Abstract
The global distribution of J2-M172 sub-haplogroups has been associated with Neolithic demic diffusion. Two branches of J2-M172, J2a-M410 and J2b-M102 make a considerable part of Y chromosome gene pool of the Indian subcontinent. We investigated the Neolithic contribution of demic dispersal from West to Indian paternal lineages, which majorly consists of haplogroups of Late Pleistocene ancestry. To accomplish this, we have analysed 3023 Y-chromosomes from different ethnic populations, of which 355 belonged to J2-M172. Comparison of our data with worldwide data, including Y-STRs of 1157 individuals and haplogroup frequencies of 6966 individuals, suggested a complex scenario that cannot be explained by a single wave of agricultural expansion from Near East to South Asia. Contrary to the widely accepted elite dominance model, we found a substantial presence of J2a-M410 and J2b-M102 haplogroups in both caste and tribal populations of India. Unlike demic spread in Eurasia, our results advocate a unique, complex and ancient arrival of J2a-M410 and J2b-M102 haplogroups into Indian subcontinent.
Population history of India has been under investigation to study the earliest settlement of anatomically modern humans Out of Africa and later to study the demographic episodes of Eurasia1,2,3. Late Pleistocene heritage of modern humans in India has been supported by archaeological findings of Middle Palaeolithic tools4. Mitochondrial genetic studies agree upon deep rooting Late Pleistocene maternal ancestry of the Indian subcontinent1,5,6. However, some Y-chromosomal studies argue that Indian subcontinent displays considerable genetic relatedness to West Eurasia corresponding to recent events7,8. First settlement of modern humans in India was 60–70 KYA (thousands years ago)6,9,10. In early Holocene, South Asia was a continent of hunter-gatherers. During and after Neolithic period, agriculturists dominated the land, especially the fertile river valleys11. Emergence of agriculture led to the major socio-cultural transition and technological development in human prehistory. The oldest evidence of agriculture comes from the Fertile Crescent (~11 KYA), the centre for demic diffusion12. However, evidences of first agriculture from South Asia indicate a timeline (~9-10 KYA) closer to the emergence of agriculture in the Fertile Crescent13.
Previous studies have established that J2a-M410 and J2b-M102 represent the Y-chromosomal component associated with demic diffusion of Neolithic farmers in North Africa and Eurasia from Mesopotamia (Iraq and Syria)14,15,16. The J2-M172 has been associated with different cultures and populations in history, such as Mediterranean/Aegan15, Greco-Anatolian, Mesopotamian and Caucasian. Presence of J2a-M410 and J2b-M102 in India has been considered a result of gene influx from Western Asia17,18. Worldwide spatial distribution of haplogroup (HG) J2a-M410 coincides with presence of archaeological records of painted pottery and ceramic figurine culture19,20. Similar material culture dating ~9 KYA has been recovered from the Neolithic sites of Mehrgarh located West of Indus Valley (now in Pakistan)21.
The population of the sub-continent shows an outstanding biological and cultural variation, which has been shaped by geographical, ecological, social and linguistic factors22. Many recent studies, particularly from the field of evolutionary genetics, have shown the extent to which each of these factors has contributed to the generation and maintenance of this diversity. It is pertinent that paternal gene pool of India comprises mainly HGs of autochthonous origin of Late Pleistocene ancestry17,18,23, and received very little gene flow from outside7,8. Many Y chromosomal studies of India addressing major demographic questions have included and broadly discussed HG J2-M17217,18,23. However, the arrival and distribution of HG J2-M172 subclades in India have not been studied comprehensively yet. Hence, the aim of this study is to address the following: (1) whether or not, the demic diffusion model stands true for the current distribution of J2-M172 in India in the backdrop of independent origin of agriculture in South Asia; (2) which contemporary populations have the closest affinities with J2-M172 in India; (3) whether distribution of J2a-M410 in India is more dominant in castes than in tribes18; and (4) what social, linguistic or geographical factors might have influenced the distribution of J2-M172 subclades. To address the above, we investigated the presence of different subclades of HG J2-M172 in diverse set of Indian populations and their affinities with rest of the world.
Results
J2-M172 composition in India: two brother clades
Out of 3023 samples, 355 chromosomes belonged to J2-M172 HG (Supplementary Table S1 and Supplementary Table S2). We could not find any J*-M304 and J2*-M172 chromosome in India. We report, a rare presence of 7 chromosomes belonging to J1-M267 from different populations which most likely entered into the Indian subcontinent in historical time24. In this study, we did not find any J2a1b-M67 Y-chromosome. This makes Indian J2a-M410 pool unique from the rest of the world, as majority of the J2a-M410 pool consists of J2a1b-M67 Y-chromosomes (Supplementary Table S1). Similarly, we could not find any J2a1a-M47, and J2a1h1-M158 chromosomes, which represent minority of J2a-M410 pool outside India. Genotyping Z2396, a newly discovered SNP, has divided the Indian J2a-M410 pool into two groups: derived and ancestral for Z2396, proving it to be polyphyletic in India. Our study shows a frequent presence of M68, a rare marker, in various tribes and groups of South India suggests its autochthonous origin. Presence of M68 was also reported in few samples of Indian origin in a study from Southeast Asia25. J2b-M102/M12 HG in India comprises largely of J2b2-M241 chromosomes with minor presence of J2b1-M205 in northwest (NW) region. Occurrence of one J2a1a-M47 and two J2a1h1-M158 chromosomes have been reported earlier in Indian samples18,23, though we could not find any in the present study.
