Abstract
Many common bacterial pathogens utilize quorum sensing to coordinate group behaviors and initiate virulence at high cell densities. The use of small molecules to block quorum sensing provides a means of abrogating pathogenic phenotypes, but many known quorum sensing modulators have limitations, including hydrolytic instability and displaying non-monotonic dose curves (indicative of additional targets and/or modes of action). To address these issues, we undertook a structure-based scaffold-hopping approach to develop new chemical modulators of the LasR quorum sensing receptor in Pseudomonas aeruginosa. We combined components from a triphenyl derivative known to strongly agonize LasR with chemical moieties known for LasR antagonism and generated potent LasR antagonists that are hydrolytically stable across a range of pH values. Additionally, many of these antagonists do not exhibit non-monotonic dose effects, delivering probes that inhibit LasR across a wider range of assay conditions relative to known lactone-based ligands.
Keywords: anti-infectives, LasR receptor, Pseudomonas aeruginosa, quorum sensing, small molecule probes, virulence
Over the past several decades, the rate at which new antimicrobial therapies have been discovered has steadily declined, while the prevalence of antibiotic resistant bacteria has simultaneously risen.1,2 The inherent bactericidal and/or bacteriostatic nature of antibiotics inevitably leads to resistance;3 therefore, considerable recent research has focused on potential approaches that diminish bacterial pathogenicity without affecting bacterial growth.4,5 By modulating bacterial cell-to-cell communication, a process termed quorum sensing (QS), many virulence phenotypes in pathogens can be attenuated without affecting bacterial viability.6−9 Our laboratory10−19 and others20−27 have become actively involved in the design of small molecule and peptidic tools capable of inhibiting QS in bacteria and exploring the roles of this signaling process in infection.
Bacteria use QS to act more as a multicellular group instead of as individual isolated cells.6−8 This communication system involves the production of diffusible chemical signals that increase in concentration proportionally to population expansion. Once a threshold signal density is reached, the molecules can productively bind to transcription factors that alter gene expression levels.6,7 In many pathogenic bacteria, QS coordinates the expression of virulence factors that are effective only when produced at high population densities on a host. The canonical QS circuit in Gram-negative bacteria is the LuxI/LuxR system, first characterized in the luminescent symbiont Vibrio fischeri.8 This system consists of a LuxI-type synthase that produces an N-acyl l-homoserine lactone (AHL) chemical signal, which in turn binds a cognate intracellular LuxR-type receptor (Figure 1). The AHL signal can passively diffuse into and out of the cell (certain bacteria also use active export).12 AHL:LuxR-type receptor binding at high cell densities typically promotes receptor homodimerization, binding to specific QS promoters, and the transcriptional regulation of group beneficial genes.
Figure 1.
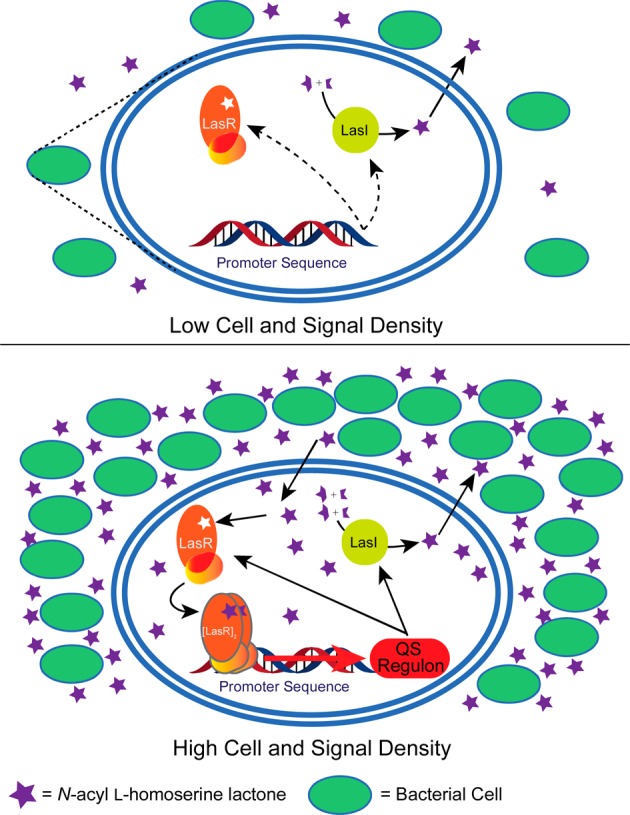
General schematic outlining the mechanism of Gram-negative quorum sensing. LasI/LasR are LuxI/LuxR homologues in P. aeruginosa. Blue double oval is an enlarged view of one bacterium.
Pseudomonas aeruginosa is associated with infections in patients with cystic fibrosis, immunocompromised individuals (e.g., patients with HIV), and chronic wounds.28,29 This Gram-negative opportunistic pathogen has a relatively complex QS network consisting of two LuxI/LuxR pairs (LasI/LasR and RhlI/RhlR), a LuxR orphan receptor (QscR, i.e., lacking an associated synthase), and the Pseudomonas quinolone signal (PQS), an alkylquinolone that binds to PqsR (a LysR-type transcriptional regulator).10,30 QS regulates about 10% of the P. aeruginosa genome,31 and these transcriptional changes enable the bacterial population to overwhelm the host’s defenses. Activation of LasR by its cognate ligand (N-(3-oxo-dodecanoyl) l-homoserine lactone (OdDHL), Figure 2A) up-regulates many virulence phenotypes (e.g., protease production and biofilm formation) and can lead to positive regulation of the Rhl and Pqs systems, placing LasR atop the QS hierarchy in a variety of environmental contexts.10 For this reason, considerable work has focused on designing small molecules and macromolecules that inhibit LasR activity10−12,17,18,25,27,32−36 and thereby inhibit associated virulence phenotypes in P. aeruginosa. Notable in this regard are contributions by Bassler,27 Greenberg,35,36 Meijler,20,21 Spring,22−25,37 and co-workers focused on novel small molecule ligands for LasR and by Janda and co-workers34,38 focused on antibodies that bind OdDHL and sequester it from LasR.
Figure 2.
