Abstract
Ribosomes are essential components of the protein synthesis machinery. The process of ribosome biogenesis is well organized and tightly regulated. Recent studies have shown that ribosomal proteins (RPs) have extraribosomal functions that are involved in cell proliferation, differentiation, apoptosis, DNA repair, and other cellular processes. The dysfunction of RPs has been linked to the development and progression of hematological, metabolic, and cardiovascular diseases and cancer. Perturbation of ribosome biogenesis results in ribosomal stress, which triggers activation of the p53 signaling pathway through RPs-MDM2 interactions, resulting in p53-dependent cell cycle arrest and apoptosis. RPs also regulate cellular functions through p53-independent mechanisms. We herein review the recent advances in several forefronts of RP research, including the understanding of their biological features and roles in regulating cellular functions, maintaining cell homeostasis, and their involvement in the pathogenesis of human diseases. We also highlight the translational potential of this research for the identification of molecular biomarkers, and in the discovery and development of novel treatments for human diseases.
Keywords: ribosomal protein, RP-MDM2-p53 pathway, ribosomopathy, cancer, drug discovery
1. INTRODUCTION
There is increasing evidence indicating that the ribosome (comprising RNA and proteins) plays a critical role in normal cellular physiology, the cellular responses to internal and external environmental stimuli, and the pathogenesis of human diseases. The synthesis of the ribosome, called ribosome biogenesis, is a highly ordered cellular process that requires a substantial expenditure of energy, therefore, it occurs primarily under nutrient-rich and growth-friendly circumstances.1 Under stress situations, the reverse phenomenon is seen, with decreased ribosome activity, reduced protein synthesis and subsequent growth arrest. Thus, ribosome biogenesis is a critical element involved in controlling cell growth and proliferation; any dysregulation of this process may result in aberrant cell proliferation and clinical manifestations of pathological states, such as cancer and metabolic disorders.2
Unraveling the mechanisms responsible for maintaining the integrity of ribosome biogenesis is critical for understanding these cellular functions, and the link between dysfunctions and the pathogenesis of diseases. In addition to being the “workshop” for ribosome biogenesis, the nucleolus is also a central hub for stress sensors.3 Disruption of ribosome biogenesis leads to nucleolar stress (also termed ribosomal stress),2 which activates the p53 signaling pathway, leading to cell cycle arrest and apoptosis.4 Additionally, ribosomal proteins (RPs) have critical roles in diverse cellular functions that are distinct from their primary role in ribosome biogenesis.5,6 These extraribosomal functions of RPs include cell growth and proliferation,7–9 apoptosis,10,11 DNA repair,12,13 cellular development14,15 and differentiation.16,17 Interestingly, a subset of RPs also acts as “watchguards” to detect the defects in ribosome biogenesis.18–22
Several human diseases have been demonstrated to be associated with defects in ribosome biogenesis, including increased cancer susceptibility.2,23 Perturbation in the extraribosomal functions of RPs is known to be involved in carcinogenesis, and aberrant ribosomal function is either a consequence or an associated feature of cancer. Diamond-Blackfan anemia (DBA)24 and 5q- syndrome25 are two clinical syndromes associated with impairments in erythropoiesis that are attributed to ribosomal gene mutations. Although previous studies demonstrated that the induction of p53 following ribosomal stress promotes extensive apoptosis in certain progenitor cell types, leading to ribosomopathies, the reason why the patients with these diseases exhibit a high incidence of malignancies remains elusive. Recent evidence suggests that the links connecting defects in ribosome biogenesis and p53 signaling pathway are very complex, and multiple factors and regulatory mechanisms are involved in this network.26,27 Additionally, it is still not well understood why the effects of ribosome dysfunction are not observed universally, but are confined to specific organ systems and cell types, such as the hematological system (erythrocytes), neurons, or skin cells.28
There is an increasing interest in further elucidating the roles of RPs in both normal physiological processes and in the pathogenesis of human diseases. Recently, several excellent reviews have been published, and interested readers are referred to those publications.3,6,18,19,28–35 In this review, we focus on the recent progress toward understanding the newly appreciated, yet still under-explored ribosomal stress pathways, with an emphasis on the extraribosomal functions of RPs and their underlying mechanisms of action. We will also highlight the roles of RPs in ribosomal anomalies and the potential of using RPs as biomarkers and molecular targets in the diagnosis and treatment of human diseases. We believe that a better understanding of the relationships between RP dysfunction and human diseases would provide new avenues for the early diagnosis of chronic diseases, such as cancer and cardiovascular diseases, and would provide novel drug targets and biomarkers for these diseases.
2. RIBOSOMAL PROTEINS AND RIBOSOME BIOGENESIS
A. Ribosome Biogenesis
The ribosome structure is complex, and considering the universal role it plays in catalyzing protein synthesis, it can be compared to a “molecular machine” composed of several distinct elements functioning as a single entity.35 Protein synthesis is a highly accurate but rapid process, and hence, ribosome biogenesis needs to be highly coordinated.
In recent years, advances in imaging technologies have unraveled the structural details of the eukaryotic ribosome and revealed more details of its interactions with messenger RNA (mRNA) and transfer RNA (tRNA).35 The full assignment of RPs in yeast and fungal 80S ribosomes have also become possible with the improved resolution.36,37 These observations led to the conclusion that the ribosome is a collection of enzymes (the ribozyme), in which ribosomal RNAs (rRNAs) function as the catalytic elements, with the RPs serving as structural ‘scaffolding’ units that organize the RNAs into appropriate configurations.35 Although the elements of the ribosome that are essential for protein synthesis have been universally conserved throughout evolution, the eukaryotic ribosomes differ from their prokaryotic counterparts in many respects, particularly regarding the presence of rRNA expansion segments and eukaryote-specific RPs, which are located on the surface of the ribosome, enveloping the evolutionarily-conserved core.34
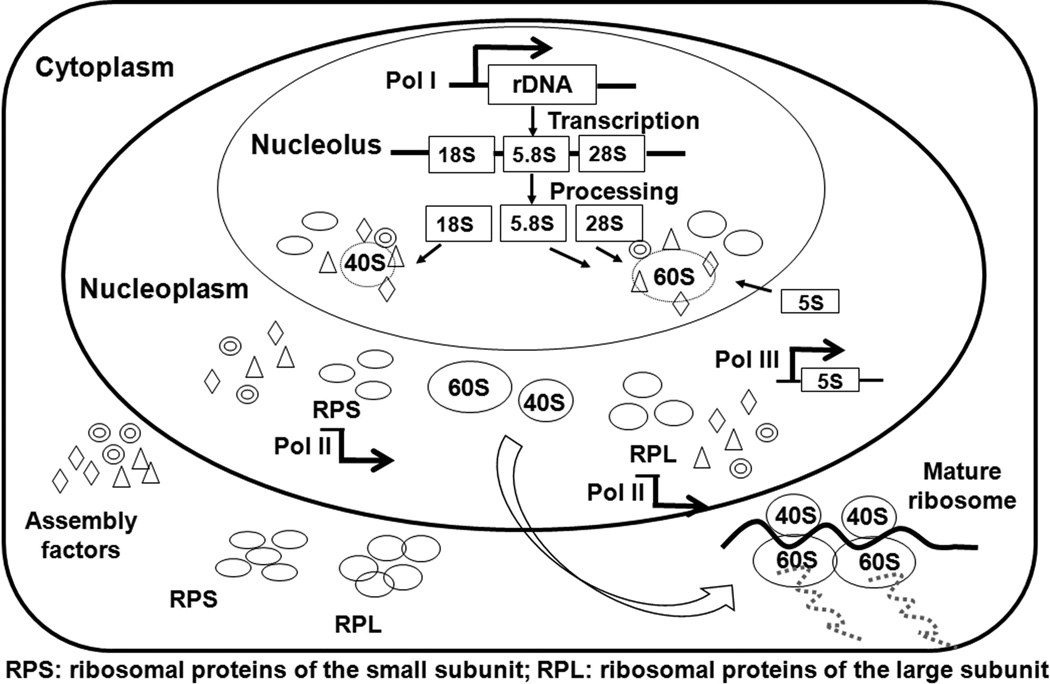
Across all evolutionary levels, ribosomes form the crux of the translational apparatus, regulating the main step in the expression of genes. Ribosomes decode genetic information, as well as form the peptide bonds during translation. The small subunit performs the former function and the large subunit catalyzes the formation of the peptide bonds.1,35 Ribosome biogenesis is a dynamic, energy-demanding, and strictly coordinated multistep process that involves the synthesis, processing, and modification of pre-rRNAs, assembly with RPs and interaction with several non-ribosomal factors, which associate with the evolving pre-ribosomal particles. In eukaryotes, ribosomes are preassembled in the nucleolus before being transferred to the cytoplasm. The process of ribosome synthesis includes the formation of pre-ribosomal particles in the nucleolus and assembly of two subunits in the cytoplasm. The full ribosome includes one large (60 S) and one small (40 S) subunit (Fig. 1).
Figure 1. The process of ribosome biogenesis.
Ribosome synthesis is a dynamic and coordinated multistep process. All three types of RNA polymerases and several hundred accessory factors participate in this process, which occurs throughout the cell. The ribosomal RNA genes are transcribed by RNA polymerase I (Pol I) into a single rRNA precursor within the nucleolus, which is subsequently cleaved and modified by several accessory factors to yield 18S, 5.8S, and 28S rRNA. The 5S rRNA gene is transcribed separately in the nucleoplasm by RNA polymerase III (Pol III). The RP genes are transcribed in the nucleoplasm by RNA polymerase II (Pol II), and these transcripts are exported to the cytoplasm for translation. The RPs and 5S RNA are imported to the nucleolus, where they assemble with rRNAs to form the small (40S) and large (60S) subunits. These preassembled subunits are then exported to the cytoplasm, where they undergo additional maturation to form the mature (80S) ribosome.
Ribosome biogenesis begins in the nucleolus, where RNA polymerase I (Pol I) first transcribes the rRNA genes into a single polycistronic transcript, which is cleaved to form 18S, 5.8S, and 28S rRNA. In yeast, Pol I transcription commences by recruiting a Pol I initiation complex at the rDNA promoter using two basal transcription factor complexes, UAF (upstream activating factor) and CF (core factor). As the transcript develops, many small nucleolar ribonucleoparticles (snoRNPs) facilitate the co-transcriptional covalent modification of numerous rRNA residues.35,38 These site-specific modifications include pseudouridylation (ψ) and methylation (M), and play an important role in ribosome function. For example, the loss of rRNA pseudouridylation decreases the translational fidelity. These RNA transcripts form ball-like structures on the 5’ end of the nascent transcripts and comprise the pre-ribosomes, corresponding to the 90S or small subunit (SSU) ‘processome’ complexes. Subsequently, the 90S–SSU processome is cleaved to form pre-40S and pre-60S particles. Meanwhile, in the nucleoplasm, RNA polymerase II (Pol II) and RNA polymerase III (Pol III) transcribe the RPs and 5S rRNA genes, respectively.39 These transcripts are then transported to the cytoplasm for translation. Upon translation, the RPs and 5S RNA are imported back into the nucleolus, where they form the pre-40S and pre-60S ribosomal subunits, along with the rRNA.40 These two subunits are then exported into the cytoplasm, where substantial structural rearrangements occur to convert the inactive pre-40S and pre-60S into functional 40S and 60S subunits (Fig. 1). Once the pre-60S has been exported into the cytoplasm, the residual large subunit ribosomal proteins associate with the non-ribosomal transacting factors, which are then recycled back to the nucleus. The process of cytoplasmic 60S ribosome maturation is essentially mediated by GTPases, such as Lsg1, and ATPases, such as Drg1.41
Functional studies of some mammalian ribosomal proteins, mostly those associated with disease states, have revealed that they are involved at various stages of pre-rRNA processing.42–45 Ribosomal proteins have been shown to be involved in the stabilization of both small and large subunit structures, rRNA processing and pre-ribosome transport, RNA folding, and/or interactions with other factors required for either ribosome synthesis or translation.46 In an elegant study by Donohue et al., the authors demonstrated that the ribosomal protein small subunit (RPS) proteins are essential for the production of the 40S ribosomal subunit, with some strictly required for initiation steps in pre-ribosome synthesis. Other RPS proteins are essentially involved in the progression of the nuclear and cytoplasmic maturation of the pre-ribosome and for nuclear export.46
Ribosomal proteins have also been speculated to stabilize secondary structures in the rRNA, to promote the formation of tertiary structures, and prevent misfolding. Ribosomal proteins also play an important role in the final large subunit structure and function. For example, L24 is essential for the proper functioning of the ribosome exit tunnel, the point of polypeptide emergence during translation, while the formation of the 60S stalk requires the incorporation of the ribosomal protein, P0.34 On the other hand, 40S cytoplasmic maturation also involves the stabilization of S3 and the endonuclease, Nob1, which mediates one of the final steps in the maturation of the small subunit i.e. the final cleavage of 20S pre-rRNA to 18S rRNA.47 In total, the two mature subunits contain four different types of rRNA, 80 different RPs and three different RNA polymerases (Pol I, Pol II, and Pol III) along with several accessory factors, all of which are apparently required for the synthesis, modification, and final assembly into the mature 80S ribosome.39
B. Ribosomal Protein Expression and Regulation
As discussed, ribosomal proteins form the basic building blocks in the ribosome assembly, playing seminal roles in the assembly and structure of ribosomes or in the initiation, elongation, or termination phases of protein translation. RPs are typically small (50–150 amino acid residues) and basic proteins with high isoelectric points (pI); their positive residues facilitate the interaction with the negatively charged phosphate groups of rRNAs or with mRNAs and tRNAs during translation.35,48 Human RPs have an average molecular weight of 18,877 Da, contain 167 amino acids and have a pI of 10.63.
Although these proteins function together, their amino acid sequences are dissimilar. Even the basic residues within the RPs are not equally distributed; instead, they occur in scattered clusters of three to four basic residues. In some RPs (e.g., S8, S9, L5, L22, L31, and the P proteins), the C-terminus contains a cluster of acidic amino acids. Although most of the RPs are basic, four RPs (the three P proteins and Sa) have acidic pI values.49 Of note, the RPs are remarkably well conserved in both structure and function, and have been throughout evolution.35
An analysis of the spatial structures of ribosomes indicates that ribosomal proteins usually contain one or several globular domains. Based on the structural packing, these proteins can be classified as α-proteins, β-barrel-containing proteins, α/β-proteins, and α+β-proteins. Most ribosomal proteins contain α-helices, β-strands that are packed into β-barrels (e.g.S12, S17, L2, L3, L14, and L24,), or α/β sandwiches (e.g. S3, S5, S6, L5, L6, L23, and L30). In addition to this compact domain, several ribosomal proteins possess elongated loops or N and C-terminal “tails” (e.g. S5, S7, S9-S14, S17, L2-L5, L15, L22, and L24), which impart substantial intramolecular mobility.49
Ribosomal proteins interact with several domains of rRNAs, acting as inter-domain clips, and help to maintain the structural integrity of the ribosomal assembly. The aforementioned elongated loops or N and C-terminal tails facilitate interactions with one or more domains of ribosomal RNA, forming inter-domain connections, as well as subunit bridges. For example, the intersubunit bridges produced by proteins S13 and L5 located on the ribosome periphery are postulated to play an important role in large-scale conformational rearrangements, e.g. in the translocation of tRNA. RPs like L24 are involved in the formation of the exit tunnel of the 60S subparticle, in addition to movement within the ribosome, thus providing translation accuracy.50 Ribosomal proteins also affect the elongation of the nascent peptide by binding to the EF-GTPase factors.
Many RPs undergo post-translational modifications. A common feature of all RP mRNAs is the 5’ terminal oligopyrimidine tract (TOP), which is made up of a cytidine (C) residue at the beginning, followed by an uninterrupted stretch of up to 5–15 pyrimidines.51 The external signals and stresses rapidly and reversibly modulate the translation of TOP mRNAs. The putative transacting factors for RP translational regulation remain elusive. For instance, it has been well documented that the La protein physically binds to TOP mRNAs, but the effects of La on their translation remain to be elucidated.52–54 It is known that the La protein is ubiquitously expressed in eukaryotic cells and associates with the 3' termini of several newly synthesized small RNAs, ultimately protecting these ends from exonucleases.55 In addition, epigenetic factors such as microRNA miR-10a has been shown to promote RP mRNAs translation by binding to the 5’ untranslated regions (UTRs).56
Post-translational modifications, such as phosphorylation, which is mediated by kinases such as phosphoinositide-3-kinase (PI3K), are involved in the regulation of TOP mRNA translation, particularly after mitogenic stimulation.57 The ribosomal protein S6, a well-established downstream target of the PI3K pathway, is phosphorylated at its C-terminus by two kinases (S6K1 and S6K2) subsequent to mitogenic stimulation. A clear correlation has been noted between the translational activation of TOP mRNAs and the hyperphosphorylation of S6, leading to the hypothesis that S6 phosphorylation is essential for recruiting TOP mRNAs to the polysomes. However, a lack of phosphorylation sites in S6 does not affect the translational control of the TOP mRNA,58 indicating that alternative signaling pathways are involved in the translational regulation of TOP mRNAs. The acidic RPs; P0, P1, and P2, which are involved in the formation of the ribosome stalk, also undergo phosphorylation.59 These post-translational modifications may be influenced by environmental factors or may be basal in nature; they may confer extraribosomal functions to these proteins.
Apart from phosphorylation, the RP production in the cell (ultimately controlling the cellular ribosome synthesis) is regulated via the proteasomal degradation of nucleolar RPs. Although the proteasome plays a prominent role in maintaining the turnover of RPs,60 in some cases, the ubiquitination of the amino acid residues of RPs can actually enhance the translational proficiency of the ribosomes.61 For example, S27a, S30, and L40 are generated as ubiquitin fusion proteins,32 but the actual function of the ubiquitin moiety remains unknown.32 An interesting phenomenon occurs in S. cerevisae, where a selective type of autophagy, known as ribophagy, which regulates the amount of ribosomes and acts as a quality control mechanism to induce the elimination of defective or wrongly assembled ribosomes, occurs under conditions of nutrient deprivation.62
C. Differences of Free and Structural Ribosomal Proteins
In eukaryotic cells, ribosomes are found either freely scattered in the cell cytoplasm or closely attached to endoplasmic reticulum (ER) membrane.63 Free and membrane-bound ribosomes are structurally and functionally identical. They differ only in their spatial distribution. Protein synthesis is usually compartmentalized with soluble proteins synthesized on free ribosomes and membrane (or secretory) proteins being synthesized on bound ribosomes.64,65 Interestingly, the proportion of the two forms of the ribosome is dependent on the physiological state and role played by the different types of tissues. Individual ribosomal subunits transform randomly between the two states (free and membrane-bound) depending upon the abundance of the particular type of mRNA molecules (those possessing ER signal sequences and those without them).66 If the protein being synthesized contains an ER signal sequence, the ribosome is directed to the ER membrane. Since several ribosomes are capable of binding to a solitary mRNA molecule, a polyribosome (also known as polysomes) is formed. The polyribosome attaches to the ER membrane following recognition of the nascent peptide chain by the signal recognition particle (SRP). As translation near the 3′ end of the mRNA molecule is completed, individual ribosomes are redirected to the cytosol.66,67 On the other hand, if the protein being encoded by the mRNA sequence lacks an ER signal sequence, the polyribosome is suspended freely in the cytosol and the protein product is formed in the cytosol itself.66,67 Both free and membrane-bound ribosomes comprise an interchangeable population and the cell can adjust their numbers on the basis of metabolic needs.
3. EXTRARIBOSOMAL FUNCTIONS OF RIBOSOMAL PROTEINS
In addition to participating in the assembly of the basal translational machinery, RPs perform additional functions in the cell, called extraribosomal functions, including regulation of the cell growth and proliferation, differentiation, apoptosis, and DNA repair.7–17 As suggested by Warner and McIntosh,6 the extraribosomal capacity of a RP can be determined based on the following criteria: 1) specific interaction(s) with some non-ribosomal component of the cell, presumably RNA or protein; 2) the interaction having an effect on cellular function; and 3) the process occurring away from the ribosome. There have been numerous reports on the extraribosomal functions of individual RPs (Table I).
Table I.
Extraribosomal Functions of Ribosomal Proteins
| RP | Extraribosomal function(s) |
Mechanism(s) | Ref. |
|---|---|---|---|
| Sa | Spleen development | Not studied | 68 |
| S2 | PRMT3 enzyme complex subunit | Not studied | 69 |
| S3 | DNA repair NFκB signaling Apoptosis Radioresistance |
Interacts with both MDM2 and p53 and stabilizes p53 Interacts with DNA base excision repair proteins (8-oxoG) Induces apoptosis via JNK activation; NF-κB complex subunit Interacts with E2F1 and upregulates Bim |
70–77 |
| S3a | Apoptosis Cell transformation Cell differentiation Drug sensitivity |
Interacts with CHOP | 69–80 |
| S4 | Cell proliferation | Cysteine protease activity | 81 |
| S5 | Cell cycle Cell differentiation |
S5 downregulates the CDK-2/4/6 levels | 82 |
| S6 | Apoptosis Cell proliferation Glucose metabolism |
S6 depletion activates the p53 pathway through L11 |
9,23,58,83,84 |
| S7 | Cell cycle Apoptosis Cellular development |
Interacts with MDM2 and stabilizes p53 S7 knockdown activates p53 in zebrafish S7 protects GADD45α from MDM2-mediated ubiquitination and degradation |
85–88 |
| S8 | Cell survival | Interacts with CDK11p46 and sensitizes cells to Fas ligand-induced apoptosis | 89 |
| S9 | Cell proliferation Cell differentiation |
S9 depletion activates the p53 pathway | 7,90 |
| S13 | Cell proliferation | S13 downregulates p27 expression and CDK2 kinase activity | 91 |
| S14 | Cell cycle Apoptosis |
S14 interacts with MDM2 and stabilizes p53 S14 depletion activates the p53 pathway S14 negatively regulates c-Myc activity |
92–95 |
| S15 | Cell cycle | Interacts with MDM2 and stabilizes p53 | 96 |
| S17 | Possible cellular development | Not studied | 97–99 |
| S19 | Cell differentiation Immunoregulation Embryonic development |
S19 knockout impairs erythropoiesis and stimulates epidermal melanocytosis S19 interacts with macrophage migration inhibitory factor (MIF) and inhibits its function Homozygous disruption causes embryonic lethality in mice, possibly related to p53 activation |
25,100–105 |
| S20 | Cell cycle Cellular development |
S20 knockout leads to epidermal melanocytosis due to p53-mediated Kit ligand expression Interacts with MDM2 and stabilizes p53 |
96, 100–103 |
| S25 | Cell cycle Apoptosis |
S25 interacts with MDM2 and stabilizes p53 | 106,107 |
| S26 | Cell cycle Apoptosis |
S26 interacts with MDM2 and stabilizes p53 S26 enhances the p53–p300 association |
108 |
| S27 | Cell proliferation Apoptosis Cell migration and invasion |
S27 interacts with MDM2 and stabilizes p53 S27 regulates NF-κB -Gadd45β signaling S27 regulates integrin β4 expression |
109–111 |
| S27a | Cell cycle Apoptosis |
S27a interacts with MDM2 and stabilizes p53 | 112 |
| S29 | Apoptosis Chemosensitization |
Downregulation of apoptosis inhibitors and upregulation of apoptosis inducers | 113,114 |
| L3 | Cell Cycle Apoptosis Cellular development |
L3 upregulates p21 through the interaction with Sp1 L3 deletion impairs the expansion of pancreatic progenitor cells in zebrafish, independent of p53 |
115,116 |
| L5 | Cell cycle Apoptosis |
Interacts with MDM2 and stabilizes p53 | 117,118 |
| L6 | Cell proliferation Chemoresistance |
Upregulates MDR proteins, GST activity/intracellular GSH, and cyclin E (gastric cancer) Interacts with MDM2 and stabilizes p53 (under ribosomal stress) Involved in normal pancreas development |
119–123 |
| L7 | Cell cycle Apoptosis |
Not studied | 124 |
| L8 | Apoptosis Cell proliferation Cellular development |
L8 deletion impairs development, inhibits cell proliferation and induces apoptosis in Drosophila | 125,126 |
| L11 | Cell cycle Apoptosis |
L11 interacts with MDM2 and stabilizes p53 Deletion of L11 activates p53 in zebrafish L11 negatively regulates the c-Myc level and activity |
4,26,127–130 |
| L13a | Immunoregulation | Inhibits the production of chemokines | 131–133 |
| L15 | Cell proliferation | Interacts with the IFN-stimulated antiviral protein, p56 | 134 |
| L17 | Cell proliferation | Inhibits vascular smooth muscle cell growth | 135 |
| L22 | Cellular development Cell transformation |
Depletion of L22 increases p53 protein synthesis in αβ T cells L22 inactivation induces Lin28B expression through NFκB |
15,136 |
| L23 | Cell cycle Apoptosis Cell invasion |
Interacts with MDM2 and stabilizes p53 L23 depletion stabilizes p53 Sequesters nucleophosmin from Miz1 |
137–139 |
| L26 | Cell cycle Apoptosis |
Interacts with MDM2 and stabilizes p53 Regulates p53 translation upon DNA damage |
140,141 |
| L29 | Cell proliferation Cellular development Angiogenesis |
L29 depletion activates p53 | 142–144 |
| L31 | Cell Proliferation | Not studied | 145,146 |
| L35a | Cell survival Drug resistance |
Not studied; overexpression contributes to drug resistance | 147,149 |
| L36a | Cell proliferation | Not studied | 8 |
| L37 | Cell cycle | L37 interacts with MDM2 and stabilizes p53 L37 depletion activates p53 through L11 |
96,150 |
| L41 | Cell survival Cell cycle Cell transformation |
L41 phosphorylates and degrades ATF4 | 151,152 |
| P1 | Cell transformation | P1 upregulates E2F1 and cyclin E | 153 |
A. Regulation of Gene Expression
It has been reported that RPs directly control gene transcription and functionally modulate transcriptional regulators, independent of their ribosomal function.5,6 For example, the translationally important K homology (KH)-domain containing S3, which is a component of the 40S ribosome subunit, also functions as a non-Rel subunit of the nuclear factor kappa B (NFκB) p65 homodimer to enhance DNA binding.73 Indeed, S3 knockdown reduces the ability of NFκB to induce selected target gene transcription.73 In addition, L7 has been identified as a co-regulator (repressor) of vitamin D receptor (VDR)-retinoid X receptor (RXR)-mediated transactivation via its interaction with the VDR.153 Similarly, L11 interacts with PPARα (peroxisome proliferator-activated receptor-α), inhibiting its ligand-dependent transcriptional activity through decreased binding to the PPAR-response element (PPRE).154
In addition to the control of specific gene transcription, RPs regulate the translation of individual proteins by a feedback mechanism. For example, S3 translation is repressed by the interaction of its C-terminal domain with its own mRNA, independent of the KH domain.155 Similarly, in response to interferon-γ, L13a is phosphorylated, released from the 60S subunit, and then specifically binds to the 3’-UTR GAIT (interferon-gamma-activated inhibitor of translation) element of ceruloplasmin (Cp) mRNA, and subsequently silences translation.131 L13a regulates the translation of specific mRNAs as part of a non-ribosomal complex, suggesting that, in addition to serving as an important part of the protein synthesis machinery, the ribosome is also a depot for proteins that modulate translation. In addition, L26 binds to the p53 mRNA 5’UTR and upregulates p53 translation after DNA damage.140
B. Cell Cycle Control
In addition to regulating gene expression,156 RPs affect cell cycle progression via various mechanisms.157–159 When expressed constitutively in Jurkat T-lymphoma cells, L7 leads to G1 arrest via the modulation of cell cycle progression-related proteins.157 In contrast, the overexpression of L15 promotes cell proliferation, while the downregulation of L15 inhibits the tumorigenicity of gastric cancer cells in nude mice.158 RPs are also required for normal cell proliferation. For example, concomitant overexpression of the nucleolar protein, nucleophosmin (NPM), facilitates the nucleolar storage of S9, facilitating ribosome biogenesis and cell proliferation.7 However, the depletion of S9 results in reduced protein synthesis and induces G1 cell cycle arrest, along with activation of p53 target genes.7 S3 is localized to the mitotic spindle and regulates the spindle dynamics by acting as a microtubule-associated protein (MAP) during mitosis.159 The depletion of S3 results in metaphase arrest, spindle abnormalities and defective chromosome movement.159
C. Regulation of Programed Cell Death
RPs have also been shown to be important in regulating apoptosis.6 S29 augments the apoptotic effects of anticancer drugs by reducing the expression of anti-apoptotic proteins and increasing the levels of pro-apoptotic proteins.113 In contrast, cancer cells overexpressing L35a exhibit reduced cell apoptosis and are more resistant to apoptosis-inducing agents than control cells, suggesting that it has a role in the response to cytotoxic damage.147 S3 induces apoptosis in response to extracellular stresses by activating JNK (c-Jun N-terminal kinases) in a caspase-dependent manner.77 This physical interaction between S3 and TRADD (tumor necrosis factor receptor (TNFR)-associated death domain) is responsible for inducing apoptosis.77 Additionally, the Akt-dependent phosphorylation of S3 inhibits its pro-apoptotic function.70 Knockdown of S3 increases the viability of HEK293 cells exposed to DNA-damaging agents, indicating that S3 is involved in DNA damage-induced cell death.71
D. Modulation of DNA Repair
There is also evidence that RPs are also involved in DNA repair.5,6 For example, S3 exhibits high binding affinity for the oxidative damage-induced 7, 8-dihydro-8-oxoguanine (8-oxoG) residues in DNA;72 it interacts with OGG1, the human base excision repair (BER) enzyme, and increases its catalytic activity towards DNA oligonucleotides containing embedded 8-oxoG residues.12 Exposure to DNA damaging agents leads to an extracellular-signal-regulated kinases (ERK)-dependent translocation of S3 to the nucleus, where it co-localizes with 8-oxoG DNA lesions.160 In addition, S3 binds to p53 and protects it from MDM2 (murine double minute 2)-mediated degradation,74 suggesting that S3 may be involved in maintaining the genomic integrity through both direct and indirect mechanisms.
