Abstract
Background
Renewed attention has been directed to the functions of the Y chromosome in the central nervous system during early human male development, due to the recent proposed involvement in neurodevelopmental diseases. PCDH11Y and NLGN4Y are of special interest because they belong to gene families involved in cell fate determination and formation of dendrites and axon.
Methods
We used RNA sequencing, immunocytochemistry and a padlock probing and rolling circle amplification strategy, to distinguish the expression of X and Y homologs in situ in the human brain for the first time. To minimize influence of androgens on the sex differences in the brain, we focused our investigation to human embryos at 8–11 weeks post-gestation.
Results
We found that the X- and Y-encoded genes are expressed in specific and heterogeneous cellular sub-populations of both glial and neuronal origins. More importantly, we found differential distribution patterns of X and Y homologs in the male developing central nervous system.
Conclusions
This study has visualized the spatial distribution of PCDH11X/Y and NLGN4X/Y in human developing nervous tissue. The observed spatial distribution patterns suggest the existence of an additional layer of complexity in the development of the male CNS.
Electronic supplementary material
The online version of this article (doi:10.1186/s13293-015-0056-4) contains supplementary material, which is available to authorized users.
Keywords: Sex differences, Human embryo development, Female, Male, Gene expression, Protocadherin, Neuroligin, X chromosome, Y chromosome, Brain, Spinal cord, Cortex, Medulla oblongata, Rolling circle amplification, PCDH11X, PCDH11Y, NLGN4X, NLGN4Y, ISLET1, OLIG2, SOX10, NeuN
Background
The current concept about the mechanism of sexual differentiation of the brain during development includes not only the action of gonadal hormones but also the function of genes encoded by the sex chromosomes [1–3]. The brain effects mediated by the actions of the Y chromosome have not attracted the same amount of interest as those mediated by genes encoded on the larger X chromosome, which is enriched in genes involved in neurodevelopment and cognition [4]. In recent years, attention to Y chromosome functions in the brain during development has increased, due to its proposed involvement in autism [5] and non-syndromic speech delay [6]. Furthermore, neuroimaging analysis within a rare cohort of humans with diverse sex chromosome aneuploidies showed that the X and Y chromosomes have opposing effects on cortical development [7] and cortical thickness asymmetry [8].
We have shown that several Y chromosome-encoded genes are expressed in the brain during human male development [9]. Of these, PCDH11Y and NLGN4Y deserve a special attention because they belong to two gene families named protocadherins and neuroligins, predominantly expressed in the nervous system and with important functions in survival, cell fate determination and formation and maturation of synapses in different neuronal populations [10, 11]. PCDH11Y is unique to humans while PCDH11X is present in our closest relatives, the chimpanzees. The duplication event that transferred the PCDH11X to the Y chromosome took place approximately six MYA ago [12]. During early investigations of PCDH11Y, when the gene was described as the only “gain of function” gene in the human genome, there was an ongoing debate regarding its human specificity [13]. The question was finally resolved in 2006 when it was shown that PCHD11Y is human-specific, and absent in both gorilla (Gorilla gorilla) and chimpanzee (Pan troglodytes) as well as other non-human primates [14].
NLGN4Y, together with its X homolog counterpart, has been proposed as candidate genes for autism spectrum disorders [5, 15]. The PCDH11X/Y gene pair has been suggested to be involved in psychosis, partly because of its putative roles in human speciation and partly due to its possible relation to structural changes in the brain [16, 17]. An important step to understand the possible functional relevance of Y-encoded genes for the development of sexual dimorphism in the brain is to investigate whether X and Y homologs are expressed in a spatial sex-specific manner.
Until now, no research has been done on the genetic contribution of X- and Y-linked genes to sex differences in the human brain prior to hormonal activation. It is therefore of great interest to investigate eventual sexual dimorphisms contributed by sex chromosome-linked genes alone with minimal influence of hormones [2]. To detect the contribution of genetic components encoded on the Y chromosome to sex differences in the brain, we used a human central nervous system (CNS) tissue acquired as early as possible during development, so that androgen hormones, such as testosterone, are not expressed or expressed at minimum levels. Indeed, in the developing Sertoli cells in the testes, androgens start to be produced around week 11 post-gestation and slowly start to influence gene expression in the brain in an activating manner after week 11 [2, 18–21].
These experiments cannot be performed using conventional in situ hybridization or immunofluorescence techniques because of cross-hybridization of probes due to high sequence similarity [15]. Indeed, different isoforms of PCDH11Y share 97 to 99 % sequence identity with the coding region of the homolog PCDH11X isoforms. Both in situ messenger RNA (mRNA) detection [22] and immunohistochemistry [23] have shown widespread expression of PCDH11X and PCDH11Y in the human brain during development, but the contribution of each gene to the observed signals could not be distinguished.
Different isoforms of NLGN4Y share 89 to 98 % sequence identity with the coding region of the homolog NLGN4X isoforms. RT-PCR experiments in individual male and female adult human brain tissues determined that both NLGN4X and NLGN4Y are expressed [24], but the spatial expression patterns of each gene, particularly during human brain development, is not known. To specifically distinguish the tissue and cellular distribution of transcripts from X and Y homolog genes, we designed a strategy using RNA sequencing technology, together with immunohistochemistry and padlock probing in combination with rolling circle amplification (RCA) [25]. As this last method relies on dual-target recognition and ligation-dependent signal amplification, it can be adapted to discriminate highly similar targets such as splice variants, mutations or members of the same gene family which may differ by only one or a few single nucleotide sequences [26]. We show here that both PCDH11Y and NLGNY are expressed in specific cellular sub-populations in the CNS that more rarely express their X homolog genes. Using a combination of immunohistochemistry and padlock probing, we also show that cells of different origin express PCDH11Y and NLGNY in the CNS during early male human development.
Results
RNA sequencing of medulla oblongata and midbrain samples from female and male human embryos
To quantify the expression of X and Y transcripts for PCDH11 and NLGN4 with high resolution and to study the expression of the other members of the neuroligin and protocadherin families, we performed RNA sequencing of CNS samples from female and male embryos, at 8–11 weeks of development. Both total RNA and polyA+ RNA were sequenced as described in the “Methods” section. Additional file 1: Table S1 shows the results of the analysis for all genes belonging to these gene families. All results are expressed in RPKM (reads per kilobase of transcript per million mapped reads). A very low number of RPKM from the Y-linked genes were detected in female tissue confirming the specificity of the technique (Fig. 1a, b). None of the X-linked genes showed significant differential expression levels in males and females in either the medulla oblongata (MO) or midbrain. To confirm the accuracy of the RNA sequencing data, we also compared the expression of PCDH11X and NLGN4X with the expression of ZFX, previously reported in adult brain as an escapee from X inactivation [27]. Figure 1c shows that ZFX had about twice the expression levels in females compared to males (adjusted p value 0.006) agreeing with previous results [27].
Fig. 1.
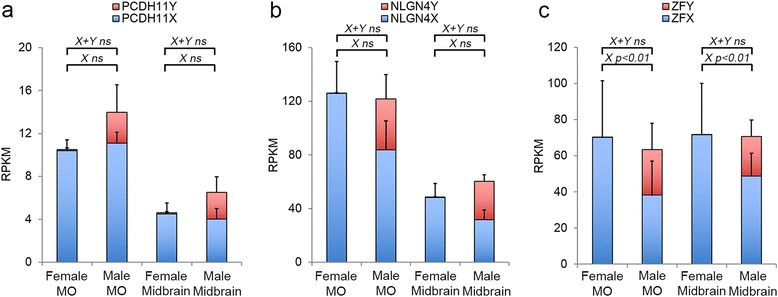
Quantification of expression levels using RNA sequencing. Staple bars show the average RPKM for both total RNA and polyA+ sequencing of female and male MO and midbrain for PCDH11X/Y (a), NLGN4X/Y (b) and ZFX/Y (c). Only ZFX showed significant expression differences between females and males in both tissues, and NLGN4X presented a non-significant increase in females
Padlock probes specifically discriminate between X and Y homologs
We used the padlock probes designed as described in the “Methods” section, as a cocktail for transcript detection by padlock probe ligation and RCA on sections prepared from male and female embryonic CNS tissues. Due to sample scarcity of female brain samples, we decided to focus on the MO and spinal cord (SC) for the in situ analysis. We observed that while the X-specific padlock probes detected transcripts in both female and male samples, the Y-specific padlock probes almost exclusively detected transcripts in male tissue sections, with very low background hybridization to females, confirming the sex specificity of the Y-specific padlock probes (Fig. 2). The low number of signals for the Y homologs, observed in the female tissues, were likely due to mis-ligation events of the Y-specific padlock probes but even more likely due to false-positive signals (autofluorescent rolling circle product like signals) identified by the image analysis software CellProfiler. In addition, we observed that both PCDH11X/Y and NLGN4X/Y mRNA expression are mainly confined to the grey matter, with very little expression in the white matter (“Wm” in Fig. 2g–j). As the samples are not comparable with respect to absolute signal counts, the counts for X and Y signals were normalized to counts per 1000 cells. When we quantified the amount of PCDH11X and PCDH11Y signals in 7 sections obtained from female SC tissues and 12 sections from male tissue, we observed that the amount of signals (per 1000 cells) for PCDH11X did not differ between male and female in both the SC and MO (Fig. 3), in accordance with the results from the RNA sequencing of the MO. On average, 50 % of the detected transcripts in the male SC corresponded to Y transcripts, and 50 % to X transcripts. In the male MO, the proportions were 27 % Y and 73 % X. When we quantified NLGN4X/Y signals in 8 sections obtained from female tissues and 14 sections from males (Table 2), we did not find differences in the number of observed signals for NLGN4X transcripts in females compared to males in both tissue types. On average, 56 % of the observed transcripts in male SC were Y transcripts, and 44 % were X transcripts. In the MO, the proportions were 51 and 49 %. These results correlated well with expression levels measured by RNA sequencing.