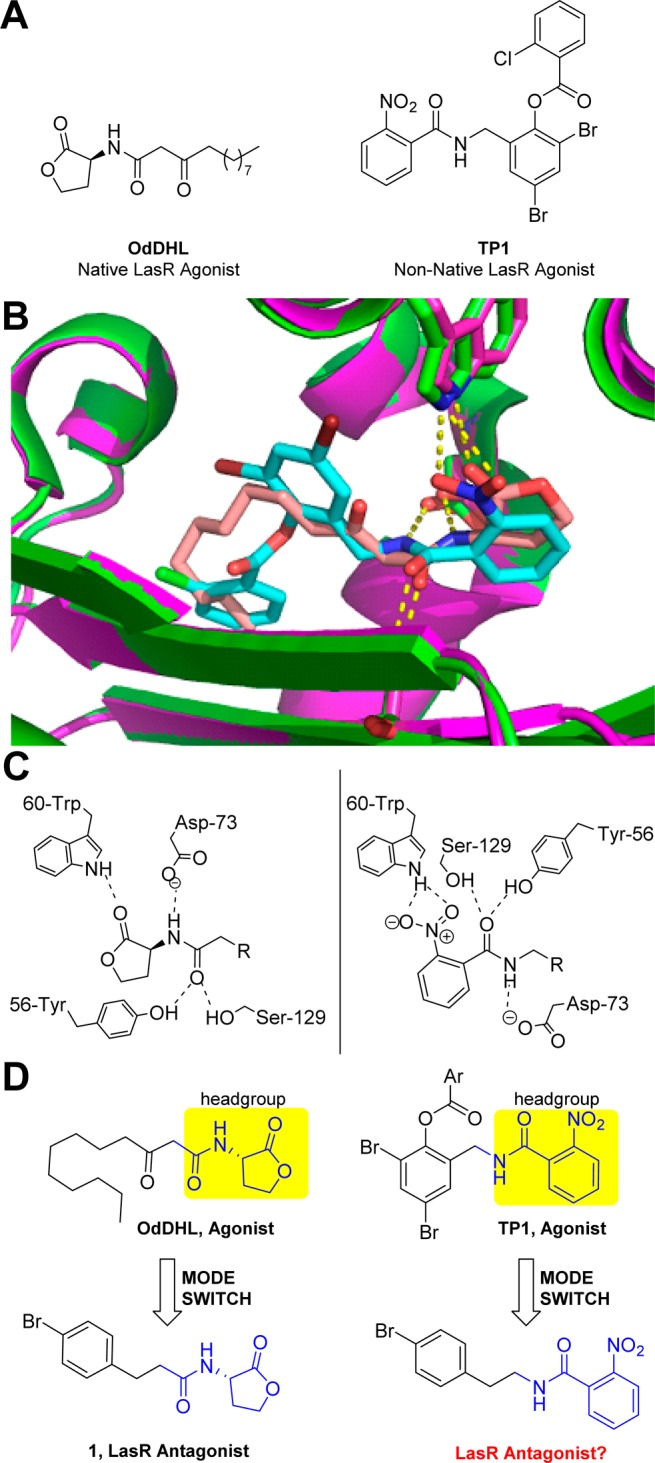
(A) Structures of the native LasR agonist OdDHL and non-native LasR agonist TP1. (B) Overlay of the ligand-binding sites in the OdDHL:LasR (pink) and TP1:LasR (green) X-ray crystal structures with many of the key hydrogen bonds indicated (dashed yellow lines). OdDHL is peach, and TP1 is cyan. Structures are from PDB IDs 3IX3 and 3IX4, respectively. (C) Two-dimensional view of the key hydrogen bonds between LasR and agonists OdDHL and TP1. (D) Illustration of our scaffold-hopping mode-switching approach, installing a p-bromo phenethyl amine on the TP1 headgroup to mimic known AHL-derived LasR antagonist 1.
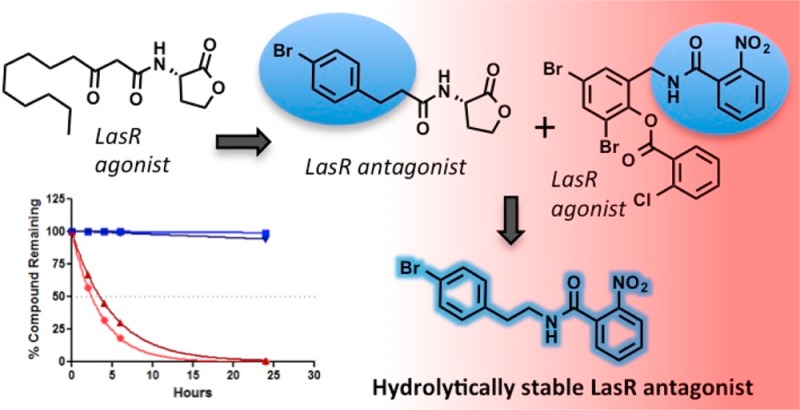
Our laboratory has developed a variety of small molecule LasR antagonists to date.10−12,17,18,32,39,40 Most of these efforts have focused on synthesizing compounds that maintain the homoserine lactone (HSL) headgroup while changing the acyl tail to various chemotypes. These modifications have resulted in the discovery of numerous non-native AHL antagonists of LasR with IC50 values from ∼0.25 to 100 μM. Although these AHLs can be used to attenuate virulence phenotypes in P. aeruginosa and represent some of the most potent LasR modulators known, they are beset by a number of shortcomings.24,25 Namely, the HSL headgroup is prone to hydrolysis in aqueous media (rendering the compounds inactive), and AHLs also can be readily degraded by bacterial and host lactonases and acylases.37,41−43 This chemotype is also actively exported from P. aeruginosa via the MexAB-OprM efflux pump.12 Further confounding their use, many of our most potent LasR antagonists exhibit a non-monotonic dose–response curve when tested in cell-based reporter assays measuring LasR transcriptional activity.18 Specifically, we observe that these compounds are capable of LasR antagonism at low concentrations, whereas at high concentrations we observe LasR agonism instead. We are currently delineating the mechanistic origins of this non-monotonic effect44 and speculate that these compounds interact with either another site on LasR or other targets at high (μM) concentrations that permit LasR activation. No matter the origin of this effect, LasR antagonists that do not display this dose–response feature are certainly desirable as research tools. Identifying such compounds, and ideally ones not prone to hydrolysis, was a main goal of the current study.