E. Regulation of Development and Differentiation
RPs play a seminal role in embryonic development.5,6 S7-deficient zebrafish embryos show development defects, including impaired hematopoiesis and abnormalities in the brain.161,162 Homozygous disruption of S19 causes embryonic lethality in mice, while mice deficient in one S19 allele show a normal growth rate and weight.105 L22 deficiency selectively stops the development of αβ-lineage T cells at the β-selection checkpoint by inducing their death, which is led by p53 induction and activation.163,164 Downregulation of the RP levels during retinoic acid-induced neuronal differentiation has also been shown.165 L29 deficiency delays osteogenesis and leads to adult bone fragility in mice.143 In addition, knockdown of L29 induces cellular differentiation.166 The depletion of S9 in glioma cells impairs 18S ribosomal RNA production, activates p53, and induces morphological differentiation in a p53-dependent manner.90 The transcriptional inactivation of S5 leads to the erythroid differentiation of murine erythroleukemia (MEL) cells, and S5 overexpression in MEL cells delays the onset of differentiation.82
F. Modulation of Cell Migration and Invasion
The differential expression of RPs in several types of metastatic cancer cells has been identified by proteomic studies using two-dimensional liquid chromatography and mass spectrometry (2D-LC/MS-MS).167 For example, overexpression of S27 (also known as metallopanstimulin-1, MPS-1) in gastric cancer tissues is correlated with metastasis. Altered S27 expression was also demonstrated to regulate gastric cancer cell migration and invasion both in vitro and in vivo.111 Integrin β4 (ITGB4) has been identified as a downstream target of S27 that mediates its effects on cell metastasis.111 L23 also plays a role in cell motility and metastasis, as demonstrated by the fact that the overexpression of L23 alters lung cancer cell morphology and enhances its invasiveness.168 The level of phosphorylated S6 is elevated in metastatic lung adenocarcinoma, which is associated with a shorter metastasis-free survival,169 indicating a role for S6 phosphorylation in cancer metastasis.
G. Regulation of Cell Transformation
Several RPs have been shown to be involved in the malignant transformation of cells.5,6 The monoallelic loss of L22 predisposes T lineage progenitors to transformation and accelerated the development of thymic lymphoma in a mouse model of T cell malignancy.136 Indeed, L22 is found to be inactivated in ~10% of human acute T cell lymphoblastic leukemia.152 In addition, S3a overexpression induces cell transformation, as assessed by the formation of cancer foci and anchorage-independent growth in vitro, and the formation of tumors in nude mice.78 P1 induces an increase in the expression of E2F1 and upregulation of cyclin E. Co-transfection of P1 with mutant rasVal12 contributed to in vitro cell transformation in NIH3T3 cells.152
H. Regulation of Angiogenesis
Angiogenesis is crucial for cancer development and progression. The loss of L29 expression markedly reduced the vascular endothelial growth factor (VEGF)-stimulated microvessel formation.124 The tumor blood vessel density in subcutaneously grown Lewis lung carcinomas was significantly reduced in L29-mutant mice, suggesting that L29 can regulate angiogenesis.142
4. RIBOSOMAL PROTEINS AND NUCLEOLAR STRESS
The nucleolus is one of the functional nuclear compartments, and is where ribosome biogenesis occurs. As discussed earlier, in response to cellular stress, the synthesis of ribosomal RNA is rapidly downregulated, probably due to the disruption of the nucleolar integrity, stabilization of p53 and induction of cell cycle arrest.3 It is interesting to note how the nucleolus transmits cellular stress signals to the p53 pathway, thus triggering cell cycle arrest and/or apoptosis based on the extent of damage and the capacity of the cell to recover.3 Disruption of the nucleolus is believed to stabilize p53 and activate the pro-apoptotic p53 signaling pathway.170 However, it has been shown that p53 stability is also regulated by an intact structure and function of the nucleolus.171
Nucleolar integrity is required to maintain ribosome biogenesis and controlling cell proliferation. As noted above, nucleolar stress, characterized by a loss of nucleolar integrity, disturbs ribosome biogenesis and halts cell proliferation.3 Nucleolar stress results in the redistribution of nucleolar proteins to the nucleoplasm, altering their interactions with MDM2 and resulting in p53 activation.18,172 These redistributed proteins include RPs, ARF (ADP-ribosylation factor), nucleophosmin (NPM/B23), nucleostemin (NS) and nucleolin, among many others. Perturbations of ribosome biogenesis, either due to impaired rRNA synthesis or RP deficiency, have been defined to cause nucleolar stress that activates p53 signaling. As would be expected given these effects, the p53 pathway is associated with many ribosomal diseases, such as DBA and the 5q- syndrome.24,25 Distinct intrinsic and extrinsic factors that disturb ribosome biogenesis and trigger nucleolar stress have been reported.1,3 This section is focused on these stresses and their consequent biological effects.
A. Impairment in rRNA Synthesis and Processing
One of the major causes of ribosomal stress is incorrect rRNA synthesis. Several proteins, such as the nucleolar protein block of proliferation 1 (Bop1), are involved in rRNA processing and ribosome assembly.173 A dominant negative Bop1 mutant inhibits ribosome biogenesis, leading to p53 activation and subsequent cell cycle arrest.20 Inactivation of p53 abrogates the mutant Bop1-induced cell cycle arrest.20 Selective inhibition of other rRNA processing factors, such as WDR3 (WD repeat 3)174 and hUTP18175, can also activate p53, reduce proliferation, and cause cell cycle arrest. The unfolded protein response (UPR) is activated in the presence of unfolded or misfolded proteins that accumulate in the endoplasmic reticulum lumen, and this also triggers p53 accumulation and activation in a PERK (PKR-like ER kinase)-dependent manner.176
The suppression of POLR1A (RNA polymerase I polypeptide A), the RNA polymerase I catalytic subunit, stabilizes p53, but upregulation of rRNA synthesis abrogates this effect,177 suggesting that the imbalance between rRNA and RPs is an inducer of ribosomal stress. On the other hand, transcription initiation factor IA (TIF-IA), a nucleolar target for the downregulation of rRNA transcription, which modulates the transcriptional activity of Pol I, mediates growth-dependent regulation of rRNA synthesis.178 The depletion of TIF-IA in mouse embryonic fibroblasts (MEFs) led to nucleolar disruption, activation of p53, cell cycle arrest and the induction of apoptosis.179 Knockdown of p53 by RNAi can overcome the cell cycle arrest and apoptosis in response to TIF-IA ablation, indicating that the nucleolus acts as a cellular stress sensor that regulates p53 stability and activity.179
Ribosome biogenesis is also a target for numerous chemotherapeutic drugs.180 Low concentrations of actinomycin D (Act D) (< 10 nM) inhibit RNA polymerase I, and consequently prevent the transcription of rRNA, which leads to nucleolar stress.117 Similar results have been seen following exposure to 5-fluorouracil (5-FU)181 or mycophenolic acid (MPA).182 In addition, p53-dependent G1 cell cycle arrest is caused by growth adverse conditions, such as serum deprivation or cell contact inhibition. This growth inhibition, possibly due to the decreased nucleolar rRNA production, facilitates L11 translocation into the nucleoplasm, inhibiting MDM2 and activating the p53 signaling pathway.128
B. Nucleolar Protein Deficiency or Malfunction
The malfunction of nucleolar proteins induces nucleolar stress, which leads to p53 activation and subsequent cell cycle arrest and apoptosis.33 The nucleolar protein nucleostemin (NS) is crucial for cell proliferation and early embryogenesis.183 NS overexpression stabilizes p53 by directly binding to MDM2. Surprisingly, knockdown of NS also leads to p53 induction and activation.183 Nucleostemin is a nucleolar GTP-binding protein and that is extensively degraded in response to GTP depletion by mycophenolic acid.184 The degradation of NS induces ribosomal stress, and activates p53 through an L11-dependent mechanism.183 On the other hand, the nucleolar protein PAK1IP1 (p21-activated protein kinase-interacting protein 1) is induced upon nucleolar disruption with 5-FU and Act D. PAK1IP1 binds to MDM2, inhibiting its ability to cause p53 ubiquitination and degradation.166 Interestingly, both PAK1IP1 overexpression and knockdown inhibit cell proliferation and induce p53-dependent G1 cell cycle arrest.185 The mechanisms underlying these effects are not fully understood.
The tumor suppressor ARF, an important player in the p53-MDM2 interaction, is known to induce both p53-dependent and -independent cell cycle arrest.186 ARF interacts with nucleophosmin B23, a multifunctional nucleolar protein implicated in ribosome synthesis, and promotes B23 polyubiquitination and degradation.186 B23 knockdown inhibits the pre-ribosomal RNA processing and induces cell death.186 Thus, ARF regulates ribosome synthesis and cell proliferation by inhibiting B23, suggesting a role for ARF in tumor surveillance. Additionally, ARF also causes the nucleoplasmic accumulation of the RNA Pol I transcription termination factor I (TTF-I) via the inhibition of B23.187 In the absence of ARF, TTF-I is targeted for ubiquitination and proteasomal degradation by MDM2.188 Moreover, ARF interacts with the upstream binding factor (UBF) and inhibits its phosphorylation, disturbing the assembly of the transcription machinery complex.189 These findings define a new pathway that regulates the cell cycle- the negative control of rRNA transcription by ARF.
The deficiency of individual RPs also causes defective ribosome biogenesis and triggers ribosomal stress, leading to p53-dependent cell cycle arrest and apoptosis.33 For example, L29 or L30 depletion disturbs 60S ribosome biogenesis and results in p53 activation.190 The effects of the hematopoietic defects in L29 mutant zebrafish depend upon p53 activation.191 L37 degradation and p53 activation are both observed in response to DNA damage.149 A deficiency of L11 in the zebrafish model also activates the p53 pathway, resulting in abnormal brain development and embryonic lethality.192 Simultaneous depletion of p53 rescues the fish from both developmental defects and apoptosis. L11 mutation in zebrafish also leads to metabolic defects, the upregulation of p53 target genes and the induction of global changes in metabolism.193
Haploinsufficiency or deficiency in small subunit ribosomal proteins can also activate proapoptotic p53 signaling pathways. For example, deletion of one allele of S6 in mouse embryos inhibits their entry into the M-phase of the cell cycle, ultimately leading to perigastrulation lethality.83 Conditional knockout of S6 in T lymphocytes suppresses their division and splenic and lymph node accumulation due to p53 activation.194 Additionally, similar erythroid phenotypes are observed in S6 knockout mice and patients with DBA and the 5q- syndrome.195 S19 deficient mice develop macrocytic anemia and bone marrow failure; p53 knockout can rescue the DBA phenotype.101 S7 knockout in zebrafish induces p53 activation and cell cycle arrest.161 In addition, aberrant movement of RPs between the nucleus and the cytoplasm also triggers nucleolar stress. Members of the β-karyopherin family, such as importin 7 (IPO7) and exportin 1 (XPO1), mediate the nuclear import of RPs and export of ribosomal subunits. Partial depletion of IPO7 induces p53 activation and p53-dependent cell growth arrest.196 Thus, defects in both the initial and later stages of ribosome synthesis cause ribosomal stress, leading to the activation of p53.
C. The RPs-MDM2-p53 Pathway
The p53 tumor suppressor regulates the expression of downstream target genes whose protein products induce cell cycle arrest, apoptosis, DNA repair, and senescence in response to various stresses,172,197–200 protecting cells from transformation and tumorigenesis. p53 is essential for maintaining the genomic stability during cell growth and division.172 Cancer cells can escape from p53 surveillance either by mutating its encoding gene, TP53, or by activating a number of proteins that suppress p53 activity.172,201–206 The predominant negative regulator of p53 is MDM2.172,201–203 MDM2 and p53 form a negative feedback loop, in which p53 activates MDM2 transcription and MDM2, in turn, inactivates p53 by targeting it for ubiquitination and proteasomal degradation.201–206 In normal cells, the p53 protein is maintained at low levels through this MDM2-p53 negative feedback loop. In response to various stress signals, the inhibitory effect of MDM2 on p53 can be circumvented by multiple cellular mechanisms.172,207–211 DNA damage can lead to the inhibition of MDM2-mediated p53 degradation by the phosphorylation of MDM2 at multiple sites207 or by the induction of SCF destruction complex-mediated MDM2 turnover.208 DNA damage signals also cause the acetylation of p53 at specific lysine residues, which activates p53 by preventing MDM2-facilitated p53 degradation.209 Oncogenic stress is another type of stress that can prevent the inhibition of p53 by MDM2. It is often associated with the overexpression of oncoproteins such as Ras (resistance to audiogenic seizures) and c-Myc (cellular myelocytomatosis oncogene). These oncogenic insults stimulate the expression of the ARF tumor suppressor, which in turn interacts with MDM2 and inhibits its ubiquitination of p53.210,211 More recently, there have been several reports supporting that the MDM2-p53 interaction is modulated by many other internal and external factors.172,212,213
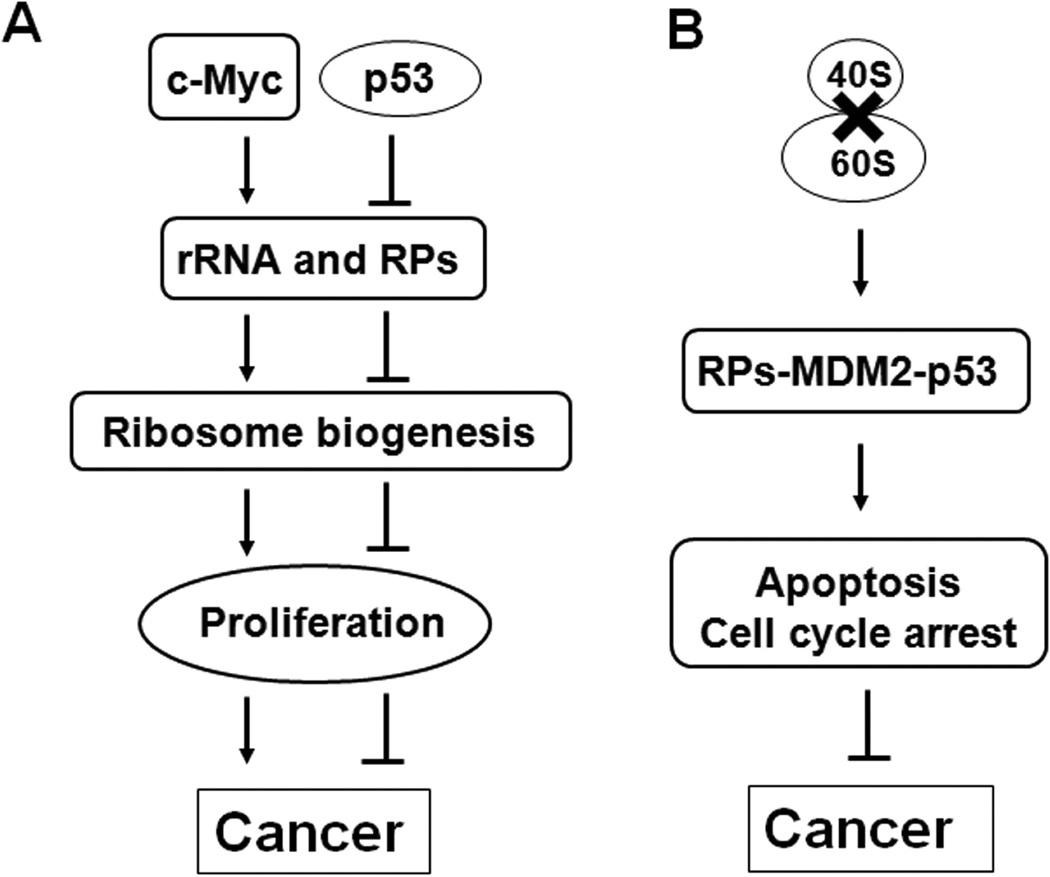
Over the past decade or so, increasing evidence has revealed the previously less-appreciated ribosomal stress-induced interactions among RPs, MDM2, p53 and related molecules, termed the RPs-MDM2-p53 pathway (Fig. 2).18 In response to nucleolar stress, several RPs translocate to the nucleoplasm, where they bind to MDM2 and inhibit the MDM2-mediated ubiquitination and degradation of p53, resulting in p53-dependent cell cycle arrest and apoptosis.18,31 The RPs-MDM2-p53 signaling pathway constitutes a “surveillance network” that monitors the integrity of ribosome biogenesis.18 In the following section, we summarize the recent findings on several RPs as regulators of the MDM2-p53 pathway.
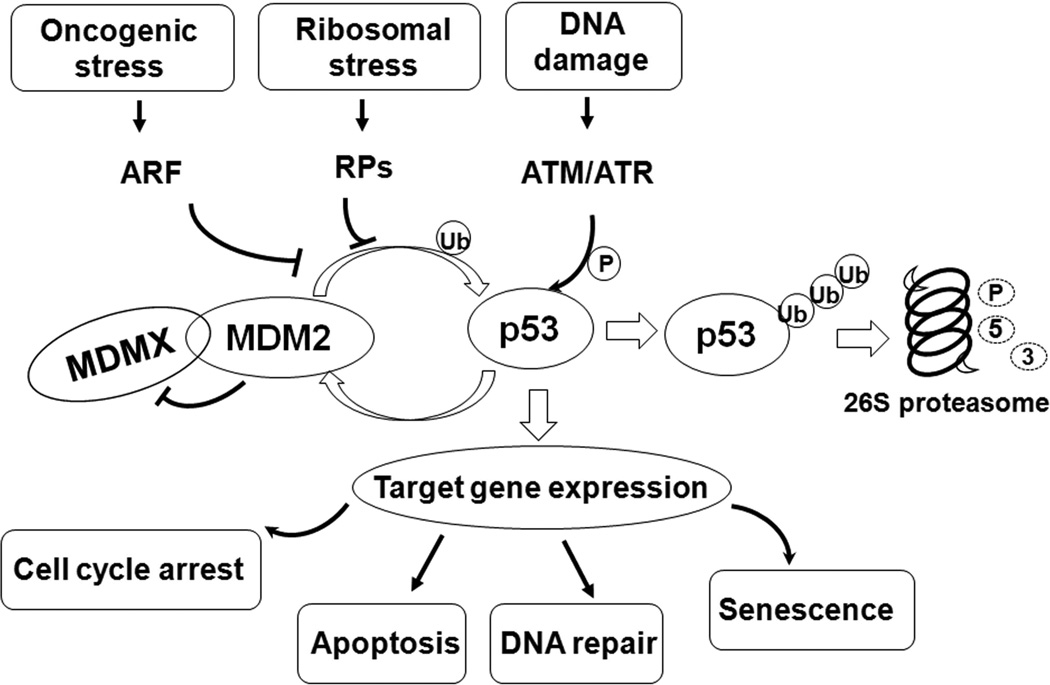
Figure 2. The MDM2-p53 signaling pathway.
The p53 tumor suppressor coordinates a complicated network of signaling pathways to prevent aberrant cell growth and proliferation. Under normal conditions, the p53 protein expression is tightly regulated and maintained at a low level by murine double minute 2 (MDM2) and MDMX. MDM2 has E3 ligase activity, and mediates the attachment of a ubiquitin (Ub) moiety to p53, which targets it for proteasomal degradation. MDM2 also binds p53 and inhibits its transcriptional activity. MDMX lacks the E3 ligase, but forms a heterodimeric complex with MDM2 to stimulate MDM2-mediated p53 degradation. MDMX also suppresses the transcriptional activity of p53 through its direct interaction with p53. In turn, p53 controls the transcription of MDM2 through a negative feedback loop. MDM2 also targets MDMX for ubiquitination and proteosomal degradation. In response to stress stimuli, the inhibitory effects on p53 are removed through distinct mechanisms, allowing p53 to be activated. For instance, exposures to radiation (ionizing and ultraviolet light) and DNA-damaging agents activate several kinases, such as ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3 related (ATR), which modify the phosphorylation states of p53, MDMX, and MDM2, leading to conformational changes in these proteins that block their interactions, resulting in p53 stabilization. Oncogenic signals stimulate the production of alternative reading frame (ARF), which binds to MDM2 and stabilizes p53. Defective ribosome biogenesis causes the release of several RPs, which bind to MDM2 and suppress its E3 ligase activity, resulting in p53 accumulation and activation. The consequence of p53 protein activation is the transactivation of several downstream genes. Depending on the cell type and the stressors, the outcome can be cell cycle arrest, apoptosis, DNA repair, or senescence.
During ribosome biogenesis, the 5S rRNA assembles with RPs L5 and L11 as a complex, before being recruited to the 60S ribosomal subunit. It was shown more than two decades ago that L5 and 5S rRNA assemble along with the MDM2 protein and the MDM2-p53 complex in murine cells,214 although the functional significance of this association was not recognized until about ten years later. L5 binds to MDM2 and inhibits p53 ubiquitination and degradation, leading to enhanced p53 transcriptional activity and p53-dependent G1 cell cycle arrest.117 The interaction of L5 with MDM2 is enhanced by treatment with a low dose of Act D; the Act D-induced p53 activation is inhibited by treatment with an siRNA (small interfering RNA) against L5. Another RP, L11, also interacts with MDM2 and inhibits MDM2 function, thus leading to p53 stabilization and activation.4,127 Further investigations showed that L11, unlike L5, inhibits the degradation of ubiquitinated MDM2, independent of its effects on p53.111 Recently, the existence of a L5-L11-5S rRNA pre-ribosomal complex has been demonstrated, and it is a part of an MDM2 inhibitory complex that stabilizes p53 in a mutually dependent manner. The complex is redirected from assembly into nascent 60S ribosomes to MDM2 inhibition as a result of defective ribosome biogenesis.215
Another ribosomal protein, L23, has also been found to activate p53 by inhibiting MDM2 function in response to ribosomal stress.137,138 However, knockdown of L23 also induces ribosomal stress and causes B23 translocation from the nucleolus to the nucleoplasm, leading to stabilization and activation of p53, suggesting that L23 functions as both an effector and a sensor in this pathway. Additionally, a synergistic effect between L5 and L11 with regard to p53 activation has been found.118 L11 cooperates with L5, resulting in a strong inhibition of MDM2’s E3 ligase activity, resulting in p53 stabilization and activation to an extent similar to that achieved by ARF.118 The capacity of L11 to bind the 5S rRNA is important for the cooperation with L5, because the mutant L11 that does not have the 5S rRNA-binding activity cannot increase the effects of L5 on MDM2.118 Preventing the degradation of both L5 and L11 is critical for p53 activation following ribosomal stress. The ribosomal stress induced by Act D results in proteasomal degradation of the newly synthesized RPs, but does not affect the ribosome-free L5 and L11, which bind to MDM2 and ubiquitinate it. Subsequent to the disruption of the nucleolus, the newly synthesized L5 and L11 continue to be imported into the nucleoli, accumulate therein, and co-localize with p53 and MDM2.216 Thus, the disrupted nucleolus, in essence, provides a “platform” for the interaction of L5 and L11 with p53/MDM2, explaining their role in p53 activation.
More recently, several investigations, including our own studies, have led to the discovery of additional RPs that modulate the MDM2-p53 pathway, including L6,122 L26,217 S3,74 S7,85,86 S14,93 S25,106 S26,108 S27,109 and S27a112 (Fig. 3). These RPs show similar, but not identical, mechanisms with regard to regulating p53 in response to ribosomal stress. For example, L6 binds to and inhibits MDM2’s E3 ubiquitin ligase activity, inhibiting MDM2-mediated p53 polyubiquitination and degradation. L6 shuttles from the nucleolus to the nucleoplasm under ribosomal stress, facilitating its binding with MDM2.122 Since all of these RPs can inhibit MDM2-mediated p53 ubiquitination, it is likely that they execute such inhibitory effects by physically interacting with MDM2 and preventing the transfer of the ubiquitin to p53. These RPs tend to bind to the central portion of MDM2, which contains the acidic domain and zinc finger domain.18,172 The acidic domain is also critical for MDM2-mediated p53 degradation,218 although how exactly the acidic domain of MDM2 contributes to the regulation of p53 stability is unclear. The binding of RPs to MDM2 may cause a conformational change in MDM2 that alters its tertiary structure within its central region, and this change might reduce its binding affinity for p53, thus weakening its ability to ubiquitinate p53. It has recently been demonstrated that the acidic and polar residues within the zinc finger domain of MDM2 are essential for its interaction with the basic residues in L11.219 However, more studies, including crystallographic studies, are needed to provide more information about the structure of the RPs-MDM2-p53 complex(es).
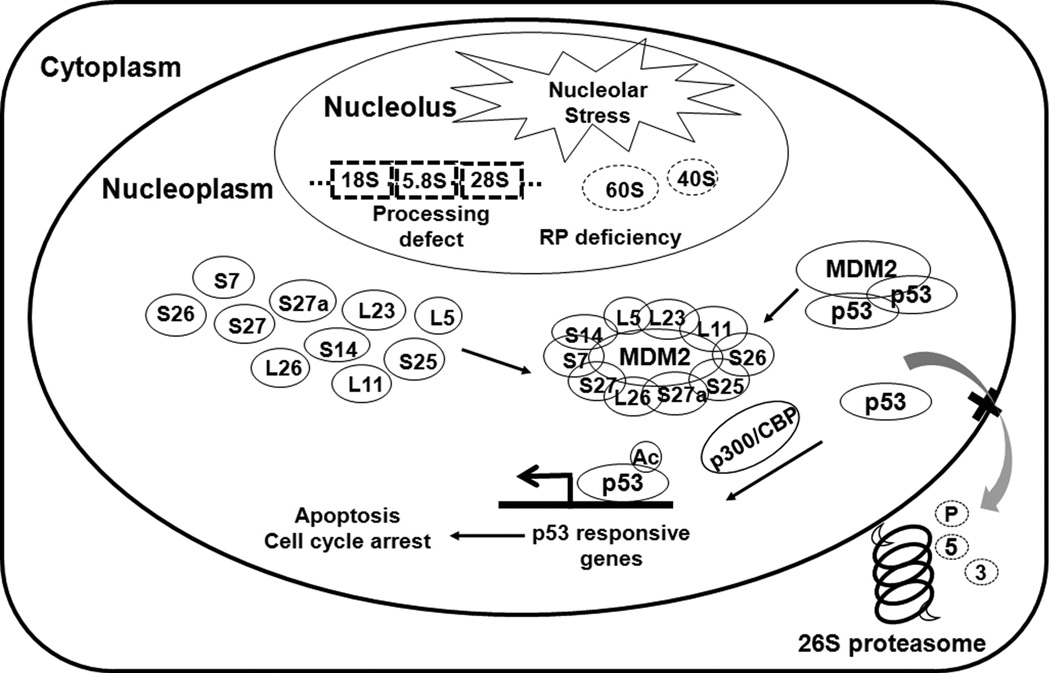
Figure 3. The RPs-MDM2-p53 interplay in nucleolar stress.