Phylogeography of J2a-M410 and J2b-M102
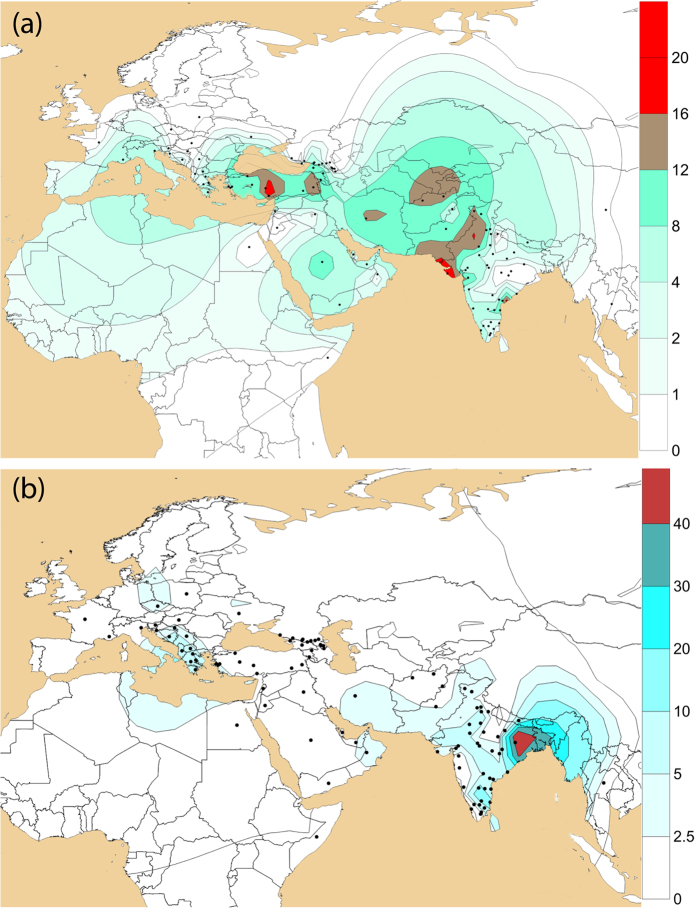
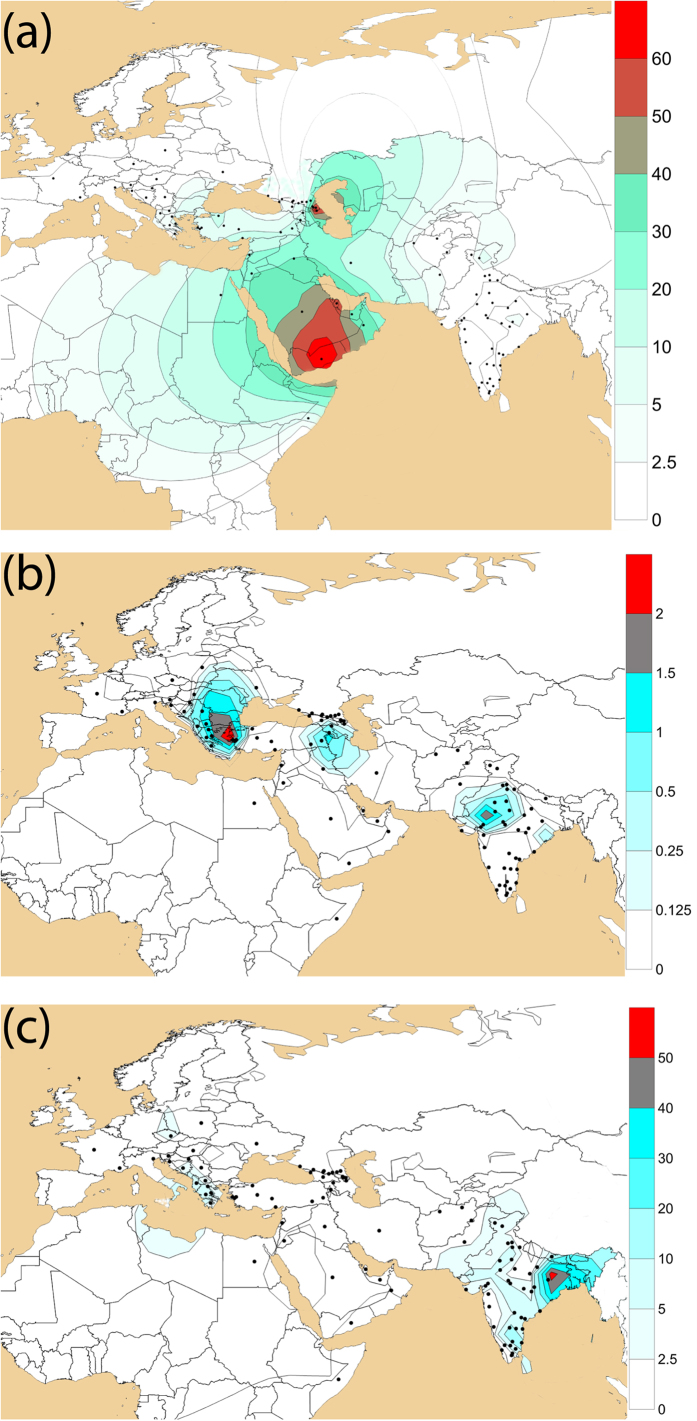
We found moderate occurrence of J2a-M410 and J2b-M102 (0–8%) in different populations inhabited in different parts of India (Supplementary Table S1). We observed substantial high frequencies of J2a-M410 (17–50% in Toda, Chenchu, Banjara, Kamboj, Lohana and Kashmiri Muslims etc.) and J2b-M102 (15–35% in Asur, Narikuravar, Pichakuntla, Shikari and Mondi, etc.) in several populations. J2a-M410 is mainly concentrated towards the NW border of India (comprising Gujarat and Rajasthan). However, high frequency and variance of J2a-M410 in PTGs (primitive tribal groups) like Toda (trasitional pastoralists) and Chenchu (hunter-gatherers and foragers) invoke interesting insights. Contrary to earlier belief, predominant presence and high variance of J2a-M410 among remote tribes dismisses any caste-specific distribution of J2a-M410 in India18. We found the distribution of both the clades geographically pronounced. From West Asia, J2a-M410 wave seems to expand West towards South-eastern Europe and East towards Central Asia and eventually to South Asia (Fig. 1a). Dense focal points of J2a-M410 can be seen along the northwest border of South Asia reaching up to South India. However, it shows a drastic decline towards East of India, consistent with our previous study26. Unlike J2a-M410, J2b-M102 is concentrated in Eastern India (Fig. 1b). Worldwide frequency patterns of J1-M267, J2a-M410, J2b-M102, J2b1-M205 and J2b2-M241 indicates their unique history of distribution (Figs 1 and 2). Frequencies and Y-STR haplotype data of different subclades of J2-M172 are given in Supplementary Table S1, Supplementary Table S3 and Supplementary Table S4.