To design new LasR antagonists that would avoid the limitations of our previous AHL leads, we utilized a structure-based scaffold-hopping approach. Triphenyl derivative TP1 (Figure 2A), discovered in a high-throughput screen by Greenberg and co-workers,35 was reported as a LasR agonist with equivalent if not enhanced potency compared to LasR’s native ligand, OdDHL (Figure 2A), in cell-based assays.45 We identified TP1 as an excellent candidate for further evaluation because of its remarkable potency, its enhanced stability to hydrolysis compared to OdDHL,35 its low MexAB-OprM efflux pump susceptibility,44 and its modular structure that is readily amenable to synthetic modification. We hypothesized that by uniting the 2-nitrophenyl “headgroup” of TP1 with various tail motifs common to our AHL-based inhibitors, we could generate novel hybrid compounds capable of LasR antagonism. Indeed, recent work by Perez and co-workers on irreversible LasR inhibitors based on TP1 is supportive of the conversion of this scaffold from LasR agonist to LasR antagonist.46 Through the synthesis and biological evaluation of a focused library based on this chemotype, we demonstrated that compounds of this new hybrid structure are capable of either agonizing or antagonizing LasR. Moreover, several new LasR antagonists with single-digit micromolar potencies were identified. Among these, one compound (17) emerged with an optimal combination of potency and maximum efficacy. More notably, this compound lacked a non-monotonic antagonism dose–response curve, demonstrating its potential utility as a tool compound. Finally, we compared the chemical stability of the new compounds to that of their HSL-containing congeners and confirmed that the hybrid compounds exhibited greatly enhanced chemical stability over a range of pH values.
Results and Discussion
We began our study by analyzing the reported X-ray crystal structures of the LasR N-terminal ligand binding domain with TP147 and with OdDHL48 (Figure 2B). Congruent with the past study by Zou and Nair,47 we determined that the 2-nitrophenyl ring of TP1 closely mimics the HSL headgroup in OdDHL, with both chemotypes making the same network of hydrogen-bonding contacts in the LasR ligand-binding site (Figure 2C). Because prior studies by our laboratory have demonstrated that the LasR agonist OdDHL can be “mode switched” to a LasR antagonist by replacing the native 3-oxo-dodecanoyl tail with alternate non-native groups (mostly aryl, such as 1 in Figure 2D),16,18 we hypothesized that the same “mode-switching” phenomenon could be possible with the nonhydrolyzable 2-nitrophenyl headgroup of TP1. Accordingly, by considering structure–activity relationships (SARs) previously determined for LasR using AHL analogues, we reasoned that combining tails from known active AHL antagonists with the 2-nitrophenyl headgroup could yield novel LasR antagonists.16−18 We note that while we have previously examined related hybrid AHL analogues, with non-native headgroups and known active aryl tails,40 these compounds had only limited activity in LasR. The structural data demonstrating the strong overlap of the TP1 2-nitrophenyl headgroup with that of the HSL strengthened our enthusiasm for exploring the activity of the new hybrid class of LasR modulators proposed here.
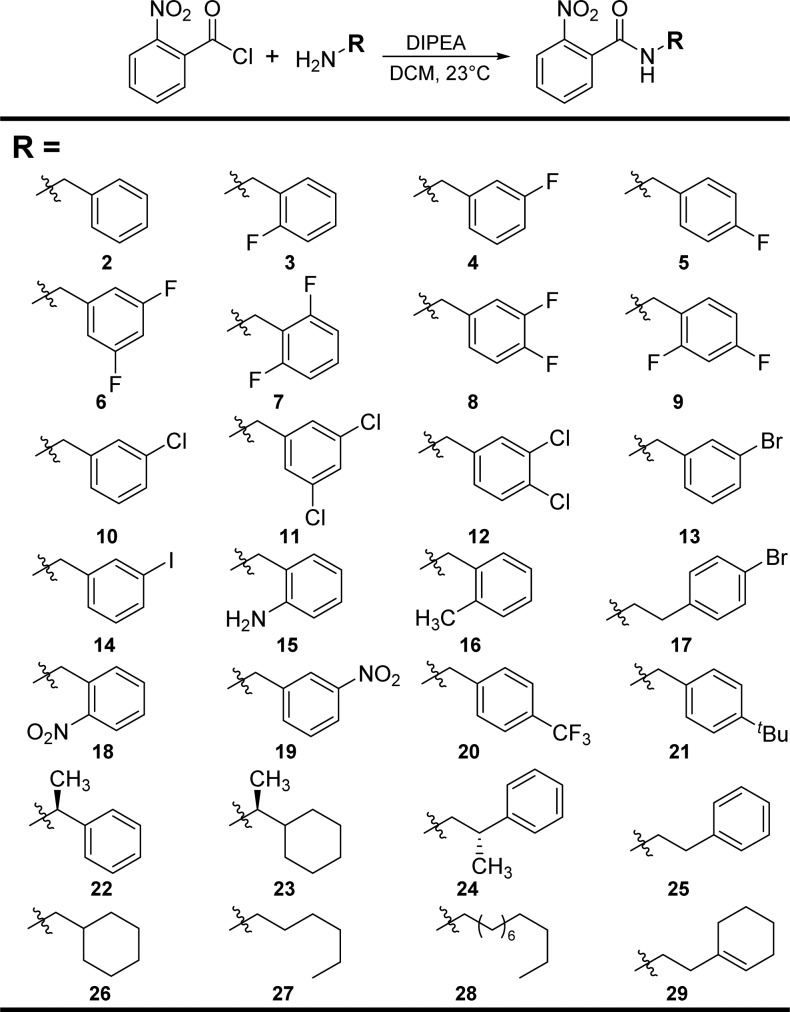
To test our strategy, we first designed a focused library of 28 hybrid compounds combining the 2-nitrophenyl headgroup with a variety of acyl groups that could presumably sample the chemical space in the LasR ligand-binding site usually occupied by the tail region of OdDHL (as judged by our analysis of the OdDHL:LasR structure) and/or have previously been shown to strongly modulate LasR when incorporated into AHLs.16−18 These compounds were readily prepared by reacting 2-nitrobenzoyl chloride with various amines in dichloromethane with Hünig’s base (Scheme 1). After compound purification, the library was evaluated for LasR agonism and antagonism using an Escherichia coli strain that contains a LasR expression plasmid and reports LasR activity via β-galactosidase production through a promoter fusion (see the Supporting Information for details).18
Scheme 1. Synthesis of the Hybrid Compound Library.