Defects in ribosome biogenesis due to impairment of rRNA synthesis or processing, nucleolar protein deficiency, or due to malfunctions trigger nucleolar stress (also called ribosomal stress). In response to nucleolar stress, a subset of RPs is released from the nucleolus to the nucleoplasm, where they bind to MDM2 and inhibit the MDM2-mediated ubiquitination and degradation of p53, leading to the stabilization and activation of p53. The RPs-MDM2-p53 interplay provides a surveillance mechanism to monitor the integrity of ribosome biogenesis and coordinate cell growth and proliferation.
Several modulators of the RPs-MDM2-p53 pathway have recently been identified, including MDMX (murine double minute 4), PICT1 (protein interacting with the C terminus 1), MYBBP1A (Myb-binding protein 1A), and hCINAP, among others.26,30,31,94 MDMX is an important negative regulator of the p53 response to ribosomal stress.26 L11 prompts MDM2-mediated ubiquitination and degradation of MDMX. Abundant MDMX in cancer cells results in decreased sensitivity to Act D due to the formation of inactive p53-MDMX complexes.26 In addition, 5S RNA binds and stabilizes the MDMX protein, and the binding between the 5S RNA and MDMX is disrupted, and MDMX is quickly degraded by MDM2, in response to ribosomal stress.220 However, the detailed role of MDMX in modulating the RPs-MDM2-p53 pathway remains unclear. For example, MDMX has been shown to facilitate the inhibition of MDM2’s E3 ligase activity by S7.86 MDMX also facilitates the S25–MDM2 interaction to modulate the E3 ubiquitin ligase activity of MDM2, but the interaction does not depend on MDMX.106
The initial description of PICT1 (also known as GLTSCR2, glioma tumor suppressor candidate region gene 2) is a tumor suppressor that interacts with and stabilizes PTEN (phosphatase and tensin homolog).221 The low levels of PICT1 in glioma tissues are associated with tumor malignancy and progression, and PICT1 overexpression enhances apoptosis in glioma cells.222 Contrarily, studies with genetic mouse models suggest PICT1 acts as an oncogene.223 Pict1−/− mice are embryonic lethal, while Pict1+/− mice develop normally. Co-depletion of p53 rescues Pict1−/− embryonic stem (ES) cells from cell cycle arrest and apoptosis.223 In a chemically-induced skin cancer model, Pict1+/− mice developed papillomas more slowly compared to their wild-type counterparts.223 Moreover, PICT1 shRNA (short hairpin RNA) induced p53-dependent growth inhibition in brain, colorectal and ovarian tumor cell lines.223 PICT1 interacts with L11 and sequesters it in the nucleolus, which inhibits the interaction between L11 and MDM2.223 Forced expression of PICT1 may protect tumor cells from nucleolar stress. Intriguingly, it has been reported that PICT1 directly binds to and stabilizes p53.224 Upon translocation to the nucleoplasm from the nucleolus following nucleolar stress, PICT1 prevents MDM2-mediated p53 degradation and induces p53 oligomerization. Thus, both deficiency and overexpression of PICT1 can result in p53 activation, which is similar to the findings observed with nucleostemin.225 PICT1 depletion during mammalian ribosome synthesis results in nucleolar stress and cell cycle arrest.225 Marginal elevations of PICT1 may cause L11 nucleolar localization, while high levels of PICT1 can allow the protein to “spill over” to the nucleoplasm and bind to p53, which may explain the different effects of PICT1 observed under different conditions.225
MYBBP1A is involved in p53 acetylation upon ribosomal stress. Acetylation is essential for p53 activation.226 As a co-factor for transcriptional regulation,227 MYBBP1A is tethered to the nucleolus through its binding to nucleolar RNA. During situations of nucleolar stress, MYBBP1A translocates from the nucleolus to the nucleoplasm, facilitates the interaction between p53 and p300, and promotes p53 acetylation.228 The depletion of L5 and L11 inhibits the translocation of MYBBP1A and the activation p53,228 indicating that there is a dynamic balance between RNA generation and export, and any disturbances (due to nucleolar stress) may alter the nucleolar RNA content and affect p53 activity through MYBBP1A.
hCINAP (human coilin-interacting nuclear ATPase protein) is an ubiquitously expressed eukaryotic nucleoplasmic enzyme that associates with Cajal bodies in the nucleoplasm, playing diverse roles in transcription and nucleotide homeostasis.229,230 It is a novel partner of S14.94 S14 stabilizes p53 by inhibiting MDM2-mediated p53 degradation,93 and this process is facilitated by S14 neddylation. hCINAP inhibits S14 neddylation, leading to reduced S14 stability and increased p53 degradation.94 Thus, hCINAP may be considered to be an important regulator of the RP-MDM2-p53 pathway.
Recent studies suggest that there is a direct link between RPs and p53 that is independent of MDM2 binding.30,31 Based on a loss-of-function genetic screening, a group of RPs was shown to directly regulate p53 function.231 The reduction of RP levels decreased the p53 levels by inhibiting p53-specific translation.30,31 RP gene mutations can cause a loss of p53 synthesis, and predispose zebrafish to the development of malignant peripheral nerve sheath tumors (MPNSTs).232 Cells extracted from MPNSTs are unable to produce p53 protein even with treated with proteasome inhibitors and γ-irradiation, which typically are strong inducers of p53.232 Interestingly, the wild-type TP53 gene is unaffected, the rates of overall protein production are normal, but the synthesis of p53 protein is not induced by the usual stimuli, indicating a potential role for RPs in the control of p53 translation.232 Supporting this idea, the specific regulation of p53 translation by individual RPs has been revealed.33 For example, in response to DNA damage, L26 binds to the p53 mRNA 5'-UTR and increases the rate of p53 protein translation,140 indicating that the direct control of p53 mRNA translation after DNA damage may represent another layer of p53 regulation by RPs. In contrast, L22 deficiency results in the selective upregulation of p53 in αβ-lineage T cells, partially through the induction of p53 synthesis,15 suggesting that L22 may have cell type-specific and stage-specific functions in T cell development.
Actively growing cells require the continuous synthesis of ribosomal RNA, RPs, and other factors to sustain cellular biosynthesis; p53 represses ribosomal gene transcription and restricts cell proliferation.233,234 In addition to rRNA, p53 also regulates the transcription of RP genes.233 For instance, p53 directly induces the expression of a RP, S27L,10,235 which is critical for DNA damage-induced cell apoptosis. We have recently demonstrated that S25 is a novel p53 downstream target.106 S25 activates p53 through its interaction with MDM2, and in turn, the activated p53 represses S25 gene transcription, forming a negative feedback loop.106 The regulation of RP gene expression is not only restricted to wild type p53; the mutant p53 (R248W) upregulates the expression of L37, P1, and S2, suggesting a mechanism for the overexpression of these RPs in human tumors.236
D. p53-Independent Pathways in Response to Ribosomal Stress
There is emerging evidence supporting that RPs are involved in the cellular response to ribosomal stress through p53-independent pathways.237 As depicted in Fig. 4, both p53-dependent and –independent mechanisms are responsible for the coordination of cell growth, proliferation, and apoptosis. These pathways can provide a basis for the development and application of biomarkers and therapeutic drugs that specifically impact ribosome biogenesis, irrespective of the p53 status.
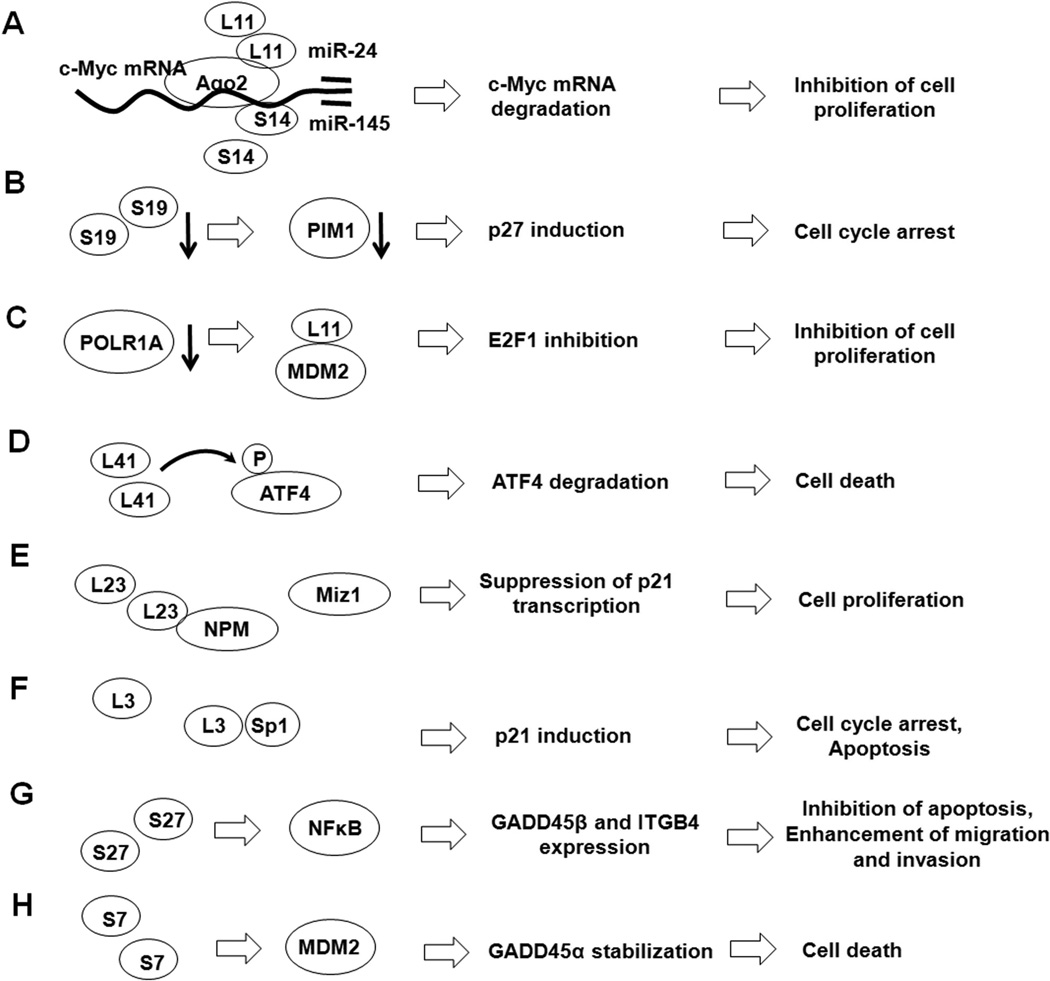
Figure 4. The p53-independent functions of RPs.
The roles of RPs in regulating cellular functions also involve p53-independent mechanisms. (A) L11 and S14 bind to c-Myc mRNA and recruit Ago-2 and miRNAs, resulting in the degradation of c-Myc mRNA. (B) S19 deficiency causes PIM1 degradation and p27 accumulation, resulting in the inhibition of cell cycle progression. (C) Knockdown of POLR1A activates the L11-MDM2 interaction and inhibits the MDM2-mediated stabilization of E2F1. (D) L41 mediates the phosphorylation and translocation of ATF4 and induces the proteosomal degradation of ATF4. (E) L23 binds to NPM, the essential co-activator of Miz1 involved in regulating p21 transcription, leading to increased cell proliferation. (F) L3 mediates p21 upregulation through its interaction with Sp1. (G) S27 regulates GADD45β and ITGB4 through NFκB signaling to inhibit apoptosis and promote cell migration and invasion. (H) The S7-MDM2 interaction stabilizes GADD45α and induces cell death.
In addition to modulating the MDM2-p53 pathway, L11 regulates c-Myc mRNA decay and protein turnover (Fig. 4A).238,239 L11 blocks the recruitment of TRRAP (transformation/transcription domain-associated protein), the co-activator of c-Myc, to the promoter regions of c-Myc target genes.130,240 Nucleolar stress can increase the binding of L11 to its targets, and decreases the TRRAP binding to c-Myc, inhibiting the expression of c-Myc downstream genes (such as E2F2 and 5S rRNA) and thus decreasing cell proliferation. Furthermore, L11 binds to the c-Myc 3’-UTR, leading to c-Myc mRNA degradation.240 Ribosomal stress inhibits c-Myc expression and activity in an L11-dependent manner.241 All of these observations suggest that L11 is a principal player in ribosome biogenesis and the cell growth process, due to its ability to modulate the functions and activities of c-Myc, a master regulator of ribosome and protein synthesis.242 Similarly, S14 has recently been suggested to be a negative regulator of c-Myc, also through the inhibition of TRRAP co-activator recruitment and c-Myc mRNA stability (Fig. 4A).92
Another protein, PIM1 (proviral integration site for Moloney murine leukemia virus 1) kinase which drives cell cycle progression, has been associated with RPs, such as S19.243 S19 deficiency dramatically destabilizes PIM1, which increases p27, inhibits cell cycle progression and reduces cell proliferation, even in the absence of p53 (Fig. 4B).243 Exogenous restoration of the PIM1 levels leads to a recovery of all of these effects, indicating that PIM1 may act as a sensor for ribosomal stress though either p53-independent or p53-dependent mechanisms.
On the other hand, siRNA knockdown of the POLR1A gene inhibits rRNA synthesis and cell cycle progression, and downregulates E2F1 in inactive p53 cells.244 The downregulation of E2F1 is due to the release of L11, which inhibits MDM2-mediated stabilization of E2F1. Thus, targeting the RNA polymerase I transcription apparatus may selectively inhibit cellular proliferation in p53-deficient environments (Fig. 4C).
The activating transcription factor 4 (ATF4) is a major coordinator of cell survival during nucleolar stress, and is commonly overexpressed in cancer. L41 induces ATF4 phosphorylation at serine 219, leading to its translocation from the nucleus to the cytoplasm for proteosomal degradation.151 L41 overexpression induces cell death and increases chemosensitivity in cancer cells (Fig. 4D).151
A negative feedback circuit between Miz-1 (Myc-associated zinc-finger protein 1) and L23 has been reported. Miz-1 inhibits cell proliferation and induces p15 and p21 expression. In the nucleolus, L23 (a direct target gene of Myc) negatively regulates Miz1-dependent transactivation by retaining B23, which is necessary for Miz1 transactivation.245 This regulatory feedback mechanism may be the link between Myc-dependent translation and Miz-1-dependent cell cycle arrest (Fig. 4E).
L3 has been suggested to be a new regulator of the cell cycle and apoptosis that positively regulates p21 expression, independent p53.246 The specific interaction between L3 and Sp1 is required for L3-mediated p21 upregulation (Fig. 4F). Furthermore, p21 overexpression leads to activation of G1/S cell cycle arrest or the induction of mitochondrial apoptotic pathways, depending on its intracellular levels.
The S27 gene is a growth factor-inducible gene. Knockdown of S27 leads to spontaneous apoptosis and growth retardation in gastric cancer cells.247 Silencing of S27 inhibits NFκB activity by reducing the phosphorylation of p65 at Ser536 and IκBα at Ser32, blocking NFκB nuclear translocation, and reducing its DNA binding activity.110 S27 knockdown-induced apoptosis is mediated by Gadd45β (growth arrest DNA damage inducible gene 45β), a direct NFκB target gene.110 Furthermore, knockdown of S27 expression inhibits invasion and migration, and reduces ITGB4 mRNA and protein expression in gastric cancer models (Fig. 4G).111 The overexpression of ITGB4 in S27 knockdown cells enhances cell invasion and migration, while knockdown of ITGB4 partially reduces these effects induced by S27 overexpression.111
S7 forms a complex with GADD45α (growth arrest and DNA damage inducible gene 45α), which regulates DNA repair, cell cycle checkpoints and apoptosis. S7 interacts with both MDM2 and GADD45α, protecting GADD45α from MDM2-mediated ubiquitination and degradation.87 However, S7 mutants lacking the ability to bind MDM2 do not stabilize GADD45α, indicating the importance of the RP-MDM2 interplay (Fig. 4H).87
E. Post-translational Modifications of Ribosomal Proteins
Post-translational modifications of RPs maintain the integrity and accuracy of the decoding machinery employed in protein translation. In the following sections, we will discuss the different post-translational modifications with respect to their stability, metabolism and biological effects.
1). RP Ubiquitination
The interaction between MDM2 and RPs modulates the protein levels and activity of RPs by post-translational modifications, including ubiquitination.19 The binding of MDM2 to L26 promotes the ubiquitination and proteasomal degradation of L26, disrupts the association of L26 with p53 mRNA, and inhibits the p53 protein synthesis mediated by L26.141 S7 is also a substrate for MDM2-mediated ubiquitination.86 The S7-ubiquitin fusion protein (S7-Ub) selectively inhibits MDM2-mediated p53 degradation and induces apoptosis better than unmodified S7.86 Of note, MDM2-mediated ubiquitination has no effect on the S7 protein level indicating that the ubiquitination of S7 by MDM2 does not target it for proteasomal degradation.86 S27a112 and S27/S27L109 are also targets of MDM2-mediated ubiquitination, but the ubiquitination of these proteins leads to their proteasomal degradation. These findings suggest that there is a feedback loop for the RPs-MDM2 interactions that regulates the magnitude and outcome of ribosomal stress.
2). RP Neddylation
The ubiquitin-like molecule, NEDD8 (Neural Precursor Cell Expressed, Developmentally Down-Regulated 8)-induced protein modification also plays an important role in regulating protein stability and activity.248 For instance, MDM2-mediated p53 neddylation inhibits its transcriptional activity.249 Intriguingly, the neddylation of RPs is required for p53 signaling in response to nucleolar stress.250 The MDM2-interacting RPs, such as S3, S7, and L11, are targets of the neddylation pathway.250 MDM2 neddylates L11 in the cytoplasm and stabilizes it, leading to its enhanced nucleolar localization. In the early stage of nucleolar stress, L11 is deneddylated by NEDD8-specific protease 1(NEDP1) and translocates from the nucleolus to the nucleoplasm, where it interacts with MDM2 and activates p53. Thus, nucleolar stress can trigger L11-mediated p53 activation that is dependent on NEDD8.27 In addition, L11 deneddylation allows it to be transiently recruited to promoter sites of p53-regulated genes, and promotes the access and binding of p53 transcriptional co-activators p300/CBP.251 However, prolonged deneddylation may induce the proteosomal degradation of L11.251 S14 is also specifically modified by NEDD8.93 hCINAP negatively regulates S14 neddylation by recruiting NEDP1.94 The decrease in S14 neddylation leads to reduced stability, incorrect localization, and an attenuated S14-MDM2 interaction, suggesting that hCINAP acts as an important regulator of the S14-MDM2-p53 pathway through the control of S14 neddylation.
3). RP Phosphorylation
Phosphorylation is one of the most common post-translational protein modifications, and is often required for functional activity of proteins. The small subunit, RPS3, is crucial for both translation initiation and the processing of DNA damage (functioning as a DNA endonuclease).13,252 Recent studies indicate that the functional switch for S3 between translation and DNA repair is regulated by its phosphorylation at different residues.253 Oxidative stress activates PKCδ (protein kinase C delta type), which phosphorylates S3 at Ser6 and Thr221, leading to its nuclear mobilization and DNA repair actvity.253,254 Upon NGF (nerve growth factor) stimulation, Akt interact with and phosphorylates S3 (Thr70), which disturbs its association with E2F1 and enhances S3 nuclear translocation, resulting in increased DNA repair activity and sustained neuronal survival.70 On the other hand, genotoxic stress induces the translocation of S3 to damage sites through ERK-mediated phosphorylation of S3 at Thr42.77,255 In addition, the phosphorylation of S3 mediates radioresistance in non-small cell lung cancer (NSCLC) cells.254 In those cells, ionizing radiation (IR) induces CK2α-mediated S3 phosphorylation (Thr221) and dissociation from TRAF2 (TNF receptor-associated factor 2), resulting in NFκB activation and upregulation of prosurvival genes. This is because S3 is a NFκB subunit and contributes to p65 DNA binding and specificity. The IKKβ (IκB kinase β) kinase-mediated phosphorylation of S3 (Ser209) is crucial for NFκB activation and anti-infective immunity.75
S6 is another example of a phospho-regulated RP.256 Mitogenic stimulation causes S6 C-terminal phosphorylation by p70 S6 kinases (S6Ks) and p90 ribosomal S6 kinases (RSKs) on Ser-235, Ser-236, Ser-240, and Ser-244.257 In addition, casein kinase 1 (CK1) mediates the phosphorylation of S6 on Ser-247.257 The phosphorylation of S6 is an important event that regulates cell growth in mammals. In quiescent mammalian cells, S6 phosphorylation is enhanced in the presence of mitogenic stimulation, growth factors, transforming agents, and carcinogenic chemicals, etc., while rapid dephosphorylation is caused by contact inhibition, nutrient starvation, heat shock or cellular stress.258 The overall outcome of S6 phosphorylation is to potentiate its mRNA cap-binding activity, but the physiological function of S6 phosphorylation remains enigmatic. Knock-in mice with mutations in all five phosphorylatable sites have been produced, and have provided further insights into the phosphorylation events.54,233 As a key player in glucose homeostasis in mice, S6 phosphorylation is necessary for regulating this process in some cell types (pancreatic β-cells and MEFs), but is expendable for the translational control of 5’-TOP mRNAs.58,259 In addition, S6 phosphorylation-deficient mice suffer from muscle weakness, which is thought to reflect impaired growth and reduced energy.260 Of note, mice lacking all five phosphorylatable sites in S6 develop fewer pancreatic cancer precursor lesions induced by DMBA (7,12-dimethylbenz(a)anthracene) or mutant K-ras.261 It was demonstrated that S6 phosphorylation attenuates K-ras-induced DNA damage and p53-mediated tumor suppression,261 indicating that the phosphorylation of S6 is important for the initiation of pancreatic cancer.
4). RP Sumoylation
The small ubiquitin-like modifier (SUMO)-mediated post-translational modification has an important regulatory role in many cellular functions, including cell cycle progression, DNA repair, and transcription.262 The nucleolar SUMO-specific protease 3, SENP3, is associated with several RPs (S4, S8, S11, and S18),263 but the functional roles of this modification are not fully understood. S3 is also a target for SUMO-1, and sumolyated S3 has increased stability compared to the unmodified protein,264 suggesting that sumoylation may represent another mechanism to protect RPs from proteolysis.
F. RPs, miRNAs and Nucleolar Stress
There is increasing evidence supporting the role of miRNAs in the regulation of the functions of RPs. It has been shown that L11 controls c-myc mRNA turnover via recruiting the miRISC (miRNA-induced silencing complex) in response to ribosomal stress.241 L11 can downregulate c-myc at the mRNA levels by binding to its 3’UTR, and recruits both miR-24 and the miRISC argonaute 2 (Ago2) core component to that region.241 The silencing of Ago2 abolishes this effect, while L11 knockdown rescues cells from miR-24-mediated c-myc mRNA reduction.241 Nucleolar stress-inducing agents, such as 5-FU or Act D, can enhance the associations of L11 with c-myc mRNA, miR-24, and Ago2. Recently, S14 has been suggested to induce c-Myc mRNA decay through the recruitment of Ago2 and miR-145, indicating that the association with the miRISC is probably a common feature of RPs that is related to regulating their mRNA stability and protein level and activity.92
Recent studies with RNA interference screening assays have suggested that reduced expression of RP genes (including L11) leads to the dissociation of miRNA complexes from target mRNAs, increasing the stability of the miRNA-targeted mRNAs, their polysome association and subsequent translation.265 These miRNAs co-sediment with ribosomes, and RP gene knockdown decreases their levels in monosomes, while increasing their target mRNAs in the polysomes. Chemical induction of nucleolar stress phenocopies RP gene knockdown, and this suggests that reduced RP gene expression regulates miRNA function through p53-dependent pathways. These findings indicate that the levels of RPs modulate miRNA-mediated repression of translation initiation, which provides a novel mechanism for the role of RPs in regulating protein translation and cell proliferation.
G. Crosstalk Between Nucleolar Stress and Other Stress Pathways
DNA damage can initiate nucleolar stress, but the mechanisms underlying this effect are not well known. A recent report suggested that genotoxic insults, such as UV (ultraviolet) irradiation and cytotoxic drugs (such as cisplatin), cause proteasome-mediated destruction of L37 in the nucleoplasm and L11-dependent stabilization of p53.149,266 These observations are validated by the fact that high levels of L37 decrease the p53-mediated DNA damage response. Thus, DNA damage-induced L37 degradation may be the link between DNA damage and the ribosomal stress pathway.149,266
In contrast, the tumor suppressor, ARF, senses oncogenic insults (such as Ras and c-Myc) and activates p53. It directly binds to L11, along with MDM2 and p53; suppressing MDM2 and enhancing the transcriptional activities of p53 and inducing cell cycle arrest.267 Silencing L11 attenuates ARF-induced cell cycle arrest and reduces p53 accumulation, demonstrating that ARF activates p53 by inducing nucleolar stress, which suggests that the ARF-MDM2-p53 and L11-MDM2-p53 pathways are functionally connected.
5. RIBOSOMAL PROTEINS AND THE PATHOGENESIS OF HUMAN DISEASES
Since RPs play a crucial role in the synthesis of proteins, which form the building blocks of tissues, disruptions in genes encoding RPs affect organogenesis, erythropoiesis and other physiological functions. Deficiencies in RPs lead to defects at distinct steps in ribosome biogenesis, resulting in a variety of disorders that affect different organs/systems. In the following sections, the discussion will be focused on the pathogenesis and therapeutic implications of these RP-associated disorders. The animal models used to study the roles of individual RPs in genetic diseases and ribosomopathies are described in Table II.
Table II.