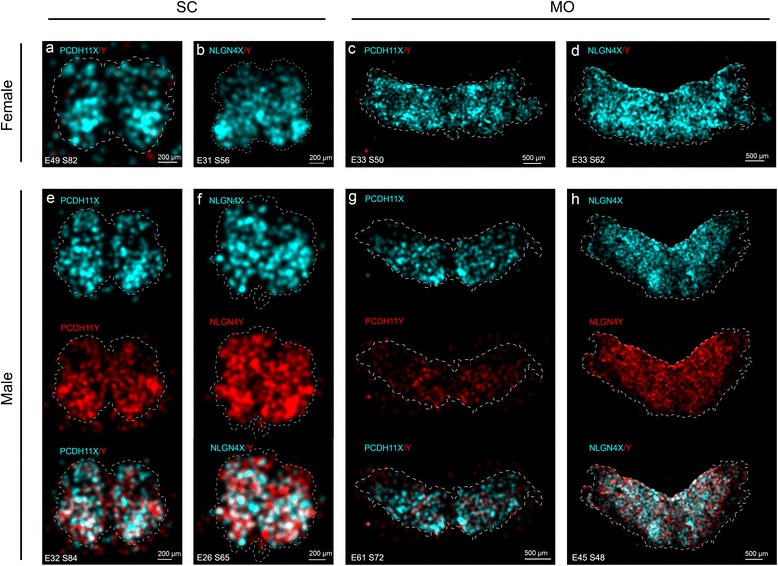
Fig. 2.
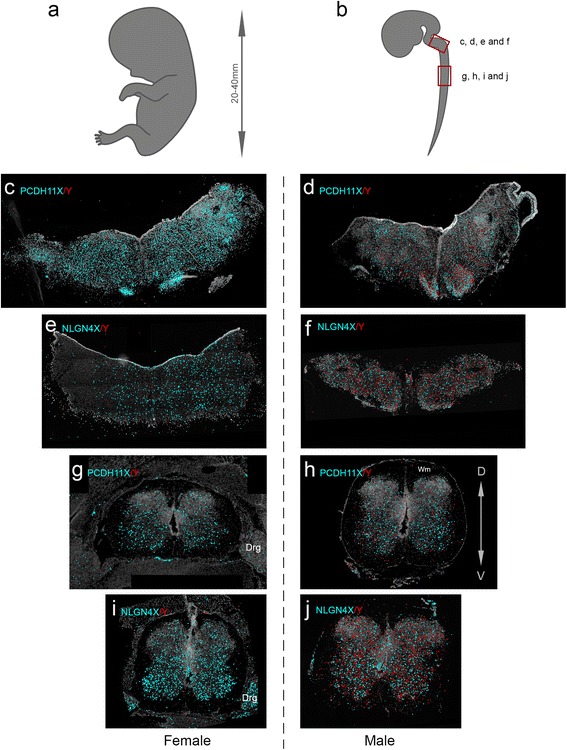
Padlock probe hybridization can distinguish between expression of X and Y homolog genes for PCDH11 and NLGN4. a, b Schematic representation of an embryo modified from Gasser [47] and the CNS of an embryo at 12 weeks of gestation modified from His [48]. The boxes in red mark the approximate position of the coronal sections shown in the rest of the figure. c Female medulla oblongata (MO) section hybridized with padlock probes for PCDH11X and Y. X signals in cyan and Y signals in red for all subfigures. d Male MO section hybridized with padlock probes for PCDH11X and Y. e Female MO section hybridized with padlock probes for NLGNX and Y. f Male MO section hybridized with padlock probes for NLGNX and Y. g Female spinal cord (SC) section hybridized with padlock probes PCDH11X and Y. h Male SC section hybridized with padlock probes for PCDH11X and Y. i Female SC section hybridized with padlock probes NLGNX and Y. j Male SC section hybridized with padlock probes for NLGNX and Y. All signals from females and males were detected using a Zeiss Axio Imager.Z2 epi-fluorescence microscope. The images were produced using the Zen software and enhanced to 15–20-pixel dots to allow visualization in ×20 magnification pictures. Drg dorsal root ganglia
Fig. 3.
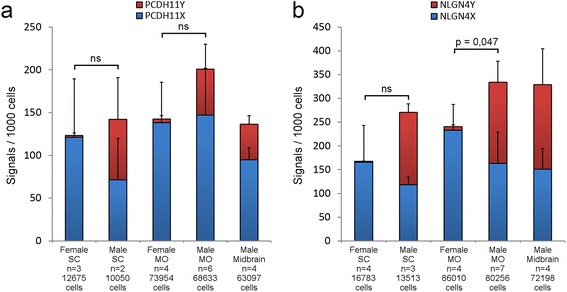
Quantification of padlock hybridization signals in SC and MO. The figure shows the average amount of signals per 1000 cells, identified by the image analysis software CellProfiler in the original microscopy images, for female and male SC, MO and midbrain. Standard errors are included for both females and males. The number of sections and the total number of cells analysed are indicated under each staple bar. a. PCDH11X/Y. b NLGN4X/Y. The clamps on top of the bars indicate the significance (or not) of the comparison between X gene expression between females and males
Table 2.
Distribution of cells with NLGN4X/Y transcripts: amount of cells with NLGN4X/Y signals
| Sample | Tissue | Gene | Gender | Cells without signals | Cells with X = 1 | Cells with Y = 1 | Cells with X + Y > = 2 | Cells with X > 1 Y = 0 | Cells with Y > 1 X = 0 | Total cell counts |
|---|---|---|---|---|---|---|---|---|---|---|
| E27_S48 | SC | NLGN4 | Female | 3098 | 364 | 3 | 4 | 45 | 0 | 3514 |
| E31_S56 | SC | NLGN4 | Female | 2955 | 493 | 3 | 0 | 94 | 1 | 3546 |
| E49_S81 | SC | NLGN4 | Female | 4225 | 350 | 4 | 0 | 47 | 0 | 4626 |
| E49_S82 | SC | NLGN4 | Female | 4767 | 297 | 9 | 1 | 23 | 0 | 5097 |
| E33_S62 | MO | NLGN4 | Female | 12,552 | 1917 | 20 | 4 | 284 | 0 | 14,777 |
| E34_S61 | MO | NLGN4 | Female | 22,028 | 3586 | 40 | 25 | 674 | 5 | 26,358 |
| E33_S63 | MO | NLGN4 | Female | 15,497 | 1652 | 49 | 14 | 177 | 1 | 17,390 |
| E34_S63 | MO | NLGN4 | Female | 24,119 | 2871 | 76 | 33 | 381 | 5 | 27,485 |
| E26_S65 | SC | NLGN4 | Male | 3165 | 200 | 303 | 36 | 26 | 25 | 3755 |
| E32_S29 | SC | NLGN4 | Male | 3917 | 319 | 447 | 75 | 25 | 36 | 4819 |
| E32_S84 | SC | NLGN4 | Male | 4120 | 310 | 381 | 69 | 28 | 31 | 4939 |
| E61_S39 | MO | NLGN4 | Male | 5283 | 379 | 414 | 58 | 19 | 47 | 6200 |
| E61_S89 | MO | NLGN4 | Male | 6123 | 633 | 710 | 117 | 56 | 75 | 7714 |
| E45_S48 | MO | NLGN4 | Male | 16,555 | 1564 | 1580 | 295 | 133 | 144 | 20,271 |
| E41_S58 | MO | NLGN4 | Male | 5719 | 563 | 584 | 421 | 50 | 49 | 7386 |
| E45_S50 | MO | NLGN4 | Male | 16,356 | 1644 | 1261 | 391 | 155 | 75 | 19,882 |
| E58_S82 | MO | NLGN4 | Male | 8219 | 1205 | 1102 | 491 | 200 | 114 | 11,331 |
| E61_S72 | MO | NLGN4 | Male | 6405 | 273 | 645 | 68 | 19 | 62 | 7472 |
| E45_S329 | Midbrain | NLGN4 | Male | 24,917 | 1623 | 2226 | 292 | 107 | 153 | 29,318 |
| E47_S77 | Midbrain | NLGN4 | Male | 5774 | 558 | 772 | 165 | 50 | 67 | 7386 |
| E45_S329 | Midbrain | NLGN4 | Male | 21,915 | 1678 | 1771 | 389 | 174 | 134 | 26,061 |
| E47_S72 | Midbrain | NLGN4 | Male | 8124 | 596 | 559 | 64 | 55 | 35 | 9433 |
Summary of signal and cell counts from the padlock probing in situ experiments. The table shows the total number of signals for each gene, and the total number of cells identified by CellProfiler, for each sample hybridized simultaneously with padlock probes for two X and Y genes
Cell classification based on transcriptional profiles
Figure 4 shows the distribution of PCDH11X/Y, NLGN4X/Y, and ACTB (housekeeping gene for tissue quality control) signals in male tissues. We observed that signal-positive cells for both PCDH11X/Y and NLGN4X/Y were in most cases positive for X or Y transcripts, but not for both. Examples of these cells are marked with arrows in Fig. 4b, d. To determine whether cells were expressing the X and the Y homologs in a mutual exclusive fashion, we classified cells with at least two assigned signals as X-specific, Y-specific or mixed. Based on this classification (Tables 1 and 2), we observed that for both gene homolog pairs, there were comparable numbers of cells in all three cell classes, indicating that cells can simultaneously express X and Y transcripts as well as exhibit a mutually exclusive fashion of expression.