Figure 1. Contour maps showing worldwide geographical frequency distribution of haplogroups.
(a) J2a-M410 and (b) J2b-M102. The maps were generated using Surfer8 of Golden Software (Golden Software Inc.), following the Kriging procedure. Black dots indicate sampling locations.
Figure 2. Contour maps showing worldwide geographical frequency distribution of haplogroups.
(a) J1-M267, (b) J2b1-M205 and (c) J2b2-M241. The maps were generated using Surfer8 of Golden Software (Golden Software Inc.), following the Kriging procedure. Black dots indicate sampling locations.
In NW and South India we found the highest variance and oldest TMRCA (time of most recent common ancestor) of J2a-M410 haplotypes, whereas in North, West and Central India the TMRCA (Table 1, Supplementary Table S5 and Supplementary Table S6) was comparatively recent. Similar to J2a-M410, J2b-M102 shows the oldest TMRCA in NW region and younger TMRCA in Central, southern, northern and eastern India. Conversely, J2b-M102 shows a slightly different distribution and younger TMRCA than its brother clade J2a-M410 in India (Table 1, Supplementary Table S5 and Supplementary S6). The J2b-M102 variance (Supplementary Table S5) is in contrast with its geographical distribution, suggesting a strong founder effect in East India.
Table 1. BATWING results of time of most recent common ancestor (TMRCA) and effective population size for groups of geographically categorized populations using (1) “observed” mutation rates66, (2) “observed” mutation rates65 (3) mutation rates predicted from logistic model65 for J2a-M410 and J2b-M102 Y-STRs.
| TMRCA (KYA) | N (Effective Population Size) | TMRCA (KYA) | N (Effective Population Size) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | 2.5% | 97.5% | Median | 2.5% | 97.5% | Median | 2.5% | 97.5% | Median | 2.5% | 97.5% | |
| J2a-M410 | J2b-M102 | |||||||||||
| Worldwide | ||||||||||||
| (1) | 6.0 | 2.9 | 12.6 | 796 | 563 | 1132 | 3.3 | 1.6 | 8.5 | 420 | 301 | 590 |
| (2) | 7.1 | 3.6 | 16.2 | 842 | 613 | 1142 | 4.5 | 2.2 | 10.9 | 579 | 428 | 784 |
| (3) | 9.9 | 4.6 | 24 | 1153 | 819 | 1609 | 6.3 | 3.1 | 15.7 | 807 | 595 | 1092 |
| Northwest India | ||||||||||||
| (1) | 3.7 | 1 | 14.6 | 179 | 96 | 346 | 3.2 | 0.7 | 15.8 | 202 | 101 | 443 |
| (2) | 5.1 | 1.4 | 19.7 | 248 | 137 | 475 | 4.5 | 1 | 21.1 | 281 | 143 | 601 |
| (3) | 7.1 | 1.9 | 27.1 | 345 | 189 | 660 | 6.2 | 1.4 | 29.5 | 389 | 197 | 835 |
| North India | ||||||||||||
| (1) | 3.1 | 0.7 | 14.3 | 236 | 119 | 509 | 2.5 | 0.4 | 16.1 | 129 | 55 | 351 |
| (2) | 4.3 | 1 | 19.5 | 331 | 169 | 703 | 3.5 | 0.6 | 21.6 | 180 | 78 | 477 |
| (3) | 6 | 1.4 | 26.5 | 457 | 233 | 956 | 4.8 | 0.8 | 30.1 | 252 | 108 | 671 |
| South India | ||||||||||||
| (1) | 4.1 | 1.6 | 12.8 | 281 | 176 | 459 | 2.3 | 0.7 | 8 | 120 | 69 | 212 |
| (2) | 5.7 | 2.2 | 17.3 | 395 | 257 | 623 | 3.2 | 1 | 10.7 | 168 | 98 | 289 |
| (3) | 8 | 3 | 23.7 | 545 | 351 | 852 | 4.4 | 1.4 | 14.9 | 233 | 136 | 403 |
| Central India | ||||||||||||
| (1) | 2.2 | 0.3 | 14.5 | 45 | 18 | 121 | 2.6 | 0.5 | 13.6 | 116 | 55 | 269 |
| (2) | 3.1 | 0.5 | 19.7 | 62 | 26 | 166 | 3.6 | 0.8 | 18.2 | 161 | 77 | 366 |
| (3) | 4.3 | 0.7 | 27.1 | 87 | 35 | 229 | 5 | 1 | 25.3 | 225 | 108 | 512 |
| East India | ||||||||||||
| (1) | — | — | — | — | — | — | 0.2 | 0 | 1.9 | 4 | 0 | 16 |
| (2) | — | — | — | — | — | — | 0.2 | 0 | 2.7 | 5 | 1 | 22 |
| (3) | — | — | — | — | — | — | 0.3 | 0 | 3.7 | 7 | 1 | 30 |
| Central Asia | ||||||||||||
| (1) | 4.9 | 1.6 | 16 | 290 | 175 | 500 | — | — | — | — | — | — |
| (2) | 6.7 | 2.3 | 21.2 | 392 | 246 | 648 | — | — | — | — | — | — |
| (3) | 9.2 | 3.2 | 29.4 | 546 | 339 | 904 | — | — | — | — | — | — |
| Caucacus | ||||||||||||
| (1) | 3.8 | 2.1 | 8.6 | 446 | 338 | 588 | — | — | — | — | — | — |
| (2) | 5.3 | 2.7 | 11.1 | 585 | 455 | 733 | — | — | — | — | — | — |
| (3) | 7.4 | 3.6 | 16.3 | 799 | 579 | 1020 | — | — | — | — | — | — |
| Europe | ||||||||||||
| (1) | 3.3 | 0.5 | 23.9 | 175 | 72 | 521 | 2.3 | 0.5 | 10.9 | 69 | 34 | 145 |
| (2) | 4.6 | 0.7 | 32.1 | 248 | 103 | 702 | 3.1 | 0.7 | 14.6 | 96 | 48 | 200 |
| (3) | 6.3 | 1 | 42.9 | 345 | 144 | 951 | 4.3 | 0.9 | 20.3 | 134 | 66 | 278 |
Among linguistic groups, our results indicate that Indo-Europeans (IE) and Dravidians (DR) have received the J2a-M410 influx approximately at the same time. However, our results indicate a very late influx and rare presence of the J2a-M410 HG in Austroasiatics (AA). In eastern region, J2b-M102 exhibits few dense focal points in contour map due to high frequency among some AA groups like Asur caused by founder effect. Comparing the TMRCA and diversity of J2b-M102 haplotypes between different linguistic groups (Table 1, Supplementary Table S5 and Supplementary Table S6) suggests that either AA received the gene flow from IE or DR very late or they have lost the diversity due to drift, followed by founder effect. Thus, the distribution of this haplogroup in India is governed primarily by geography. Apart from geographical separation, linguistic affiliation seems to be responsible to shape J2-M172 pool in India. Male effective population sizes for J2a-M410 show rapid expansion when moving from NW region to Gangetic plains while it drops rapidly in Central India and again expands in South India (Table 1, Supplementary Table S6 and Supplementary Fig. S1). However, J2b male effective population sizes remain more or less constant throughout the subcontinent except for East India, where it declines sharply (Table 1, Supplementary Table S6 and Supplementary Fig. S1).