Animal Models Used to Investigate RPs and Ribosomopathies
| RP | Species | Alteration | Phenotype | Mechanism | Ref. |
|---|---|---|---|---|---|
| S6 | Mouse | Conditional deletion (+/−) in liver cells | Cell cycle blockade in hepatocytes | Inhibits cyclin E mRNA expression | 9 |
| S6 | Mouse | Conditional deletion (+/−) in T cells | Cell cycle arrest in T cells | p53 pathway activation | 194 |
| S6 | Mouse | Deletion (+/−) in growing oocytes | Perigastrulation lethality | p53 pathway activation | 83 |
| S6 | Mouse | Phosphorylation site mutation | Smaller size, decreased pancreatic insulin secretion, and impaired glucose tolerance | Not studied | 58 |
| S7 | Mouse | ENU screening for mutation | Decreased body size, skeletal anomalies, midventral white spotting, and eye and central nervous system malformations | p53 pathway activation | 88 |
| S14 | Mouse | Conditional deletion (+/−) in hematopoietic progenitor cells | Hematopoietic progenitor cell deficiencies | p53 pathway activation | 102 |
| S19 | Mouse | Deletion (−/−) | Zygotes do not develop to normal blastocysts | Not studied | 105 |
| S20 | Mouse | Mutation (Dsk+/−) |
Increased pigmentation, erythrocyte hypoplasia | p53 stabilization stimulates Kit ligand expression | 103 |
| S7 | Zebrafish | Morpholino knockdown | Apoptosis, cell cycle arrest, impaired hematopoiesis | Activation of the p53 pathway and MMP family genes | 161 |
| S19 | Zebrafish | Morpholino knockdown | Erythropoietic failure | Activation of p53 family members (p53/ΔNp63/TAp73) | 191 |
| S29 | Zebrafish | Morpholino knockdown | Erythropoietic failure | Not studied | 162 |
| L22 | Mouse | Deletion (−/−) | Impaired αβ T cell development | Induction of p53 protein synthesis | 15 |
| L24 | Mouse | Mutation in C57BLKS Bst/+ mice | p53 pathway activated | p53 pathway activation | 268,269 |
| L27a | Mouse | Mutation in sooty foot ataxia (SFA) mice | Epidermal hyperpigmentation | p53 pathway activation | 270 |
| L29 | Mouse | Deletion (−/−) | Mild growth retardation | Decrease in global protein synthesis | 271 |
| L11 | Zebrafish | Morpholino knockdown | Defects in the development of hematopoietic stem cells (HSCs) and the maintenance of erythroid cells | p53 pathway activation | 192 |
| L38 | Zebrafish | Morpholino knockdown | Shorter body trunk | Not studied | 162 |
A. Developmental Disorders
Proper ribosome biogenesis maintains normal protein translation, which is essential for normal cellular functions. Therefore, the proteins required for ribosome assembly are vigorously and ubiquitously expressed. During times of rapid proliferation, such as embryonic development, the demand for functional ribosomes becomes even greater. Indeed, several studies in model organisms have underscored the importance of RPs during development.272 For instance, the disruptions of RP genes in Drosophila contribute to the “Minute” phenotype, which is characterized by slow development, absent or short bristles, poor fertility, and recessive lethality; these gene deficiencies lead to growth retardation by impairing the overall protein synthesis capacity of the ribosome.272 Knockdown of a large cohort of RP genes in developing zebrafish has demonstrated a variety of axis defects and abnormalities in the central nervous system (CNS) that can be attributed to the loss of individual RPs.162 For example, the knockdown of S15 leads to an enlarged fourth ventricle, and knockdown of S3, S5, S29, and L35a lead to brain deformities, while extreme body trunk abnormalities are seen in organisms with S29 knockdown.162 Meanwhile, in S7-deficient zebrafish embryos, p53 is activated, leading to apoptosis, cell cycle arrest, and impaired hematopoiesis.162 The matrix metalloproteinase (MMP) family genes are also activated in S7 mutants, indicating that there might also be improper migration of cells, further causing abnormal development.162 Concomitant knockdown of the p53 protein only partially reversed the abnormal phenotype, indicating that both p53-dependent and -independent mechanisms are involved in the onset of these developmental disorders.162
Homozygous mutations in RPs cause gross developmental defects and generally result in embryonic lethality.273 In zebrafish with RP mutations, defects in the endoderm-derived tissues are frequently noted in homozygous embryos.232 L23a and L6 are expressed in exocrine pancreatic progenitor cells and are essential for normal pancreas development. The haploinsufficiency of RP genes disrupts ribosome assembly, triggering ribosomal stress and inducing a p53-dependent cellular stress response.123 Surprisingly, in both L23 and L6 mutant embryos, the simultaneous knockdown of p53 is unable to restore the normal growth of pancreatic progenitor cells, suggesting that L23a and L6 have p53-independent roles in the physiological development of the pancreas. Altered liver and gut development in RPL mutants have also been reported, suggesting that developing endodermal tissues express high levels of RPL genes, and are very sensitive to the disruption of RPL gene function.123 It has been suggested that high levels of proliferation among pancreatic, liver, and gut progenitor cells may render these tissues highly sensitive to RPL disruption.123 Each of these affected tissues ultimately generate large numbers of adult cells, which are characterized by high-level protein synthesis and secretion, and the associated requirement for well-developed rough endoplasmic reticulum in these adult cell types, further making them susceptible to dysfunction in association with ribosome defects.123
In higher organisms such as mammals, mutations in RPs are typically associated with a wide array of specific abnormalities.274 One example comprises the tail-short (Ts) laboratory mice, which possess short, kinky tails and numerous skeletal abnormalities.274 L38 located within the Ts locus is altered in these mutants.274 Similarly, a deletion within L24 has been identified in mouse “Belly spot and tail (Bst)” mutants, which exhibit a kinked tail with a ventral midline spot.246 The expression of wild-type transgenes of L24 rescue these abnormalities.268
Heterozygous targeted mutant S6 mice exhibit embryonic lethality,83 while the mice with heterozygous targeted mutations of L22 or L29 can survive.15,271 On the other hand, homozygous targeted mutations of L22 cause αβ T-cell-specific developmental defects.15 Mice heterozygous for mutations in S19, S20, or L27a exhibit epidermal hyperpigmentation and anemia, along with decreases in body size, while the heterozygosity of L27a also causes cerebellar ataxia.103,275 The abnormal phenotypes of these heterozygous mutations can be rescued by the concomitant removal of one copy of TP53.162 S19-null mice are embryonic lethal prior to implantation, but mice heterozygous for the disrupted S19 allele have normal growth and organ development.105 The disruption of S7 results in reduced body size, skeletal malformations, mid-ventral white spotting (from severe developmental abnormalities in melanoblasts), and eye malformations, which can be attenuated by simultaneous TP53 deletion.88 Interestingly, S7 mutant mice do not show anemia,88 an abnormality commonly associated with RP mutations in humans. These observations support the existence of distinct functions of the RPs, and possible species-associated differences in their functions.6 S6, S7, S19, S20, and L27a are all critical for epidermal cell development;88,103,268,269,270 S7, L24, and L38 play essential roles in skeletal and neural development,88,269,276 and S14, S19, and S20 are necessary for hematopoiesis.95,103
RPs may also play an important role in the maintenance of cognitive functions.277–279 High L10 levels are observed in the murine hippocampus, a key site in the brain for promoting memory and learning. Interestingly, in humans, this gene is located at chromosome Xq28, an autism candidate region.278 Autism comprises a complex group of behavioral disorders characterized by impaired language and social skills. Indeed, two non-synonymous mutations of L10, L206M and H213Q have been detected in four males with autism.278 However, a larger study in human subjects has not found any causative mutations of L10 with respect to the susceptibility to autism.277–279
In model organisms, the phenotypes associated with RP mutation(s) appear to occur via three distinct mechanisms: 1) global protein synthesis suppression; 2) suppression of specific protein synthesis; and 3) extraribosomal functions. Diminished global protein synthesis has been identified in the L24Bst/+ phenotype, while S7Mtu/+ mice present with defective 18S rRNA preprocessing in the brain and liver, without any decrease in global protein synthesis.276 In S7Mtu/+ mice, there is impaired rRNA preprocessing along with p53 activation, indicating the intimidate association of p53 with the genesis and progression of RP-related disorders.88 Due to the essential and ubiquitous nature of RPs, it is believed that even slight disruptions in their functions can result in a wide range of developmental disorders.280
B. Cardiovascular and Metabolic Disorders
Several RPs have been implicated in the development and progression of cardiovascular diseases.281–283 For instance, the Minute syndrome in Drosophila, which is associated with RP haploinsufficiency, also affects the heart.281–283 The cardiomyopathy associated with the Minute syndrome is caused by haploinsufficiency for genes encoding cytoplasmic RPs. Specifically, the heart-specific knockdown of S15a severely impairs the cardiac function in adult Drosophila.283 RPs also have a role in the vascular system. For example, L17 acts as a vascular smooth muscle cell growth inhibitor and limits carotid intima thickening in mice.135 L17 expression inhibits the growth of vascular smooth muscle cells by arresting the cells at the G0/G1 phase.135 L17 is paralogous to the L23 protein, and L23 is known to activate p53 by inhibiting MDM2 in response to nucleolar stress. Thus, L17 may also act as a cell cycle inhibitor through p53 activation, although no evidence of its role in the p53 pathway has been reported.
Transgenic knock-in mice in which all five phosphorylatable serines for the S6 protein were substituted with alanines exhibit impaired pancreatic β-cell function and glucose homeostasis, along with muscle weakness.58,257,259 Pancreatic insufficiency is also manifested as an inability to secrete enough insulin, and thus, these mice have faulty glucose utilization. Interestingly, the global protein synthesis rate in MEFs derived from S6 P−/− mice is significantly higher than that in cells derived from wild-type mice.58 These observations suggest that S6 phosphorylation may be a positive regulator of the cell size and whole body glucose homeostasis, while being a negative regulator of global protein synthesis. However, the underlying mechanisms are not fully understood.
S6K, a S6 kinase, is intimately associated with several cardiovascular and metabolic functions. S6K is a major downstream target of mTOR (mammalian target of rapamycin).284 The mTOR signaling network regulates cell proliferation, growth, and survival, and is involved in metabolic regulation and tumor transformation. S6K is known to be rapidly and strongly activated after myocardial infarction, leading to pathological cardiac remodeling through myocardial infarction-enhanced Akt signaling.285 S6K1 (S6 kinase 1) is implicated in the pathogenesis of Type 2 diabetes, as S6K1 knockdown in mice leads to hypoinsulinemia.286 Similarly, rats treated with S6K1 antisense oligonucleotides exhibit reduced body weight gain and appetite, improved glucose utilization, and reduced fasting insulin levels.286 The depletion of hepatic S6K1 in db/db mice using S6K1 shRNA results in the downregulation of SREBP1c (sterol regulatory element binding protein-1c) gene expression in the liver, along with reduced hepatic and serum triglyceride concentrations.287 The deletion of S6K1 leads to an increased life span and resistance to aging-related disorders, such as motor dysfunction, bone disorders, and loss of insulin sensitivity.288 S6K1 inhibition also reduces adiposity, which inversely correlates with longevity in mice. Regular exercise in adolescent rats reduces the basal phosphorylation levels of S6 via a decrease in S6K1.288 However, as discussed earlier, S6K is also implicated in the mitogenic and nutrient-responsive PI3K-mTOR pathway, and S6K controls cell growth by regulating ribosome biogenesis at the translational level. Several of its effects may therefore not be directly related to S6 phosphorylation, but to a more general control of global protein synthesis via its effects on ribosome synthesis.
The disruption of one L13a allele confers resistance to lipid-induced oxidative apoptosis in CHO (Chinese hamster ovary) cells, indicating that L13a may be a potential therapeutic target for the treatment of hyperlipidemia-associated conditions.289 A recent study demonstrated that knockdown of L13a in macrophages abolished the translational termination of inflammatory chemokines.133 Interestingly, unlike S19 or S24, the depletion of L13a does not impair ribosome biogenesis or overall protein synthesis in human monocytes.290 Considering the fact that inflammation and lipidemic conditions are intimately linked, it is worthwhile to further investigate the specific role(s) of L13a in the progression of inflammatory conditions such as atherosclerosis.
C. Ribosomopathies
Ribosome dysfunction causes specific pathological conditions called ribosomopathies, a collection of rare genetic disorders characterized by macrocytic anemia in association with growth retardation and developmental abnormalities.23 Although ribosomes are active in all cell types, the predominant phenotype of ribosomopathy is a failure of erythropoiesis.29 A better understanding of the pathogenesis of ribosomopathies would help develop approaches to their early diagnosis and treatment.
1). Diamond Blackfan Anemia (DBA)
DBA is a congenital erythroid aplasia characterized by defective erythropoiesis, physical abnormalities such as short stature and cardiac defects, as well as an increased risk for cancer.24,291 It usually presents in infancy, with lethargy and pallor being the most common clinical symptoms.24 Approximately half of the DBA patients carry mutations and deletions in RP genes, such as those encoding S19,24 S24,292 S17,97–99 L5,293,294 L11,293,294 L26,295 L35a,148 S7,294 S10,296 S26,296 or S15297. The mutations observed in the RPs include nonsense, missense, frameshift, and splice site mutations. Mutations in S19, the most common mutations found in DBA, lead to defective pre-rRNA processing of the 18S rRNA, and reduce the 40S ribosomal subunit production.298 S19 deficiency in zebrafish results in a phenotype resembling DBA, with the suppression of neural differentiation through p53 and ∆Np63 deregulation.299 ∆Np63 is required for the specification of the non-neural ectoderm; its upregulation leads to craniofacial abnormalities during gastrulation. The induction of p53 causes p21 accumulation and subsequent cell cycle arrest in erythroid progenitor cells, leading to hypoproliferative anemia.102 Clinical evidence also suggests that mutations in the large ribosomal unit proteins are linked to specific physical abnormalities observed in DBA patients.148,293,294 L5 is associated with higher incidences of cleft lip/palate and cardiac anomalies associated with DBA, while isolated thumb abnormalities may be linked to L11 mutations.293,294 The haploinsufficiency of RPL proteins reduces the amount of 60S ribosome and the mature 80S ribosome, probably contributing to the bone marrow failure and potential cancer predisposition in DBA patients.148 The deletion of L11 in the zebrafish model causes defects in hematopoiesis193 and embryonic development192 through a p53-depedent mechanism.
2). 5q- Syndrome
The 5q- syndrome, a unique subtype of myelodysplastic syndrome (MDS) with del (5q) as the sole cytogenetic abnormality, is manifested as severe macrocytic anemia, normal/high platelets with dysplastic micromegakaryocytes, and rare progression to acute myeloid leukemia (AML), in contrast to other types of MDS.300 S14 haploinsufficiency has been identified as the major genetic abnormality in 5q-syndrome patients.25 S14 deficiency causes erythroid defects, with relative preservation of the other lineages. Conversely, the overexpression of S14 in bone marrow CD34+ cells from 5q- syndrome patients rescues their erythroid differentiation.25 S14 deficiency blocks the processing of 18S rRNA in human erythroleukaemia cells25 and recapitulates the macrocytic anemia of 5q- syndrome in mice.95 The deletion of miR-145/146a is responsible for other hematological phenotypes, such as thrombocytosis and megakaryocytic defects.301 The loss of S14 induces and activates p53, leading to p21-mediated cell cycle arrest and destruction of erythroid progenitor cells.95,102,302
3). Treacher Collins Syndrome
Treacher Collins syndrome (TCS) is a congenital autosomal dominant disorder associated with craniofacial development, including hypoplasia of the facial bones, particularly the lower mandible and cheek; abnormalities of the eyes; and alterations of the external ears.303 To date, more than 200 mutations, including deletions, insertions, nonsense mutations, and alternative splicing have been reported,304 all of them leading to a truncated Treacle protein.304 The Treacle protein is a putative nucleolar phosphoprotein that plays a role in rDNA transcription and 18S pre-rRNA methylation. All TCS patients are heterozygous for a Tcof1 (Treacher Collins-Franceschetti syndrome 1) mutation; and the disease manifestations are due to haploinsufficiency, not dominant negative effects. Intensive investigations in murine models have shown that Tcof1 is essential for the formation and proliferation of neural crest cells.305 In Tcof1+/− mice, the haploinsufficiency of Tcof1 disturbs ribosome biogenesis, resulting in p53-dependent apoptosis and cell cycle arrest in neuroepithelial cells.303 The surviving population proliferates more slowly than Tcof1 wildtype cells, further decreasing the number of neural crest cells, which leads to the characteristic hypoplasia seen in TCS.303 Since the Treacle protein is involved in ribosome synthesis and processing, a non-functional variants implies that there is likely a lack of mature ribosomes. This may be the reason for both the apoptosis and the slow growth rate. Supporting this idea, fewer mature ribosomes are also observed in the neuroepithelium of Tcof1+/− mice.303 However, the reason for the specific defects in the neuroepithelium is still unclear, since TCOF1 is expressed in various other tissues.
4). Shwachman Diamond Syndrome
Shwachman diamond syndrome (SDS) is a rare autosomal recessive disorder manifested as bone marrow failure, skeletal deformities, and exocrine pancreatic hypoplasia.306–308 Patients exhibit a predisposition to leukemia, and the symptoms are typically manifested in early infancy, including fat malabsorption, growth failure, and fat soluble vitamin deficiency (vitamins A, D, E and K).306–308 The most common hematological aberration observed is neutropenia. Up to 90% of the SDS patients exhibit biallelic mutations in the SBDS gene (named after Drs. Shwachman, Bodian and Diamond, who first described the syndrome) which is involved in ribosome synthesis and processing.308–310 Low expression of many RP genes, including S9, S20, L6, L15, L22, L23 and L29, and genes involved in rRNA and mRNA processing, is observed in SDS patients.23 An increase in Fas-mediated apoptosis is also seen, presumably leading to bone marrow failure.311 A recent study demonstrated the possibility that pancreatic tumors may be associated with SDS, thereby broadening the clinical phenotype of the disease.312 In vivo SDS models based on mammals, insects, and fish replicate the genetic and/or developmental aspects of ribosomopathies, and have led to the identification of pathways and candidate molecules that are important in the pathogenesis of the diseases.
D. Cancer
Impairments of ribosome biogenesis and protein translation have been shown to be associated with cancer.2,18,29,304 Disruption of one or several steps that control ribosome and protein synthesis can affect cell cycle progression and cell growth, leading to malignant transformation.2 In various cellular, animal, and clinical models, ribosome protein synthesis, as well as ribosome translation initiation, has been shown to be regulated by both oncogenes and tumor suppressor genes.2,18,29,304 Although clinical studies on the relationship between RP expression and human cancers are still limited, the available data indicate that the up- or downregulation of RPs can be seen in a variety of human cancers. In this section, we will focus on several representative pathways that demonstrate the importance of the RPs in cancer development and progression (Fig. 5).
Figure 5. Proposed models of the roles of RPs in cancer.
The ribosomal and extraribosomal functions of RPs are involved in carcinogenesis, cancer progression, and metastasis. (A) Aberrant ribosome biogenesis due to oncogene activation (such as c-Myc) causes uncontrolled cell growth and proliferation, increasing the risk of malignant transformation and carcinogenesis. On the contrary, p53 negatively regulates rRNA synthesis and ribosome biogenesis, protecting the cells from transformation and carcinogenesis. (B) The impairment of RPs-MDM2-p53 is linked to cancer progression. A MDM2 mutation in the central zinc finger (C305F) disrupts its interaction with L11, and significantly accelerates Eμ-Myc-induced lymphomagenesis in mice.
The oncogene and transcription factor Myc modifies several genes that are necessary for ribosomal assembly.29,313 Its overexpression drives the ribosome and protein biosynthesis in tumor cells, thereby initiating tumorigenesis via increased cell growth and proliferation.241,314 Myc facilitates rRNA transcription315 and processing316 and the production of other tralslation appartus compontents, including RPs.241 The direct link between Myc and RPs in carcinogenesis has been demonstrated using transgenic mouse models, which showed that the loss of one allele of L24 or L38 decreases the incidence of Eμ-Myc-driven lymphomagenesis and delays tumor onset.317 B lymphocytes heterozygous for L24 and overexpressing Myc show normal rates of total protein synthesis.317 It has been speculated that the haploinsufficiency of the L24 gene antagonizes Myc’s ability to induce tumors by inhibiting the translation of cyclin-dependent kinase 11 during mitosis and blocking the switch from CAP to IRES (internal ribosomal entry site)–dependent translation. However, when two alleles of L24 are present along with Myc overexpression, there is a considerable increase in protein synthesis during mitosis. Thus, even a moderate decrease in the expression of a single RP gene can hinder the ability of Myc to initiate tumorigenesis.317 In contrast, Pol I and Pol III transcription are repressed by p53 and retinoblastoma (Rb).234,318 In cancer cells that harbor inactivating mutations in p53 and Rb, deregulation of Pol I and Pol III activity can lead to tumorigenesis (Fig. 5A).318
The extraribosomal functions of RPs may be critically invovled in carcinogenesis. Many RPs were identified as haploinsufficient tumor suppressors in a cancer screening study in a zebrafish model.319 Heterozygous mutations in several RP genes (those encoding L35, S15a, S8, L36, L7, S7, L13, S29, and L23a) led to an increased risk of developing MPNSTs, a rare tumor type, in laboratory strains of zebrafish. These nerve sheath tumors are unable to efficiently synthesize p53 protein, even in the presence of p53 mRNA. Interestingly, a homozygous loss-of-function point mutation in the TP53 gene also leads to the same tumor type in zebrafish.232 Therefore, it is possible that an appropriate amount of RP expression is necessary for p53 activation and signaling;232 and haploinsufficiency of RPs may lead to disruption of this tumor suppressor pathway, ultimately leading to carcinogenesis.
How these RPs act as tumor suppressors, or in certain cases, as tumor-causing or promoting genes is unknown. The main function of the RPs is ribosome assembly and the maintenance of the efficiency and accuracy of translation. Defects in the RPs themselves will lead to impaired protein synthesis, and as such, the level of critical tumor suppressors may be decreased below a threshold level. This may lead to the cell not being protected from genotoxic and other insults, which can ultimately cause malignant transformation. Mutations or loss of certain RPs may dramatically affect the level or function of their binding partners (either tumor suppressors or oncogenes). A recent study provided direct evidence of the extraribosomal function of RPs in preventing cancer, demonstarting that inactivation of L22 predisposes cells to transformmation in vitro and leukemogenesis in vivo, and that the loss of L22 induces stemness factor Lin28B expression through a NFκB-dependent mechanism.136 Lin28B has been shown to increase cell proliferation and promote tumor growth,320,321 suggesting the involvement of a novel L22-NFκB-Lin28B signaling pathway in the development of T-ALL (T-acute lymphoblastic leukemia).320,321
The involvement of RPs in the regulation of the MDM2-p53 pathway is also linked to carcinogenesis and cancer progression (Fig. 5B). For example, the C305F missense mutation in the central zinc finger domain of MDM2 disrupts the interaction of MDM2 with L11 and L5, inhibiting its nuclear transport and proteasomal degradation, and promoting p53 degradation.322 In fact, transgenic mice with the MDM2 C305F mutant retain the normal p53 response to DNA damage, but do not show the p53 response to nucleolar stress.323 Indeed, perturbations in the RP-MDM2 interaction significantly increase the rate of Eμ-Myc-induced lymphomagenesis.323 Furthermore, the p53 response induced by nucleolar stress does not require p19ARF, suggesting that an RPs-MDM2-p53 interaction mediates an ARF-independent c-Myc-activated tumor suppression pathway.323
In addition to the p53 signaling pathway, RPs also influence tumorigenesis via the regulation of other important molecules. For example, S13 overexpression promotes multidrug resistance and decreases drug-induced apoptosis in gastric cancer cells.91 S13 downregulates p27 expression and CDK2 kinase activity, thus promoting the G1 to S phase transition, whereas S13 knockdown leads to the G1 arrest of gastric cancer cells.91 Similarly, L6 and L23 enhance the resistance to multiple chemotherapeutic agents. Downregulation of L6 reverses multidrug resistance (MDR) and sensitizes cells to adriamycin-induced apoptosis.121 Additionally, L23 enhances glutathione S transferase (GST) activity and the intracellular glutathione content in the cells.324
RPs have also been shown to contribute to radioresistance.76 In non-small cell lung cancer cells, ionizing radiation (IR) led to casein kinase 2α (CK2α)-mediated phosphorylation of S3, which induced the dissociation of the S3-TRAF2 complex and led to NFκB activation, resulting in a significant upregulation of prosurvival genes (cIAP1, cIAP2, and survivin).76 Another interesting observation is that the phosphorylation of S6 is increased in pancreatic acinar cells upon challenge with DMBA, a carcinogen, or transgenic expression of mutant K-ras.261 The development of pancreatic cancer precursor lesions is greatly reduced in S6P−/− mice, which are incapable of S6 phosphorylation.261
6. THE POTENTIAL VALUE OF RIBOSOMAL PROTEINS IN THE DIAGNOSIS AND TREATMENT OF HUMAN DISEASES
RPs constitute the functionally active part of the ribosome, in addition to acting as a scaffold that keeps proteins in position for optimal functioning. However, it is now clear that the RPs often perform auxiliary functions and act as “sentinels for self-evaluation of cellular health”.6 Thus, it is not surprising that even slight disruptions in RPs can lead to a wide array of pathologies. Due to their involvement in pathways that are tightly linked to cell growth, proliferation, and metabolism, RPs may serve as promising diagnostic and prognostic biomarkers and therapeutic targets for human diseases. In the following section, we focus on the implications of RPs in various disease states and their potential for diagnosing disease and as targets for treatment.
A. Ribosomal Proteins as Cancer Biomarkers
Differential expression of several RP genes is observed in different human disorders, especially in genetic diseases and cancer (Table III). It is known that aberrant ribosome synthesis contributes to increased cellular proliferation, but whether the differential expression of RPs is a causative factor or simply an associated feature in enhanced cell proliferation is unclear.
Table III.
Ribosomal Protein Expression and Human Cancers
| Cancer Type | RP(s) | Alteration | Implications for disease | Ref. |
|---|---|---|---|---|
| Liver cancer | S8, L12, L23a, L27, L30, L36, L36a | Upregulation | Increased L36 expression is associated with better survival | 8,325,326 |
| Gastric cancer | L15, S13, L6, L13 | Upregulation | Upregulation is associated with increased cell proliferation, drug resistance, and poor survival | 91,120,158,327 |
| Colorectal cancer | L13, S11, L7, L10a, L44, S19, L19 (feces), S27L (cancer tissues and feces) | Upregulation | Fecal L19 expression is associated with advanced disease while elevated S27L expression correlates with a better prognosis | 327–333 |
| Sa, S8, S12, S18, S24, L13a, L18, L28, L32, and L35a | Downregulation | Downregulation is associated with the ribosomes of the mucosal epithelia | ||
| Prostate cancer | S2, L19 | Upregulation | Elevated L19 expression is associated with advanced disease | 334–337 |
| Esophageal cancer | L14, L15, pS6 | Downregulation (L14); Upregulation (L15, pS6) |
Elevated levels of pS6 are associated with shorter survival and an adverse prognosis | 338–340 |
| Lung cancer | L22, pS6 | Downregulation (L22); Upregulation (pS6) |
Higher pS6 expression is associated with a shorter metastasis-free survival | 169,341 |
| Breast cancer | L41 | Downregulation | L41 downregulation is related to malignant transformation | 150 |
| Osteosarcoma | L7a | Downregulation | Downregulation of L7a is associated with poor survival of osteosarcoma patients with lung metastasis | 342 |
| Leukemia Lymphoma | S6, L23 | Upregulation | Elevated levels of L23 are associated with poor survival | 343,344 |
| Ovarian cancer | S4X | Upregulation | High expression of RPS4X is associated with a lower risk of death and later disease progression | 345 |
Differential display (DD) analyses demonstrated that the S8, L12, L23a, L27 and L30 mRNA levels are enhanced in human hepatocellular carcinoma (HCC) tissue samples and cell lines.325 Immunohistochemical analyses suggested that L36 is expressed in 75% of HCC patients, but is not detected in corresponding non-tumor liver tissues. The L36 level is higher in the early stages of cancer, and L36 expression is associated with a better overall survival, suggesting that it may represent an independent prognostic factor for HCC.326 L36a mRNA is preferentially overexpressed in 85% of HCC tissues and in all eight HCC cell lines in a previous study.8 The overexpression of L36a enhances colony formation and cell proliferation.8
L15 expression is markedly upregulated in gastric cancer cell lines and tissues.158 Inhibition of L15 expression suppresses gastric cancer cell growth in vitro and reduces tumorigenicity in vivo.158 S13 expression is also upregulated in human gastric cancer.91 S13 knockdown inhibits gastric cancer cell growth, with significant G1 cell cycle arrest, probably through the upregulation of p27.91 L6 is overexpressed in adriamycin-resistant SGC7901 gastric cancer cells.120 Similarly, L6 is also upregulated in gastric cancer tissues compared to normal gastric mucosa,120 and contributes to increased cell growth via cyclin E upregulation.120 Patients with low levels of L6 have better survival than those with higher levels, suggesting that L6 may be a potential prognostic biomarker for gastric cancer patients.120 The upregulation of L13 mRNA expression was observed in 10 of 36 (28%) gastric cancer tissue samples, and the upregulation of the L13 gene was associated with the clinical stage of the disease.327
The expression of RPs in colorectal cancer (CRC) differs from that seen in the normal mucosa.328 The RP genes overexpressed in CRC include the large subunit protein, L5, and the small subunit proteins, S3, S6, S8, and S12.329 Another study demonstrated differential expression of 12 RPs (Sa, S8, S11, S12, S18, S24, L7, L13a, L18, L28, L32, and L35a) in CRC compared with paired normal colonic mucosa.330 S11 and L7 were highly expressed, and all other ribosomal proteins were markedly decreased, in CRC tissue samples.330 Fecal L19 expression is associated with advanced tumor stages and addictive to serum CEA (carcinoembryonic antigen), and can be used to predict the prognosis of CRC patients.331 A fluorescent mRNA differential display analysis also identified several RPs (L10a, L44, S11, and S19) that are remarkably upregulated in human colon carcinoma.332 The upregulation of L13 mRNA expression was observed in 19 of 46 (41%) colorectal cancer tissue samples compared to adjacent normal tissue samples. Knocking down L13 expression using small interfering RNA (siRNA) results in a dramatic attenuation of colon cancer cell growth in vitro and decreased tumorigenicity in vivo.327 In contrast, recent evidence suggests that increased levels of S27L in either feces or cancer tissue are associated with a better prognosis. Thus, assessing the RP levels can be useful as a predictive index for disease progression and can enable the personalization of therapies, particularly in intermediate-stage CRC patients.333
Another tumor type exhibiting differential RP expression is prostate cancer, wherein the mRNA levels of L19 are elevated, contributing to reduced patient survival.334 The siRNA-mediated L19 knockdown abolishes aggressive phenotypes of the disease.335 Thus, L19 may serve as both a prognostic marker and a valid therapeutic target for prostate cancer. S2 is overexpressed in malignant prostate cancer cell lines and tissues, and has been reported to be a novel diagnostic marker for prostate cancer.336,337
Decreased expression of L14338 and overexpression of L15339 in esophageal cancer have previously been suggested. Additionally, hyperphosphorylation of S6 and an increased phosphorylated S6/S6 (pS6/S6) ratio were seen in esophageal squamous cell carcinoma patients, and were associated with a reduced disease-free survival.340 Importantly, knockdown of S6 and S6K1 increased cell death via the downregulation of cyclin D1, and also suppressed cell migration and invasion via the downregulation of ERK/JNK phosphorylation in esophageal cancer cells.340 These findings suggest that pS6 and the ratio of pS6/S6 are closely related to tumor progression and have prognostic significance in esophageal squamous cell carcinoma.340
The RPs appear to have roles in a wide variety of other cancer types as well. For example, the expression levels of L22 mRNA and protein have been shown to be significantly downregulated in NSCLC patients.341 In addition, high pS6 expression was suggested to be a negative prognostic factor for lung adenocarcinoma, because it was associated with the time-to-metastasis in patients with early stage lung adenocarcinoma.169 pS6 is also overexpressed in brain metastatic lesions of lung cancer.169 The downregulation of L41 mRNA was detected in 75% of primary breast cancers.150 Downregulation of L7a is associated with a poorer survival of osteocarcinoma patients with lung metastasis.342 The S6 protein is highly expressed in diffuse large B cell lymphomas (DLBCL), and knockdown of S6 leads to decreased proliferation of these cells in culture.343 L23 mRNA is overexpressed in higher-risk MDS patients, and elevated L23 expression is inversely associated with apoptosis in CD34+ bone marrow cells. The L23 level can serve as an independent prognostic indicator, regardless of the patient age, IPSS (international prognostic scoring system) score, or hemoglobin level. Thus, a higher level of L23 predicts disease progression and poor survival.344 A recent study indicated that high levels of X-linked RPS4 (RPS4X) correlated with reduced disease progression and a decreased risk of death in patients with high-grade serous epithelial ovarian cancer. Indeed, depletion of RPS4X reduces the cell growth rate and increases the resistance to cisplatin in ovarian cancer cell lines.345
B. Ribosomal Proteins as Molecular Targets for Drug Discovery and Development
Considering that aberrant ribosome biogenesis and protein translation are associated with cell growth and cell proliferation, hematological and neurological disorders, and an increased risk of cancer and other chronic diseases, it would be reasonable to target the components of the ribosome biogenesis and the translation machinery that are deregulated in specific disease states in order to develop novel targeted therapies. Below, we will discuss a few examples to illustrate the potential of RPs as novel drug targets.