Fig. 4.
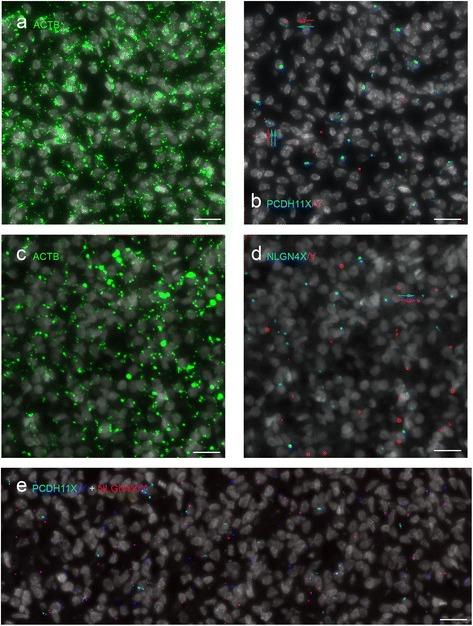
Padlock hybridization with ACTB, PCDH11X/Y and NLGN4X/Y. a, b. Male MO sections simultaneously hybridized with one padlock probe for ACTB, five probes for PCDH11X and five for PCDH11Y. a, b The same region of the section with staining for nuclei and ACTB (a) and PCDH11X (cyan) and PCDH11Y (red) (b). Most stained cells express either PCDH11X or PCDH11Y. Rarely, some cells express both X and Y transcripts (marked with arrows). c, d Male MO section simultaneously hybridized with one padlock probe for ACTB, four probes for NLGN4X and four for NLGN4Y. Nuclei and ACTB staining are shown in c, and NLGN4X (cyan) and NLGN4Y (red) staining in d. e Male SC section simultaneously hybridized with five probes for PCDH11X (cyan), five for PCDH11Y (blue), four probes for NLGN4X (red) and four for NLGN4Y (purple)
Table 1.
Distribution of cells with PCDH11X/Y transcripts: amount of cells with PCDH11X/Y signals
| Sample | Tissue | Gene | Gender | Cells without signals | Cells with X = 1 | Cells with Y = 1 | Cells with X + Y > = 2 | Cells with X > 1 Y = 0 | Cells with Y > 1 X = 0 | Total cell counts |
|---|---|---|---|---|---|---|---|---|---|---|
| E27_S44 | SC | PCDH11 | Female | 3442 | 470 | 1 | 0 | 89 | 0 | 4002 |
| E31_S91 | SC | PCDH11 | Female | 3358 | 190 | 4 | 0 | 23 | 1 | 3576 |
| E49_S82 | SC | PCDH11 | Female | 4793 | 252 | 18 | 0 | 34 | 0 | 5097 |
| E33_S50 | MO | PCDH11 | Female | 13,517 | 1479 | 10 | 6 | 321 | 0 | 15,333 |
| E34_S20 | MO | PCDH11 | Female | 12,485 | 1069 | 0 | 0 | 192 | 0 | 13,746 |
| E33_S63 | MO | PCDH11 | Female | 16,189 | 1042 | 45 | 1 | 113 | 0 | 17,390 |
| E34_S63 | MO | PCDH11 | Female | 25,784 | 1452 | 85 | 13 | 151 | 0 | 27,485 |
| E32_S53 | SC | PCDH11 | Male | 4915 | 106 | 61 | 10 | 5 | 14 | 5111 |
| E32_S84 | SC | PCDH11 | Male | 4319 | 257 | 269 | 45 | 31 | 18 | 4939 |
| E45_S75 | MO | PCDH11 | Male | 14,214 | 895 | 427 | 108 | 87 | 28 | 15,759 |
| E61_S49 | MO | PCDH11 | Male | 5944 | 636 | 78 | 21 | 122 | 2 | 6803 |
| E41_S58 | MO | PCDH11 | Male | 6824 | 409 | 99 | 13 | 41 | 0 | 7386 |
| E45_S50 | MO | PCDH11 | Male | 16,881 | 1553 | 948 | 240 | 175 | 85 | 19,882 |
| E58_S82 | MO | PCDH11 | Male | 9437 | 1122 | 356 | 160 | 241 | 15 | 11,331 |
| E61_S72 | MO | PCDH11 | Male | 6848 | 373 | 194 | 17 | 34 | 6 | 7472 |
| E47_S71 | Midbrain | PCDH11 | Male | 9635 | 683 | 172 | 58 | 53 | 9 | 10,610 |
| E45_S330 | Midbrain | PCDH11 | Male | 15,907 | 595 | 369 | 43 | 62 | 17 | 16,993 |
| E47_S72 | Midbrain | PCDH11 | Male | 8758 | 468 | 135 | 18 | 47 | 7 | 9433 |
| E45_S329 | Midbrain | PCDH11 | Male | 24,012 | 1259 | 555 | 70 | 137 | 28 | 26,061 |
Summary of signal and cell counts from the padlock probing in situ experiments. The table shows the total number of signals for each gene, and the total number of cells identified by CellProfiler, for each sample hybridized simultaneously with padlock probes for two X and Y genes
To evaluate how PCDH11X/Y and NLGN4X/Y were expressed in relation to each other, we hybridized sections simultaneously with the four sets of padlock probes using a four colour detection system as described in the “Methods” section (Fig. 4e). The analysis of single-cell expression profiles shows all possible combinations of the four transcripts (Table 3).
Table 3.
Distribution of cells with simultaneous detection of PCDH11X/Y and NLGN4X/Y transcripts
| Sample ID | Tissue | Gene | Gender | Total cell counts | Cells with PX > = 1 | Cells with PY > = 1 | Cells with NX > = 1 | Cells with NY > = 1 | Cells with PX + Y > = 2 | Cells with NX + Y > = 2 | Cells with PX + NX > = 2 | Cells with PY + NY > = 2 |
| E49_S82 | SC | Dual | Female | 5097 | 247 | 16 | 282 | 6 | 0 | 1 | 38 | 2 |
| E33_S63 | MO | Dual | Female | 17,390 | 993 | 36 | 1671 | 39 | 1 | 12 | 153 | 4 |
| E34_S63 | MO | Dual | Female | 27,485 | 1292 | 71 | 2940 | 68 | 5 | 25 | 302 | 2 |
| E32_S84 | SC | Dual | Male | 4939 | 208 | 241 | 278 | 353 | 32 | 49 | 31 | 16 |
| E41_S58 | MO | Dual | Male | 7386 | 318 | 75 | 566 | 573 | 8 | 367 | 39 | 15 |
| E58_S82 | MO | Dual | Male | 11,331 | 887 | 241 | 1108 | 977 | 100 | 361 | 207 | 50 |
| E61_S72 | MO | Dual | Male | 7472 | 322 | 155 | 245 | 628 | 13 | 60 | 31 | 27 |
| E45_S329 | Midbrain | Dual | Male | 26,061 | 1084 | 447 | 1625 | 1714 | 46 | 335 | 142 | 42 |
| E47_S72 | Midbrain | Dual | Male | 9433 | 421 | 107 | 591 | 530 | 15 | 56 | 42 | 16 |
| Sample ID | Tissue | Gene | Gender | Cells with PX + NY > = 2 | Cells with PY + NX > = 2 | Cells with PX + NX + NY > = 3 | Cells with PY + NX + NY > = 3 | Cells with PX + PY + NX > = 3 | Cells with PX + PY + NY > = 3 | Cells with PXandY + NXandY > = 4 | Cells with no signals | |
| E49_S82 | SC | Dual | Female | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4504 | |
| E33_S63 | MO | Dual | Female | 7 | 5 | 2 | 0 | 2 | 0 | 0 | 14,467 | |
| E34_S63 | MO | Dual | Female | 5 | 9 | 4 | 3 | 4 | 3 | 1 | 22,751 | |
| E32_S84 | SC | Dual | Male | 38 | 23 | 11 | 7 | 11 | 7 | 2 | 3639 | |
| E41_S58 | MO | Dual | Male | 43 | 5 | 50 | 4 | 50 | 4 | 0 | 5318 | |
| E58_S82 | MO | Dual | Male | 169 | 58 | 100 | 22 | 100 | 22 | 8 | 6991 | |
| E61_S72 | MO | Dual | Male | 50 | 15 | 4 | 3 | 4 | 3 | 1 | 5915 | |
| E45_S329 | Midbrain | Dual | Male | 141 | 74 | 29 | 20 | 29 | 20 | 5 | 20,338 | |
| E47_S72 | Midbrain | Dual | Male | 46 | 17 | 6 | 2 | 6 | 2 | 0 | 7581 |
The table shows the total number of signals for each gene, and the total number of cells identified by CellProfiler, for each sample hybridized simultaneously with padlock probes for the four X or Y genes: PCDH11X (PX), PCDH11Y (PY), NLGN4X (NX) and NLGNY (NY)
Differential spatial distribution of X and Y expressing cells in the CNS
To conduct an analysis of the spatial distribution patterns and objectively investigate whether transcripts for PCDH11X, PCDH11Y, NLGN4X and NLGN4Y are differentially distributed in the CNS as well as to visualize the extent of overlap of areas with X and Y signals, we compared our image data with a randomized image data set. As a first step, with these three goals in mind, we generated kernel density estimation plots as described in the “Methods” section and plotted the X and Y signal densities onto images of the SC and MO tissue sections (Fig. 5). In the SC, the signal density of both NLGN4X and PCDH11X varied across each tissue section, with clearly higher signal density in the ventral horns than in the dorsal horns in both male and female tissues (Fig. 5a, b, e–f). In contrast to the X homologs, NLGN4Y presented an even distribution over the whole SC, suggesting that there are cells which are more prone to express the Y homolog than the X homolog in some parts of the sections, namely in the dorsal horns (Fig. 5f). PCDH11Y did not show any consistent signal distribution differences between the ventral and dorsal regions (Fig. 5e). Different expression patterns were also observed when we analysed sections from the MO (Fig. 5c, d, g–h). Indeed, density plots showed how PCDH11X appeared to be expressed patch-wise in both male and female MO tissues while PCDH11Y showed a more evenly distributed expression pattern, to a larger extent than for PCDH11X confined to the periphery of the tissue. Similarly, the NLGN4Y transcripts appeared to be homogeneously distributed over the tissue sections, while the density of NLGN4X transcripts varied across the tissues.