Affinities with world population
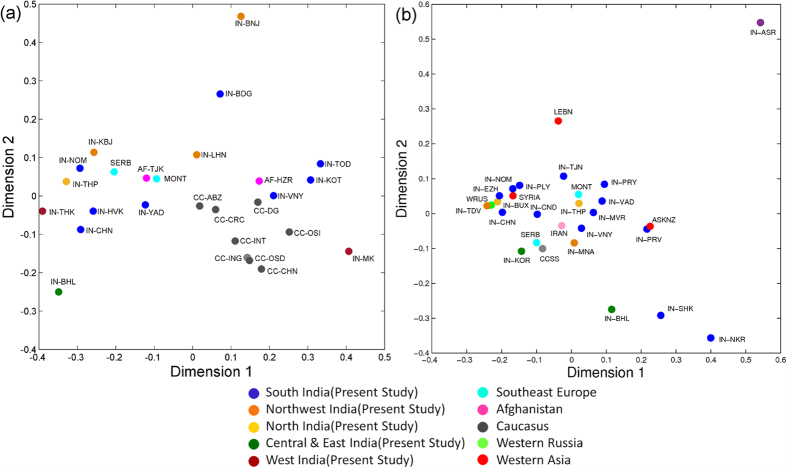
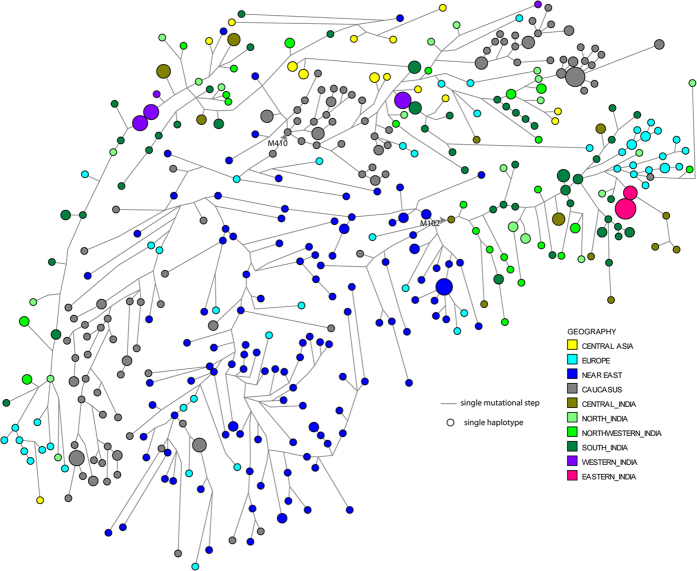
MDS (Multidimensional Scaling) plot based on Rst values for HG J2a-M410 shows that Indian populations do not form a single cluster but are closer to the Central Asian and European populations, while Caucasians (populations from Caucasus) form a separate cluster (Fig. 3a). However, MDS for HG J2b-M102 shows that only few Indian nomadic tribes and East Indian populations lie outside the major Indian cluster (Fig. 3b). MJ (median joining) tree (Fig. 4) depicts geographically pronounced clustering of Y-STR haplotypes. No sharing of STR haplotypes for J2a-M410 and J2b-M102 along with geographical clustering has been observed. High diversities of J2a-M410 and J2b-M102 haplotypes are evident from the MJ and RM (reduced Median) networks (Fig. 4, Supplementary Fig. S2 and Supplementary Fig. S3).
Figure 3. Multidimensional scaling (MDS) plot based on Rst values of Y-STRs for haplogroups.
(a) J2a-M410 and (b) J2b-M102. Dots represent different populations worldwide. CC = Caucasian, IN = Indian, AF = Afghanistani, ABZ = Abkhaz, BDG = Baduga, BNJ = Banjara, BHL = Bahelia, CHN = Chechen (Chechnya), DG = Chechen (Dagestan), INT = Chechen Ingushetia, CHN = Chenchu, CRC = Circassians, HVK = Havik, HZR = Hazara, ING = Ingush, KBJ = Kamboj, KOT = Kota, LHN = Lohana, MK = Mahadev Koli, MONT = Montenegrin, OSD = Ossets (Digor), OSI = Ossets (Iron), NOM = Indian Nomads (Pichakuntla & Mondi), SERB = Serbian, TJK = Tajik, THK = Thakar, THP = Thapa, TOD = Toda, VNY = Vanniyar, YAD = Yadav, IRAN = Iranian, LEBN = Lebanese, MONT = Montenegrin, SERB = Serbian, SYRIA = Syrian, WRUS = WesternRussia, CCSS = Caucasus, ASR = Asur, BHL = Bahelia, BUX = Buxas, CND = CapeNadar, CHN = Chenchu, EZH = Ezhava, KOR = Korku, MVR = Maravar, MNA = Meena, NKR = Narikuravar, PLY = Paliyan, PRV = Paravar, PRY = Parayar, NOM = Indian Nomads (Pichakuntla & Mondi), SHK = Shikari, TDV = Tadvi, TJN = Tamil Jains, THP = Thapa, VNY = Vanniyar, VAD = Yadhava, ASKNZ = Ashkenazi.