1). Gene Therapy and Pharmacotherapy for Ribosomopathies
Because many DBA patients exhibit a deficiency in S19, gene therapy has been attempted using lentiviral vectors containing the S19 gene transduced into the bone marrow CD34+ cells from DBA patients.346 The enforced expression of the S19 transgene improves the proliferation of S19 mutant CD34+ cells and enhances the erythroid development of S19-deficient hematopoietic progenitors, suggesting the feasibility of gene therapy for S19-deficient DBA. Furthermore, a high level of S19 expression is required for correction of the erythroid development, and transplantation of unsorted S19-transduced CD34+ cells from mobilized peripheral blood led to successful gene therapy effects in vivo.346 These results indicate that targeted therapies modulating specific RPs in disease states may represent a new approach to treatment of this type of ribosomopathy.
The induction and activation of the p53 pathway are common in human disorders of defective ribosome biogenesis, such as DBA102 and 5q- syndrome.302 Lenalidomide (Len), a thalidomide derivative, stabilizes MDM2 and accelerates p53 degradation.347 Len treatment leads to increased phosphorylation of MDM2 on Ser-166/186, and disturbs the binding of S14, resulting in suppressed MDM2 autoubiquitination, restoring the function of MDM2 in 5q- syndrome, and overcoming the p53 activation.347 Thus, the inhibition of p53 in patients with ribosomopathies seems to be a promising therapeutic concept, since the typical disease phenotype arises due to improper p53 activation, which leads to the apoptosis of hematopoietic progenitor cells. However, it is important to note that long-term p53 inactivation may lead to an increased risk of various cancers. The inactivation of upstream regulators of p53, such as MDM2-interacting RPs (i.e., L11) could be possible alternatives to reduce this risk.348
2). Treatment of Neurological Disorders
S3 plays crucial roles in oxidative stress and DNA repair. Based on the protein transduction domain, PEP-1, researchers have developed a novel construct, PEP-1-S3, which has been shown to protect against experimental cerebral ischemic damage.349 In addition, the PEP-1-S3 construct could protect dopaminergic neurons from oxidative stress in an MPTP (1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine)-induced mouse model of Parkinsonism.350 PEP-1-S3 could be efficiently delivered to the substantia nigra, where it significantly inhibited the generation of reactive oxygen species (ROS) and DNA fragmentation, and improved cell survival. This observed neuroprotection was related to the antiapoptotic activity of PEP-1-S3, which suggests that it might represent a novel therapy for Parkinson’s disease.
3). Therapy for Inflammatory Diseases
Considering that S19 plays a role in the inflammatory response by binding to the pro-inflammatory cytokine macrophage migration inhibitory factor (MIF),104 researchers have sought to establish whether recombinant S19 can exert anti-inflammatory effects in a mouse model of anti-GBM (glomerular basement membrane) glomerulonephritis (GN), in which MIF is known to play an important role. GBM-GN mice treated with S19 showed the absence of glomerular crescents, glomerular necrosis, renal dysfunction and/or proteinuria. Interestingly, S19 was shown to block the upregulation of MIF and CD74 and inactivate ERK and NFκB signaling, thereby inhibiting macrophage and T-cell infiltration, Th1 and Th17 responses, and the expression of pro-inflammatory cytokines.351
4). Inhibition of rRNA Transcription and RP Expression for Cancer Therapy
Uncontrolled ribosome synthesis due to increased rRNA transcription by Pol I leads to increased cellular proliferation and cancer. An orally active small molecule, CX-5461, targets rRNA transcription and selectively kills B-lymphoma cells with little effect on the wild-type population.352 CX-5461 selectively inhibits only Pol I-driven transcription, causing nucleolar disruption and subsequent activation of the p53-dependent apoptotic signaling pathway. Additionally, CX-5461 inhibits rRNA synthesis and induces both autophagy and senescence, but not apoptosis, via a p53-independent mechanism in solid tumor cells.353 Thus, selectively inhibiting rRNA transcription could serve as a promising therapeutic strategy for cancer, in both hematological and solid tumors.353 Another small molecule compound, CX-3543, also inhibited Pol I transcription and induced apoptosis in cancer cells by causing the selective disruption of the nucleolin/rDNA complex.354
Silencing RP expression by RNAi techniques has been shown to inhibit the proliferation of human pancreatic,144 gastric,119 and prostate335 cancer cells both in vitro and in vivo. DNAZYM-1P, a 'ribozyme-like' oligonucleotide, has recently been developed to target the overexpression of S2. DNAZYM-1P decreased the S2 expression in malignant prostate cells with little effect on normal cells. DNAZYM-1P inhibited cell growth and induced apoptosis in prostate cancer cells. In vivo, DNAZYM-1P blocked tumor growth and metastasis, and eventually eradicated tumors. Moreover, DNAZYM-1P improved the disease-free survival of tumor-bearing mice in a dose-dependent manner.337
Another interesting study reported that recombinant L23a and L31 proteins cloned from the giant panda possessed anticancer activities, as shown by their ability to retard the growth and proliferation of human laryngeal carcinoma, Hep-2, and human hepatoma, HepG-2, cells.145,146
5). Modulation of the RPs-MDM2-p53 Pathway for Cancer Therapy
More recently, experimental cancer therapy targeting the S7-MDM2-p53 pathway has been reported.328 This study employed recombinant LZ-8 (rLZ-8) to inhibit precursor ribosomal RNA synthesis, and was shown to reduce polysome formation, triggering nucleolar stress and resulting in an increased binding of S7 to MDM2, leading to p53 activation.355 LZ-8, an immunomodulatory protein, was derived from the medicinal mushroom Ganoderma lucidum (Reishi or Ling Zhi). As noted above, rLZ-8 treatment activates p53, leading to p21 expression, G1 arrest, and the inhibition of cell growth in a p53-dependent manner. It also suppressed the growth of xenograft tumors in mice.355 In another report,356 adenovirus-mediated delivery of L23 was shown to inhibit the proliferation of gastric cancer cells harboring wildtype p53 in vitro and in vivo. Exogenous L23 stabilizes and activates p53 though the inhibition of the MDM2-p53 interaction, inducing cell cycle arrest and apoptosis.356
7. MITOCHONDRIAL RIBOSOMAL PROTEINS –A BRIEF DISCUSSION
This review has focused mainly on the cytoplasmic ribosomal proteins. However, the mitochondria have their own protein synthesis machinery. Mammalian mitochondrial ribosomes (55S) consist of small (28S) and large (39S) subunits.357,358 The 55S ribosome contains more than 75 mitochondrial ribosomal proteins (MRPs), encoded by nuclear genes that are imported into the mitochondria, where they are assembled into the mitochondrial ribosome. The small subunit of this ribosome contains 29 proteins, whereas the large subunit has about 50 proteins.
As expected, MRPs play key roles in the ribosomal structure and assembly, and also in mitochondrial protein translation. Knockdown of MRPs caused mitonuclear protein imbalances and impaired mitochondrial respiration. Similar to the cytoplasmic RPs, MRPs also possess several extra-ribosomal functions unrelated to mitochondrial translation. Several MRPs (MRPS29,359,360 MRPS30,361,362 MRPL37,363 MRPL41,364) have also been reported play a role in apoptosis. For example, MRPL41 physically interacts with Bcl-2, which may contribute to its pro-apoptotic activities.364 Studies also reported that MRPL41 stabilizes the p53 protein to induce apoptosis.365 Interestingly, MRPL41 also causes p53-independent G1 phase cell cycle arrest via stabilization of p27.365 MRPL41 has also been shown to stabilize p21 and arrest cells in the G1 phase under conditions of serum starvation.366
MRPL41 is downregulated in breast and kidney cancer cell lines/tissues, suggesting that it may have tumor-suppressor properties.367 The overexpression of the MRPS36 retards cell proliferation via the induction of p21 expression and phosphorylation of p53.368 Contrarily, the overexpression of the MRPS18-2 protein leads to the immortalization and dedifferentiation of primary rat embryonic fibroblasts, thus indicating that it has a potential role in tumorigenesis.369 MRPL59 (CRIF1, CR6-interacting factor 1) has been identified as a transcription co-factor that interacts with Gadd45 and negatively regulates cell growth and cell cycle progression.370 MRPL58 (ICT1, immature colon carcinoma transcript 1) is essential for cell viability. Knockdown MRPL58 inhibits cell proliferation and decreases the mitochondrial membrane potential and mass, and also decreases the cytochrome c oxidase activity.371
Recently, Chinese researchers have established a link between the overexpression of MRPS12 and the proliferation and migration of gastric cancer cells.372 Studies have also identified MRPs with major mutations leading to mitochondrial translation deficiencies and lethality. Abnormal expression of MRPs is also observed in various cancers, such as gliomas, breast cancer, squamous cell carcinoma, and osteosarcoma.370,373,374 For example, the expression of MRPL59 is dramatically reduced in adrenal adenoma and papillary thyroid cancer.370 SNPs in the MRPS30 gene were shown to correlate with breast cancer in both Caucasian and Chinese populations, especially in the older subjects.373,374
8. CONCLUSIONS AND FUTURE DIRECTIONS
Our understanding of the ribosome and RPs based on information obtained during the past two decades has considerably improved. RPs were initially thought to be “housekeeping” genes, but are now considered to have diverse extraribosomal functions, including roles in cellular functions ranging from cell cycle progression to cell death, to malignant transformation and cellular metabolism. Another important extraribosomal function includes the surveillance of ribosome synthesis. Importantly, an aberration or deregulation in any of these processes may drive malignant transformation and lead to an abnormal cellular phenotype.375 The discovery of extraribosomal functions of the ribosomal components adds an additional layer of complexity to the relationship between dysregulated ribosome biogenesis and disease states such as cancer. In fact, it is difficult to distinguish the primary from secondary effects of mutant RPs. Critically, one must appreciate that the nucleolar proteome is not static, but exhibits dynamic responses when stimulated with physiological and pathological stresses, such as nutrient and growth factor starvation. These environmental factors regulate the nucleolar localization and trafficking of many proteins through controlled sequestration and release. Disturbances in the normal nucleolar function and structure lead to disruption of this regulation, and as a consequence, affect multiple cellular functions. In fact, several pathways that modulate Pol I transcription during ribosome biogenesis/protein synthesis are subject to regulation by the nucleolus. Thus, the nucleolus is both a target of cancer signaling, and also an upstream regulator of pathways important for normal cell growth and function. There is frequent cross-talk between the components of the ribosome, especially the ribosomal proteins, and other signaling pathways; these proteins undergo numerous modifications in response to stress and physiological demands.376
The disruption of ribosome biogenesis is an important causal factor in the induction of ribosomal stress. It affects the ultimate fate of the cell, and may result in cell cycle arrest or apoptosis via a p53-mediated pathway. Indeed, numerous reports have emphasized the importance of the RPs-MDM2-p53 interplay, which helps to maintain the integrity of ribosome biogenesis, and ultimately, normal cellular development and function. Studies on human ribosomopathies, as well as animal models, have highlighted the role of the p53 pathway in the clinical manifestations of many human ribosomal disorders, such as DBA and 5q-syndrome. It appears that p53 activation is a common cellular response, but the consequences of this response vary in different cell types, probably due to the differences in the tissue-specific activation of p53’s downstream targets.
The fact that the dysregulation of p53 is an important causal factor in more than half of human cancers, and the involvement of ribosomal stress in the activation and regulation of p53 in relation to cell cycle control, makes studies of the RP-MDM2-p53 pathways pivotal to understanding the mechanisms of carcinogenesis, and for the development of novel treatments for cancer. We have already discussed how several RPs, such as L11, L5, L23, L26, L6, etc., modulate the MDM2-p53 pathway, in some cases playing redundant roles. Owing to the large number of RPs and their similar functions, one should be careful when evaluating the roles of RPs as molecular biomarkers and therapeutic targets for human disease.
Further investigations will be required to reveal the regulatory network associated with this signaling cascade. For instance, the exact mechanisms that control RP shuttling between the nucleolus and nucleoplasm after ribosomal stress are not clear. The interacting partners for MDM2-binding RPs have also not yet been identified. In addition, RPs are short-lived proteins that need to be post-translationally modified to ensure their stability and nuclear transport. The modifications of RPs that are involved in the RPs-MDM2-p53 pathway deserve further exploration. Interestingly, the HBx (Hepatitis B virus X) oncoprotein confers nucleolar stress-induced p53 expression by disrupting the interaction between L11 and MDM2,377 suggesting the possible inactivation of the L11-MDM2-p53 pathway by the HBx oncoprotein in hepatocellular carcinogenesis. However, the relationship between defective ribosome biogenesis and cancer is complicated and not fully understood. The altered expression of some RPs seems to be a common feature in many cancers, however, it is unclear whether increased ribosome activity due to increased cellular proliferation leads to RP overexpression, or whether these RPs are actually responsible for the cell transformation. Although changes in RP levels have been suggested as potential biomarkers for cancer prognosis, most of these studies were descriptive and lacked mechanistic insights. Further studies will therefore be needed to reveal the functional roles of distinct sets of RPs that are deregulated in specific cancer types.
Furthermore, it is not clear how the MDM2-binding RPs modulate MDM2’s E3 ubiquitin ligase activity. For example, it is unclear whether MDM2-binding RPs function individually or as a subribosomal complex in response to nucleolar stress. The emerging findings indicate that the pathways linking ribosome defects and p53 signaling are more complicated than previously considered, and it is most likely that multiple players are involved in these interactions. Finally, how individual RPs perform non-redundant roles in each stage of the ribosomal stress response is also a topic that needs to be addressed.
More recent evidence suggests that the RP-associated p53 induction in response to ribosomal stress is independent of the RPs-MDM2 interaction. A direct link between RP deficiency and impaired p53 protein synthesis has been established in the absence of MDM2.123,228 In addition, p53-independent cell cycle arrest mechanisms in response to ribosome biogenesis have also been described. Moreover, the role of RPs in regulating proteins other than p53 may reflect their diverse extraribosomal functions, and represent distinct outcomes of ribosomal stress. Therefore, future studies are needed to explore the putative extraribosomal functions of RPs in regulating p53-independent pathways.
The differential expression of RPs in several cancer types makes them attractive candidates that may serve as noninvasive biomarkers for cancer. Ribosome biogenesis appears to be an attractive target for cancer prevention or therapy. Supporting this possibility, the activation of p53 by nucleolar stress has been shown during the treatment of B-cell lymphoma in mice. The newly appreciated RPs-MDM2-p53 pathway also offers an opportunity for anticancer drug development. For example, it is now recognized that targeting both MDM2 and MDMX presents a more effective approach towards p53 activation and subsequent tumor growth suppression.378 An ideal anticancer drug candidate would be a small molecule that could bind to the zinc finger domain of MDM2 and prevent its interaction with p53 in a manner similar to L11. This hypothetical molecule could potentially activate p53 by deactivating both MDM2 and MDMX, since turning on the nucleolar stress-RP-p53 pathway can simultaneously deactivate both MDM2 and MDMX. Thus, designing lead compounds that mimic the ability of RPs to bind to MDM2 could open a door for the future development of novel therapeutic agents.
In addition, RPs have a role in the metabolic control of the cell, mainly via the S6 kinase/mTOR signaling pathways. RPs are also critical in organogenesis; it will be important to investigate their roles in organ growth, metabolic control, and interactions with growth factors in order to obtain a better understanding of their roles in maintaining homeostasis.
Considering that RPs are associated with numerous signaling pathways and regulate, as well as are regulated by, various genetic and epigenetic factors, future studies are needed to explore the role of RPs under normal and pathological conditions with respect to both the p53-dependent and –independent pathways. It is hoped that a better understanding of these intricate pathways and the complex interplay between RPs and various signaling pathways will provide new strategies for the diagnosis, prevention, treatment and monitoring of RP-associated human diseases.
ACKNOWLEDGEMENTS
The contents of the paper are solely the responsibility of the authors, and do not necessarily represent the official views of the National Institutes of Health or other funding agencies. R.Z. was supported by NIH grants R01 CA112029, R01 CA121211, and R01 CA186662. We thank the current and former members of our laboratories for their contributions to the publications cited in this work. We thank Dr. Jiang-Jiang Qin and Mr. Sukesh Voruganti for helpful discussions during the preparation of this manuscript. The research field of ribosomal proteins reviewed in this article is rapidly expanding; we apologize for not being able to cite all of the references published in the recent years, due to space limitations.
Biographies
Wei Wang is an Assistant Professor in the Department of Pharmaceutical Sciences, School of Pharmacy, Texas Tech University Health Sciences Center (TTIHSC), Amarillo, TX. USA. She obtained her M.D. in Clinical Medicine from Changzhi Medical College, Changzhi, China and her Ph.D. in Molecular Pharmacology from Fudan University, Shanghai, China. Prior to joining the faculty at TTUHSC, she completed her post-doctoral training and also worked as a Research Associate at the University of Alabama School of Medicine, Birmingham, AL, USA. With over 10 years of post-doctoral experience in the field of cancer research, particularly in the area of cancer biology and experimental therapeutics, her research has been continuously focused on molecular cancer therapy. Her particular interests include cancer cell signaling pathways, the development of experimental therapeutics and the evaluation of their mechanisms of action.
Subhasree Nag is a Ph.D. candidate in the Department of Pharmaceutical Sciences, School of Pharmacy, Texas Tech University Health Sciences Center, Amarillo, TX. USA. She completed her master’s degree at the Institute of Chemical Technology, Mumbai, India. Her research focuses on the preclinical pharmacology of MDM2 inhibitors as anticancer agents.
Xu Zhang is a Post-doctoral Research Associate in the Department of Pharmaceutical Sciences, School of Pharmacy, Texas tech University Health Sciences Center (TTUHSC), Amarillo, TX. USA. He obtained his M.D. and Ph.D. in Clinical Laboratory Medicine from Jiangsu University Zhenjiang, China. During his graduate studies, he was an international scholar at TTUHSC. His research focuses on the signaling pathways in cancer cells and their roles in carcinogenesis and cancer progression.
Ming-Hai Wang is Professor and Amarillo Community Endowed Chair, Department of Biomedical Sciences, and the Director of Cancer Biology Center, School of Pharmacy, Texas Tech University Health Sciences Center, Amarillo, TX. USA. He obtained his M.D. from Zhejiang University School of Medicine, Hangzhou, China, and his Ph.D. in Immunology from Zhejiang University School of Medicine, Hangzhou, China/Forschung-sinstitut of Borstel, the Medical University of Luebeck, Luebeck, F.R. Germany (a joint Ph.D. program). He completed his postdoctoral fellowship in the Department of Immunology and Cell Biology, the Medical University of Luebeck, Luebeck, F.R. Germany and was a Visiting Associate in the Laboratory of Immunobiology at the National Cancer Institute, National Institutes of Health, Bethesda, MD. USA. Prior to joining TTUHSC, he was an Assistant Professor at Wayne State University School of Medicine, Detroit, Michigan, USA and Associate Professor of Medicine at the University of Colorado School of Medicine, Denver, CO. USA. His research interest is in the field of cancer biology and carcinogenesis, with a major focus on receptor tyrosine kinases and antibody targeted therapy.
Hui Wang is a Principal Investigator, Professor, the Head of the Laboratory of Molecular Pharmacology and Toxicology, and the Director of Food Safety Research Center at the Institute for Nutritional Sciences (INS), Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, China. She obtained her Ph.D. from Tianjin Medical University/School of Medicine, the University of Alabama at Birmingham (a joint PhD program). She continued her post-doctoral fellowship at the University of Alabama School of Medicine, Birmingham, AL, USA, where she joined the faculty in the Department of Pharmacology and Toxicology, Division of Clinical Pharmacology as an Instructor and was promoted to Assistant Professor. She joined the INS in October 2005. Her research interests include cancer biology, cancer prevention, drug discovery and molecular pharmacology and toxicology, as well as nutrition, environmental health and disease prevention, and applied nutriceuticals and food safety.
Jianwei Zhou is a Professor in the Department of Molecular Cell Biology and Toxicology, School of Public Health, and Dean of the Graduate School, Nanjing Medical University, Nanjing, China. He obtained his M.D. in Public Health and his Ph.D. in Occupational and Environmental Health at Nanjing Medical College, Nanjing, China. He was an exchange scholar at the University of California, Davis, CA, USA between 1996 and 1998. His research interests include molecular and cellular biology, cancer biology, environmental toxicology and experimental therapy.
Ruiwen Zhang is a Professor in the Department of Pharmaceutical Sciences, School of Pharmacy, Texas Tech University Health Sciences Center (TTUHSC), Amarillo, TX. USA. He obtained his M.D. and Ph.D. in Toxicology from Fudan University Shanghai Medical College, Shanghai, China. He completed his Postdoctoral/Clinical Pharmacology Fellowship at the University of Alabama School of Medicine, Birmingham, AL. USA, where he then joined the faculty in Clinical Pharmacology in the Department of Pharmacology and Toxicology as an Assistant Professor and was promoted to Professor and Director of the Cancer Pharmacology Laboratory. He is certified by the American Board of Toxicology (D.A.B.T.) and was on the Board of Directors from 2009 until 2013. Dr. Zhang was elected as a Fellow of the American Association for the Advancement of Science (AAAS) in 2009. His major research interests include translational medicine, cancer biology, gene silencing, drug discovery, experimental therapy, and clinical pharmacology and toxicology, focusing on oncogenes and tumor suppressor genes.