Fig. 5.
Signal density maps in the spinal cord and medulla oblongata sections from female and male embryos hybridized with PCDH11X/Y and NLGNX/Y. The distributions of Y and X signals in different tissue sections are displayed as kernel density estimation plots where Y signal density is displayed in red and X signal density in cyan. The intensity of the colour reflects the density of the signal. Overlap in signal densities results in a mixed colour, whereas equal proportion of Y and X results in white colour. The outline of the tissue is indicated by a dashed white line. Orientation of the SC tissues is ventral down and dorsal up. The signal density in the female spinal cord is displayed in a for PCDH11X and in b for NLGN4X. c, d Signal density in the female medulla oblongata for PCDH11X and NLGN4X, respectively. e Signal density of PCDH11X/Y in the male spinal cord is shown individually for the two homologs as well as a plot of the two transcript densities combined. NLGN4X/Y is displayed in the same way on f. Signal densities in the male medulla oblongata are shown in g and h for PCDH11X/Y and NLGN4X/Y
We then compared the observed data for male samples with a randomized data set and plotted the pixels whose colour is over-represented by more than 3 standard deviations compared to the mean value from 100 randomizations as described in the “Methods” section. For NLGN4X and Y, we see a deviation in the representation of Y-dominant pixels from the random data. X-dominant pixels are generally under-represented while Y-dominant pixels are over-represented in our data (Fig. 6a) in comparison to the randomized data set. This is consistent with the observation that the expression of NLGN4X is confined to some regions while NLGN4Y is more universally expressed. We also see tendencies to an under-representation of mixed pixels. When plotting the pixels deviating more than +3 SD, we observed a higher prevalence of NLGN4Y transcripts in the peripheral parts of the SC (Fig. 6b). An additional example is given in Additional file 2: Figure S1. For PCDH11 in the MO, we observe high signal density for both PCDH11X and Y in the olivary nucleus (OL), but we also see an under-representation of pixels of mixed intensities, pointing towards a compartmentalized expression of PCDH11X and PCDH11Y. When comparing the analysed samples to their respective randomized data with 95 % confidence intervals (corresponding to a significance level of 0.05), we clearly see an over-representation of NLGN4Y-dominant pixels in the SC and brain samples, while in the MO, the trend is less clear and the difference between the data and randomized simulations is not statistically significant (Fig. 6c). We do not observe any consistent pattern for NGLN4X across all samples. For PCDH11X/Y, a majority of the samples show a strong deviation from randomness, and again, Y-dominant pixels are over-represented and no stable pattern is found for X-dominant pixels (Fig. 6d).
Fig. 6.
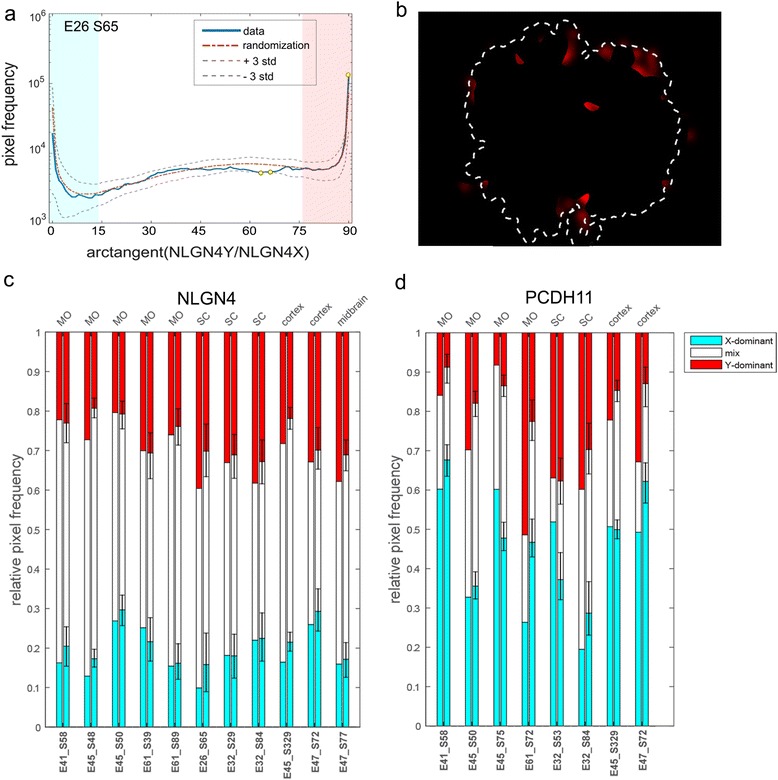
Comparison of the observed spatial signal distribution to randomized signal distributions in male MO for NLGN4X/Y. The frequency of pixels of varying intensity ratios of X and Y for NLGN4X/Y in male SC (E32_S84) are displayed in the histogram in a. A cutoff of 80 % colour purity was set to classify pixels as X dominant and Y dominant, displayed in the histogram as a blue field and a pink field, respectively. Pixels deviating more than 3 SD are shown as yellow dots on the blue line and then plotted in b visualize the location of the pixels over-represented in the sample tissue compared to the randomized data set. The proportions of X-dominant, Y-dominant and mixed pixels in the analysed samples are shown for all samples next to their respective randomized data set in c for NLGN4X/Y and in d for PCDH11X/Y. Relative pixel frequency from the random distributions for all samples is shown in c and d.
When comparing the ventral and dorsal counts for the homolog transcripts in spinal cord as the ratio between X and Y transcripts (Additional file 3: Table S2), we found that NLGN4X in females showed higher expression in the ventral SC (n = 5, t test, one tail, p = 0.045) compared to the dorsal. PCDH11X did not exhibit any significant differences in regional expression in females (n = 2, t test, one tail, p = 0.08). Neither NLGN4Y nor PCDH11Y presented significant differences in the dorsal to ventral expression patterns in males. This confirms the results from the analysis of the Kernel density estimation (KDE) plots, as the over-representation of Y-dominant pixels (located in the dorsal region) is expected when the Y homolog transcripts are more homogeneously distributed and the X homolog transcripts largely confined to the ventral region. Also, under-representation of X-dominant pixels is expected for the clustered expression pattern of the X homolog transcripts as these to a large extent overlap with the Y transcript signals resulting in mixed pixels in the ventral region. These observations suggested to us that there exist different populations of cells in the CNS with respect to the expression of X or Y homologs. In other words, our results indicate that spatial sexual dimorphism in the expression of these two genes exists.
Determination of the cellular identity of Y expressing cells using a combination of immunohistochemistry with padlock probing
To determine the precursor, glial or neuronal identity of positive cells for PCDH11X/Y and NLGN4X/Y in the MO, we combined padlock probe hybridizations with immuno-histochemical labelling as described in the “Methods” section. We used NeuN antibodies, known to label neurons [28], Olig2 antibodies, which stain precursor cells for oligodendrocytes [29], and Sox10 antibodies, which stain neural progenitor cells which will give rise to both neural and glial cells in the CNS [30]. Figure 7a shows that both NeuN+ and NeuN− cells express PCDH11X and PCDH11Y transcripts. For example, the olivary nucleus (ON) region contains PCDH11X and Y signals but no NeuN staining. Similarly, Fig. 7b shows also a partial overlap between NLGNX and Y signals and NeuN staining. Interestingly, NLGN4X and Y signals were observed in the ependymal layer (“Ep” in Fig. 7b) while signals for PCDH11X and Y were sparse in this region (not shown). Ep is the layer from which glial cells originate and migrate [31]. Olig2 staining is shown in Fig. 7c, d. Only a small subset of Olig2+ cells and Sox10+ cells contained signals for PCDH11X, PCDH11Y, NLGN4X or NLGN4Y, suggesting partial expression of X and Y gametolog genes in neuronal cell precursors (Fig. 7e, f). In addition to labelling precursors for oligodendrocytes, Olig2 is known to be expressed in motor neurons [29]. Furthermore, as described in the section related to density map analysis above, we observed increased expression of PCDH11X and NLGN4X in the ventral horn of the SC, a region known to contain several nuclei for motor neurons. Therefore, we also performed combined padlock probing with immunostaining for Islet-1 antibodies, known to label motor neurons, in the middle and ventral horn of the SC [32]. Additional file 4: Figure S2 shows that we found co-localization of PCDH11X and Y signals and Islet-1 in some positive cells. This suggests that PCDH11X and Y are expressed more in motor neuron differentiating cells during early stages of development than in sensory neurons, known to occupy the dorsal horn of the SC [33]. In conclusion, both PCDH11X and Y and NLGNX and Y are expressed in heterogeneous sub-populations of cells at different developmental states, including developing neurons and glial precursors.