All compounds had purities ≥95%; see the Supporting Information for full characterization data.
We began by screening the library for LasR agonism at 50 μM. Among the various chemotypes represented, we anticipated that compound 28 would show LasR agonism, because it contained a tail similar in length to that of OdDHL. The primary screen revealed m-nitro compound 19 and, as anticipated, 28 as LasR agonists, whereas the remaining compounds were inactive. Dose–response analyses using the E. coli reporter strain revealed that 19 and 28 agonized LasR with EC50 values of 25 and 0.36 μM, respectively (see the Supporting Information). We note that these agonists are considerably less active than TP1 and OdDHL (each with EC50 values of <5 nM in this strain). Nevertheless, this result demonstrated that hybrid compounds that combine the 2-nitrophenyl headgroup with chemical moieties distinct from TP1 can act as LasR modulators. It is interesting that the other library members with structures very similar to 19 and 28 failed to agonize LasR, which suggests that the tail groups in 19 and 28 may be capable of making specific contacts with LasR that serve to stabilize the active protein. LuxR-type proteins are known to be highly selective for agonist-type ligands, whether native or non-native,19 and these data for LasR match this trend.
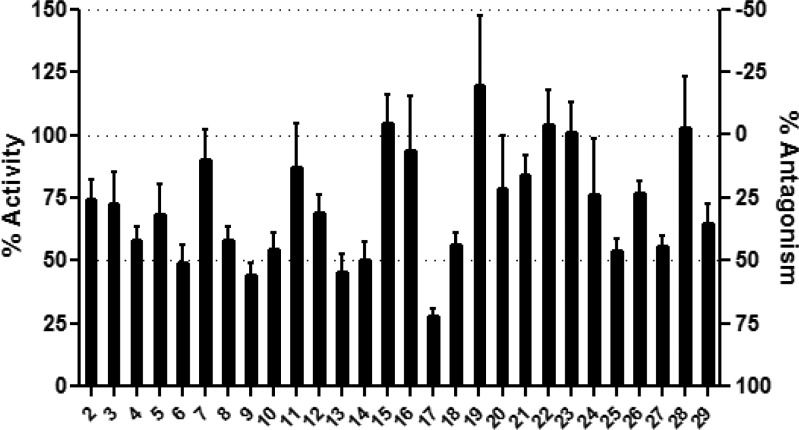
We next evaluated the library in a primary LasR antagonism screen at 50 μM in competition against OdDHL (at 5 nM; see the Supporting Information). Many of the compounds showed strong antagonism of LasR at this concentration (Figure 3), demonstrating that this hybrid scaffold was poised for LasR antagonism. Examination of the assay data revealed some interesting SAR trends for LasR antagonism by compounds with benzylamine-derived tails. Specifically, halogen substitution at the meta position of the aryl tail engendered efficacious LasR inhibition at 50 μM (e.g., compounds 4, 6, 8, 10, 13, and 14). Compounds that contained this functionality had an average LasR antagonism of 48%, whereas those lacking it averaged only 23% antagonism. This SAR trend mirrors our previous observation that meta-substitution on the aryl groups of phenylacetyl HSL analogues (i.e., PHLs) engendered strong LasR antagonism.18 In addition, we found that the one-carbon longer hybrid homologues derived from phenethylamine, most notably p-bromo derivative 17, showed LasR inhibitory activity. These single-point assay data encouraged us to evaluate hybrid compounds with ∼40% LasR antagonism for potency.
Figure 3.
Primary antagonism data for the hybrid compound library in the E. coli LasR reporter strain. Compounds were evaluated at 50 μM in the presence of an EC70 of OdDHL (5 nM). Culture treated with only 5 nM OdDHL was set to 0% antagonism/100% activity. Culture treated with only DMSO was set to 100% antagonism/0% activity (see the Supporting Information for full assay details).
After generating antagonism dose–response curves, we noted that IC50 values for the selected compounds ranged from 4.8 to 52 μM and maximum antagonism (i.e., efficacy) ranged from 41 to 78% (Table 1). These data further highlighted the SAR describing the heightened activity of compounds containing meta-halogenated phenyl rings linked to the 2-nitrophenyl headgroup (e.g., 4, 6, 8, 10, 13, and 14). Indeed, these compounds exhibited an average IC50 of 12 μM. Compounds without this structural moiety, on the other hand, had an average IC50 of 31 μM. Interestingly, the o-nitro compound (18), a regioisomer of the m-nitro LasR agonist 19 noted above, showed strong LasR antagonism instead (IC50 = 9.7 μM). Many of the most potent compounds, however, displayed a slight to moderate non-monotonic dose–response that is usually characteristic of potent AHL-derived LuxR-type receptor antagonists.18 Specifically, compounds 6, 10, and 13 showed substantial curve inversion to LasR agonism at concentrations >100 μM, whereas compound 14 exhibited a very slight LasR agonism upturn at 250 μM (see curves in the Supporting Information). Furthermore, maximum efficacy for 6, 10, 13, and 14 was modest and averaged 56% antagonism. However, p-bromo compound 17, based on aryl AHL analogue 1 (Figure 2D), defied these two trends. In fact, 17 displayed no non-monotonic characteristics, was the most potent LasR antagonist identified in our focused library (IC50 = 4.8 μM), and had a maximum efficacy that was among the best (72%). As AHL 1 is one of our most potent reported LasR antagonists (in this E. coli reporter44 and others18), it follows that this scaffold-hopping approach on TP1 involving the same tail region could produce a potent LasR antagonist.