Figure 4. Network connecting Y-STR haplotypes within J2-M172.
The network was constructed using a median joining with maximum parsimony (MP) algorithm as implemented in the Network 4.612 program. The size of the circle is proportional to the numbers of the samples.
Discussion
J2-M172 is a predominant HG in West and Central Asia. Populations living West from India show high frequency, subclade variation and presence of paragroups. Worldwide subclade diversity and distribution of J2a-M410 suggest its spread from West and Central Asia into India through NW corridor (Fig. 1a). The spatial distribution of J2a-M410 throughout Middle East and Central Asia is overlapped by presence of Neolithic artifacts such as painted pottery and ceramic culture19. The earliest precursor known of Indus Valley civilization, Mehrgarh (NW of Indian subcontinent, now in Pakistan), provides one of the oldest (~9KYA) evidences of origin of agriculture and plant domestication suited by early Holocene climate27. Additionally, these Neolithic sites of Mehrgarh showed the earliest evidence of transformation of subsistence from hunting-gathering to settled agriculture owing to the idea that the first farmers from Indus were agro-pastoral, and semi-nomadic people28. It is interesting to note that the concentration of J2a-M410 over the geography largely mimics the agricultural centres29. J2a-M410 in India peaks at NW region and shows a clinal pattern towards Central and East, however, again rises considerably in South. J2b-M102 has been found in all parts of India in low to moderate frequency, but it is significantly frequent among some nomadic PTGs of South India. From eastern region, some of the Austroasiatic tribes carry a high frequency of J2b2-M241. In the present study, we also see high frequency of J2a-M410 and J2b-M102 in remote undisturbed foragers with recent history of hunting gathering (eg. Asur, Chenchu), pastorals (eg. Toda tribe with high J2a-M68) and nomadic tribes (eg. Banjara, Bahelia etc). Most of the nomadic tribes were from NW region or had recently migrated from the region towards South India (eg. Narikruwar, Shikari, Mondi, Pichakuntla). Considering these facts and arguments, one can deduce that these groups could be the relic of agro-pastoral communities spreading from the NW region of the subcontinent in the past.
Various studies have given evidences to support the influence of Neolithic from Near East on Indian subcontinent (in Mehrgarh) dated around 10.5 KYA 30 and references therein. Noted similarities between Mehrgarh and Near East are domesticated wheat varieties, early phases of farming, pottery, other archaeological artefacts, some domesticated plants and herd animals30. A vast arid region of Iran and Afghanistan lies in between Near East and Indus Valley, leaving possibility of rainfall agriculture only in the foothills and cul-de-sac valleys31. Yet, the area was not an undefeatable geographical barrier for Neolithic spread. Some sections of the Silk Road (route South of the Caspian sea) connecting Badakhshan (north-eastern Afghanistan and south-eastern Tajikistan) with West Asia, Egypt and India were in use by 5 KYA32. Other section of Silk Road connecting Badakhshan to the Mesopotamian plains (the Great Khorasan Road) was in use by 6 KYA31. Archaeological evidences support similarities among widely separated Neolithic sites in these regions33 and plausibility of migration of population34.
The AMOVA results found that geographically distant populations have higher Fst values (Table 2). Lower Fst values between NW India and geographical regions West from India show them to be less differentiated. Higher Fst values between NW and South India suggest their high level of differentiation. TMRCAs of J2a-M410 and J2b-M102 of NW and South India advocate an early arrival of these lineages to Indian subcontinent. Genetic relatedness of NW Indians with population to West from NW border and remarkable presence of J2-M172 HGs in remote Indian tribes along with other social strata, may represent the early Holocene expansion in NW India (including the Indus Valley) diffusing towards Central Asia and spreading agriculture eastwards to the Gangetic plains during pre-Harappan times (6-7 KYA). However, presence of J2-M172 subclades in India can not only be substantiated by Neolithic spread. Firstly, lack of any sublineage of J2a-M410 (M67, M47 and M92) representing majority of the pool outside India, implies towards an older and unique history of this HG into the subcontinent. J2a1b-M67 and J2a-M92 lineages have been well correlated temporally and spatially with the spread of earliest farmers and Bronze Age cultures in Anatolia, south-eastern Europe and Mediterranean15. Secondly, in indigenous Indian populations, negligible presence of other Y-HGs like R1b1b2-M26935, G2a-P15 and E1b1b1a1b-V1316,36,37, which are associated with demographic spread of Neolithic. Thirdly, in addition to prominent absence of J2a1b-M67 and J2a-M92 in southwestern Asia, practical absence of HGs J1-M267 and G-M201 in India, respectively, occur at 9% and 10.9% in Turkey38, 33.1% and 2.2% in Iraq14, and 3.4% and 6% in Pakistan18, indicate towards different dispersal events from Middle East to southeastern Europe and southwestern Asia. Complete absence of any paragroup of J clade in India reinforces the established theory of J2 subclades entering into the subcontinent from NW/West to India. Given all that, and granted their exogenous origin, J2a-M410 and J2b-M102 in the subcontinent may reflect any combination of unknown and known movements. Though, the genealogical ages for Indian J2a-M410 and J2b-M102 are correlating with appearance of agriculture in Indus Valley (~6KYA) and Mehrgarh (~9KYA) and falling well within the Neolithic range, differential presence and distribution of J2-M172 sublineages and other associated HGs depict a complex picture. Most likely events responsible for the current distribution of J2-M172 sublineages into Indian subcontinent could be any combination of 1) entry of herders from West and Central Asia/Middle East during late glacial maximum (LGM) of Holocene, 2) Neolithic demic diffusion from the West, and 3) Bronze and Iron age migration/admixtures.