Fig. 7.
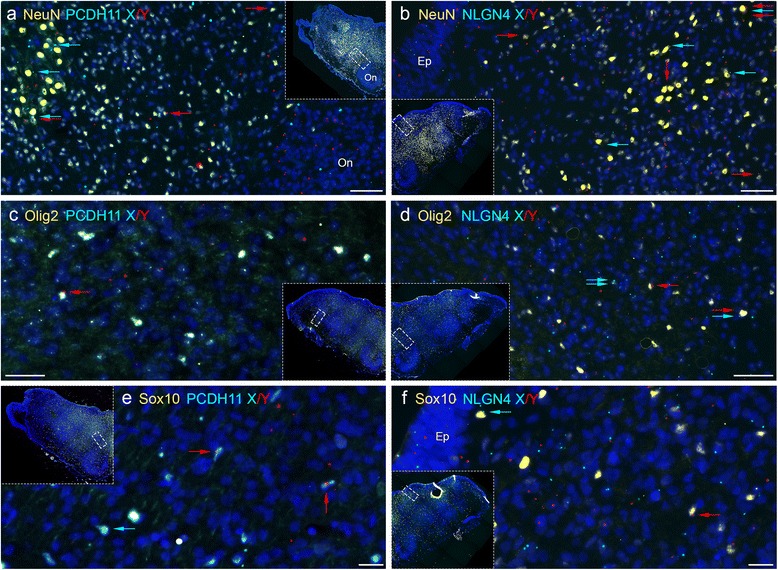
Combined padlock hybridization and immunohistochemistry for PCDH11X/Y and NLGNX/Y in MO samples from human male embryos. a, b Padlock probe hybridizations with probes for PCDH11X and PCDH11Y (a) or NLGN4X and NLGN4Y (b) were combined with immunohistochemistry using NeuN antibodies. NeuN-positive cells are stained in yellow, PCDH11X signals in sky blue and PCDH11Y in red. DAPI staining in dark blue allows visualization of nuclei. Each subfigure is an enlargement of the MO tissue showed in each inset picture. Some NeuN+ cells contain signals for PCDH11X (marked with blue arrows), PCDH11Y (red arrows) or both X and Y transcripts (blue and red arrows). c, d Padlock probe hybridization with probes for PCDH11X/Y (a) or NLGN4X/Y (b) was combined with immunohistochemistry using Olig2 antibodies. d, f Padlock probe hybridization with probes for PCDH11X/Y (a) or NLGN4X/Y (b) was combined with immunohistochemistry using Sox10 antibodies. Ep ependymal layer
Discussion
This study provides the first evidence of sexually dimorphic spatial expression patterns of Y chromosome genes and their X-encoded homologs in the CNS tissues of human embryos during early development. We also show that X and Y transcripts can be simultaneously expressed in single cells and that a fraction of cells express the X or the Y homolog alone. Furthermore, the combined results from padlock probe hybridizations and RNA sequencing showed that the sum of gametolog gene expression in males is similar to the expression of the X homolog gene in females. Taking both main observations together, we can conclude that while the total expression of the gametolog genes is similar among the sexes, sex specificity in the developing male SC and MO may be due to the fact that Y transcripts are expressed in different sub-populations of cells (possibly cellular networks) than those expressing X transcripts in females. However, our results suggest but do not prove that X and Y transcripts can be expressed in different cells or that there exist different cell populations expressing the X and the Y homologs in different proportions.
The PCDH11X and Y genes are not the only members of the protocadherin family that showed combinatorial expression in different cells. In fact, eight protocadherins and five classical cadherins were expressed in a combinatorial way in cells of the primary somatosensory cortex of the adult mouse [34]. The authors showed that most single neurons express more than one cadherin in different combinations in all layers of cerebral cortex. Also, as in our study, some of the cadherin-positive cells were NeuN positive while others were not, indicating both neuronal and possibly glial expression of these family members. They proposed that the combinatorial expression of multiple cadherin genes provides a molecular code that contributes to the molecular specification of the vast complexity of cellular networks and connectivity in the brain. Our results suggest that an additional male-specific code may act in combination with the above, resulting in a subset of male-specific cellular networks or connectivity.
The specific functions of PCDH11Y and NLGN4Y cannot be studied by knockout experiments in mice, since rodents do not have these genes encoded in their genomes. On the other hand, the homolog X genes are present on rodents. Knockout mice for Pcdh11x have not yet been produced. However, in utero electroporation of embryos with short hairpin RNA (shRNA) or overexpression vectors against Pcdh11x, as well as transient transfection of neural stem cells collected from E13.5 mice embryos has been done recently [35]. These experiments showed that overexpression of Pcdh11x in embryonic neural stem cells leads to reduced neural stem cell differentiation, as well as enhanced neural proliferation, both in vivo and in vitro, while reduction of Pcdh11x resulted in increased cell migration and premature neuronal differentiation in mice. Very recently, it was shown that overexpression of Pcdh11x in primary cultures of cortical neurons derived from embryonic day 16 mice reduced dendritic complexity, whereas knockdown increased dendritic branching [36]. Assuming that the human PCDH11Y has similar functions as the X gene has in mice, then the expression of this gene only in human males could result in enhanced neural cell proliferation only in males and only in a subset of cell populations. A fundamental question for the future is whether the newly discovered subset of human male cells that express PCDH11Y (and not PCDH11X) are also present in females or whether they form a male-specific cellular network produced only in males during CNS development.
The effect of the removal of the homolog X-encoded gene has also been studied for Nlgn4x. Mice with a loss-of-function mutation in the murine ortholog Nlgn4x, which encodes the synaptic cell adhesion protein neuroligin-4, exhibited highly selective deficits in reciprocal social interactions and communication [37], although later generations of the same line of Nlgn4 mutant mice did not show these behaviours [38]. Also, absence of Nlgn4x caused a decreased network response to stimulation in both excitatory and inhibitory circuits in somatosensory cortical slices of juvenile mice [39].
In humans, multiple genetic studies have implicated a significant role for NLGN4X in the susceptibility to several neurodevelopmental disorders. For example, Xp22.3 deletions including NLGN4X have long been associated with autism [40]. Also, a frameshift mutation in NLGN4X causing a premature stop codon was observed in all affected individuals in a large family, in which 10 males had X-linked mental retardation, 2 had autism, and 1 had pervasive developmental disorder [41]. PCDH11X is also proposed to affect neurodevelopment in humans. For example, deletion of this gene was found in a patient with developmental dyslexia [42], and deletion of both PCDH11X and PCDH11Y was discovered in a child with non-syndromic language delay [6]. Despite these prior human genetic studies, it is still not known what biological processes and cellular mechanisms are compromised when mutations in PCDH11X or NLGN4X occur. As mentioned before, functional studies of these genes can only partially be addressed in animal models. Therefore, it seems that functional insight in humans, particularly during neurodevelopment, will be best addressed by the use of human pluripotent stem cell (hiPSC) approaches [43]. Interestingly, neural stem cell (NSC) models derived from both hiPSCs and human embryonic stem cells have recently been used to study the function of NLGN4X [44]. The authors used shRNAmir to repress NLGNX expression. They observed that decreased NLGN4X levels during differentiation of NSCs into neurons caused delayed neuronal development, decreased neurite formation and less cell–cell connections.
Conclusions
The main findings of our investigations were that the X and Y homolog genes for PCDH11 and NLGN4 were mostly expressed in different glial and neuronal cell populations in the CNS during human male development. Also, we found differential distribution patterns of X and Y homologs in the male developing central nervous system. We propose that future experiments on NSC models derived both from females and males in which X and Y homolog genes are differentially modulated will shed light on the possible involvement of these genes in the development, maturation or connectivity of sex-specific cellular networks in the human brain.
Methods
Human embryonic CNS samples and sample preparation
Samples were obtained from human embryos between the 7 and 11 gestational weeks. The gestational age was estimated from the last menstrual period and from an ultrasound scan in which crown-rump length of the foetus was measured. Samples were dissected after surgical terminations of pregnancies performed at the University Hospital in Uppsala and after maternal written consent and approval from the regional Ethics Committee in Uppsala (number 2011/329). All procedures were made in general anaesthesia by intravenous administration of 1 ml alfentanil 0.5 mg/ml and 15–20 ml propofol 10 mg/ml. No gases for inhalation were used during the process, except for a mix of air and oxygen. The short procedure for evacuating the uterine cavity was performed by a gynaecologist with vacuum aspiration. The tissue specimens were disrupted by the surgical procedure, and the tissue types most frequently intact were, in declining order, the MO, SC and midbrain. Prior to sectioning, tissue samples were fixed for at least 16 h at 4 °C in 0.1 M phosphate-buffered saline (PBS) containing 4 % paraformaldehyde (PFA; Sigma-Aldrich). The samples were then cryoprotected in 30 % sucrose/PBS. Twelve-micrometre-thick sections were cut on a freezing microtome after mounting in TissueTec® (check name), collected on SuperFrost® Plus microscope slides (VWR International), and stored at −80 °C prior to hybridizations. The samples for RNA sequencing were snap frozen on dry ice - chilled Eppendorf tubes and stored at −80 °C prior to RNA extractions. To determine the sex of each embryo, DNA was extracted from arm tissue using DNasy® Blood & Tissue Kit (Qiagen). PCR on DNA samples was done using male-specific primers for STS sY14 (SRY), (5′-GAATATTCCCGCTCTCCGGA-3′, 5′-GCTGGTGCTCCATTCTTGAG-3). Complete information about the samples used for each experiment can be found in Additional file 5: Table S3.