Table 1. Potency and Maximum Efficacy Values for Selected Compounds in the E. coli LasR Reporter.
| compd | IC50 (μM)a | 95% CI (μM)b | max % antagonismc |
|---|---|---|---|
| 4 | 21 | 10–43 | 60 |
| 6 | 6.6d | 4.1–10 | 55 |
| 8 | 16 | 10–25 | 58 |
| 9 | 45 | 27–74 | 78 |
| 10 | 9.0d | 4.1–10 | 50 |
| 13 | 9.6d | 4.5–20 | 57 |
| 14 | 6.5d | 3.6–11 | 62 |
| 17 | 4.8 | 3.2–7.0 | 72 |
| 18 | 9.7 | 4.8–20 | 59 |
| 20 | 35 | 21–59 | 41 |
| 24 | 37 | 26–53 | 71 |
| 25 | 21 | 12–37 | 72 |
| 27 | 40 | 30–55 | 78 |
| 29 | 52 | 32–84 | 72 |
Calculated by testing the compound’s ability to abrogate LasR based activation of lasI-lacZ over a range of concentrations in the presence of 5 nM OdDHL.
CI = 95% confidence interval for IC50 value.
Denotes the lowest amount of LasR activity seen for individual compounds at any concentration throughout the curve. See the Supporting Information for individual dose–response curves.
Dose–response antagonism curve shows inversion to agonism (i.e., non-monotonic behavior) at high compound concentrations. Concentrations at which LasR agonism was observed were excluded from IC50 calculations.
Looking more broadly at the library, when we compared the activities of these new hybrid compounds to our previously reported non-native AHL analogues that contain the same tail structures,16−18 the hybrid compounds consistently have slightly reduced potencies relative to the parent AHL analogues. To obtain more quantitative comparative data, we submitted the HSL-containing congeners of compounds 14, 17, and 25 to identical LasR antagonism assay conditions and found that the AHL congeners were 11-, 18-, and 2-fold more potent, respectively (see the Supporting Information for dose curves of AHL compounds). However, the hybrid compounds displayed other beneficial qualities for use as probe molecules that offset this reduction in potency; we delineate these features below for compounds 14 and 17.
In contrast to other potent hybrid compounds identified herein (e.g., 6, 10, and 13), compounds 14 and 17 display only slight or no non-monotonic effects in their LasR antagonism curves, respectively. Accordingly, these compounds largely maintained their LasR inhibitory activities at high concentrations, an extremely desirable trait for probe compounds intended for use as QS inhibitors (e.g., if used at ∼10–50 times their IC50). In contrast, the AHL congeners of 14 and 17 elicit a strong non-monotonic effect that induces LasR agonism at higher compound concentrations (in both E. coli and P. aeruginosa reporter strains).44 The dose–response curves of AHL analogue 1 and hybrid compound 17 explicitly illustrate this phenomenon (Figure 4, panel A vs panel B). Again, we are currently working to fully understand the mechanistic origins of such non-monotonic dose curves (for AHLs and certain hybrid compounds). With or without these mechanistic data, if one seeks to probe a system by antagonizing LasR and observing the downstream effects, AHL analogue 1 must be dosed carefully to ensure that LasR is not being unintentionally agonized (Figure 4A). In contrast, antagonist 17 unambiguously causes LasR antagonism (Figure 4B), and this compound’s maximum efficacy surpasses that of AHL analogue 1. These features serve to showcase the value of 17 as a chemical tool to study LasR relative to AHL 1 and that one must look at characteristics beyond an IC50 value to best ascertain a compound’s utility.
Figure 4.
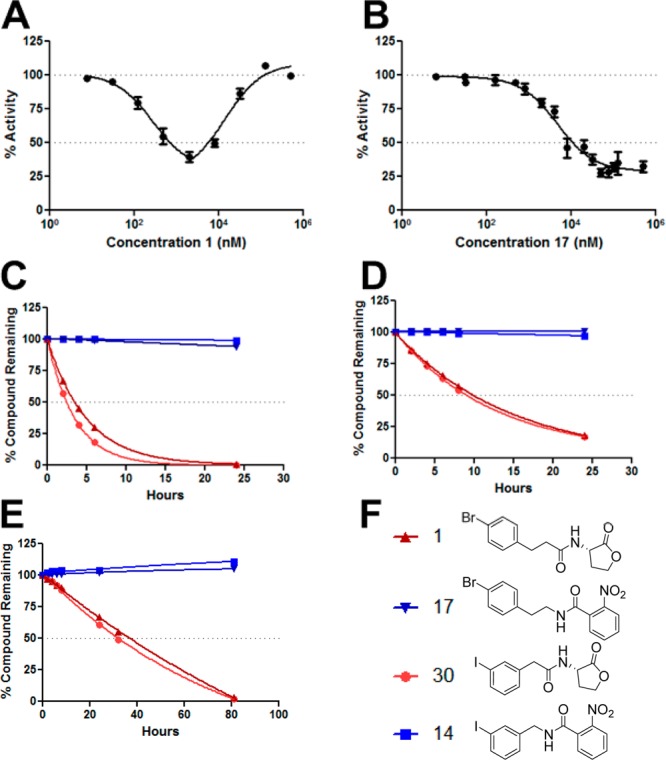
LasR antagonism dose–response curves for AHL analogue 1 (A) and hybrid compound 17 (B) in the E. coli reporter. AHL analogues 1 and 30 decompose in aqueous buffers at pH 8 (C), 7 (D), and 6 (E), whereas hybrid compounds 14 and 17 are stable throughout the assay. Compound structures are shown in panel F. Compound concentrations were determined using HPLC referenced to an internal standard (see the Supporting Information).
As highlighted above, another chemical liability of AHLs relative to other QS probe molecules is their hydrolytic instability,37,41−43 and we reasoned that hybrid compounds such as 14 and 17, lacking the HSL headgroup, should be highly robust in aqueous media. The rate of AHL hydrolysis will of course be dependent on the medium type and assay conditions, and this can be affected by variable pH throughout the length of assays. Of direct interest to LasR, P. aeruginosa is known to preferentially metabolize weak carbon acids, which has been shown to increase the pH of standard culture (i.e., LB) media to approximately 8.5 over 48 h.37 Therefore, we decided to evaluate the hydrolytic stabilities of compounds 14 and 17 over time in buffer solutions at pH 6, 7, and 8 and compare their stabilities to those of their AHL congeners (30 and 1, respectively; Figure 4C–F). The compound solutions were incubated at 37 °C with shaking, and aliquots were examined every 2 h using high-performance liquid chromatography (HPLC).