Table 2. Analysis of Molecular Variance (AMOVA) using (a) J2a and (b) J2b Y-STR between groups of geographically categorized populations.
| Geographical Group | Fst |
|---|---|
| (a) | |
| Northern & North-western India vs Europe | 0.1546 |
| Northern & North-western India vs Central Asia | 0.18 |
| Northern & North-western India vs Caucasus | 0.3747 |
| Northern & North-western India vs Southern India | 0.3493 |
| Southern India vs Central Asia | 0.304 |
| Southern India vs Europe | 0.3315 |
| Southern India vs Caucasus | 0.4104 |
| (b) | |
| Northern & North-western India vs Europe | 0.5008 |
| Northern & North-western India vs Near East | 0.5591 |
| Northern & North-western India vs Central India | 0.5463 |
| Northern & North-western India vs Southern India | 0.5763 |
| Northern & North-western India vs Eastern India | 0.695 |
| Southern India vs Europe | 0.5738 |
| Southern India vs Near East | 0.6201 |
Conclusion
Absence or negligible presence of classical markers of Eurasian demic diffusion in India advocates against it to be the sole explanation of J2a-M410 and J2b-M102 distribution in the subcontinent. High variance, haplotype diversity with no sharing haplotype, geographically pronounced phylogeny and seemingly autochthonous origin of sublineage J2a-M68, suggest towards the antiquity of the HGs. Dispersal of J2a-M410 and J2b-M102 from Near East to NW region and further eastwards of the subcontinent seems to have unique and complex history of various known and unknown possible events. Regardless of the complexity of dispersal, NW region appears to be the corridor for entry of these haplogroups into India. Remarkable presence of J2a-M410 among tribal groups inhabited in remote geographical regions strongly dismisses the earlier belief of it to be caste-specific.
Material and Methods
Sample composition
We studied a total of 3023 Y- chromosomes belonging to 77 different Indian populations. Of these, 1102 individuals were from different primitive tribes, 362 individuals were from nomadic tribes, 856 individuals were from different castes and 541 individuals were from different tribes. These individuals were affiliated to different linguistic groups (~1249 Indo-Europeans, ~1163 Dravidians, ~407 Austroasiatics and ~78 Tibeto-Burman) distributed all over India. In addition to Indian samples, we also analysed 39 samples of Thapa population from Nepal. Details of populations studied are given in the Supplementary Table S2. Prior to the sample collection, informed written consents were obtained from all the subjects participating in this study. This study was approved by the Institutional Ethical Committee of the CSIR-Centre for Cellular and Molecular Biology, Hyderabad, India. The methods were carried out in accordance with approved guidelines.
Y chromosome analysis
Biallelic Y chromosome markers defining HG J-M304 and its subclades (M304, M267, M172, M410, M102, M47, M67, M68, M158, M205, M241, Z2396 and Z1827) were genotyped following the hierarchy of the Y chromosome phylogeny. Samples were genotyped through Sanger sequencing using BigDye™ Terminator cycle sequencing kit (Applied Biosystems, USA). For Y-STR genotyping, every second or third sample from each population was randomly selected based on the frequency of the subclades of HG J-M304 in the respective population. Out of 355 J-M304 possessing Y chromosomes, 158 were genotyped for 17 Y-STR markers with AmpFlSTR® PCR amplification kit (Applied Biosystems, USA). Detailed method of genotyping is given in Text S1. J2-M172 Individuals were resolved for downstream biallelic markers and assigned to the HG J2-M172 subclades (ISOGG, http://www.isogg.org/tree/).
Haplotype analysis
We obtained haplogroup frequency data for 6966 individuals from different studies, representing various geographical regions and ethnicity worldwide14,18,38,39,40,41,42,43,44,45,46,47,48,49,50,51. We also retrieved worldwide dataset of 17 Y-STR markers for 1157 individuals belonging to HG J2-M172 for comparison23,26,39,51,52,53,54,55,56,57. Details of the dataset are given in Supplementary Table S1 and Supplementary Table S4. Median joining (MJ) and reduced median (RM), resolved with the maximum parsomony (MP) algorithm, were created using the 15 Y-STR data (Supplementary Table S3 and Supplementary Table S4) and Network 4.612 software (www.fluxus-engineering.com). Equal weights were given to each locus. One Steiner tree was selected and shown (Fig. 1, Supplementary Fig. S2 and Supplementary Fig. S3). The age estimates (Supplementary Table S5) based on 15 Y-STR loci variations (Supplementary Table S3, Supplementary Table S4 and Supplementary Table S7) were calculated using the method described in Zhivotovsky et al. 58 and updated in Sengupta et al. 18. The TMRCA of Y chromosomal HGs have been derived from ρ statistic, assuming evolutionary mutation rate 6.9 × 10−4 per locus per generation58 as well as TD statistics, assuming genealogical mutation rate 2.1 × 10−3 per locus per generation59,60,61.
Population diversity indices, Rst values and AMOVA (Analysis of Molecular Variance) based Fst were calculated using Arlequin 3.5 software62. MDS (Multi Dimensional Scaling) plots of Rst values were drawn using MATLAB 8.03 (http://www.mathworks.nl/products/matlab/). HG isofrequency maps were generated using Surfer v 8 (Golden Software Inc., Golden, Colorado), following the Kriging procedure. We excluded two DYS385 loci from all current analyses due to technical issue. We have also filtered out some highly deviating haplotype samples from any age estimation analyses. The deviating samples were detected by determining their high mutational distance from the modal haplotypes. The selected samples used for TMRCA analyses are listed in Supplementary Table S7.
BATWING analysis
TMRCAs and the times of individual population splits were calculated using the Bayesian approach implemented in BATWING, considering exponential growth from a constant-size population model63. We used broad prior distributions: gamma (2, 400) for population growth rate per generation (α), gamma (1, 200) for the time in coalescent units when exponential growth began, normal (2000, 1000) for effective population size (N)35. To achieve more plausible time estimates for Neolithic coalescent sublineage like J2-M17264, we used genealogical rates. We considered three different sets of mutation rates and prior distributions for each marker65,66 (Supplementary Table S8). For each BATWING simulation, we performed 300 million MCMC cycles. We discarded first 50 million MCMC iterations as burn-ins and sampled 1 in every 500 cycle for estimation. Thus, the results are inferred from 500,000 samples taken from 250 million MCMC iterations. The TMRCA was calculated as the product of ‘N’, generation time (30 years) and the height of the tree ‘T’ estimated by BATWING (Table 1 and Supplementary Table S6). The statistical analysis was performed using R package 3.2.0 (http://www.R-project.org/).
