 (2012). [DOI] [PubMed] [Google Scholar]
- Shen W. et al. Chlorogenic acid inhibits LPS-induced microglial activation and improves survival of dopaminergic neurons. Brain Res Bull 88, 487–494, 10.1016/j.brainresbull.2012.04.010 (2012). [DOI] [PubMed] [Google Scholar]
- Wang G. F. et al. Anti-hepatitis B virus activity of chlorogenic acid, quinic acid and caffeic acid in vivo and in vitro. Antiviral Res 83, 186–190, 10.1016/j.antiviral.2009.05.002 (2009). [DOI] [PubMed] [Google Scholar]
- Xu X., Zhu X., Dwek R. A., Stevens J. & Wilson I. A. Structural characterization of the 1918 influenza virus H1N1 neuraminidase. J Virol 82, 10493–10501, 10.1128/JVI.00959-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWilliam H. et al. Analysis Tool Web Services from the EMBL-EBI. Nucleic Acids Res 41, W597–600, 10.1093/nar/gkt376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger L. L. C. The PyMOL Molecular Graphics System, Version 1.3r1 (2010).
- Lagorce D. et al. The FAF-Drugs2 server: a multistep engine to prepare electronic chemical compound collections. Bioinformatics 27, 2018–2020, 10.1093/bioinformatics/btr333 (2011). [DOI] [PubMed] [Google Scholar]
- Leeson P. Drug discovery: Chemical beauty contest. Nature 481, 455–456, 10.1038/481455a (2012). [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W. & Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46, 3–26 (2001). [DOI] [PubMed] [Google Scholar]
- Johansson B. E., Grajower B. & Kilbourne E. D. Infection-permissive immunization with influenza virus neuraminidase prevents weight loss in infected mice. Vaccine 11, 1037–1039 (1993). [DOI] [PubMed] [Google Scholar]
- Kiefer F., Arnold K., Kunzli M., Bordoli L. & Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res 37, D387–392, 10.1093/nar/gkn750 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J. & Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 (2006). [DOI] [PubMed] [Google Scholar]
- Schwede T., Kopp J., Guex N. & Peitsch M. C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31, 3381–3385 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S. & Coleman R. G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52, 1757–1768, 10.1021/ci3001277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. Molecular Docking of Potential Inhibitors for Influenza H7N9. Comput Math Methods Med 2015, 480764, 10.1155/2015/480764 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30, 2785–2791, 10.1002/jcc.21256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19, 1639–1662 (1998). [Google Scholar]
- Huey R. & Morris G. M. Using AutoDock 4 with AutoDockTools: A Tutorial. The Scripps Research Institute, USA, 54–56 (2008). [Google Scholar]
- Huey R., Morris G. M., Olson A. J. & Goodsell D. S. A semiempirical free energy force field with charge–based desolvation. J Comput Chem 28, 1145–1152 (2007). [DOI] [PubMed] [Google Scholar]
- Mitrasinovic P. M. Advances in the structure-based design of the influenza A neuraminidase inhibitors. Curr Drug Targets 11, 315–326 (2010). [DOI] [PubMed] [Google Scholar]
- Gong J., Xu W. & Zhang J. Structure and functions of influenza virus neuraminidase. Curr Med Chem 14, 113–122 (2007). [DOI] [PubMed] [Google Scholar]
- Monajjemi M., Mousavi M., Rezaei S. & Mollaamin F. Molecular dynamic and monte carlo study on nanoenergetic binding sites of neuraminidase in different media. Afr J Microbiol Res 6, 1713–1717 (2012). [Google Scholar]
- Li X. et al. Essential sequence of influenza neuraminidase DNA to provide protection against lethal viral infection. DNA Cell Biol 25, 197–205, 10.1089/dna.2006.25.197 (2006). [DOI] [PubMed] [Google Scholar]
- Ritz C. & Streibig J. C. Bioassay analysis using R. J Stat Softw 12, 1–22 (2005). [Google Scholar]
- Cheung C.-L. et al. Distribution of amantadine-resistant H5N1 avian influenza variants in Asia. J Infect Dis 193, 1626–1629 (2006). [DOI] [PubMed] [Google Scholar]
- Hayden F. G. et al. Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N Engl J Med 321, 1696–1702 (1989). [DOI] [PubMed] [Google Scholar]
- Suzuki H. et al. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother 9, 195–200 (2003). [DOI] [PubMed] [Google Scholar]
- Hurt A. C., Holien J. K., Parker M., Kelso A. & Barr I. G. Zanamivir-resistant influenza viruses with a novel neuraminidase mutation. J Virol 83, 10366–10373 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies B. E. Pharmacokinetics of oseltamivir: an oral antiviral for the treatment and prophylaxis of influenza in diverse populations. J Antimicrob Chemother 65, ii5–ii10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day A. J. et al. Dietary flavonoid and isoflavone glycosides are hydrolysed by the lactase site of lactase phlorizin hydrolase. FEBS Lett 468, 166–170 (2000). [DOI] [PubMed] [Google Scholar]
- Chen X., Yin O. Q., Zuo Z. & Chow M. S. Pharmacokinetics and modeling of quercetin and metabolites. Pharm Res 22, 892–901 (2005). [DOI] [PubMed] [Google Scholar]
- Gonthier M.-P., Verny M.-A., Besson C., Rémésy C. & Scalbert A. Chlorogenic acid bioavailability largely depends on its metabolism by the gut microflora in rats. J Nutr 133, 1853–1859 (2003). [DOI] [PubMed] [Google Scholar]
- Azuma K. et al. Absorption of chlorogenic acid and caffeic acid in rats after oral administration. J Agric Food Chem 48, 5496–5500 (2000). [DOI] [PubMed] [Google Scholar]