Preparation of RNA-seq libraries from total and polyA+ RNA
Frozen tissues ranging from 70–190 mg were homogenized in 2 ml TRIzol® reagent (Ambion) using an Ultratorrax T25 homogenizator (Labortechnik). Total RNA was extracted using RiboPure kit (Ambion) according to the manufacturers’ instructions. PolyA RNA was enriched from 1 μg total RNA using MicroPoly (A) Purist kit (Ambion) according to the manufacturer’s instructions. The quantity and quality of the input RNA was controlled using a RNA 6000 Pico chip on a Bioanalyzer (Agilent Technologies) and only RIN values above 7 were used in the analysis.
Complementary DNA (cDNA) library preparation was conducted at the Uppsala Genome Centre (SciLifeLab). Briefly, a ribosomal RNA (rRNA) depletion step was performed with 56 mg as input amount for all samples, using the RiboMinus Eukaryote kit (Life Technologies). Whole transcriptome libraries were then constructed using the SOLiD total RNA-Seq kit (rev B, July 2011, Life Technologies). Emulsion PCR was performed using the SOliD EZ Bead System (Life Technologies) and the libraries were then sequenced on three lanes with the SOLiD 5500xl System (Life Technologies).
Sequence alignment and data analysis
All sequencing reads were aligned to the hg19 build of the human genome using the LifeScope software (version 2.5, Life Technologies). Reads mapping to exonic regions were counted using the HTSeq software (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html) only counting reads having the quality of 30 or higher (Additional file 6: Figure S3) using the hg19 gene list (gff file) provided from the iGenomes project (Illumina) [45]. Differential gene expression was calculated using the DESeq2 software (version 1.6.0) [46]. DESeq2 approximates the null distribution using a negative binomial model under the assumption that the two conditions have the same read abundance, in order to calculate the probability of genes having a differential expression between two samples. Correction for multiple testing was done by DESeq2 using the procedure of Benjamini and Hochberg [45]. When counting reads aligned to specific exons used for design of padlock probes, the HTseq software was used as previously described, using the specific exons as input. RPKM calculations were performed using the R RPKM library. Reads overlapping each padlock probe were counted using SAM tools together with in-house perl scripts.
Sample pre-treatment for in situ experiments
Tissue samples were dried at room temperature for 1 h prior to two washes in 0.1 % diethylpyrocarbonate (DEPC)-treated PBS. The tissue was then permeabilized in 0.2 mg/ml pepsin (Sigma) in 0.1 M HCl at 37 °C for 2 min. After permeabilization, the slides were washed twice in DEPC-treated PBS. All of the following reactions were performed in Secure Seal hybridization chambers (Invitrogen).
Padlock probe design strategy
Alignments of transcripts for PCDH11X and PCDH11Y showed very high levels of sequence identity between these genes (Additional file 7: Figure S4a), with 97 to 99 % sequence identity for each exon (Additional file 8: Table S4a). Similarly, alignments for NLGN4X and NLGN4Y presented 89 to 98 % identity at the exon level (Additional file 7: Figure S4b and Additional file 8: Table S4b). We selected two positions in exon 5 of PCDH11X/Y to design padlock probes P1 and P2 (Additional file 7: Figure S4a). These positions differ by one nucleotide in all X transcripts compared to all Y transcripts. In addition, we selected three positions in exon 9 which differ between all PCDH11X long transcripts (NM_032968, NM_032969, NM_001168360, NM_001138361, NM_001138362, NM_001138363), compared to the only known PCDH11Y long transcript (NM_032973), and we designed probes P3, P4 and P5. Similarly, for NLGN4X/Y, we selected one position in exon 3, one in exon 5 and 2 positions in exon 6, all of them present in all X and Y transcripts, to design padlock probes N1 to N4 (Additional file 7: Figure S4b). The complete sequences of all padlock probes including cDNA primers and detection sequences are listed in Additional file 9: Table S5. All padlock probes were 5′-phosphorylated at a concentration of 2 μM with 0.2 U μl−1 T4 polynucleotide kinase (Fermentas) in the manufacturer’s buffer A plus 1 mM ATP for 30 min at 37 °C, followed by enzyme inactivation for 10 min at 65 °C.
In situ cDNA detection using padlock probes
To start the reverse transcription step, 1 μM of each cDNA primer was added to the slides with 20 U μl−1 of RevertAid H minus M-MuLV reverse transcriptase (Fermentas), 500 nM dNTPs (Fermentas), 0.2 μg μl−1 BSA (NEB) and 1 U μl−1 RiboLock RNase Inhibitor (Fermentas) in the M-MuLV reaction buffer. The slides were incubated for 3 h at 37 °C. After incubation, the slides were washed briefly in PBS-T (DEPC-PBS with 0.05 % Tween-20 (Sigma)), followed by a post-fixation step in 3.7 % (w/v) paraformaldehyde in DEPC–PBS for 45 min at room temperature. Subsequently, the samples were washed twice in PBS-T.
To make the target cDNA strands available for padlock probe hybridization, the RNA portion of the created RNA–DNA hybrids was degraded with ribonuclease H. This was performed in the same step as the padlock probe hybridization and ligation. Ligation was carried out with 100 nM of each padlock probe in a mixture of 0.5 U μl−1 Tth DNA Ligase (Gene Craft Germany), 0.4 U μl−1 RNase H (Fermentas), 1 U μl−1 RiboLock RNase Inhibitor, Tth ligase buffer, 50 mM KCl and 20 % formamide. Incubation was performed first at 37 °C for 30 min, followed by 45 min at 45 °C. RCA was performed with 1 U μl−1 Φ29 DNA polymerase (Fermentas) in the supplied reaction buffer, 1 U μl−1 RiboLock RNase Inhibitor, 250 μM dNTPs, 0.2 μg μl−1 BSA and 5 % glycerol. Incubation was carried out for 16 h at 37 °C. The incubation was followed by a wash in PBS-T.
RCA products were visualized using 100 nM of each corresponding detection probe (Additional file 9: Table S5) in 2× SSC and 20 % formamide at 37 °C for 30 min. Each detection oligonucleotide is available labelled with FITC, Texas Red (TR), Cy3, Cy5 or AF750. For two colour detection of X and Y transcripts, we used Cy3 and TR, respectively. ACTB transcripts were detected using Cy5-labelled detection oligonucleotide. For simultaneous four colour detection of transcripts from four genes, we used Cy3-labelled detection oligonucleotides for PCDH11X, TR for PCDH11Y, AF750 for NLGN4X and Cy5 for NLGN4Y. For simultaneous four colour detection of splice variants, we used Cy3 for PCDH11X-PAN, Cy5 for PCDH11X-Long, FITC for PCDH11Y-PAN and TR for PCDH11Y-Long. The slides were then washed in PBS-T, and nuclei staining was done using Hoechst 33343 (Life Technologies) 50 μg μl−1 for 12 min at room temperature. Finally, the slides were washed with PBS-T followed by removal of the secure-seals, and the slides were dehydrated using a series of 70, 85 and 99.5 % ethanol for 2 min in each. The dry slides were mounted with SlowFade® Gold antifade reagent (Life Technologies).
Image acquisition and processing
The tissue slides were scanned using a Zeiss Axio Imager.Z2 epi-fluorescence microscope (filter cubes in Additional file 10: Table S6) and the original tiles (each one field of view of the camera) exported as grayscale images for image processing using the image analysis tool CellProfiler (http://www.cellprofiler.org/). A customized pipeline was set up for each of the samples for identification of transcript signals and nuclei based on the fluorescent signals. The nuclei were segmented based on global Otsu thresholding and the cell borders defined by a fixed distance of 20 pixels from the nuclei border (Additional file 11: Figure S5). Furthermore, the transcript signals were identified by manual thresholding subsequent to top-hat filtering of the images and then assigned to a parent cell based on in which identified cell they were detected. The transcript signals were counted in the whole tissue to obtain total counts for comparison to the RNA sequencing data. The numbers of detected transcript signals were summarized as well as the data for each of the segmented cells and their assigned children signals to classify cells based on expression of X, Y or both X and Y. The coordinates of the transcript signals were used to construct density plots of the tissues for investigation of the regional distribution of X and Y transcripts of NLGN4 and PCDH11.
Image analysis
To statistically analyse whether differences in signal density distributions of X and Y transcripts were of biological relevance, we compared the observed data for male samples with a randomized data set. An image of a randomized distribution of signals was generated for each sample using the same positions of the signal and the same number of X and Y transcript counts as in the observed data. KDE plots were generated for each sample and randomized image data. The KDE plots were generated from binary images where each signal is represented by 1 pixel. The pixels are convoluted with a rotational symmetric 2D Gaussian kernel of 150 pixels with a standard deviation of 25 pixels. The Y and X pixel intensity ratio was calculated for the density plots, and the frequency of these pixel ratios for each sample data was plotted in a histogram together with the Y and X pixel intensity ratio frequency from the corresponding randomized data set (±3 SD; 100 randomizations). Since we were particularly interested to see whether we could find indications of cell populations having preferential expression of either X or Y transcripts, we decided to divide our data into three ratio bins. A dominant X or Y pixel was defined as a pixel with 80 % colour purity for either the X or Y homolog, leaving the rest of the pixels in a mixed bin.