REFERENCES
- 1.Lafontaine DL, Tollervey D. The function and synthesis of ribosomes. Nat Rev Mol Cell Biol. 2001;2(7):514–520. doi: 10.1038/35080045. [DOI] [PubMed] [Google Scholar]
- 2.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3(3):179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 3.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40(2):216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3(6):577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 5.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem Sci. 1996;21(5):164–165. [PubMed] [Google Scholar]
- 6.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34(1):3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindstrom MS, Zhang Y. Ribosomal protein S9 is a novel B23/NPM-binding protein required for normal cell proliferation. J Biol Chem. 2008;283(23):15568–15576. doi: 10.1074/jbc.M801151200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH, You KR, Kim IH, Cho BH, Kim CY, Kim DG. Over-expression of the ribosomal protein L36a gene is associated with cellular proliferation in hepatocellular carcinoma. Hepatology. 2004;39(1):129–138. doi: 10.1002/hep.20017. [DOI] [PubMed] [Google Scholar]
- 9.Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288(5473):2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 10.He H, Sun Y. Ribosomal protein S27L is a direct p53 target that regulates apoptosis. Oncogene. 2007;26(19):2707–2716. doi: 10.1038/sj.onc.1210073. [DOI] [PubMed] [Google Scholar]
- 11.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560(1–3):81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 12.Hegde V, Wang M, Deutsch WA. Human ribosomal protein S3 interacts with DNA base excision repair proteins hAPE/Ref-1 and hOGG1. Biochemistry. 2004;43(44):14211–14217. doi: 10.1021/bi049234b. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Chubatsu LS, Admon A, Stahl J, Fellous R, Linn S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J Biol Chem. 1995;270(23):13620–13629. doi: 10.1074/jbc.270.23.13620. [DOI] [PubMed] [Google Scholar]
- 14.Flygare J, Kiefer T, Miyake K, Utsugisawa T, Hamaguchi I, Da Costa L, Richter J, Davey EJ, Matsson H, Dahl N, Wiznerowicz M, Trono D, Karlsson S. Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia. Blood. 2005;105(12):4627–4634. doi: 10.1182/blood-2004-08-3115. [DOI] [PubMed] [Google Scholar]
- 15.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26(6):759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Da Costa L, Narla G, Willig TN, Peters LL, Parra M, Fixler J, Tchernia G, Mohandas N. Ribosomal protein S19 expression during erythroid differentiation. Blood. 2003;101(1):318–324. doi: 10.1182/blood-2002-04-1131. [DOI] [PubMed] [Google Scholar]
- 17.Zhan Y, Melian NY, Pantoja M, Haines N, Ruohola-Baker H, Bourque CW, Rao Y, Carbonetto S. Dystroglycan and mitochondrial ribosomal protein L34 regulate differentiation in the Drosophila eye. Plos One. 2010;5(5):e10488. doi: 10.1371/journal.pone.0010488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Lu H. Signaling to p53: Ribosomal proteins find their way. Cancer Cell. 2009;16(5):369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Liao JM, Liao WJ, Lu H. Scission of the p53-MDM2 Loop by Ribosomal Proteins. Genes Cancer. 2012;3(3–4):298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21(13):4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G. Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol. 2009;11(4):501–508. doi: 10.1038/ncb1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun XX, Wang YG, Xirodimas DP, Dai MS. Perturbation of 60 S ribosomal biogenesis results in ribosomal protein L5- and L11-dependent p53 activation. J Biol Chem. 2010;285(33):25812–25821. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115(16):3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, Tentler D, Mohandas N, Carlsson B, Dahl N. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21(2):169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 25.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451(7176):335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilkes DM, Chen L, Chen J. MDMX regulation of p53 response to ribosomal stress. EMBO J. 2006;25(23):5614–5625. doi: 10.1038/sj.emboj.7601424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundqvist A, Liu G, Mirsaliotis A, Xirodimas DP. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009;10(10):1132–1139. doi: 10.1038/embor.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13(6):355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shenoy N, Kessel R, Bhagat TD, Bhattacharyya S, Yu Y, McMahon C, Verma A. Alterations in the ribosomal machinery in cancer and hematologic disorders. J Hematol Oncol. 2012;5:32. doi: 10.1186/1756-8722-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29(30):4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 31.Miliani de Marval PL, Zhang Y. The RP-Mdm2-p53 pathway and tumorigenesis. Oncotarget. 2011;2(3):234–238. doi: 10.18632/oncotarget.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shcherbik N, Pestov DG. Ubiquitin and ubiquitin-like proteins in the nucleolus: multitasking tools for a ribosome factory. Genes Cancer. 2010;1(7):681–689. doi: 10.1177/1947601910381382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakraborty A, Uechi T, Kenmochi N. Guarding the 'translation apparatus': defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip Rev RNA. 2011;2(4):507–522. doi: 10.1002/wrna.73. [DOI] [PubMed] [Google Scholar]
- 34.Thomson E, Ferreira-Cerca S, Hurt E. Eukaryotic ribosome biogenesis at a glance. J Cell Sci. 2013;126(Pt 21):4815–4821. doi: 10.1242/jcs.111948. [DOI] [PubMed] [Google Scholar]
- 35.Wilson DN, Doudna Cate JH. The structure and function of the eukaryotic ribosome. Cold Spring Harb Perspect Biol. 2012;4(5):a011536. doi: 10.1101/cshperspect.a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science. 2011;334(6058):941–948. doi: 10.1126/science.1211204. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334(6062):1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- 38.Doudna JA, Rath VL. Structure and function of the eukaryotic ribosome: the next frontier. Cell. 2002;109(2):153–156. doi: 10.1016/s0092-8674(02)00725-0. [DOI] [PubMed] [Google Scholar]
- 39.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14(3):313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 40.Ciganda M, Williams N. Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA. 2011;2(4):523–533. doi: 10.1002/wrna.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39(2):196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24(16):2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choesmel V, Fribourg S, Aguissa-Toure AH, Pinaud N, Legrand P, Gazda HT, Gleizes PE. Mutation of ribosomal protein RPS24 in Diamond-Blackfan anemia results in a ribosome biogenesis disorder. Hum Mol Genet. 2008;17(9):1253–1263. doi: 10.1093/hmg/ddn015. [DOI] [PubMed] [Google Scholar]
- 44.Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, Karlsson S, Ellis SR. Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood. 2007;109(3):980–986. doi: 10.1182/blood-2006-07-038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Idol RA, Robledo S, Du HY, Crimmins DL, Wilson DB, Ladenson JH, Ladenson JH, Bessler M, Mason PJ. Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production. Blood Cells Mol Dis. 2007;39(1):35–43. doi: 10.1016/j.bcmd.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 46.O'Donohue MF, Choesmel V, Faubladier M, Fichant G, Gleizes PE. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J Cell Biol. 2010;190(5):853–866. doi: 10.1083/jcb.201005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35(5):260–266. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gould HJ, Martini OH, King HS. 80S ribosomal proteins. Biochem J. 1972;129(3):31P–32P. doi: 10.1042/bj1290031p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korobeinikova AV, Garber MB, Gongadze GM. Ribosomal proteins: structure, function, and evolution. Biochemistry (Mosc) 2012;77(6):562–574. doi: 10.1134/S0006297912060028. [DOI] [PubMed] [Google Scholar]
- 50.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289(5481):920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 51.Barth-Baus D, Stratton CA, Parrott L, Myerson H, Meyuhas O, Templeton DJ, Landreth GE, Hensold JO. S6 phosphorylation-independent pathways regulate translation of 5'-terminal oligopyrimidine tract-containing mRNAs in differentiating hematopoietic cells. Nucleic Acids Res. 2002;30(9):1919–1928. doi: 10.1093/nar/30.9.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwartz EI, Intine RV, Maraia RJ. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5'-terminal oligopyrimidine mRNA metabolism. Mol Cell Biol. 2004;24(21):9580–9591. doi: 10.1128/MCB.24.21.9580-9591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brenet F, Socci ND, Sonenberg N, Holland EC. Akt phosphorylation of La regulates specific mRNA translation in glial progenitors. Oncogene. 2009;28(1):128–139. doi: 10.1038/onc.2008.376. [DOI] [PubMed] [Google Scholar]
- 54.Caldarola S, De Stefano MC, Amaldi F, Loreni F. Synthesis and function of ribosomal proteins--fading models and new perspectives. FEBS J. 2009;276(12):3199–3210. doi: 10.1111/j.1742-4658.2009.07036.x. [DOI] [PubMed] [Google Scholar]
- 55.Wolin SL, Cedervall T. The La protein. Ann Rev Biochem. 2002;71:375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- 56.Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30(4):460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 57.Vogt PK. PI 3-kinase, mTOR, protein synthesis and cancer. Trends Mol Med. 2001;7(11):482–484. doi: 10.1016/s1471-4914(01)02161-x. [DOI] [PubMed] [Google Scholar]
- 58.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19(18):2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasler P, Brot N, Weissbach H, Parnassa AP, Elkon KB. Ribosomal proteins P0, P1, and P2 are phosphorylated by casein kinase II at their conserved carboxyl termini. J Biol Chem. 1991;266(21):13815–13820. [PubMed] [Google Scholar]
- 60.Lam YW, Lamond AI, Mann M, Andersen JS. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol. 2007;17(9):749–760. doi: 10.1016/j.cub.2007.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spence J, Gali RR, Dittmar G, Sherman F, Karin M, Finley D. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell. 2000;102(1):67–76. doi: 10.1016/s0092-8674(00)00011-8. [DOI] [PubMed] [Google Scholar]
- 62.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 63.Blobel G, Potter VR. Studies on free and membrane-bound ribosomes in rat liver. I. Distribution as related to total cellular RNA. J Mol Biol. 1967;26(2):279–292. doi: 10.1016/0022-2836(67)90297-5. [DOI] [PubMed] [Google Scholar]
- 64.Andrews TM, Tata JR. Protein synthesis by membrane-bound and free ribosomes of secretory and non-secretory tissues. Biochem J. 1971;121(4):683–694. doi: 10.1042/bj1210683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lerner RS, Seiser RM, Zheng T, Lager PJ, Reedy MC, Keene JD, Nicchitta CV. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 2003;9(9):1123–1137. doi: 10.1261/rna.5610403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 67.Stephens SB, Dodd RD, Brewer JW, Lager PJ, Keene JD, Nicchitta CV. Stable ribosome binding to the endoplasmic reticulum enables compartment-specific regulation of mRNA translation. Mol Biol Cell. 2005;16(12):5819–5831. doi: 10.1091/mbc.E05-07-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bolze A, Mahlaoui N, Byun M, Turner B, Trede N, Ellis SR, Abhyankar A, Itan Y, Patin E, Brebner S, Sackstein P, Puel A, Picard C, Abel L, Quintana-Murci L, Faust SN, Williams AP, Baretto R, Duddridge M, Kini U, Pollard AJ, Gaud C, Frange P, Orbach D, Emile JF, Stephan JL, Sorensen R, Plebani A, Hammarstrom L, Conley ME, Selleri L, Casanova JL. Ribosomal protein SA haploinsufficiency in humans with isolated congenital asplenia. Science. 2013;340(6135):976–978. doi: 10.1126/science.1234864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi S, Jung CR, Kim JY, Im DS. PRMT3 inhibits ubiquitination of ribosomal protein S2 and together forms an active enzyme complex. Biochim Biophys Acta. 2008;1780(9):1062–1069. doi: 10.1016/j.bbagen.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 70.Lee SB, Kwon IS, Park J, Lee KH, Ahn Y, Lee C, Kim J, Choi SY, Cho SW, Ahn JY. Ribosomal protein S3, a new substrate of Akt, serves as a signal mediator between neuronal apoptosis and DNA repair. J Biol Chem. 2010;285(38):29457–29468. doi: 10.1074/jbc.M110.131367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hegde V, Yadavilli S, Deutsch WA. Knockdown of ribosomal protein S3 protects human cells from genotoxic stress. DNA Repair (Amst) 2007;6(1):94–99. doi: 10.1016/j.dnarep.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 72.Hegde V, Wang M, Deutsch WA. Characterization of human ribosomal protein S3 binding to 7,8-dihydro-8-oxoguanine and abasic sites by surface plasmon resonance. DNA Repair (Amst) 2004;3(2):121–126. doi: 10.1016/j.dnarep.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 73.Wan F, Anderson DE, Barnitz RA, Snow A, Bidere N, Zheng L, Hegde V, Lam LT, Staudt LM, Levens D, Deutsch WA, Lenardo MJ. Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell. 2007;131(5):927–939. doi: 10.1016/j.cell.2007.10.009. [DOI] [PubMed] [Google Scholar]
- 74.Yadavilli S, Mayo LD, Higgins M, Lain S, Hegde V, Deutsch WA. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst) 2009;8(10):1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wan F, Weaver A, Gao X, Bern M, Hardwidge PR, Lenardo MJ. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat Immunol. 2011;12(4):335–343. doi: 10.1038/ni.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang HJ, Youn H, Seong KM, Jin YW, Kim J, Youn B. Phosphorylation of Ribosomal Protein S3 and Antiapoptotic TRAF2 Protein Mediates Radioresistance in Non-small Cell Lung Cancer Cells. J Biol Chem. 2013;288(5):2965–2975. doi: 10.1074/jbc.M112.385989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jang CY, Kim HD, Kim J. Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem Biophys Res Commun. 2012;421(3):474–478. doi: 10.1016/j.bbrc.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 78.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141(3):741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cui K, Coutts M, Stahl J, Sytkowski AJ. Novel interaction between the transcription factor CHOP (GADD153) and the ribosomal protein FTE/S3a modulates erythropoiesis. J Biol Chem. 2000;275(11):7591–7596. doi: 10.1074/jbc.275.11.7591. [DOI] [PubMed] [Google Scholar]
- 80.Hu ZB, Minden MD, McCulloch EA, Stahl J. Regulation of drug sensitivity by ribosomal protein S3a. Blood. 2000;95(3):1047–1055. [PubMed] [Google Scholar]
- 81.Yadaiah M, Sudhamalla B, Rao PN, Roy KR, Ramakrishna D, Hussain Syed G, Ramaiah KV, Bhuyan AK. Arrested cell proliferation through cysteine protease activity of eukaryotic ribosomal protein S4. FASEB J. 2013;27(2):803–810. doi: 10.1096/fj.12-217752. [DOI] [PubMed] [Google Scholar]
- 82.Matragkou CN, Papachristou ET, Tezias SS, Tsiftsoglou AS, Choli-Papadopoulou T, Vizirianakis IS. The potential role of ribosomal protein S5 on cell cycle arrest and initiation of murine erythroleukemia cell differentiation. J Cell Biochem. 2008;104(4):1477–1490. doi: 10.1002/jcb.21722. [DOI] [PubMed] [Google Scholar]
- 83.Panic L, Tamarut S, Sticker-Jantscheff M, Barkic M, Solter D, Uzelac M, Grabusić K, Volarević S. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol Cell Biol. 2006;26(23):8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jeon YJ, Kim IK, Hong SH, Nan H, Kim HJ, Lee HJ, Masuda ES, Meyuhas O, Oh BH, Jung YK. Ribosomal protein S6 is a selective mediator of TRAIL-apoptotic signaling. Oncogene. 2008;27(31):4344–4352. doi: 10.1038/onc.2008.73. [DOI] [PubMed] [Google Scholar]
- 85.Chen D, Zhang Z, Li M, Wang W, Li Y, Rayburn ER, Hill DL, Wang H, Zhang R. Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene. 2007;26(35):5029–5037. doi: 10.1038/sj.onc.1210327. [DOI] [PubMed] [Google Scholar]
- 86.Zhu Y, Poyurovsky MV, Li Y, Biderman L, Stahl J, Jacq X, Prives C. Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell. 2009;35(3):316–326. doi: 10.1016/j.molcel.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gao M, Li X, Dong W, Jin R, Ma H, Yang P, Hu M, Li Y, Hao Y, Yuan S, Huang J, Song L. Ribosomal protein S7 regulates arsenite-induced GADD45alpha expression by attenuating MDM2-mediated GADD45alpha ubiquitination and degradation. Nucleic Acids Res. 2013;41(10):5210–5222. doi: 10.1093/nar/gkt223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Watkins-Chow DE, Cooke J, Pidsley R, Edwards A, Slotkin R, Leeds KE, Mullen R, Baxter LL, Campbell TG, Salzer MC, Biondini L, Gibney G, Phan Dinh Tuy F, Chelly J, Morris HD, Riegler J, Lythgoe MF, Arkell RM, Loreni F, Flint J, Pavan WJ, Keays DA. Mutation of the diamond-blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 2013;9(1):e1003094. doi: 10.1371/journal.pgen.1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hao Y, Kong X, Ruan Y, Gan H, Chen H, Zhang C, Ren S, Gu J. CDK11p46 and RPS8 associate with each other and suppress translation in a synergistic manner. Biochem Biophys Res Commun. 2011;407(1):169–174. doi: 10.1016/j.bbrc.2011.02.132. [DOI] [PubMed] [Google Scholar]
- 90.Lindstrom MS, Nister M. Silencing of ribosomal protein S9 elicits a multitude of cellular responses inhibiting the growth of cancer cells subsequent to p53 activation. PLoS One. 2010;5(3):e9578. doi: 10.1371/journal.pone.0009578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo X, Shi Y, Gou Y, Li J, Han S, Zhang Y, Huo J, Ning X, Sun L, Chen Y, Sun S, Fan D. Human ribosomal protein S13 promotes gastric cancer growth through down-regulating p27(Kip1) J Cell Mol Med. 2011;15(2):296–306. doi: 10.1111/j.1582-4934.2009.00969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou X, Hao Q, Liao JM, Liao P, Lu H. Ribosomal Protein S14 Negatively Regulates c-Myc Activity. J Biol Chem. 2013;288(30):21793–21801. doi: 10.1074/jbc.M112.445122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32(3):388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang J, Bai D, Ma X, Guan J, Zheng X. hCINAP is a novel regulator of ribosomal protein-HDM2-p53 pathway by controlling NEDDylation of ribosomal protein S14. Oncogene. 2014;33(2):246–254. doi: 10.1038/onc.2012.560. [DOI] [PubMed] [Google Scholar]
- 95.Barlow JL, Drynan LF, Hewett DR, Holmes LR, Lorenzo-Abalde S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ, Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat Med. 2010;16(1):59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Daftuar L, Zhu Y, Jacq X, Prives C. Ribosoal Proteins RPL37, RPS15 and RPS20 Regulate the Mdm2-p53-MdmX Network. PLoS One. 2013;8(7):e68667. doi: 10.1371/journal.pone.0068667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28(12):1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 98.Farrar JE, Vlachos A, Atsidaftos E, Carlson-Donohoe H, Markello TC, Arceci RJ, Ellis SR, Lipton JM, Bodine DM. Ribosomal protein gene deletions in Diamond-Blackfan anemia. Blood. 2011;118(26):6943–6951. doi: 10.1182/blood-2011-08-375170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Song MJ, Yoo EH, Lee KO, Kim GN, Kim HJ, Kim SY, Kim SH. A novel initiation codon mutation in the ribosomal protein S17 gene (RPS17) in a patient with Diamond-Blackfan anemia. Pediatr Blood Cancer. 2010;54(4):629–631. doi: 10.1002/pbc.22316. [DOI] [PubMed] [Google Scholar]
- 100.Moniz H, Gastou M, Leblanc T, Hurtaud C, Cretien A, Lecluse Y, Raslova H, Larghero J, Croisille L, Faubladier M, Bluteau O, Lordier L, Tchernia G, Vainchenker W, Mohandas N, Da Costa L. DBA Group of Société d'Hématologie et d'Immunologie Pédiatrique-SHIP. Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis. 2012;3:e356. doi: 10.1038/cddis.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jaako P, Flygare J, Olsson K, Quere R, Ehinger M, Henson A, Ellis S, Schambach A, Baum C, Richter J, Larsson J, Bryder D, Karlsson S. Mice with ribosomal protein S19 deficiency develop bone marrow failure and symptoms like patients with Diamond-Blackfan anemia. Blood. 2011;118(23):6087–6096. doi: 10.1182/blood-2011-08-371963. [DOI] [PubMed] [Google Scholar]
- 102.Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A, Berliner N, Kutok JL, Ebert BL. Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood. 2011;117(9):2567–2576. doi: 10.1182/blood-2010-07-295238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McGowan KA, Li JZ, Park CY, Beaudry V, Tabor HK, Sabnis AJ, Zhang W, Fuchs H, de Angelis MH, Myers RM, Attardi LD, Barsh GS. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat Genet. 2008;40(8):963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Filip AM, Klug J, Cayli S, Frohlich S, Henke T, Lacher P, Eickhoff R, Bulau P, Linder M, Carlsson-Skwirut C, Leng L, Bucala R, Kraemer S, Bernhagen J, Meinhardt A. Ribosomal protein S19 interacts with macrophage migration inhibitory factor and attenuates its pro-inflammatory function. J Biol Chem. 2009;284(12):7977–7985. doi: 10.1074/jbc.M808620200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsson H, Davey EJ, Draptchinskaia N, Hamaguchi I, Ooka A, Leeven P, Forsberg E, Karlsson S, Dahl N. Targeted disruption of the ribosomal protein S19 gene is lethal prior to implantation. Mol Cell Biol. 2004;24(9):4032–4037. doi: 10.1128/MCB.24.9.4032-4037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang X, Wang W, Wang H, Wang MH, Xu W, Zhang R. Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene. 2013;32(22):2782–2791. doi: 10.1038/onc.2012.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Adilakshmi T, Laine RO. Ribosomal protein S25 mRNA partners with MTF-1 and La to provide a p53-mediated mechanism for survival or death. J Biol Chem. 2002;277(6):4147–4151. doi: 10.1074/jbc.M109785200. [DOI] [PubMed] [Google Scholar]
- 108.Cui D, Li L, Lou H, Sun H, Ngai SM, Shao G, Tang J. The ribosomal protein S26 regulates p53 activity in response to DNA damage. Oncogene. 2013;33(17):2225–2235. doi: 10.1038/onc.2013.170. [DOI] [PubMed] [Google Scholar]
- 109.Xiong X, Zhao Y, He H, Sun Y. Ribosomal protein S27-like and S27 interplay with p53-MDM2 axis as a target, a substrate and a regulator. Oncogene. 2011;30(15):1798–1811. doi: 10.1038/onc.2010.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yang ZY, Qu Y, Zhang Q, Wei M, Liu CX, Chen XH, Yan M, Zhu ZG, Liu BY, Chen GQ, Wu YL, Gu QL. Knockdown of metallopanstimulin-1 inhibits NF-kappaB signaling at different levels: the role of apoptosis induction of gastric cancer cells. Int J Cancer. 2012;130(12):2761–2770. doi: 10.1002/ijc.26331. [DOI] [PubMed] [Google Scholar]
- 111.Yang ZY, Jiang H, Qu Y, Wei M, Yan M, Zhu ZG, Liu BY, Chen GQ, Wu YL, Gu QL. Metallopanstimulin-1 Regulates Invasion and Migration of Gastric Cancer Cells Partially Through Integrin beta4. Carcinogenesis. 2013;34(12):2851–2860. doi: 10.1093/carcin/bgt226. [DOI] [PubMed] [Google Scholar]
- 112.Sun XX, DeVine T, Challagundla KB, Dai MS. Interplay between ribosomal protein S27a and MDM2 protein in p53 activation in response to ribosomal stress. J Biol Chem. 2011;286(26):22730–22741. doi: 10.1074/jbc.M111.223651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Khanna N, Sen S, Sharma H, Singh N. S29 ribosomal protein induces apoptosis in H520 cells and sensitizes them to chemotherapy. Biochem Biophys Res Commun. 2003;304(1):26–35. doi: 10.1016/s0006-291x(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 114.Khanna N, Reddy VG, Tuteja N, Singh N. Differential gene expression in apoptosis: identification of ribosomal protein S29 as an apoptotic inducer. Biochem Biophys Res Commun. 2000;277(2):476–486. doi: 10.1006/bbrc.2000.3688. [DOI] [PubMed] [Google Scholar]
- 115.Russo A, Esposito D, Catillo M, Pietropaolo C, Crescenzi E, Russo G. Human rpL3 induces G(1)/S arrest or apoptosis by modulating p21 (waf1/cip1) levels in a p53-independent manner. Cell Cycle. 2013;12(1):76–87. doi: 10.4161/cc.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Provost E, Wehner KA, Zhong X, Ashar F, Nguyen E, Green R, Parsons MJ, Leach SD. Ribosomal biogenesis genes play an essential and p53-independent role in zebrafish pancreas development. Development. 2012;139(17):3232–3241. doi: 10.1242/dev.077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J Biol Chem. 2004;279(43):44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 118.Horn HF, Vousden KH. Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene. 2008;27(44):5774–5784. doi: 10.1038/onc.2008.189. [DOI] [PubMed] [Google Scholar]
- 119.Wu Q, Gou Y, Wang Q, Jin H, Cui L, Zhang Y, He L, Wang J, Nie Y, Shi Y, Fan D. Downregulation of RPL6 by siRNA inhibits proliferation and cell cycle progression of human gastric cancer cell lines. PLoS One. 2011;6(10):e26401. doi: 10.1371/journal.pone.0026401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gou Y, Shi Y, Zhang Y, Nie Y, Wang J, Song J, Jin H, He L, Gao L, Qiao L, Wu K, Fan D. Ribosomal protein L6 promotes growth and cell cycle progression through upregulating cyclin E in gastric cancer cells. Biochem Biophys Res Commun. 2010;393(4):788–793. doi: 10.1016/j.bbrc.2010.02.083. [DOI] [PubMed] [Google Scholar]
- 121.Du J, Shi Y, Pan Y, Jin X, Liu C, Liu N, Han Q, Lu Y, Qiao T, Fan D. Regulation of multidrug resistance by ribosomal protein l6 in gastric cancer cells. Cancer Biol Ther. 2005;4(2):242–247. doi: 10.4161/cbt.4.2.1477. [DOI] [PubMed] [Google Scholar]
- 122.Bai D, Zhang J, Xiao W, Zheng X. Regulation of the HDM2-p53 pathway by ribosomal protein L6 in response to ribosomal stress. Nucleic Acids Res. 2014;42(3):1799–1811. doi: 10.1093/nar/gkt971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Provost E, Weier CA, Leach SD. Multiple ribosomal proteins are expressed at high levels in developing zebrafish endoderm and are required for normal exocrine pancreas development. Zebrafish. 2013;10(2):161–169. doi: 10.1089/zeb.2013.0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Neumann F, Krawinkel U. Constitutive expression of human ribosomal protein L7 arrests the cell cycle in G1 and induces apoptosis in Jurkat T-lymphoma cells. Exp Cell Res. 1997;230(2):252–261. doi: 10.1006/excr.1996.3417. [DOI] [PubMed] [Google Scholar]
- 125.Li H, Pan L, Gou K. Depletion of ribosomal protein L8 impairs Drosophila development and is associated with apoptosis. Sci China Life Sci. 2010;53(9):1092–1097. doi: 10.1007/s11427-010-4059-4. [DOI] [PubMed] [Google Scholar]
- 126.Swoboda RK, Somasundaram R, Caputo L, Ochoa EM, Gimotty PA, Marincola FM, Van Belle P, Barth S, Elder D, Guerry D, Czerniecki B, Schuchter L, Vonderheide RH, Herlyn D. Shared MHC class II-dependent melanoma ribosomal protein L8 identified by phage display. Cancer Res. 2007;67(8):3555–3559. doi: 10.1158/0008-5472.CAN-06-2763. [DOI] [PubMed] [Google Scholar]
- 127.Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol. 2003;23(23):8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bhat KP, Itahana K, Jin A, Zhang Y. Essential role of ribosomal protein L11 in mediating growth inhibition-induced p53 activation. EMBO J. 2004;23(12):2402–2412. doi: 10.1038/sj.emboj.7600247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dai MS, Shi D, Jin Y, Sun XX, Zhang Y, Grossman SR, Lu H. Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem. 2006;281(34):24304–24313. doi: 10.1074/jbc.M602596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dai MS, Arnold H, Sun XX, Sears R, Lu H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 2007;26(14):3332–3345. doi: 10.1038/sj.emboj.7601776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115(2):187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 132.Mukhopadhyay R, Ray PS, Arif A, Brady AK, Kinter M, Fox PL. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol Cell. 2008;32(3):371–382. doi: 10.1016/j.molcel.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Poddar D, Basu A, Baldwin WM, 3rd, Kondratov RV, Barik S, Mazumder B. An extraribosomal function of ribosomal protein L13a in macrophages resolves inflammation. J Immunol. 2013;190(7):3600–3612. doi: 10.4049/jimmunol.1201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hsu YA, Lin HJ, Sheu JJ, Shieh FK, Chen SY, Lai CH, Tsai FJ, Wan L, Chen BH. A novel interaction between interferon-inducible protein p56 and ribosomal protein L15 in gastric cancer cells. DNA Cell Biol. 2011;30(9):671–679. doi: 10.1089/dna.2010.1149. [DOI] [PubMed] [Google Scholar]