As predicted from the literature,35,37,41−43 the AHLs decomposed at the fastest rate in a mildly alkaline buffer (Figure 4C), at a moderate rate in neutral buffer (Figure 4D), and slowest under slightly acidic conditions (Figure 4E). The HSL half-lives for AHLs 1 and 30 at neutral pH were between 8.5 and 11 h. Because many assays routinely last longer than 12 h, these data demonstrate that the concentration of active AHL will vary widely throughout an assay, providing an additional layer of complexity to designing the assay, analyzing data, and drawing conclusions. Furthermore, if the pH becomes slightly alkaline, which occurs regularly with P. aeruginosa cultures, HSL decomposition accelerates. The decomposition of 1 and 30 at pH 8 illustrates this point, as their half-lives decrease to between 2 and 3.5 h (Figure 4C). In contrast, hybrid compounds 14 and 17 were highly stable under each buffer condition tested (blue lines, Figure 4C–E), showing no decomposition. These results further emphasize the worth of such hybrid chemotypes as hydrolytically stable LasR modulators that can be used in a range of medium types, including slightly acidic or alkaline conditions. Although not evaluated here, we also predict that these non-AHL derived compounds should display enhanced stability toward bacteria- and host-derived enzymes known to degrade AHLs such as lactonases and acylases.37,41−43
In summary, we report herein our efforts toward the design of new QS modulators in P. aeruginosa with enhanced stabilities and activities for use as chemical tools. We outline a structure-based scaffold-hopping approach for the design of new synthetic ligands to modulate the P. aeruginosa QS receptor, LasR. We replaced the HSL headgroup with a 2-nitrophenyl fragment derived from the known LasR agonist, TP1, and combined that component with various tail regions of non-native AHL analogues known for LasR antagonism to generate a 28-member focused library. Screening of this library in an E. coli LasR reporter strain led to the discovery of many new LasR modulators, some of which displayed strong potency and efficacy as LasR antagonists. Compound 17 was the most valuable tool compound emerging from this study, with an IC50 of 4.8 μM and a maximum efficacy of 72%. Importantly, 17 does not display the non-monotonic LasR modulatory activity exhibited by known potent AHL analogues and other hybrid compounds identified in this library. Furthermore, this new hybrid chemotype also had greatly enhanced hydrolytic stability compared to HSL containing congeners. Ongoing studies will focus on improving compound potency, replacing the nitro group with an isosteric substitute resulting in compounds with greater in vivo tolerance,49 and evaluation of these compounds in other LuxR-type receptors. These experiments will be reported in due course.
Acknowledgments
Financial support for this work was provided by the NIH (R01 GM109403). M.C.O. acknowledges support from the Arnold and Mabel Beckman Foundation through an Arnold O. Beckman Postdoctoral Fellowship. NMR and MS instrumentation at UW—Madison is supported by the NSF (CHE-1048642 and CHE-0342998) and the NIH (1S10 0D020022), respectively, and a generous gift from Paul J. Bender.
Glossary
Abbreviations
- QS
quorum sensing
- AHL
N-acyl l-homoserine lactone
- HSL
homoserine lactone
- PQS
Pseudomonas quinolone signal
- SAR
structure–activity relationship
- OdDHL
N-(3-oxo-dodecanoyl) l-homoserine lactone
- HPLC
high-performance liquid chromatography
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.5b00112.
Synthetic protocols and full compound characterization data, biological assay protocols, dose–response curves, and full details of compound stability studies. (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Boucher H. W.; Talbot G. H.; Benjamin D. K.; Bradley J.; Guidos R. J.; Jones R. N.; Murray B. E.; Bonomo R. A.; Gilbert D. (2013) 10 × ’20 Progress—Development of New Drugs Active against Gram-Negative Bacilli: An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 56, 1685–1694. 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B.; Guidos R.; Gilbert D.; Bradley J.; Boucher H. W.; Scheld W. M.; Bartlett J. G.; Edwards J. (2008) The Epidemic of Antibiotic-Resistant Infections: A Call to Action for the Medical Community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 155–164. 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- Wright G. D. (2007) The Antibiotic Resistome: The Nexus of Chemical and Genetic Diversity. Nat. Rev. Microbiol. 5, 175–186. 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- Cegelski L.; Marshall G. R.; Eldridge G. R.; Hultgren S. J. (2008) The Biology and Future Prospects of Antivirulence Therapies. Nat. Rev. Microbiol. 6, 17–27. 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatworthy A. E.; Pierson E.; Hung D. T. (2007) Targeting Virulence: A New Paradigm for Antimicrobial Therapy. Nat. Chem. Biol. 3, 541–548. 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- Rutherford S. T.; Bassler B. L. (2012) Bacterial Quorum Sensing: Its Role in Virulence and Possibilities for Its Control. Cold Spring Harbor Perspect. Med. 2, 1–26. 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilli A.; Bassler B. L. (2006) Bacterial Small-Molecule Signaling Pathways. Science 311, 1113–1116. 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua C.; Greenberg E. P. (2002) Listening in on Bacteria: Acyl-Homoserine Lactone Signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- Njoroge J.; Sperandio V. (2009) Jamming Bacterial Communication: New Approaches for the Treatment of Infectious Diseases. EMBO Mol. Med. 1, 201–210. 