Analysis of dorsal vs. ventral SC signal distribution
The definition of dorsal and ventral border in SC tissue was done by dividing the total distance from dorsal to ventral horn at one position on each side of the SC. These two points acted as basis for a horizontal border separating the dorsal from the ventral region. Signals in each part of the tissue were counted using ImageJ Cell Counter plugin. Statistical evaluation was done using two-sample t test and one tail and assuming unequal variances.
Combined padlock hybridization and immunohistochemistry
To combine the padlock detection and immunohistochemistry, we first hybridized slides with padlock probes as described above. After that, the slides were washed three times for 10 min in PBS and blocked for 1 h in 0.25 % bovine serum albumin in PBS with 0.25 % Triton X-100. Each of the following primary antibodies: anti-NeuN (mouse, 1:1000; Abcam), anti-Olig2 (goat, 1:20; R&D Systems), anti-Sox10 (goat, 1:20; R&D Systems) and anti-Islet 1 (goat, 1:20; R&D Systems) was incubated overnight at 4 °C. After washing with PBS, the corresponding secondary antibodies were applied for 4 h at room temperature: Alexa 594 goat anti-rabbit, FITC goat anti-mouse, Alexa 488 donkey anti-goat and Alexa 597 goat anti-chicken (1:400; Life Technologies). Immunolabelled cells were mounted with Dako ultramount aqueous permanent mounting medium (Dako). Cell imaging was performed using epi-fluorescence microscope Ziess (Zeiss Axio Imager.Z2 epi-fluorescence microscope, filter cubes in Additional file 10: Table S6). The images were assembled in Photoshop CS6 (Adobe).
Acknowledgements
We are grateful to Drs. Art Arnold and Harold Zakon for critically reviewing parts of this manuscript. Also, I (EJ) greatly acknowledge Dr. Arnold for inspiration for this work since we met for the first time at a Gordon Conference in 2010. We would also like to thank Ammar Zaghlool for preparation of polyA RNA for the RNA sequencing. This study was funded by the Swedish Research Foundation (EJ, LF, ED) grant name: Sex determination factors in the brain encoded in the Y chromosome (Project number K2012-61X-22089-01-3) and the Swedish Research Council (MN).
Additional file
RNA sequencing results for all genes belonging to protocadherin and neuroligin families. The table shows expression values as base mean values for females and males, the position of each gene in the genome, fold change in positive values for male-biased genes and negative values for female-biased genes and p values adjusted for multiple testing as described in the “Methods” section. (DOCX 27 kb)
Example of the objective analysis of spatial expression image data. Kernel density estimation (KDE) plots of PCDH11X/Y transcript signals detected in embryonic brain tissue are shown in a. At the top, the KDE plot of PCDH11X (cyan) has been merged with the one of PCDH11Y (red). From the separate KDE plots, two clear features can be distinguished; the Y homolog is more evenly expressed over the whole tissue while the X homolog shows higher expression in confined regions, namely along the lower edge and at the tip to the right. The observed spatial distribution of X and Y signals is compared to a random distribution in the histogram b, based on the relative contribution of X and Y signal intensity in each pixel in the KDE plot. The distribution of pixels in the KDE plot is shown as a blue line together with the average of 100 randomized data sets as a red line. Pixels deviating in number from what would be expected by chance (±3 SD from averaged random) are shown as a yellow dots on the blue line. The bar chart in b shows that we have a higher number of pixels classified as Y-dominant than we have X-dominant pixels, confirming our observations in the KDE plots in a that some regions express the Y homolog to a higher extent than the X homolog. Pixels in the observed signal distribution deviating more than 3 standard deviations from the random signal distribution, are plotted back onto the tissue in c and further confirms the patterns of X and Y homolog expression observed in a. The Y homolog shows dominant expression along the upper edge of the tissue section while the lower edge of the tissue section is dominated by mixed pixels (signal contribution from both the homologs). (TIF 623 kb)
Signal counts and statistical analysis for the ventral and dorsal part of the spinal cord. The table shows signal counts in dorsal vs. ventral SC sections acquired by using ImageJ software (Cell Counter plugin). Distributions for PCDH11X, PCDH11Y, NLGN4X and NLGN4Y are shown for slides obtained from seven female and four male embryos. Percentage distribution shows the ratio between X and Y homologs. (DOCX 16 kb)
Combined padlock hybridization for PCDH11X and Y and Immunohistochemistry with Islet-1 antibodies in SC samples from human male embryos. a, b Padlock probe hybridization with probes for PCDH11X and PCDH11Y was combined with immunohistochemistry using islet-1 antibodies. Islet-1-positive cells are stained in yellow, PCDH11X signals are in sky blue and PCDH11Y in red. DAPI staining in dark blue allows visualization of nuclei. Part b is a detail enlargement of the complete SC section shown in a. (TIF 4288 kb)
Description of the embryos and tissues used for each experiment. The table shows all embryonal sections and tissues used for RNA-seq, padlock probe and immunohistochemistry experiments. Embryos named E57, E49, E55 and E54 were used for RNA sequencing. The slides named E32_S62, E45_S71, E45_S72 and E45_S73 were included in the combined experiments of padlock probing and immunohistochemistry. Remaining embryonal sections were used for padlock probing experiments only. Experiments in which simultaneous detection of PCDH11X, PCDH11Y, NLGN4X and NLGN4Y was done are indicated by “PCDH11X/Y + NLGN4X/Y” in the column named “Gene”. (DOCX 19 kb)
Filtration of RNA sequencing data. The figure shows the reads mapped to the two last exons of NLGN4Y. Sequences mapping to more than one location in the genome are removed by filtration as described in the “Methods” section. After filtration, Y-specific sequences are not detected in females, indicating that the filtration process was effective in separating X- and Y-specific sequences. (TIF 1198 kb)
Alignment of transcripts for PCDH11X/Y and NLGN4X/Y. The colours represent percentage of identity in each region with dark blue corresponding to 100 % identity. The name of each transcript is given according to UCSC nomenclature with accession numbers in parenthesis. The limits between exon sequences are marked with red vertical lines. The exact position of each exon in the gene (hg 19) is given in Additional file 8: Table S4. The positions for padlock probes P1 to P5 are indicated at the bottom. In these positions, all X transcripts differ from all Y transcripts by one nucleotide. The exact positions (hg 19) of the nucleotide differences and the complete padlock probe sequences are shown in Additional file 9: Table S5. Alignments for transcripts of NLGN4X/Y. The positions of padlock probes N1 to N4 are indicated at the bottom. These probes are located in exons 3, 5 and 6. (TIF 869 kb)
Percentage of sequence identity between exons of PCDH11X/Y and NLGNX/Y. Top. For each exon, the start and stop positions in the genome (hg 19) is given for NLGN4X and NLGN4Y, the size in base pairs, the percent of identity for X and Y exons, and the name of transcripts that contain the particular exon. Shaded in grey are those exons used for padlock probe design. Bottom. Same as above for exons contained in PCDH11X and PCDH11Y. The percentage of identity for X and Y exons was very high, ranging from 97 to 99 %. Notes: *This exon present in all transcripts except Y transcript uc004fte.2. **this exon also shares 87 % sequence identity to X position 5922232 to 5922326; ***repeated sequence with high sequence identity to sequences located in several chromosomes. ****high sequence identity also on chromosome 12. *****except Y transcript uc010nw.1. (DOCX 20 kb)
Padlock probe sequences. The name and complete sequence for each probe is shown. In lowercase are the sequences included in the X and Y homologs for PCDH11, NLGN4 and ACTB. In uppercase are the sequences used for hybridization to detection oligos. Each PCDH11X/Y probe can be detected by two alternative detection oligos, allowing experiments in which only long transcripts or all transcripts are stained. In red colour are the base changes between all X and Y transcripts followed by the position of these bases in the genome. (DOCX 18 kb)
Filters used for fluorescent imaging with the Zeiss Axio Imager.Z2 epi-fluorescence microscope. (DOCX 13 kb)
Cell segmentation using CellProfiler. The outlines of the cells defined by CellProfiler by nuclei segmentation. The outlines of the cells were defined at a 20-pixel distance from the outlines of the nuclei, which were identified by Global Otsu classification based on the DAPI nuclear staining. (TIF 2666 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MMJ, EL and MM performed the experiments. MMJ, EL, JH, and XQ analysed the data. MMJ, EJ, MN and EL wrote the paper with comments from all co-authors. MN, EL, EJ, LF and MMJ conceived of the study and planned the experiments. MN, ED, LF and EJ contributed material and reagents. All authors read and approved the final manuscript.
Contributor Information
Mats Nilsson, Email: mats.nilsson@scilifelab.se.
Elena Jazin, Email: elena.jazin@ebc.uu.se.