- 135.Smolock EM, Korshunov VA, Glazko G, Qiu X, Gerloff J, Berk BC. Ribosomal protein L17, RpL17, is an inhibitor of vascular smooth muscle growth and carotid intima formation. Circulation. 2012;126(20):2418–2427. doi: 10.1161/CIRCULATIONAHA.112.125971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rao S, Lee SY, Gutierrez A, Perrigoue J, Thapa RJ, Tu Z, Jeffers JR, Rhodes M, Anderson S, Oravecz T, Hunger SP, Timakhov RA, Zhang R, Balachandran S, Zambetti GP, Testa JR, Look AT, Wiest DL. Inactivation of ribosomal protein L22 promotes transformation by induction of the stemness factor, Lin28B. Blood. 2012;120(18):3764–3773. doi: 10.1182/blood-2012-03-415349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dai MS, Zeng SX, Jin Y, Sun XX, David L, Lu H. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol Cell Biol. 2004;24(17):7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jin A, Itahana K, O'Keefe K, Zhang Y. Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol Cell Biol. 2004;24(17):7669–7680. doi: 10.1128/MCB.24.17.7669-7680.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M. A ribosomal protein L23-nucleophosmin circuit coordinates Mizl function with cell growth. Nat Cell Biol. 2008;10(9):1051–1061. doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 140.Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123(1):49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- 141.Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M. Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell. 2008;32(2):180–189. doi: 10.1016/j.molcel.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jones DT, Lechertier T, Reynolds LE, Mitter R, Robinson SD, Kirn-Safran CB, Hodivala-Dilke KM. Endogenous ribosomal protein L29 (RPL29): a newly identified regulator of angiogenesis in mice. Dis Model Mech. 2013;6(1):115–124. doi: 10.1242/dmm.009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Oristian DS, Sloofman LG, Zhou X, Wang L, Farach-Carson MC, Kirn-Safran CB. Ribosomal protein L29/HIP deficiency delays osteogenesis and increases fragility of adult bone in mice. J Orthop Res. 2009;27(1):28–35. doi: 10.1002/jor.20706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li C, Ge M, Yin Y, Luo M, Chen D. Silencing expression of ribosomal protein L26 and L29 by RNA interfering inhibits proliferation of human pancreatic cancer PANC-1 cells. Mol Cell Biochem. 2012;370(1–2):127–139. doi: 10.1007/s11010-012-1404-x. [DOI] [PubMed] [Google Scholar]
- 145.Sun B, Hou YL, Hou WR, Zhang SN, Ding X, Su XL. cDNA Cloning, Overexpression, Purification and Pharmacologic Evaluation for Anticancer Activity of Ribosomal Protein L23A Gene (RPL23A) from the Giant Panda. Int J Mol Sci. 2012;13(2):2133–2147. doi: 10.3390/ijms13022133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Su XL, Hou YL, Yan XH, Ding X, Hou WR, Sun B, Zhang SN. Expression, purification, and evaluation for anticancer activity of ribosomal protein L31 gene (RPL31) from the giant panda (Ailuropoda melanoleuca) Mol Biol Rep. 2012;39(9):8945–8954. doi: 10.1007/s11033-012-1763-0. [DOI] [PubMed] [Google Scholar]
- 147.Lopez CD, Martinovsky G, Naumovski L. Inhibition of cell death by ribosomal protein L35a. Cancer Lett. 2002;180(2):195–202. doi: 10.1016/s0304-3835(02)00024-1. [DOI] [PubMed] [Google Scholar]
- 148.Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr, Meltzer P, Esposito D, Beggs AH, Schneider HE, Grabowska A, Ball SE, Niewiadomska E, Sieff CA, Vlachos A, Atsidaftos E, Ellis SR, Lipton JM, Gazda HT, Arceci RJ. Abnormalities of the large ribosomal subunit protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 2008;112(5):1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Llanos S, Serrano M. Depletion of ribosomal protein L37 occurs in response to DNA damage and activates p53 through the L11/MDM2 pathway. Cell Cycle. 2010;9(19):4005–4012. doi: 10.4161/cc.9.19.13299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wang S, Huang J, He J, Wang A, Xu S, Huang SF, Xiao S. RPL41, a small ribosomal peptide deregulated in tumors, is essential for mitosis and centrosome integrity. Neoplasia. 2010;12(3):284–293. doi: 10.1593/neo.91610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang A, Xu S, Zhang X, He J, Yan D, Yang Z, Xiao S. Ribosomal protein RPL41 induces rapid degradation of ATF4, a transcription factor critical for tumour cell survival in stress. J Pathol. 2011;225(2):285–292. doi: 10.1002/path.2918. [DOI] [PubMed] [Google Scholar]
- 152.Artero-Castro A, Kondoh H, Fernandez-Marcos PJ, Serrano M, Ramon y Cajal S, Lleonart ME. Rplp1 bypasses replicative senescence and contributes to transformation. Exp Cell Res. 2009;315(8):1372–1383. doi: 10.1016/j.yexcr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 153.Berghofer-Hochheimer Y, Zurek C, Wolfl S, Hemmerich P, Munder T. L7 protein is a coregulator of vitamin D receptor retinoid X receptor-mediated transactivation. J Cell Biochem. 1998;69(1):1–12. doi: 10.1002/(sici)1097-4644(19980401)69:1<1::aid-jcb1>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 154.Gray JP, Davis JW, 2nd, Gopinathan L, Leas TL, Nugent CA, Vanden Heuvel JP. The ribosomal protein rpL11 associates with and inhibits the transcriptional activity of peroxisome proliferator-activated receptor-alpha. Toxicol Sci. 2006;89(2):535–546. doi: 10.1093/toxsci/kfj040. [DOI] [PubMed] [Google Scholar]
- 155.Kim HD, Kim TS, Joo YJ, Shin HS, Kim SH, Jang CY, Lee CE, Kim J. RpS3 Translation Is Repressed by Interaction With Its Own mRNA. J Cell Biochem. 2010;110(2):294–303. doi: 10.1002/jcb.22537. [DOI] [PubMed] [Google Scholar]
- 156.Lindström MS. Emerging functions of ribosomal proteins in gene-specific transcription and translation. Biochem Biophys Res Commun. 2009;379(2):167–170. doi: 10.1016/j.bbrc.2008.12.083. 6. [DOI] [PubMed] [Google Scholar]
- 157.Neumann F, Krawinkel U. Constitutive expression of human ribosomal protein L7 arrests the cell cycle in G(1) and induces apoptosis in Jurkat T-lymphoma cells. Exp Cell Res. 1997;230(2):252–261. doi: 10.1006/excr.1996.3417. [DOI] [PubMed] [Google Scholar]
- 158.Wang H, Zhao LN, Li KZ, Ling R, Li XJ, Wang L. Overexpression of ribosomal protein L15 is associated with cell proliferation in gastric cancer. BMC Cancer. 2006;6:91. doi: 10.1186/1471-2407-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Jang CY, Kim HD, Zhang X, Chang JS, Kim J. Ribosomal protein S3 localizes on the mitotic spindle and functions as a microtubule associated protein in mitosis. Biochem Biophys Res Commun. 2012;429(1–2):57–62. doi: 10.1016/j.bbrc.2012.10.093. [DOI] [PubMed] [Google Scholar]
- 160.Yadavilli S, Hegde V, Deutsch WA. Translocation of human ribosomal protein S3 to sites of DNA damage is dependant on ERK-mediated phosphorylation following genotoxic stress. DNA Repair (Amst) 2007;6(10):1453–1462. doi: 10.1016/j.dnarep.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Duan J, Ba Q, Wang ZL, Hao M, Li XG, Hu PT, Zhang D, Zhang R, Wang H. Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. Int J Biochem Cell Biol. 2011;43(8):1218–1227. doi: 10.1016/j.biocel.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 162.Uechi T, Nakajima Y, Nakao A, Torihara H, Chakraborty A, Inoue K, Kenmochi N. Ribosomal protein gene knockdown causes developmental defects in zebrafish. PLoS One. 2006;1:e37. doi: 10.1371/journal.pone.0000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Anderson SJ, Lauritsen JPH, Hartman MG, Foushee AMD, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alpha beta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26(6):759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 164.Stadanlick JE, Zhang Z, Lee SY, Hemann M, Biery M, Carleton MO, Zambetti GP, Anderson SJ, Oravecz T, Wiest DL. Developmental arrest of T cells in Rpl22-deficient mice is dependent upon multiple p53 effectors. J Immunol. 2011;187(2):664–675. doi: 10.4049/jimmunol.1100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bevort M, Leffers H. Down regulation of ribosomal protein mRNAs during neuronal differentiation of human NTERA2 cells. Differentiation. 2000;66(2–3):81–92. doi: 10.1046/j.1432-0436.2000.660203.x. [DOI] [PubMed] [Google Scholar]
- 166.Liu JJ, Huang BH, Zhang JQ, Carson DD, Hooi SC. Repression of HIP/RPL29 expression induces differentiation in colon cancer cells. J Cell Physiol. 2006;207(2):287–292. doi: 10.1002/jcp.20589. [DOI] [PubMed] [Google Scholar]
- 167.Kreunin P, Yoo C, Urquidi V, Lubman DM, Goodison S. Differential expression of ribosomal proteins in a human metastasis model identified by coupling 2-D liquid chromatography and mass spectrometry. Cancer Genomics Proteomics. 2007;4(5):329–339. [PMC free article] [PubMed] [Google Scholar]
- 168.Liu F, Li Y, Yu Y, Fu S, Li P. Cloning of novel tumor metastasis-related genes from the highly metastatic human lung adenocarcinoma cell line Anip973. J Genet Genomics. 2007;34(3):189–195. doi: 10.1016/S1673-8527(07)60020-4. [DOI] [PubMed] [Google Scholar]
- 169.McDonald JM, Pelloski CE, Ledoux A, Sun M, Raso G, Komaki R, Wistuba II, Bekele BN, Aldape K. Elevated phospho-S6 expression is associated with metastasis in adenocarcinoma of the lung. Clin Cancer Res. 2008;14(23):7832–7837. doi: 10.1158/1078-0432.CCR-08-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22(22):6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Boyd MT, Vlatkovic N, Rubbi CP. The nucleolus directly regulates p53 export and degradation. J Cell Biol. 2011;194(5):689–703. doi: 10.1083/jcb.201105143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nag S, Qin JJ, Srivenugopal K, Wang MH, Zhang R. The MDM2-p53 Pathway Revisited. J Biomed Res. 2013;27(4):254–271. doi: 10.7555/JBR.27.20130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Strezoska Z, Pestov DG, Lau LF. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5. 8S RRNA processing and 60S ribosome biogenesis. Mol Cell Biol. 2000;20(15):5516–5528. doi: 10.1128/mcb.20.15.5516-5528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.McMahon M, Ayllon V, Panov KI, O'Connor R. Ribosomal 18 S RNA Processing by the IGF-I-responsive WDR3 Protein Is Integrated with p53 Function in Cancer Cell Proliferation. J Biol Chem. 2010;285(24):18309–18318. doi: 10.1074/jbc.M110.108555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Holzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Längst G, Eick D. Defects in 18 S or 28 S rRNA Processing Activate the p53 Pathway. J Biol Chem. 2010;285(9):6364–6370. doi: 10.1074/jbc.M109.054734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang F, Hamanaka RB, Bobrovnikova-Marjon E, Gordan JD, Dai MS, Lu H, Simon MC, Diehl JA. Ribosomal stress couples the unfolded protein response to p53-dependent cell cycle arrest. J Biol Chem. 2006;281(40):30036–30045. doi: 10.1074/jbc.M604674200. [DOI] [PubMed] [Google Scholar]
- 177.Donati G, Bertoni S, Brighenti E, Vici M, Trere D, Volarevic S, Montanaro L, Derenzini M. The balance between rRNA and ribosomal protein synthesis up- and downregulates the tumour suppressor p53 in mammalian cells. Oncogene. 2011;30(29):3274–3288. doi: 10.1038/onc.2011.48. [DOI] [PubMed] [Google Scholar]
- 178.Schnapp A, Pfleiderer C, Rosenbauer H, Grummt I. A growth-dependent transcription initiation factor (TIF-IA) interacting with RNA polymerase I regulates mouse ribosomal RNA synthesis. EMBO J. 1990;9(9):2857–2863. doi: 10.1002/j.1460-2075.1990.tb07475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yuan XJ, Zhou YG, Casanova E, Chai MQ, Kiss E, Grone HJ, Schütz G, Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell. 2005;19(1):77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 180.Burger K, Muhl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, Eick D. Chemotherapeutic Drugs Inhibit Ribosome Biogenesis at Various Levels. J Biol Chem. 2010;285(16):12416–12425. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sun XX, Dai MS, Lu H. 5-fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem. 2007;282(11):8052–8059. doi: 10.1074/jbc.M610621200. [DOI] [PubMed] [Google Scholar]
- 182.Sun XX, Dai MS, Lu H. Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J Biol Chem. 2008;283(18):12387–12392. doi: 10.1074/jbc.M801387200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dai MS, Sun XX, Lu H. Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol Cell Biol. 2008;28(13):4365–4376. doi: 10.1128/MCB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lo D, Dai MS, Sun XX, Zeng SX, Lu H. Ubiquitin- and MDM2 E3 ligase-independent proteasomal turnover of nucleostemin in response to GTP depletion. J Biol Chem. 2012;287(13):10013–10020. doi: 10.1074/jbc.M111.335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yu WS, Qiu ZW, Gao N, Wang LR, Cui HX, Qian Y, Jiang L, Luo J, Yi Z, Lu H, Li D, Liu M. PAK1IP1, a ribosomal stress-induced nucleolar protein, regulates cell proliferation via the p53-MDM2 loop. Nucleic Acids Res. 2011;39(6):2234–2248. doi: 10.1093/nar/gkq1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Itahana K, Bhat KP, Jin AW, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol Cell. 2003;12(5):1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 187.Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF Tumor Suppressor Controls Ribosome Biogenesis by Regulating the RNA Polymerase I Transcription Factor TTF-I. Mol Cell. 2010;38(4):539–550. doi: 10.1016/j.molcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 188.Lessard F, Stefanovsky V, Tremblay MG, Moss T. The cellular abundance of the essential transcription termination factor TTF-I regulates ribosome biogenesis and is determined by MDM2 ubiquitinylation. Nucleic Acids Res. 2012;40(12):5357–5367. doi: 10.1093/nar/gks198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ayrault O, Andrique L, Fauvin D, Eymin B, Gazzeri S, Seite P. Human tumor suppressor p14(ARF) negatively regulates rRNA transcription and inhibits UBF1 transcription factor phosphorylation. Oncogene. 2006;25(58):7577–7586. doi: 10.1038/sj.onc.1209743. [DOI] [PubMed] [Google Scholar]
- 190.Sun XX, Wang YG, Xirodimas DP, Dai MS. Perturbation of 60 S Ribosomal Biogenesis Results in Ribosomal Protein L5-and L11-dependent p53 Activation. J Biol Chem. 2010;285(33):25812–25821. doi: 10.1074/jbc.M109.098442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Uechi T, Nakajima Y, Chakraborty A, Torihara H, Higa S, Kenmochi N. Deficiency of ribosomal protein S19 during early embryogenesis leads to reduction of erythrocytes in a zebrafish model of Diamond-Blackfan anemia. Hum Mol Genet. 2008;17(20):3204–3211. doi: 10.1093/hmg/ddn216. [DOI] [PubMed] [Google Scholar]
- 192.Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N. Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One. 2009;4(1):e4152. doi: 10.1371/journal.pone.0004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Danilova N, Sakamoto KM, Lin S. Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. Br J Haematol. 2011;152(2):217–228. doi: 10.1111/j.1365-2141.2010.08396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sulic S, Panic L, Barkic M, Mercep M, Uzelac M, Volarevic S. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19(24):3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.McGowan KA, Pang WW, Bhardwaj R, Perez MG, Pluvinage JV, Glader BE, Malek R, Mendrysa SM, Weissman IL, Park CY, Barsh GS. Reduced ribosomal protein gene dosage and p53 activation in low-risk myelodysplastic syndrome. Blood. 2011;118(13):3622–3633. doi: 10.1182/blood-2010-11-318584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Golomb L, Bublik DR, Wilder S, Nevo R, Kiss V, Grabusic K, Volarevic S, Oren M. Importin 7 and Exportin 1 Link c-Myc and p53 to Regulation of Ribosomal Biogenesis. Mol Cell. 2012;45(2):222–232. doi: 10.1016/j.molcel.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 198.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 199.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6(12):909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 201.Juven T, Barak Y, Zauberman A, George DL, Oren M. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene. 1993;8(12):3411–3416. [PubMed] [Google Scholar]
- 202.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7(7A):1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 203.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12(2):461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature. 1993;362(6423):857–860. doi: 10.1038/362857a0. [DOI] [PubMed] [Google Scholar]
- 205.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387(6630):296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 206.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387(6630):299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 207.Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, Kastan MB, Katzir E, Oren M. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15(9):1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Inuzuka H, Tseng A, Gao D, Zhai B, Zhang Q, Shaik S, Wan L, Ang XL, Mock C, Yin H, Stommel JM, Gygi S, Lahav G, Asara J, Xiao ZX, Kaelin WG, Jr, Harper JW, Wei W. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCF(beta-TRCP) ubiquitin ligase. Cancer Cell. 2010;18(2):147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Li M, Luo J, Brooks CL, Gu W. Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem. 2002;277(52):50607–50611. doi: 10.1074/jbc.C200578200. [DOI] [PubMed] [Google Scholar]
- 210.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92(6):725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 211.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1(1):20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 212.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol Cancer Res. 2003;1(14):1001–1008. [PubMed] [Google Scholar]
- 213.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14(17):5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Marechal V, Elenbaas B, Piette J, Nicolas JC, Levine AJ. The ribosomal L5 protein is associated with mdm-2 and mdm-2-p53 complexes. Mol Cell Biol. 1994;14(11):7414–7420. doi: 10.1128/mcb.14.11.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Donati G, Peddigari S, Mercer CA, Thomas G. 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 2013;4(1):87–98. doi: 10.1016/j.celrep.2013.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bursac S, Brdovcak MC, Pfannkuchen M, Orsolic I, Golomb L, Zhu Y, Katz C, Daftuar L, Grabušić K, Vukelić I, Filić V, Oren M, Prives C, Volarevic S. Mutual protection of ribosomal proteins L5 and L11 from degradation is essential for p53 activation upon ribosomal biogenesis stress. Proc Natl Acad Sci U S A. 2012;109(50):20467–20472. doi: 10.1073/pnas.1218535109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhang Y, Wang J, Yuan Y, Zhang W, Guan W, Wu Z, Jin C, Chen H, Zhang L, Yang X, He F. Negative regulation of HDM2 to attenuate p53 degradation by ribosomal protein L26. Nucleic Acids Res. 2010;38(19):6544–6554. doi: 10.1093/nar/gkq536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Kawai H, Wiederschain D, Yuan ZM. Critical contribution of the MDM2 acidic domain to p53 ubiquitination. Mol Cell Biol. 2003;23(14):4939–4947. doi: 10.1128/MCB.23.14.4939-4947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhang Q, Xiao H, Chai SC, Hoang QQ, Lu H. Hydrophilic residues are crucial for ribosomal protein L11 (RPL11) interaction with zinc finger domain of MDM2 and p53 protein activation. J Biol Chem. 2011;286(44):38264–38274. doi: 10.1074/jbc.M111.277012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Li M, Gu W. A critical role for noncoding 5S rRNA in regulating Mdmx stability. Mol Cell. 2011;43(6):1023–1032. doi: 10.1016/j.molcel.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yim JH, Kim YJ, Ko JH, Cho YE, Kim SM, Kim JY, Lee S, Park JH. The putative tumor suppressor gene GLTSCR2 induces PTEN-modulated cell death. Cell Death Differ. 2007;14(11):1872–1879. doi: 10.1038/sj.cdd.4402204. [DOI] [PubMed] [Google Scholar]
- 222.Kim YJ, Cho YE, Kim YW, Kim JY, Lee S, Park JH. Suppression of putative tumour suppressor gene GLTSCR2 expression in human glioblastomas. J Pathol. 2008;216(2):218–224. doi: 10.1002/path.2401. [DOI] [PubMed] [Google Scholar]
- 223.Sasaki M, Kawahara K, Nishio M, Mimori K, Kogo R, Hamada K, Itoh B, Wang J, Komatsu Y, Yang YR, Hikasa H, Horie Y, Yamashita T, Kamijo T, Zhang Y, Zhu Y, Prives C, Nakano T, Mak TW, Sasaki T, Maehama T, Mori M, Suzuki A. Regulation of the MDM2-P53 pathway and tumor growth by PICT1 via nucleolar RPL11. Nat Med. 2011;17(8):944–951. doi: 10.1038/nm.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Lee S, Kim JY, Kim YJ, Seok KO, Kim JH, Chang YJ, Kang HY, Park JH. Nucleolar protein GLTSCR2 stabilizes p53 in response to ribosomal stresses. Cell Death Differ. 2012;19(10):1613–1622. doi: 10.1038/cdd.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lo D, Lu H. Nucleostemin: Another nucleolar"Twister" of the p53-MDM2 loop. Cell Cycle. 2010;9(16):3227–3232. doi: 10.4161/cc.9.16.12605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133(4):612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Owen HR, Elser M, Cheung E, Gersbach M, Kraus WL, Hottiger MO. MYBBP1a is a novel repressor of NF-kappaB. J Mol Biol. 2007;366(3):725–736. doi: 10.1016/j.jmb.2006.11.099. [DOI] [PubMed] [Google Scholar]
- 228.Yamauchi T, Keough RA, Gonda TJ, Ishii S. Ribosomal stress induces processing of Mybbp1a and its translocation from the nucleolus to the nucleoplasm. Genes Cells. 2008;13(1):27–39. doi: 10.1111/j.1365-2443.2007.01148.x. [DOI] [PubMed] [Google Scholar]
- 229.Drakou CE, Malekkou A, Hayes JM, Lederer CW, Leonidas DD, Oikonomakos NG, Lamond AI, Santama N, Zographos SE. hCINAP is an atypical mammalian nuclear adenylate kinase with an ATPase motif: structural and functional studies. Proteins. 2012;80(1):206–220. doi: 10.1002/prot.23186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Malekkou A, Lederer CW, Lamond AI, Santama N. The nuclear ATPase/adenylate kinase hCINAP is recruited to perinucleolar caps generated upon RNA pol.II inhibition. FEBS Lett. 2010;584(22):4559–4564. doi: 10.1016/j.febslet.2010.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Castro ME, Leal JF, Lleonart ME, Ramon YCS, Carnero A. Loss-of-function genetic screening identifies a cluster of ribosomal proteins regulating p53 function. Carcinogenesis. 2008;29(7):1343–1350. doi: 10.1093/carcin/bgm302. [DOI] [PubMed] [Google Scholar]
- 232.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A. 2008;105(30):10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18(4):1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 234.Zhai W, Comai L. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol. 2000;20(16):5930–5938. doi: 10.1128/mcb.20.16.5930-5938.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li J, Tan J, Zhuang L, Banerjee B, Yang X, Chau JF, Lee PL, Hande MP, Li B, Yu Q. Ribosomal protein S27-like, a p53-inducible modulator of cell fate in response to genotoxic stress. Cancer Res. 2007;67(23):11317–11326. doi: 10.1158/0008-5472.CAN-07-1088. [DOI] [PubMed] [Google Scholar]
- 236.Loging WT, Reisman D. Elevated expression of ribosomal protein genes L37, RPP-1, and S2 in the presence of mutant p53. Cancer Epidemiol Biomarkers Prev. 1999;8(11):1011–1016. [PubMed] [Google Scholar]
- 237.Donati G, Montanaro L, Derenzini M. Ribosome biogenesis and control of cell proliferation: p53 is not alone. Cancer Res. 2012;72(7):1602–1607. doi: 10.1158/0008-5472.CAN-11-3992. [DOI] [PubMed] [Google Scholar]
- 238.Dai MS, Lu H. Crosstalk between c-Myc and ribosome in ribosomal biogenesis and cancer. J Cell Biochem. 2008;105(3):670–677. doi: 10.1002/jcb.21895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dai MS, Sears R, Lu H. Feedback regulation of c-Myc by ribosomal protein L11. Cell Cycle. 2007;6(22):2735–2741. doi: 10.4161/cc.6.22.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Dai MS, Sun XX, Lu H. Ribosomal Protein L11 Associates with c-Myc at 5 S rRNA and tRNA Genes and Regulates Their Expression. J Biol Chem. 2010;285(17):12587–12594. doi: 10.1074/jbc.M109.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Challagundla KB, Sun XX, Zhang X, DeVine T, Zhang Q, Sears RC, Dai MS. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol Cell Biol. 2011;31(19):4007–4021. doi: 10.1128/MCB.05810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer. 2010;10(4):301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- 243.Iadevaia V, Caldarola S, Biondini L, Gismondi A, Karlsson S, Dianzani I, Loreni F. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene. 2010;29(40):5490–5469. doi: 10.1038/onc.2010.279. [DOI] [PubMed] [Google Scholar]
- 244.Donati G, Brighenti E, Vici M, Mazzini G, Trere D, Montanaro L, Derenzini M. Selective inhibition of rRNA transcription downregulates E2F-1: a new p53-independent mechanism linking cell growth to cell proliferation. J Cell Sci. 2011;124(17):3017–3028. doi: 10.1242/jcs.086074. [DOI] [PubMed] [Google Scholar]
- 245.Wanzel M, Russ AC, Kleine-Kohlbrecher D, Colombo E, Pelicci PG, Eilers M. A ribosomal protein L23-nucleophosmin circuit coordinates Miz1 function with cell growth. Nat Cell Biol. 2008;10(9):1051–1061. doi: 10.1038/ncb1764. [DOI] [PubMed] [Google Scholar]
- 246.Russo A, Esposito D, Catillo M, Pietropaolo C, Crescenzi E, Russo G. Human rpL3 induces G(1)/S arrest or apoptosis by modulating p21(waf1/cip1) levels in a p53-independent manner. Cell Cycle. 2013;12(1):76–87. doi: 10.4161/cc.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Wang YW, Qu Y, Li JF, Chen XH, Liu BY, Gu QL, Zhu ZG. In vitro and in vivo evidence of metallopanstimulin-1 in gastric cancer progression and tumorigenicity. Clin Cancer Res. 2006;12(16):4965–4973. doi: 10.1158/1078-0432.CCR-05-2316. [DOI] [PubMed] [Google Scholar]
- 248.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 249.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118(1):83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 250.Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008;9(3):280–286. doi: 10.1038/embor.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Mahata B, Sundqvist A, Xirodimas DP. Recruitment of RPL11 at promoter sites of p53-regulated genes upon nucleolar stress through NEDD8 and in an Mdm2-dependent manner. Oncogene. 2012;31(25):3060–3071. doi: 10.1038/onc.2011.482. [DOI] [PubMed] [Google Scholar]
- 252.Kim SH, Lee JY, Kim J. Characterization of a wide range base-damage-endonuclease activity of mammalian rpS3. Biochem Biophys Res Commun. 2005;328(4):962–967. doi: 10.1016/j.bbrc.2005.01.045. [DOI] [PubMed] [Google Scholar]
- 253.Kim TS, Kim HD, Kim J. PKCdelta-dependent functional switch of rpS3 between translation and DNA repair. Biochimica Et Biophysica Acta. 2009;1793(2):395–405. doi: 10.1016/j.bbamcr.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 254.Kim TS, Kim HD, Shin HS, Kim J. Phosphorylation status of nuclear ribosomal protein S3 is reciprocally regulated by protein kinase C{delta} and protein phosphatase 2A. J Biol Chem. 2009;284(32):21201–21208. doi: 10.1074/jbc.M109.018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kim HD, Lee JY, Kim J. Erk phosphorylates threonine 42 residue of ribosomal protein S3. Biochem Biophys Res Commun. 2005;333(1):110–115. doi: 10.1016/j.bbrc.2005.05.079. [DOI] [PubMed] [Google Scholar]
- 256.Krieg J, Hofsteenge J, Thomas G. Identification of the 40 S ribosomal protein S6 phosphorylation sites induced by cycloheximide. J Biol Chem. 1988;263(23):11473–11477. [PubMed] [Google Scholar]
- 257.Hutchinson JA, Shanware NP, Chang H, Tibbetts RS. Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J Biol Chem. 2011;286(10):8688–8696. doi: 10.1074/jbc.M110.141754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Meyuhas O. Physiological roles of ribosomal protein S6: one of its kind. Int Rev Cell Mol Biol. 2008;268:1–37. doi: 10.1016/S1937-6448(08)00801-0. [DOI] [PubMed] [Google Scholar]
- 259.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31(6):342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 260.Ruvinsky I, Katz M, Dreazen A, Gielchinsky Y, Saada A, Freedman N, Mishani E, Zimmerman G, Kasir J, Meyuhas O. Mice deficient in ribosomal protein S6 phosphorylation suffer from muscle weakness that reflects a growth defect and energy deficit. PLoS One. 2009;4(5):e5618. doi: 10.1371/journal.pone.0005618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Khalaileh A, Dreazen A, Khatib A, Apel R, Swisa A, Kidess-Bassir N, Maitra A, Meyuhas O, Dor Y, Zamir G. Phosphorylation of Ribosomal Protein S6 Attenuates DNA Damage and Tumor Suppression during Development of Pancreatic Cancer. Cancer Res. 2013;73(6):1811–1820. doi: 10.1158/0008-5472.CAN-12-2014. [DOI] [PubMed] [Google Scholar]
- 262.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18(1):1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 263.Haindl M, Harasim T, Eick D, Muller S. The nucleolar SUMO-specific protease SENP3 reverses SUMO modification of nucleophosmin and is required for rRNA processing. EMBO Rep. 2008 Mar;9(3):273–279. doi: 10.1038/embor.2008.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Jang CY, Shin HS, Kim HD, Kim JW, Choi SY, Kim J. Ribosomal protein S3 is stabilized by sumoylation. Biochem Biophys Res Commun. 2011;414(3):523–527. doi: 10.1016/j.bbrc.2011.09.099. [DOI] [PubMed] [Google Scholar]