10.1002/emmm.200900032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. A.; Eibergen N. R.; Moore J. D.; Blackwell H. E. (2015) Small Molecule Disruption of Quorum Sensing Cross-Regulation in Pseudomonas aeruginosa Causes Major and Unexpected Alterations to Virulence Phenotypes. J. Am. Chem. Soc. 137, 1510–1519. 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdt J. P.; McInnis C. E.; Schell T. L.; Rossi F. M.; Blackwell H. E. (2014) Mutational Analysis of the Quorum-Sensing Receptor LasR Reveals Interactions That Govern Activation and Inhibition by Nonlactone Ligands. Chem. Biol. 21, 1361–1369. 10.1016/j.chembiol.2014.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. D.; Gerdt J. P.; Eibergen N. R.; Blackwell H. E. (2014) Active Efflux Influences the Potency of Quorum Sensing Inhibitors in Pseudomonas aeruginosa. ChemBioChem 15, 435–442. 10.1002/cbic.201300701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal-Gan Y.; Stacy D. M.; Foegen M. K.; Koenig D. W.; Blackwell H. E. (2013) Highly Potent Inhibitors of Quorum Sensing in Staphylococcus aureus Revealed through a Systematic Synthetic Study of the Group-III Autoinducing Peptide. J. Am. Chem. Soc. 135, 7869–7882. 10.1021/ja3112115. [DOI] [PubMed] [Google Scholar]
- Praneenararat T.; Palmer A. G.; Blackwell H. E. (2012) Chemical Methods to Interrogate Bacterial Quorum Sensing Pathways. Org. Biomol. Chem. 10, 8189. 10.1039/c2ob26353j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattmann M. E.; Blackwell H. E. (2010) Small Molecules That Modulate Quorum Sensing and Control Virulence in Pseudomonas aeruginosa. J. Org. Chem. 75, 6737–6746. 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; Mattmann M. E.; Blackwell H. E. (2008) Evaluation of a Focused Library of N-Aryl l-Homoserine Lactones Reveals a New Set of Potent Quorum Sensing Modulators. Bioorg. Med. Chem. Lett. 18, 5978–5981. 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; O’Neill J. C.; Miller D. M.; Wezeman R. J.; Mattmann M. E.; Lin Q.; Blackwell H. E. (2008) Comparative Analyses of N-Acylated Homoserine Lactones Reveal Unique Structural Features That Dictate Their Ability to Activate or Inhibit Quorum Sensing. ChemBioChem 9, 389–400. 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; O’Neill J. C.; Miller D. M.; Mattmann M. E.; Blackwell H. E. (2007) Modulation of Bacterial Quorum Sensing with Synthetic Ligands: Systematic Evaluation of N-Acylated Homoserine Lactones in Multiple Species and New Insights into Their Mechanisms of Action. J. Am. Chem. Soc. 129, 13613–13625. 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; O’Neill J. C.; Blackwell H. E. (2007) N-Phenylacetanoyl-l-Homoserine Lactones Can Strongly Antagonize or Superagonize Quorum Sensing in Vibrio fischeri. ACS Chem. Biol. 2, 315–320. 10.1021/cb700036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayo J.; Amara N.; Krief P.; Meijler M. M. (2011) Live Cell Labeling of Native Intracellular Bacterial Receptors Using Aniline-Catalyzed Oxime Ligation. J. Am. Chem. Soc. 133, 7469–7475. 10.1021/ja200455d. [DOI] [PubMed] [Google Scholar]
- Amara N.; Mashiach R.; Amar D.; Krief P.; Spieser S. A. H.; Bottomley M. J.; Aharoni A.; Meijler M. M. (2009) Covalent Inhibition of Bacterial Quorum Sensing. J. Am. Chem. Soc. 131, 10610–10619. 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- Morkunas B.; Galloway W. R. J. D.; Wright M.; Ibbeson B. M.; Hodgkinson J. T.; O’Connell K. M. G.; Bartolucci N.; Della Valle M.; Welch M.; Spring D. R. (2012) Inhibition of the Production of the Pseudomonas aeruginosa Virulence Factor Pyocyanin in Wild-Type Cells by Quorum Sensing Autoinducer-Mimics. Org. Biomol. Chem. 10, 8452. 10.1039/c2ob26501j. [DOI] [PubMed] [Google Scholar]
- Hodgkinson J. T.; Galloway W. R. J. D.; Wright M.; Mati I. K.; Nicholson R. L.; Welch M.; Spring D. R. (2012) Design, Synthesis and Biological Evaluation of Non-Natural Modulators of Quorum Sensing in Pseudomonas aeruginosa. Org. Biomol. Chem. 10, 6032. 10.1039/c2ob25198a. [DOI] [PubMed] [Google Scholar]
- Galloway W. R. J. D.; Hodgkinson J. T.; Bowden S.; Welch M.; Spring D. R. (2012) Applications of Small Molecule Activators and Inhibitors of Quorum Sensing in Gram-Negative Bacteria. Trends Microbiol. 20, 449–458. 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Galloway W. R. J. D.; Hodgkinson J. T.; Bowden S. D.; Welch M.; Spring D. R. (2011) Quorum Sensing in Gram-Negative Bacteria: Small-Molecule Modulation of AHL and AI-2 Quorum Sensing Pathways. Chem. Rev. 111, 28–67. 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- Hentzer M.; Wu H.; Andersen J. B.; Riedel K.; Rasmussen T. B.; Bagge N.; Kumar N.; Schembri M. A.; Song Z.; Kristoffersen P.; Manefield M.; Costerton J. W.; Molin S.; Eberl L.; Steinberg P.; Kjelleberg S.; Høiby N.; Givskov M. (2003) Attenuation of Pseudomonas aeruginosa Virulence by Quorum Sensing Inhibitors. EMBO J. 22, 3803–3815. 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin C. T.; Miller L. C.; Siryaporn A.; Drescher K.; Semmelhack M. F.; Bassler B. L. (2013) A Quorum-Sensing Inhibitor Blocks Pseudomonas aeruginosa Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. U. S. A. 110, 17981–17986. 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadikot R. T.; Blackwell T. S.; Christman J. W.; Prince A. S. (2005) Pathogen-Host Interactions in Pseudomonas aeruginosa Pneumonia. Am. J. Respir. Crit. Care Med. 171, 1209–1223. 10.1164/rccm.200408-1044SO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak J. B.; Cannon C. L.; Pier G. B. (2000) Establishment of Pseudomonas aeruginosa Infection: Lessons from a Versatile Opportunist. Microbes Infect. 2, 1051–1060. 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- Schuster M.; Greenberg E. P.. LuxR-Type Proteins in Pseudomonas aeruginosa Quorum Sensing: Distinct Mechanisms with Global Implications. In Chemical Communication among Bacteria; Williams S. C., Bassler B. L., Eds.; ASM Press: Washington, DC, 2008; pp 133−144. [Google Scholar]