References
- 1.McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14(6):677–83. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jazin E, Cahill L. Sex differences in molecular neuroscience: from fruit flies to humans. Nat Rev Neurosci. 2010;11(1):9–17. doi: 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- 3.Cahill L. Why sex matters for neuroscience. Nat Rev Neurosci. 2006;7(6):477–84. doi: 10.1038/nrn1909. [DOI] [PubMed] [Google Scholar]
- 4.Kopsida E, Stergiakouli E, Lynn PM, Wilkinson LS, Davies W. The role of the Y chromosome in brain function. Open Neuroimaging J. 2009;2:20–30. doi: 10.2174/1876528900902010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross JL, Tartaglia N, Merry DE, Dalva M, Zinn AR. Behavioral phenotypes in males with XYY and possible role of increased NLGN4Y expression in autism features. Genes Brain Behav. 2015;14(2):137–44. doi: 10.1111/gbb.12200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Speevak MD, Farrell SA. Non-syndromic language delay in a child with disruption in the Protocadherin11X/Y gene pair. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(4):484–9. doi: 10.1002/ajmg.b.31186. [DOI] [PubMed] [Google Scholar]
- 7.Raznahan A, Lee NR, Greenstein D, Wallace GL, Blumenthal JD, Clasen LS et al. Globally divergent but locally convergent X- and Y-chromosome influences on cortical development. Cerebral cortex. 2014. doi:10.1093/cercor/bhu174. [DOI] [PMC free article] [PubMed]
- 8.Lin A, Clasen L, Lee NR, Wallace GL, Lalonde F, Blumenthal J, et al. Mapping the stability of human brain asymmetry across five sex-chromosome aneuploidies. J Neurosci. 2015;35(1):140–5. doi: 10.1523/JNEUROSCI.3489-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reinius B, Jazin E. Prenatal sex differences in the human brain. Mol Psychiatry. 2009;14(11):987. doi: 10.1038/mp.2009.114. [DOI] [PubMed] [Google Scholar]
- 10.Morishita H, Yagi T. Protocadherin family: diversity, structure, and function. Curr Opin Cell Biol. 2007;19(5):584–92. doi: 10.1016/j.ceb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Bottos A, Rissone A, Bussolino F, Arese M. Neurexins and neuroligins: synapses look out of the nervous system. Cell Mol Life Sci. 2011;68(16):2655–66. doi: 10.1007/s00018-011-0664-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams NA, Close JP, Giouzeli M, Crow TJ. Accelerated evolution of Protocadherin11X/Y: a candidate gene-pair for cerebral asymmetry and language. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(6):623–33. doi: 10.1002/ajmg.b.30357. [DOI] [PubMed] [Google Scholar]
- 13.Gagneux P. A Pan-oramic view: insights into hominoid evolution through the chimpanzee genome. Trends Ecol Evol. 2004;19(11):571–6. doi: 10.1016/j.tree.2004.09.002. [DOI] [Google Scholar]
- 14.Wilson ND, Ross LJ, Crow TJ, Volpi EV. PCDH11 is X/Y homologous in Homo sapiens but not in Gorilla gorilla and Pan troglodytes. Cytogenet Genome Res. 2006;114(2):137–9. doi: 10.1159/000093329. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Yu S, Fu Y, Li X. Synaptic proteins and receptors defects in autism spectrum disorders. Front Cell Neurosci. 2014;8:276. doi: 10.3389/fncel.2014.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crow TJ. The ‘big bang’ theory of the origin of psychosis and the faculty of language. Schizophr Res. 2008;102(1-3):31–52. doi: 10.1016/j.schres.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Crow TJ. The XY, gene hypothesis of psychosis: origins and current status. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(8):800–24. doi: 10.1002/ajmg.b.32202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Falgueras A, Swaab DF. Sexual hormones and the brain: an essential alliance for sexual identity and sexual orientation. Endocr Dev. 2010;17:22–35. doi: 10.1159/000262525. [DOI] [PubMed] [Google Scholar]
- 19.Hanley NA, Hagan DM, Clement-Jones M, Ball SG, Strachan T, Salas-Cortes L, et al. SRY, SOX9, and DAX1 expression patterns during human sex determination and gonadal development. Mech Dev. 2000;91(1-2):403–7. doi: 10.1016/S0925-4773(99)00307-X. [DOI] [PubMed] [Google Scholar]
- 20.Nugent BM, McCarthy MM. Epigenetic underpinnings of developmental sex differences in the brain. Neuroendocrinology. 2011;93(3):150–8. doi: 10.1159/000325264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sekido R. The potential role of SRY in epigenetic gene regulation during brain sexual differentiation in mammals. Adv Genet. 2014;86:135–65. doi: 10.1016/B978-0-12-800222-3.00007-3. [DOI] [PubMed] [Google Scholar]
- 22.Helbig I, Matigian NA, Vadlamudi L, Lawrence KM, Bayly MA, Bain SM, et al. Gene expression analysis in absence epilepsy using a monozygotic twin design. Epilepsia. 2008;49(9):1546–54. doi: 10.1111/j.1528-1167.2008.01630.x. [DOI] [PubMed] [Google Scholar]
- 23.Priddle TH, Crow TJ. Protocadherin 11X/Y a human-specific gene pair: an immunohistochemical survey of fetal and adult brains. Cerebral cortex (New York, N Y : 1991) 2013;23(8):1933–41. doi: 10.1093/cercor/bhs181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34(1):27–9. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary BP, Landegren U. Padlock probes: circularizing oligonucleotides for localized DNA detection. Science (New York, N Y) 1994;265(5181):2085–8. doi: 10.1126/science.7522346. [DOI] [PubMed] [Google Scholar]
- 26.Nilsson M, Barbany G, Antson DO, Gertow K, Landegren U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat Biotechnol. 2000;18(7):791–3. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- 27.Trabzuni D, Ramasamy A, Imran S, Walker R, Smith C, Weale ME, et al. Widespread sex differences in gene expression and splicing in the adult human brain. Nat Commun. 2013;4:2771. doi: 10.1038/ncomms3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan W, Zhang YP, Hou Z, Huang C, Zhu H, Zhang CQ et al. Novel insights into NeuN: from neuronal marker to splicing regulator. Molecular neurobiology. 2015. doi:10.1007/s12035-015-9122-5. [DOI] [PubMed]
- 29.Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157–63. doi: 10.1016/S0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 30.Drerup CM, Wiora HM, Topczewski J, Morris JA. Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development. 2009;136(15):2623–32. doi: 10.1242/dev.030577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregg C, Weiss S. Generation of functional radial glial cells by embryonic and adult forebrain neural stem cells. J Neurosci. 2003;23(37):11587–601. doi: 10.1523/JNEUROSCI.23-37-11587.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu Q, Li D, Louis KR, Li X, Yang H, Sun Q, et al. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat Commun. 2014;5:3449. doi: 10.1038/ncomms4449. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 2008;11(11):1283–93. doi: 10.1038/nn.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna-K K, Hertel N, Redies C. Cadherin expression in the somatosensory cortex: evidence for a combinatorial molecular code at the single-cell level. Neuroscience. 2011;175:37–48. doi: 10.1016/j.neuroscience.2010.11.056. [DOI] [PubMed] [Google Scholar]
- 35.Zhang P, Wu C, Liu N, Niu L, Yan Z, Feng Y, et al. Protocadherin 11 x regulates differentiation and proliferation of neural stem cell in vitro and in vivo. J Mol Neurosci. 2014;54(2):199–210. doi: 10.1007/s12031-014-0275-x. [DOI] [PubMed] [Google Scholar]
- 36.Wu C, Niu L, Yan Z, Wang C, Liu N, Dai Y et al. Pcdh11x negatively regulates dendritic branching. Journal of molecular neuroscience : MN. 2015. doi:10.1007/s12031-015-0515-8. [DOI] [PubMed]
- 37.Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci U S A. 2008;105(5):1710–5. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ey E, Yang M, Katz AM, Woldeyohannes L, Silverman JL, Leblond CS et al. Absence of deficits in social behaviors and ultrasonic vocalizations in later generations of mice lacking neuroligin4. Genes, brain, and behavior. 2012. doi:10.1111/j.1601-183X.2012.00849.x. [DOI] [PMC free article] [PubMed]
- 39.Delattre V, La Mendola D, Meystre J, Markram H, Markram K. Nlgn4 knockout induces network hypo-excitability in juvenile mouse somatosensory cortex in vitro. Sci Rep. 2013;3:2897. doi: 10.1038/srep02897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas NS, Sharp AJ, Browne CE, Skuse D, Hardie C, Dennis NR. Xp deletions associated with autism in three females. Hum Genet. 1999;104(1):43–8. doi: 10.1007/s004390050908. [DOI] [PubMed] [Google Scholar]
- 41.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard M-P, et al. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74(3):552–7. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veerappa AM, Saldanha M, Padakannaya P, Ramachandra NB. Genome-wide copy number scan identifies disruption of PCDH11X in developmental dyslexia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(8):889–97. doi: 10.1002/ajmg.b.32199. [DOI] [PubMed] [Google Scholar]
- 43.Zhu H, Lensch MW, Cahan P, Daley GQ. Investigating monogenic and complex diseases with pluripotent stem cells. Nat Rev Genet. 2011;12(4):266–75. doi: 10.1038/nrg2951. [DOI] [PubMed] [Google Scholar]
- 44.Shi L, Chang X, Zhang P, Coba MP, Lu W, Wang K. The functional genetic link of NLGN4X knockdown and neurodevelopment in neural stem cells. Hum Mol Genet. 2013;22(18):3749–60. doi: 10.1093/hmg/ddt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasser RF. Atlas of human embryos. Baltimore: Lippincott Williams and Wilkins; 1975.
- 48.His W. Die anotomische nomenclatur. Nomina Anatomica. Der von der Anatomischen Gesellschaft auf ihrer IX. Versammlung in Basel angenommenen Namen. Versammlung in Basel angenommenen Namen. Leipzig: Verlag von Veit&Comp; 1895.