- 265.Janas MM, Wang E, Love T, Harris AS, Stevenson K, Semmelmann K, Shaffer JM, Chen PH, Doench JG, Yerramilli SV, Neuberg DS, Iliopoulos D, Housman DE, Burge CB, Novina CD. Reduced expression of ribosomal proteins relieves microRNA-mediated repression. Mol Cell. 2012;46(2):171–186. doi: 10.1016/j.molcel.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 266.Frum RA, Zhang YP. The RP-p53-Mdm2 pathway: A new link to genetic integrity? Cell Cycle. 2010;9(22):4427. doi: 10.4161/cc.9.22.13606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Dai MS, Challagundla KB, Sun XX, Palam LR, Zeng SX, Wek RC, Lu H. Physical and Functional Interaction between Ribosomal Protein L11 and the Tumor Suppressor ARF. J Biol Chem. 2012;287(21):17120–17129. doi: 10.1074/jbc.M111.311902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Oliver ER, Saunders TL, Tarle SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Barkic M, Crnomarkovic S, Grabusic K, Bogetic I, Panic L, Tamarut S, Cokaric M, Jeric I, Vidak S, Volarevic S. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29(10):2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Terzian T, Dumble M, Arbab F, Thaller C, Donehower LA, Lozano G, Justice MJ, Roop DR, Box NF. Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J Pathol. 2011;224(4):540–552. doi: 10.1002/path.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kirn-Safran CB, Oristian DS, Focht RJ, Parker SG, Vivian JL, Carson DD. Global growth deficiencies in mice lacking the ribosomal protein HIP/RPL29. Dev Dyn. 2007;236(2):447–460. doi: 10.1002/dvdy.21046. [DOI] [PubMed] [Google Scholar]
- 272.Saeboe-Larssen S, Lyamouri M, Merriam J, Oksvold MP, Lambertsson A. Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics. 1998;148(3):1215–1224. doi: 10.1093/genetics/148.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101(35):12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ishijima J, Yasui H, Morishima M, Shiroishi T. Dominant lethality of the mouse skeletal mutation Tail-short (Ts) is determined by the Ts allele from mating partners. Genomics. 1998;49(3):341–350. doi: 10.1006/geno.1998.5277. [DOI] [PubMed] [Google Scholar]
- 275.Terzian T, Dumble M, Arbab F, Thaller C, Donehower LA, Lozano G, Justice MJ, Roop DR, Box NF. Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J Pathol. 2011;224(4):540–552. doi: 10.1002/path.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145(3):383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Chiocchetti A, Pakalapati G, Duketis E, Wiemann S, Poustka A, Poustka F, Klauck SM. Mutation and expression analyses of the ribosomal protein gene RPL10 in an extended German sample of patients with autism spectrum disorder. Am J Med Genet A. 2011;155A(6):1472–1475. doi: 10.1002/ajmg.a.33977. [DOI] [PubMed] [Google Scholar]
- 278.Gong X, Delorme R, Fauchereau F, Durand CM, Chaste P, Betancur C, Goubran-Botros H, Nygren G, Anckarsäter H, Rastam M, Gillberg IC, Kopp S, Mouren-Simeoni MC, Gillberg C, Leboyer M, Bourgeron T. An investigation of ribosomal protein L10 gene in autism spectrum disorders. BMC Med Genet. 2009;10:7. doi: 10.1186/1471-2350-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Klauck SM, Felder B, Kolb-Kokocinski A, Schuster C, Chiocchetti A, Schupp I, Wellenreuther R, Schmötzer G, Poustka F, Breitenbach-Koller L, Poustka A. Mutations in the ribosomal protein gene RPL10 suggest a novel modulating disease mechanism for autism. Mol Psychiatry. 2006;11(12):1073–1084. doi: 10.1038/sj.mp.4001883. [DOI] [PubMed] [Google Scholar]
- 280.Terzian T, Box N. Genetics of ribosomal proteins: "curiouser and curiouser". PLoS Genet. 2013;9(1):e1003300. doi: 10.1371/journal.pgen.1003300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Alexander SJ, Woodling NS, Yedvobnick B. Insertional inactivation of the L13a ribosomal protein gene of Drosophila melanogaster identifies a new Minute locus. Gene. 2006;368:46–52. doi: 10.1016/j.gene.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 282.Van Beest M, Mortin M, Clevers H. Drosophila RpS3a, a novel Minute gene situated between the segment polarity genescubitus interruptus and dTCF. Nucleic Acids Res. 1998;26(19):4471–4475. doi: 10.1093/nar/26.19.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Casad ME, Abraham D, Kim IM, Frangakis S, Dong B, Lin N, Wolf MJ, Rockman HA. Cardiomyopathy is associated with ribosomal protein gene haplo-insufficiency in Drosophila melanogaster. Genetics. 2011;189(3):861–870. doi: 10.1534/genetics.111.131482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18(16):1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 285.Di R, Wu X, Chang Z, Zhao X, Feng Q, Lu S, Luan Q, Hemmings BA, Li X, Yang Z. S6K inhibition renders cardiac protection against myocardial infarction through PDK1 phosphorylation of Akt. Biochem J. 2012;441(1):199–207. doi: 10.1042/BJ20110033. [DOI] [PubMed] [Google Scholar]
- 286.Treins C, Alliouachene S, Hassouna R, Xie Y, Birnbaum MJ, Pende M. The combined deletion of S6K1 and Akt2 deteriorates glycemic control in a high-fat diet. Mol Cell Biol. 2012;32(19):4001–4011. doi: 10.1128/MCB.00514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Li S, Ogawa W, Emi A, Hayashi K, Senga Y, Nomura K, Hara K, Yu D, Kasuga M. Role of S6K1 in regulation of SREBP1c expression in the liver. Biochem Biophys Res Commun. 2011;412(2):197–202. doi: 10.1016/j.bbrc.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 288.Um SH, Frigerio F, Watanabe M, Picard F, Joaquin M, Sticker M, Fumagalli S, Allegrini PR, Kozma SC, Auwerx J, Thomas G. Absence of S6K1 protects against age- and diet-induced obesity while enhancing insulin sensitivity. Nature. 2004;431(7005):200–205. doi: 10.1038/nature02866. [DOI] [PubMed] [Google Scholar]
- 289.Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, Behlke MA, Ory DS, Schaffer JE. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14(1):33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Chaudhuri S, Vyas K, Kapasi P, Komar AA, Dinman JD, Barik S, Mazumder B. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA. 2007;13(12):2224–2237. doi: 10.1261/rna.694007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Flygare J, Karlsson S. Diamond-Blackfan anemia: erythropoiesis lost in translation. Blood. 2007;109(8):3152–3154. doi: 10.1182/blood-2006-09-001222. [DOI] [PubMed] [Google Scholar]
- 292.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, Niewiadomska E, Da Costa L, Tchernia G, Niemeyer C, Meerpohl JJ, Stahl J, Schratt G, Glader B, Backer K, Wong C, Nathan DG, Beggs AH, Sieff CA. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79(6):1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Petrtylova K, Mihal V, Stary J, Cerna Z, Jabali Y, Pospisilova D. Identification of mutations in the ribosomal protein L5 (RPL5) and ribosomal protein L11 (RPL11) genes in Czech patients with Diamond-Blackfan anemia. Hum Mutat. 2009;30(3):321–327. doi: 10.1002/humu.20874. [DOI] [PubMed] [Google Scholar]
- 294.Gazda HT, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Zaucha JM, Glader B, Niemeyer C, Meerpohl JJ, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Ribosomal protein L5 and L11 mutations are associated with cleft palate and abnormal thumbs in Diamond-Blackfan anemia patients. Am J Hum Genet. 2008;83(6):769–780. doi: 10.1016/j.ajhg.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Gazda HT, Preti M, Sheen MR, O'Donohue MF, Vlachos A, Davies SM, Kattamis A, Doherty L, Landowski M, Buros C, Ghazvinian R, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Glader B, Atsidaftos E, Lipton JM, Gleizes PE, Beggs AH. Frameshift mutation in p53 regulator RPL26 is associated with multiple physical abnormalities and a specific pre-ribosomal RNA processing defect in diamond-blackfan anemia. Hum Mutat. 2012;33(7):1037–1044. doi: 10.1002/humu.22081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Doherty L, Sheen MR, Vlachos A, Choesmel V, O'Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE, Ball SE, Niewiadomska E, Matysiak M, Glader B, Arceci RJ, Farrar JE, Atsidaftos E, Lipton JM, Gleizes PE, Gazda HT. Ribosomal protein genes RPS10 and RPS26 are commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2010;86(2):222–228. doi: 10.1016/j.ajhg.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Landowski M, O'Donohue MF, Buros C, Ghazvinian R, Montel-Lehry N, Vlachos A, Sieff CA, Newburger PE, Niewiadomska E, Matysiak M, Glader B, Atsidaftos E, Lipton JM, Beggs AH, Gleizes PE, Gazda HT. Novel deletion of RPL15 identified by array-comparative genomic hybridization in Diamond-Blackfan anemia. Hum Genet. 2013;132(11):1265–1274. doi: 10.1007/s00439-013-1326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Choesmel V, Bacqueville D, Rouquette J, Noaillac-Depeyre J, Fribourg S, Cretien A, Leblanc T, Tchernia G, Da Costa L, Gleizes PE. Impaired ribosome biogenesis in Diamond-Blackfan anemia. Blood. 2007;109(3):1275–1283. doi: 10.1182/blood-2006-07-038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Danilova N, Sakamoto KM, Lin S. Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood. 2008;112(13):5228–5237. doi: 10.1182/blood-2008-01-132290. [DOI] [PubMed] [Google Scholar]
- 300.Barlow JL, Drynan LF, Trim NL, Erber WN, Warren AJ, McKenzie AN. New insights into 5q- syndrome as a ribosomopathy. Cell Cycle. 2010;9(21):4286–4293. doi: 10.4161/cc.9.21.13742. [DOI] [PubMed] [Google Scholar]
- 301.Starczynowski DT, Kuchenbauer F, Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R, Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16(1):49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 302.Pellagatti A, Marafioti T, Paterson JC, Barlow JL, Drynan LF, Giagounidis A, Pileri SA, Cazzola M, McKenzie AN, Wainscoat JS, Boultwood J. Induction of p53 and up-regulation of the p53 pathway in the human 5q- syndrome. Blood. 2010;115(13):2721–2723. doi: 10.1182/blood-2009-12-259705. [DOI] [PubMed] [Google Scholar]
- 303.Sakai D, Trainor PA. Treacher Collins syndrome: unmasking the role of Tcof1/treacle. Int J Biochem Cell Biol. 2009;41(6):1229–1232. doi: 10.1016/j.biocel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst. 2010;6(3):481–493. doi: 10.1039/b919670f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat Med. 2008;14(2):125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Ganapathi KA, Shimamura A. Ribosomal dysfunction and inherited marrow failure. Br J Haematol. 2008;141(3):376–387. doi: 10.1111/j.1365-2141.2008.07095.x. [DOI] [PubMed] [Google Scholar]
- 307.Burroughs L, Woolfrey A, Shimamura A. Shwachman-Diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol Oncol Clin North Am. 2009;23(2):233–248. doi: 10.1016/j.hoc.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Boocock GR, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat Genet. 2003;33(1):97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 309.Ganapathi KA, Austin KM, Lee CS, Dias A, Malsch MM, Reed R, Shimamura A. The human Shwachman-Diamond syndrome protein, SBDS, associates with ribosomal RNA. Blood. 2007;110(5):1458–1465. doi: 10.1182/blood-2007-02-075184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Rujkijyanont P, Adams SL, Beyene J, Dror Y. Bone marrow cells from patients with Shwachman-Diamond syndrome abnormally express genes involved in ribosome biogenesis and RNA processing. Br J Haematol. 2009;145(6):806–815. doi: 10.1111/j.1365-2141.2009.07692.x. [DOI] [PubMed] [Google Scholar]
- 311.Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107(12):4583–4588. doi: 10.1182/blood-2005-12-4831. [DOI] [PubMed] [Google Scholar]
- 312.Dhanraj S, Manji A, Pinto D, Scherer SW, Favre H, Loh ML, Chetty R, Wei AC, Dror Y. Molecular characteristics of a pancreatic adenocarcinoma associated with Shwachman-Diamond syndrome. Pediatr Blood Cancer. 2013;60(5):754–760. doi: 10.1002/pbc.24453. [DOI] [PubMed] [Google Scholar]
- 313.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, Roobeek I, Weis I, Voute PA, Schwab M, Versteeg R. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20(6):1383–1393. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69(23):8839–8843. doi: 10.1158/0008-5472.CAN-09-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlén S, Hydbring P, Söderberg O, Grummt I, Larsson LG, Wright AP. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol. 2005;7(3):303–310. doi: 10.1038/ncb1225. [DOI] [PubMed] [Google Scholar]
- 316.Schlosser I, Holzel M, Murnseer M, Burtscher H, Weidle UH, Eick D. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 2003;31(21):6148–6156. doi: 10.1093/nar/gkg794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature. 2008;456(7224):971–975. doi: 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Felton-Edkins ZA, Kenneth NS, Brown TR, Daly NL, Gomez-Roman N, Grandori C, Eisenman RN, White RJ. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2(3):181–184. [PubMed] [Google Scholar]
- 319.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2(5):E139. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O'Sullivan M, Lu J, Phillips LA, Lockhart VL, Shah SP, Tanwar PS, Mermel CH, Beroukhim R, Azam M, Teixeira J, Meyerson M, Hughes TP, Llovet JM, Radich J, Mullighan CG, Golub TR, Sorensen PH, Daley GQ. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.King CE, Wang L, Winograd R, Madison BB, Mongroo PS, Johnstone CN, Rustgi AK. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene. 2011;30(40):4185–4193. doi: 10.1038/onc.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Lindstrom MS, Jin A, Deisenroth C, White Wolf G, Zhang Y. Cancer-associated mutations in the MDM2 zinc finger domain disrupt ribosomal protein interaction and attenuate MDM2-induced p53 degradation. Mol Cell Biol. 2007;27(3):1056–1068. doi: 10.1128/MCB.01307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Macias E, Jin A, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang Y. An ARF-independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 Interaction. Cancer Cell. 2010;18(3):231–243. doi: 10.1016/j.ccr.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Shi Y, Zhai H, Wang X, Han Z, Liu C, Lan M, Du J, Guo C, Zhang Y, Wu K, Fan D. RPsS13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp Cell Res. 2004;296(2):337–346. doi: 10.1016/j.yexcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 325.Kondoh N, Shuda M, Tanaka K, Wakatsuki T, Hada A, Yamamoto M. Enhanced expression of S8, L12, L23a, L27 and L30 ribosomal protein mRNAs in human hepatocellular carcinoma. Anticancer Res. 2001;21(4A):2429–2433. [PubMed] [Google Scholar]
- 326.Song MJ, Jung CK, Park CH, Hur W, Choi JE, Bae SH, Choi JY, Choi SW, Han NI, Yoon SK. RPL36 as a prognostic marker in hepatocellular carcinoma. Pathol Int. 2011;61(11):638–644. doi: 10.1111/j.1440-1827.2011.02716.x. [DOI] [PubMed] [Google Scholar]
- 327.Kobayashi T, Sasaki Y, Oshima Y, Yamamoto H, Mita H, Suzuki H, Toyota M, Tokino T, Itoh F, Imai K, Shinomura Y. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int J Mol Med. 2006;18(1):161–170. [PubMed] [Google Scholar]
- 328.Lai MD, Xu J. Ribosomal proteins and colorectal cancer. Curr Genomics. 2007;8(1):43–49. doi: 10.2174/138920207780076938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Pogue-Geile K, Geiser JR, Shu M, Miller C, Wool IG, Meisler AI, Pipas JM. Ribosomal protein genes are overexpressed in colorectal cancer: isolation of a cDNA clone encoding the human S3 ribosomal protein. Mol Cell Biol. 1991;11(8):3842–3849. doi: 10.1128/mcb.11.8.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, Nakayama J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51(5):567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 331.Huang CJ, Chien CC, Yang SH, Chang CC, Sun HL, Cheng YC, Liu CC, Lin SC, Lin CM. Faecal ribosomal protein L19 is a genetic prognostic factor for survival in colorectal cancer. J Cell Mol Med. 2008;12(5B):1936–1943. doi: 10.1111/j.1582-4934.2008.00253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ojala P, Sundstrom J, Gronroos JM, Virtanen E, Talvinen K, Nevalainen TJ. mRNA differential display of gene expression in colonic carcinoma. Electrophoresis. 2002;23(11):1667–1676. doi: 10.1002/1522-2683(200206)23:11<1667::AID-ELPS1667>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 333.Huang CJ, Yang SH, Lee CL, Cheng YC, Tai SY, Chien CC. Ribosomal protein s27-like in colorectal cancer: a candidate for predicting prognoses. PLoS One. 2013;8(6):e67043. doi: 10.1371/journal.pone.0067043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bee A, Ke Y, Forootan S, Lin K, Beesley C, Forrest SE, Foster CS. Ribosomal protein l19 is a prognostic marker for human prostate cancer. Clin Cancer Res. 2006;12(7 Pt 1):2061–2065. doi: 10.1158/1078-0432.CCR-05-2445. [DOI] [PubMed] [Google Scholar]
- 335.Bee A, Brewer D, Beesley C, Dodson A, Forootan S, Dickinson T, Gerard P, Lane B, Yao S, Cooper CS, Djamgoz MB, Gosden CM, Ke Y, Foster CS. siRNA knockdown of ribosomal protein gene RPL19 abrogates the aggressive phenotype of human prostate cancer. PLoS One. 2011;6(7):e22672. doi: 10.1371/journal.pone.0022672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Ohkia A, Hu Y, Wang M, Garcia FU, Stearns ME. Evidence for prostate cancer-associated diagnostic marker-1: immunohistochemistry and in situ hybridization studies. Clin Cancer Res. 2004;10(7):2452–2458. doi: 10.1158/1078-0432.ccr-03-0170. [DOI] [PubMed] [Google Scholar]
- 337.Wang M, Hu YJ, Stearns ME. RPS2: a novel therapeutic target in prostate cancer. J Exp Clin Cancer Res. 2009;28:6. doi: 10.1186/1756-9966-28-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Huang XP, Zhao CX, Li QJ, Cai Y, Liu FX, Hu H, Xu X, Han YL, Wu M, Zhan QM, Wang MR. Alteration of RPL14 in squamous cell carcinomas and preneoplastic lesions of the esophagus. Gene. 2006;366(1):161–168. doi: 10.1016/j.gene.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 339.Wang Q, Yang C, Zhou J, Wang X, Wu M, Liu Z. Cloning and characterization of full-length human ribosomal protein L15 cDNA which was overexpressed in esophageal cancer. Gene. 2001;263(1–2):205–209. doi: 10.1016/s0378-1119(00)00570-9. [DOI] [PubMed] [Google Scholar]
- 340.Kim SH, Jang YH, Chau GC, Pyo S, Um SH. Prognostic significance and function of phosphorylated ribosomal protein S6 in esophageal squamous cell carcinoma. Mod Pathol. 2013;26(3):327–335. doi: 10.1038/modpathol.2012.161. [DOI] [PubMed] [Google Scholar]
- 341.Yang M, Sun H, Wang H, Zhang S, Yu X, Zhang L. Down-regulation of ribosomal protein L22 in non-small cell lung cancer. Med Oncol. 2013;30(3):646. doi: 10.1007/s12032-013-0646-0. [DOI] [PubMed] [Google Scholar]
- 342.Zheng SE, Yao Y, Dong Y, Lin F, Zhao H, Shen Z, Sun YJ, Tang LN. Down-regulation of ribosomal protein L7A in human osteosarcoma. J Cancer Res Clin Oncol. 2009;135(8):1025–1031. doi: 10.1007/s00432-008-0538-4. [DOI] [PubMed] [Google Scholar]
- 343.Hagner PR, Mazan-Mamczarz K, Dai B, Balzer EM, Corl S, Martin SS, Zhao XF, Gartenhaus RB. Ribosomal protein S6 is highly expressed in non-Hodgkin lymphoma and associates with mRNA containing a 5' terminal oligopyrimidine tract. Oncogene. 2011;30(13):1531–1541. doi: 10.1038/onc.2010.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Wu LY, Li X, Xu F, Chang CK, He Q, Zhang Z, Zhng Y. Over-expression of RPL23 in myelodysplastic syndromes is associated with apoptosis resistance of CD34(+) cells and predicts poor prognosis and distinct response to CHG chemotherapy or decitabine. Ann Hematol. 2012;91(10):1547–1554. doi: 10.1007/s00277-012-1486-2. [DOI] [PubMed] [Google Scholar]
- 345.Tsofack SP, Meunier L, Sanchez L, Madore J, Provencher D, Mes-Masson AM, Lebbel M. Low expression of the X-linked ribosomal protein S4 in human serous epithelial ovarian cancer is associated with a poor prognosis. BMC cancer. 2013;13:303. doi: 10.1186/1471-2407-13-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Flygare J, Olsson K, Richter J, Karlsson S. Gene therapy of Diamond Blackfan anemia CD34(+) cells leads to improved erythroid development and engraftment following transplantation. Exp Hematol. 2008;36(11):1428–1435. doi: 10.1016/j.exphem.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 347.Wei S, Chen X, McGraw K, Zhang L, Komrokji R, Clark J, Caceres G, Billingsley D, Sokol L, Lancet J, Fortenbery N, Zhou J, Eksioglu EA, Sallman D, Wang H, Epling-Burnette PK, Djeu J, Sekeres M, Maciejewski JP, List A. Lenalidomide promotes p53 degradation by inhibiting MDM2 auto-ubiquitination in myelodysplastic syndrome with chromosome 5q deletion. Oncogene. 2013;32(9):1110–1120. doi: 10.1038/onc.2012.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Olausson KH, Nistér M, Lindström MS. p53-Dependent and -Independent Nucleolar Stress Responses. Cells. 2012;1:774–798. doi: 10.3390/cells1040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hwang IK, Yoo KY, Kim DW, Kim SY, Park JH, Ryoo ZY, Kim J, Choi SY, Won MH. Ischemia-induced ribosomal protein S3 expressional changes and the neuroprotective effect against experimental cerebral ischemic damage. J Neurosci Res. 2008;86(8):1823–1835. doi: 10.1002/jnr.21621. [DOI] [PubMed] [Google Scholar]
- 350.Ahn EH, Kim DW, Shin MJ, Kim YN, Kim HR, Woo SJ, Kim SM, Kim DS, Kim J, Park J, Eum WS, Hwang HS, Choi SY. PEP-1-ribosomal protein S3 protects dopaminergic neurons in an MPTP-induced Parkinson's disease mouse model. Free Radic Biol Med. 2013;55:36–45. doi: 10.1016/j.freeradbiomed.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 351.Lv J, Huang XR, Klug J, Frohlich S, Lacher P, Xu A, Meinhardt A, Lan HY. Ribosomal protein S19 is a novel therapeutic agent in inflammatory kidney disease. Clin Sci (Lond) 2013;124(10):627–637. doi: 10.1042/CS20120526. [DOI] [PubMed] [Google Scholar]
- 352.Bywater MJ, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe MK, Ryckman DM, Rice WG, Schmitt C, Lowe SW, Johnstone RW, Pearson RB, McArthur GA, Hannan RD. Inhibition of RNA Polymerase I as a Therapeutic Strategy to Promote Cancer-Specific Activation of p53. Cancer Cell. 2012;22(1):51–65. doi: 10.1016/j.ccr.2012.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Drygin D, Lin A, Bliesath J, Ho CB, O'Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71(4):1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 354.Drygin D, Siddiqui-Jain A, O'Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JK, Von Hoff D, Anderes K, Rice WG. Anticancer Activity of CX-3543: A Direct Inhibitor of rRNA Biogenesis. Cancer Res. 2009;69(19):7653–7661. doi: 10.1158/0008-5472.CAN-09-1304. [DOI] [PubMed] [Google Scholar]
- 355.Wu CT, Lin TY, Hsu HY, Sheu F, Ho CM, Chen EIT. Ling Zhi-8 mediates p53-dependent growth arrest of lung cancer cells proliferation via the ribosomal protein S7-MDM2-p53 pathway. Carcinogenesis. 2011;32(12):1890–1896. doi: 10.1093/carcin/bgr221. [DOI] [PubMed] [Google Scholar]
- 356.Zhang YF, Shi YQ, Li XH, Du WQ, Luo GH, Gou YW, Wang X, Guo X, Liu J, Ding J, Wu K, Fan D. Inhibition of the p53-MDM2 interaction by adenovirus delivery of ribosomal protein L23 stabilizes p53 and induces cell cycle arrest and apoptosis in gastric cancer. J Gene Med. 2010;12(2):147–156. doi: 10.1002/jgm.1424. [DOI] [PubMed] [Google Scholar]
- 357.Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol Biol Rep. 1994;19(3):183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 358.O'Brien TW. Properties of human mitochondrial ribosomes. IUBMB Life. 2003;55(9):505–513. doi: 10.1080/15216540310001626610. [DOI] [PubMed] [Google Scholar]
- 359.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276(35):33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- 360.Kissil JL, Cohen O, Raveh T, Kimchi A. Structure-function analysis of an evolutionary conserved protein, DAP3, which mediates TNF-alpha- and Fas-induced cell death. EMBO J. 1999;18(2):353–362. doi: 10.1093/emboj/18.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Carim L, Sumoy L, Nadal M, Estivill X, Escarceller M. Cloning, expression, and mapping of PDCD9, the human homolog of Gallus gallus pro-apoptotic protein p52. Cytogenet Cell Genet. 1999;87(1–2):85–88. doi: 10.1159/000015397. [DOI] [PubMed] [Google Scholar]
- 362.Cavdar Koc E, Ranasinghe A, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL. A new face on apoptosis: death-associated protein 3 and PDCD9 are mitochondrial ribosomal proteins. FEBS Lett. 2001;492(1–2):166–170. doi: 10.1016/s0014-5793(01)02250-5. [DOI] [PubMed] [Google Scholar]
- 363.Levshenkova EV, Ukraintsev KE, Orlova VV, Alibaeva RA, Kovriga IE, Zhugdernamzhilyn O, Frolova EI. The structure and specific features of the cDNA expression of the human gene MRPL37. Bioorg khim. 2004;30(5):499–506. doi: 10.1023/b:rubi.0000043788.86955.ea. [DOI] [PubMed] [Google Scholar]
- 364.Chintharlapalli SR, Jasti M, Malladi S, Parsa KV, Ballestero RP, Gonzalez-Garcia M. BMRP is a Bcl-2 binding protein that induces apoptosis. J Cell Biochem. 2005;94(3):611–626. doi: 10.1002/jcb.20292. [DOI] [PubMed] [Google Scholar]
- 365.Yoo YA, Kim MJ, Park JK, Chung YM, Lee JH, Chi SG, Kim JS, Yoo YD. Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol. 2005;25(15):6603–6616. doi: 10.1128/MCB.25.15.6603-6616.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kim MJ, Yoo YA, Kim HJ, Kang S, Kim YG, Kim JS, Yoo YD. Mitochondrial ribosomal protein L41 mediates serum starvation-induced cell-cycle arrest through an increase of p21(WAF1/CIP1) Biochem Biophys Res Comm. 2005;338(2):1179–1184. doi: 10.1016/j.bbrc.2005.10.064. [DOI] [PubMed] [Google Scholar]
- 367.Charafe-Jauffret E, Ginestier C, Iovino F, Wicinski J, Cervera N, Finetti P, Hur MH, Diebel ME, Monville F, Dutcher J, Brown M, Viens P, Xerri L, Bertucci F, Stassi G, Dontu G, Birnbaum D, Wicha MS. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009;69(4):1302–1313. doi: 10.1158/0008-5472.CAN-08-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Chen YC, Chang MY, Shiau AL, Yo YT, Wu CL. Mitochondrial ribosomal protein S36 delays cell cycle progression in association with p53 modification and p21(WAF1/CIP1) expression. J Cell Biochem. 2007;100(4):981–990. doi: 10.1002/jcb.21079. [DOI] [PubMed] [Google Scholar]
- 369.Kashuba E, Pavan Yenamandra S, Darekar SD, Yurchenko M, Kashuba V, Klein G, Szekely L. MRPS18-2 protein immortalizes primary rat embryonic fibroblasts and endows them with stem cell-like properties. Proc Natl Acad Sci. 2009;106(47):19866–19871. doi: 10.1073/pnas.0911545106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Chung HK, Yi YW, Jung NC, Kim D, Suh JM, Kim H, Park KC, Song JH, Kim DW, Hwang ES, Yoon SH, Bae YS, Kim JM, Bae I, Shong M. CR6-interacting factor 1 interacts with Gadd45 family proteins and modulates the cell cycle. J Biol Chem. 2003;278(30):28079–28088. doi: 10.1074/jbc.M212835200. [DOI] [PubMed] [Google Scholar]
- 371.Handa Y, Hikawa Y, Tochio N, Kogure H, Inoue M, Koshiba S, Güntert P, Inoue Y, Kigawa T, Yokoyama S, Nameki N. Solution structure of the catalytic domain of the mitochondrial protein ICT1 that is essential for cell vitality. J Mol Biol. 2010;404(2):260–273. doi: 10.1016/j.jmb.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 372.Chen D, Zhang R, Shen W, Fu H, Liu S, Sun K, Sun X. RPS12-specific shRNA inhibits the proliferation, migration of BGC823 gastric cancer cells with S100A4 as a downstream effector. Int J Oncol. 2013;42(5):1763–1769. doi: 10.3892/ijo.2013.1872. [DOI] [PubMed] [Google Scholar]
- 373.Li H, Beeghly-Fadiel A, Wen W, Lu W, Gao YT, Xiang YB, Cai Q, Long J, Shi J, Chen K, Zheng Y, Shu XO, Zheng W. Gene-environment interactions for breast cancer risk among Chinese women: a report from the Shanghai Breast Cancer Genetics Study. Am J Epidemiol. 2013;177(2):161–170. doi: 10.1093/aje/kws238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.O'Brien KM, Cole SR, Poole C, Bensen JT, Herring AH, Engel LS, Millikan RC. Replication of breast cancer susceptibility loci in whites and African Americans using a Bayesian approach. Am J Epidemiol. 2014;179(3):382–394. doi: 10.1093/aje/kwt258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Bursac S, Brdovcak MC, Donati G, Volarevic S. Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim Biophys Acta. 2014;1842(6):817–830. doi: 10.1016/j.bbadis.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 376.Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD. Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. 2014;1842(6):802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 377.Kapoor NR, Ahuja R, Shukla SK, Kumar V. The HBx protein of hepatitis B virus confers resistance against nucleolar stress and anti-cancer drug-induced p53 expression. FEBS Lett. 2013;587(9):1287–1292. doi: 10.1016/j.febslet.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 378.Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7(1):1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]