Additional Information
How to cite this article: Singh, S. et al. Dissecting the influence of Neolithic demic diffusion on Indian Y-chromosome pool through J2-M172 haplogroup. Sci. Rep. 6, 19157; doi: 10.1038/srep19157 (2016).
Supplementary Material
Acknowledgments
We thank all the donors for participating in this study. KT was supported by the Network project (GENESIS-BSC0121) of Council of Scientific and Industrial Research (CSIR), India. KT also acknowledges support from the UK-India Education and Research Initiative grant (RG-4772). LS was supported by the Bhatnagar Fellowship of Council of Scientific and Industrial Research (CSIR), India and JC. Bose Fellowship of Department of Science and Technology (DST), India. GC is supported by the European Union European Regional Development Fund through the Centre of Excellence in Genomics to Estonian Biocentre, by Estonian Personal grants PUT-766 and Estonian Institutional Grant IUT24-1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions S.S., K.T. and G.C. conceived and designed the experiments. S.S., K.S.K., S.K.S., S.N., L.S. and K.T. collected samples. S.S., A.S., R.R., K.S.K., S.K.S., A.S.-2, S.A.S., V.P., V.K. and P.V. performed the experiments. S.S. and G.C. analyzed the data. A.P. did BATWING analysis. L.S. and K.T. contributed reagents/materials/tools. S.S. prepared first draft of the manuscript. G.C. and K.T. provided substantial inputs for manuscript writing. All authors read and approved the final manuscript.
References
- Thangaraj K. et al. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup ‘M’ in India. BMC Genomics 7, 151 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Thangaraj K., Patterson N., Price A. L. & Singh L. Reconstructing Indian population history. Nature 461, 489–94 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellars P., Gori K. C., Carr M., Soares P. A. & Richards M. B. Genetic and archaeological perspectives on the initial modern human colonization of southern Asia. Proc. Natl. Acad. Sci. USA. 110, 10699–10704 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V. N. Prehistoric human colonisation of India. J. Biosci. 26, 491–531 (2001). [DOI] [PubMed] [Google Scholar]
- Kivisild T. et al. In Genomic Diversity: Applications in Human Population Genetics (eds Papiha S. S., Deka R. & Chakraborty R.) Ch. 11, 135–152 (Kluwer, New York, 1999). [Google Scholar]
- Metspalu M. et al. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 5, 26 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A. et al. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 13, 2277–2290 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I. et al. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 7, 42 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kivisild T. et al. The genetic heritage of earliest settlers persist in both the Indian tribal and caste populations. Am. J. Hum. Genet. 72, 313–332 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K. et al. Reconstructing the origin of Andaman Islanders. Science 308, 996 (2005). [DOI] [PubMed] [Google Scholar]
- Fuller D. Q. & Murphy C. Overlooked but not forgotten: India as a center for agricultural domestication. Gen. Anthropol. 21, 1–8 (2014). [Google Scholar]
- Diamond J. & Bellwood P. Farmers and their languages: the first expansions. Science 300, 597–603 (2003). [DOI] [PubMed] [Google Scholar]
- Fuller D. Q. Agricultural origins and frontiers in South Asia: a working synthesis. J. World Prehist. 20, 1–86 (2006). [Google Scholar]
- Al-Zahery N. et al. Y-chromosome and mtDNA polymorphisms in Iraq, a crossroad of the early human dispersal and of post-Neolithic migrations. Mol. Phylogenet. Evol. 28, 458–472 (2003). [DOI] [PubMed] [Google Scholar]
- Di Giacomo F. et al. Y chromosomal haplogroup J as a signature of the post-neolithic colonization of Europe. Hum. Genet. 115, 357–371 (2004). [DOI] [PubMed] [Google Scholar]
- Semino O. et al. Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the neolithization of Europe and later migratory events in the Mediterranean area. Am. J. Hum. Genet. 74, 1023–1034 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo S. et al. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc. Natl. Acad. Sci. USA. 103, 843–848 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S. et al. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenousand exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am. J. Hum. Genet. 78, 202–221 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauvin J. The Birth Of The Gods And The Origins Of Agriculture (Cambridge University Press, Cambridge 2000). [Google Scholar]
- King R. & Underhill P. A. Congruent distribution of Neolithic painted pottery and ceramic figurines with Y-chromosome lineages. Antiquity 76, 707–714 (2002). [Google Scholar]
- Possehl G. L. Indus Age: The Beginnings (Univ. Pennsylvania Press, Philadelphia, 1999). [Google Scholar]
- Chaubey G., Metspalu M., Kivisild T. & Villems R. Peopling of South Asia: Investigating the caste-tribe continuum in India. Bioessays 29, 91–100 (2007). [DOI] [PubMed] [Google Scholar]
- Arunkumar G. et al. Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system. PLoS One 7, e50269 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaaswarkhanth M. et al. Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations. Eur. J. Hum. Genet. 18, 354–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. M., Perumal R., Keat P. Y. & Kuehn D. L. Haplotype diversity of 16 Y-chromosomal STRs in three main ethnic populations (Malays, Chinese and Indians) in Malaysia. Forensic Sci. Int. 167, 70–76 (2007). [DOI] [PubMed] [Google Scholar]
- Thangaraj K. et al. The influence of natural barriers in shaping the genetic structure of Maharashtra populations. PloS one 5, e15283 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. K. Origin of agriculture and domestication of plants and animals linked to early Holocene climate amelioration. Curr. Sci. 87, 54–59 (2004). [Google Scholar]
- Allchin R. & Allchin B. Origins of a Civilization: The Prehistory and Early Archaeology of South Asia (New Delhi, Viking, 1997). [Google Scholar]
- Fuller D. Q. Non-human genetics, agricultural origins and historical linguistics in South Asia In The Evolution And History Of Human Populations In South Asia (eds Petraglia M. D. & Allchin B.) 393–443 (Springer: Netherlands, 2007). [Google Scholar]