- Wagner V. E.; Bushnell D.; Passador L.; Brooks A. I.; Iglewski B. H. (2003) Microarray Analysis of Pseudomonas Aeruginosa Quorum-Sensing Regulons: Effects of Growth Phase and Environment. J. Bacteriol. 185, 2080–2095. 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; Wezeman R. J.; Siegel A. P.; Blackwell H. E. (2005) Small Molecule Inhibitors of Bacterial Quorum Sensing and Biofilm Formation. J. Am. Chem. Soc. 127, 12762–12763. 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- Smith K. M.; Bu Y.; Suga H. (2003) Induction and Inhibition of Pseudomonas Aeruginosa Quorum Sensing by Synthetic Autoinducer Analogs. Chem. Biol. 10, 81–89. 10.1016/S1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Amara N.; Krom B. P.; Kaufmann G. F.; Meijler M. M. (2011) Macromolecular Inhibition of Quorum Sensing: Enzymes, Antibodies, and beyond. Chem. Rev. 111, 195–208. 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
- Müh U.; Hare B. J.; Duerkop B. A.; Schuster M.; Hanzelka B. L.; Heim R.; Olson E. R.; Greenberg E. P. (2006) A Structurally Unrelated Mimic of a Pseudomonas aeruginosa Acyl-Homoserine Lactone Quorum-Sensing Signal. Proc. Natl. Acad. Sci. U. S. A. 103, 16948–16952. 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müh U.; Schuster M.; Heim R.; Singh A.; Olson E. R.; Greenberg E. P. (2006) Novel Pseudomonas aeruginosa Quorum-Sensing Inhibitors Identified in an Ultra-High-Throughput Screen. Antimicrob. Agents Chemother. 50, 3674–3679. 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glansdorp F. G.; Thomas G. L.; Lee J. K.; Dutton J. M.; Salmond G. P. C.; Welch M.; Spring D. R. (2004) Synthesis and Stability of Small Molecule Probes for Pseudomonas aeruginosa Quorum Sensing Modulation. Org. Biomol. Chem. 2, 3329–3336. 10.1039/b412802h. [DOI] [PubMed] [Google Scholar]
- Kaufmann G. F.; Sartorio R.; Lee S.-H.; Mee J. M.; Altobell L. J.; Kujawa D. P.; Jeffries E.; Clapham B.; Meijler M. M.; Janda K. D. (2006) Antibody Interference with N-Acyl Homoserine Lactone-Mediated Bacterial Quorum Sensing. J. Am. Chem. Soc. 128, 2802–2803. 10.1021/ja0578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis C. E.; Blackwell H. E. (2011) Thiolactone Modulators of Quorum Sensing Revealed through Library Design and Screening. Bioorg. Med. Chem. 19, 4820–4828. 10.1016/j.bmc.2011.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnis C. E.; Blackwell H. E. (2011) Design, Synthesis, and Biological Evaluation of Abiotic, Non-Lactone Modulators of LuxR-Type Quorum Sensing. Bioorg. Med. Chem. 19, 4812–4819. 10.1016/j.bmc.2011.06.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates E. A.; Philipp B.; Buckley C.; Atkinson S.; Chhabra S. R.; Sockett R. E.; Goldner M.; Dessaux Y.; Cámara M.; Smith H.; Williams P. (2002) N-Acylhomoserine Lactones Undergo Lactonolysis in a pH-, Temperature-, and Acyl Chain Length-Dependent Manner during Growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect. Immun. 70, 5635–5646. 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F.; Wang L.-H.; Wang J.; Dong Y.-H.; Hu J. Y.; Zhang L.-H. (2005) Quorum Quenching Enzyme Activity Is Widely Conserved in the Sera of Mammalian Species. FEBS Lett. 579, 3713–3717. 10.1016/j.febslet.2005.05.060. [DOI] [PubMed] [Google Scholar]
- Ozer E. A.; Pezzulo A.; Shih D. M.; Chun C.; Furlong C.; Lusis A. J.; Greenberg E. P.; Zabner J. (2005) Human and Murine Paraoxonase 1 Are Host Modulators of Pseudomonas aeruginosa Quorum-Sensing. FEMS Microbiol. Lett. 253, 29–37. 10.1016/j.femsle.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Moore J. D.; Rossi F. M.; Welsh M. A.; Nyffeler K. E.; Blackwell H. E.. A Comparative Analysis of Synthetic Quorum Sensing Modulators in Pseudomonas aeruginosa: New Insights Into Mechanism, Active Efflux Susceptibility, Phenotypic Response, and Next-Generation Ligand Design. J. Am. Chem. Soc., 2015, in press, DOI: 10.1021/jacs.5b06728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhari J. S.; Kinoyama I.; Struss A. K.; Pullanikat P.; Lowery C. A.; Lardy M.; Janda K. D. (2011) Synthesis and Molecular Modeling Provide Insight into a Pseudomonas aeruginosa Quorum Sensing Conundrum. J. Am. Chem. Soc. 133, 3840–3842. 10.1021/ja111138y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. T.; Noto J. G.; Nichols-O’Neill L.; Perez L. J. (2015) Potent Irreversible Inhibitors of LasR Quorum Sensing in Pseudomonas aeruginosa. ACS Med. Chem. Lett. 6, 162–167. 10.1021/ml500459f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y.; Nair S. K. (2009) Molecular Basis for the Recognition of Structurally Distinct Autoinducer Mimics by the Pseudomonas aeruginosa LasR Quorum-Sensing Signaling Receptor. Chem. Biol. 16, 961–970. 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley M. J.; Muraglia E.; Bazzo R.; Carfì A. (2007) Molecular Insights into Quorum Sensing in the Human Pathogen Pseudomonas aeruginosa from the Structure of the Virulence Regulator LasR Bound to Its Autoinducer. J. Biol. Chem. 282, 13592–13600. 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Meanwell N. A. (2011) Synopsis of Some Recent Tactical Application of Bioisosteres in Drug Design. J. Med. Chem. 54, 2529–2591. 10.1021/jm1013693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.