- Gangal K., Sarson G. R. & Shukurov A. The Near-Eastern Roots of the Neolithic in South Asia. PloS one 9, e95714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiebert F. T. & Dyson R. H. Prehistoric Nishapur and frontier between Central Asia and Iran. Iranica Antiqua 37, 113–149 (2002). [Google Scholar]
- Kuzmina E. E. & Mair V. H. The Prehistory Of The Silk Road (Univ. Pennsylvania Press, Philadelphia, 2008). [Google Scholar]
- Alizadeh A. Excavations at the prehistoric mound of Chogha Bonut, Khuzestan, Iran: Technical report (University of Chicago, Illinois, 2003). [Google Scholar]
- Harris D. R. Origins Of Agriculture In Western Central Asia: An Environmental-Archaeological Study (Univ. Pennsylvania Press, Philadelphia, 2010). [Google Scholar]
- Balaresque P. et al. A predominantly Neolithic origin for European paternal lineages. PLoS Boil. 8, e1000285 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M. F. et al. Out of Africa and back again: nested cladistic analysis of human Y chromosome variation. Mol. Biol. Evol. 15, 427–441 (1998). [DOI] [PubMed] [Google Scholar]
- Lacan M. et al. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc. Natl. Acad. Sci. USA. 108, 18255–18259 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnioğlu C. et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum. Genet. 14, 127–148 (2004). [DOI] [PubMed] [Google Scholar]
- Balanovsky O. et al. Parallel evolution of genes and languages in the Caucasus region. Mol. Biol. Evol. 28, 2905–2920 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis J. R. et al. The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am. J. Hum. Genet. 74, 532–544 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores C. et al. Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan. J. Hum. Genet. 50, 435–41 (2005). [DOI] [PubMed] [Google Scholar]
- Sanchez J. J., Hallenberg C., Børsting C., Hernandez A. & Morling N. High frequencies of Y chromosome lineages characterized by E3b1, DYS19-11, DYS392-12 in Somali males. Eur. J. Hum. Genet. 13, 856–866 (2005). [DOI] [PubMed] [Google Scholar]
- Regueiro M., Cadenas A. M., Gayden T., Underhill P. A. & Herrera R. J. Iran: tricontinental nexus for Y-chromosome driven migration. Hum. Hered. 61, 132–43 (2006). [DOI] [PubMed] [Google Scholar]
- Cadenas A. M., Zhivotovsky L. A., Cavalli-Sforza L. L., Underhill P. A. & Herrera R. J. Y-chromosome diversity characterizes the Gulf of Oman. Eur. J. Hum. Genet. 16, 374–386 (2007). [DOI] [PubMed] [Google Scholar]
- Battaglia V. et al. Y-chromosomal evidence of the cultural diffusion of agriculture in Southeast Europe. Eur. J. Hum. Genet. 17, 820–830 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalloua P. A. et al. Y-chromosomal diversity in Lebanon is structured by recent historical events. Am. J. Hum. Genet. 82, 873–882 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Amero K. K. et al. Saudi Arabian Y-Chromosome diversity and its relationship with nearby regions. BMC Genet. 10, 59 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R. J. et al. The coming of the Greeks to Provence and Corsica: Y-chromosome models of archaic Greek colonization of the western Mediterranean. BMC Evol. Biol. 11, 69 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regueiro M. et al. High levels of Paleolithic Y-chromosome lineages characterize Serbia. Gene 498, 59–67 (2012). [DOI] [PubMed] [Google Scholar]
- Herrera K. et al. Neolithic patrilineal signals indicate that the Armenian plateau was repopulated by agriculturalists. Eur. J. Hum. Genet. 20, 313–320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. Afghanistan’s ethnic groups share a Y-chromosomal heritage structured by historical events. PloS One 7, e34288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roewer L. et al. Analysis of Y chromosome STR haplotypes in the European part of Russia reveals high diversities but non-significant genetic distances between populations. Int. J. Legal Med. 122, 219–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sibai M. et al. Geographical Structure of the Y-chromosomal Genetic Landscape of the Levant: A coastal-inland contrast. Ann. Hum. Genet. 73, 568–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabal S. et al. Human Y‐chromosome short tandem repeats: A tale of acculturation and migrations as mechanisms for the diffusion of agriculture in the Balkan Peninsula. Am. J. Phys. Anthropol. 142, 380–390 (2010). [DOI] [PubMed] [Google Scholar]
- Dulik M. C. et al. Mitochondrial DNA and Y chromosome variation provides evidence for a recent common ancestry between native Americans and Indigenous Altaians. Am. J. Hum. Genet. 90, 229–246 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber M. et al. Influences of history, geography, and religion on genetic structure: the Maronites in Lebanon. Eur. J. Hum. Genet. 19, 334–340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robino C. et al. Analysis of Y-chromosomal SNP haplogroups and STR haplotypes in an Algerian population sample. Int. J. Legal Med. 122, 251–255 (2008). [DOI] [PubMed] [Google Scholar]
- Zhivotovsky L. A. et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am. J. Hum. Genet. 74, 50–61 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmao L. et al. Mutation rates at Y chromosome specific microsatellites. Hum. Mutat. 26, 520–528 (2005). [DOI] [PubMed] [Google Scholar]
- Sanchez-Diz P. et al. Population and segregation data on 17 Y-STRs: results of a GEP-ISFG collaborative study. Int. J. Legal Med. 122, 529–533 (2008). [DOI] [PubMed] [Google Scholar]
- Ge J., Budowle B. & Aranda X. G. Mutation rates at Y chromosome short tandem repeats in Texas populations. Forensic Sci. Int. Genet. 3, 179–184 (2009). [DOI] [PubMed] [Google Scholar]
- Excoffier L., Laval G. & Schneider S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005). [PMC free article] [PubMed] [Google Scholar]
- Wilson I. J., Weale M. E. & Balding D. J. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J. R. Stat. Soc. Ser. A Stat. Soc. 166, 155–188 (2003). [Google Scholar]
- Wang C. C. & Li H. Comparison of Y-chromosomal lineage dating using either evolutionary or genealogical Y-STR mutation rates. bioRxiv, http://dx.doi.org/10.1101/004705 (2014). [Google Scholar]
- Burgarella C. & Navascue´ s, M. Mutation rate estimates for 110 Y-chromosome STRs combining population and father-son pair data. Eur. J. Hum. Genet. 19, 70–75 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W. et al. A worldwide survey of human male demographic history based on Y-SNP and Y-STR data from the HGDP-CEPH populations. Mol. Biol. Evol. 27, 385–393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.