Abstract
A new member of the Orthomyxoviridae family, influenza D virus (IDV), was first reported in swine in the Midwest region of the United States. This study aims to extend our knowledge on the IDV epidemiology and to determine the impact of bovine production systems on virus spread. A total of 15 isolates were recovered from surveillance of bovine herds in Mississippi, and two genetic clades of viruses co-circulated in the same herd. Serologic assessment from neonatal beef cattle showed 94% seropositive, and presumed maternal antibody levels were substantially lower in animals over six months of age. Active IDV transmission was shown to occur at locations where young, weaned, and comingled calves were maintained. Serological characterization of archived sera suggested that IDV has been circulating in the Mississippi cattle populations since at least 2004. Continuous surveillance is needed to monitor the evolution and epidemiology of IDV in the bovine population.
Keywords: Influenza D virus, Surveillance, Beef cattle, Influenza virus, Serological surveillance, Maternal antibody, Mississippi
Highlights
-
•
A total of 10 influenza D viruses (IDVs) were recovered from beef cattle at Mississippi.
-
•
2 antigenically distinct clades of viruses co-circulated in the same herd.
-
•
Active IDV transmission occurs among young, weaned, and comingled calves.
-
•
Serological surveillance suggests that IDV has been present in the Mississippi cattle population at least since 2004.
Introduction
Influenza viruses belong to the Orthomyxoviridae family and comprise three of its genera: Influenza virus A, B, and C (IAV, IBV, and ICV, respectively). ICV is commonly associated with fever, coughing, and rhinorrhea in children, and occasionally lower respiratory tract infections in infants (Matsuzaki et al., 2006). In addition to humans, swine infection with human ICV has been reported in China (Guo et al., 1983). In 2011, a novel influenza virus was reported in swine from Oklahoma (Hause et al., 2013) with 50% protein sequence identity with ICV but no cross-reactivity with human ICV generated serum. This virus was provisionally defined as influenza D virus (IDV) (Hause et al., 2014). IDV is a single-strand, negative sense RNA virus with 7 genome segments that are predicted to encode 9 proteins, including glycoprotein hemagglutinin–esterase fusion (HE), polymerases PB2, PB1, and P3, nucleoprotein, matrix proteins (M1 and CM2), and nonstructural proteins (NS1 and NEP). To date, bovine IDV has been reported in United States, France, and China (Ducatez et al., 2015, Hause et al., 2013, Jiang et al., 2014). In the United States, IDV has been reported in cattle population in Texas, California, and in Midwestern states, such as Kansas, Minnesota, Nebraska, and Oklahoma, suggesting, based on serological investigation, that cattle are the natural host reservoir of this new virus (Hause et al., 2014). Furthermore, metagenomic analysis showed that IDV is one of the common microbes identified via metagenomics sequencings of Californian dairy calves diagnosed as bovine respiratory diseases (BRD) from single farm (Ng et al., 2015), suggesting a correaltion of IDV with BRD.
The objectives of this study were to further examine the presence of IDV in Mississippi and to determine the impact of beef production management practices on virus infection and maintenance. Our results showed that IDV has been present in the Mississippi cattle population since at least 2004 and that weaned, comingled calves likely play an important role in maintenance and transmission of IDV.
Results
Virus recovered from Mississippi Cattle
Of the 55 sick calves, 18 (32.7%) were seropositive for IDV. The GMT for IDV of the 55 sick calves was 172.8 (±215.7) ( Table 1 ). Each sick calf had been held at the facility for an average of 24.0 (±15.7) days at time of sampling, and had received 1.9 (±1.0) treatments for respiratory disease. Respiratory swabs testing positive by quantitative RT-PCR for IDV were obtained from 16 of 55 (29.1%) sick calves and 2 of 82 (2.4%) healthy calves. Thirteen of the 16 positive specimens from sick calves and both of the positive specimens from healthy calves were succesfully isolates in cell culture. Two samples had signs of myotic contamination due to lack of anti-fungal in the transport media, but filtration after primary passage made it possible to isolate one of these two contaminated samples.
Table 1.
Summary of the serum samples collected in Mississippi cattle.
| Dataset | Year | Management style | Clinical signs | Age | Seropositive rate (n) | GMT |
|---|---|---|---|---|---|---|
| I | 2014 | Order-buyer | No | 6–8 m | 22.5% (89a) | 1:91.9 (±56.0) |
| 2014 | Order-buyer | Yes | 6–8 m | 32.7% (55) | 1:172.8 (±215.7) | |
| II | 2013 | Cow–calf operation | No | 24–36 h | 95.1 (284) | 1:345.6 (±491.6) |
| 2014 | Cow–calf operation | No | 24–36 h | 92.1 (164) | 1:557.7 (±498.6) | |
| III | 2004 | Cow–calf operation | No | 6 m–12 y | 18.3% (241) | 1:146.9 (±45.9) |
| 2005 | Cow–calf operation | No | 6–8 m | 14.8% (223) | 1:63.5 (±30.2) | |
| 2006 | Cow–calf operation | No | 6 m–14 y | 13.5% (141) | 1:83.0 (±53.9) |
These 89 sera were collected from 82 individual calves, and seven of these calves were sampled twice.
For the 45 sick calves which had both nasal and nasopharyngeal specimens taken, our results showed that 8 of the 45 (17.8%) were nasally positive and 7 of 45 (15.6%) were nasopharyngeally positive, however only 3 of the 45 (6.7%) were both nasally and nasopharyngeally positive.
Among these fifteen isolates, four isolates, including D/bovine/Mississippi/C00046N/2014, D/bovine/Mississippi/C00013N/2014, D/bovine/Mississippi/C00030P/2014, and D/bovine/Mississippi/C00014N/2014, were selected to determine whether multiple viruses were once isolated. Results showed that these four isolates were negative against BVDV, PI3, adenovirus, coronavirus, BHV, and BRSV after isolation in HRT-18G cells.
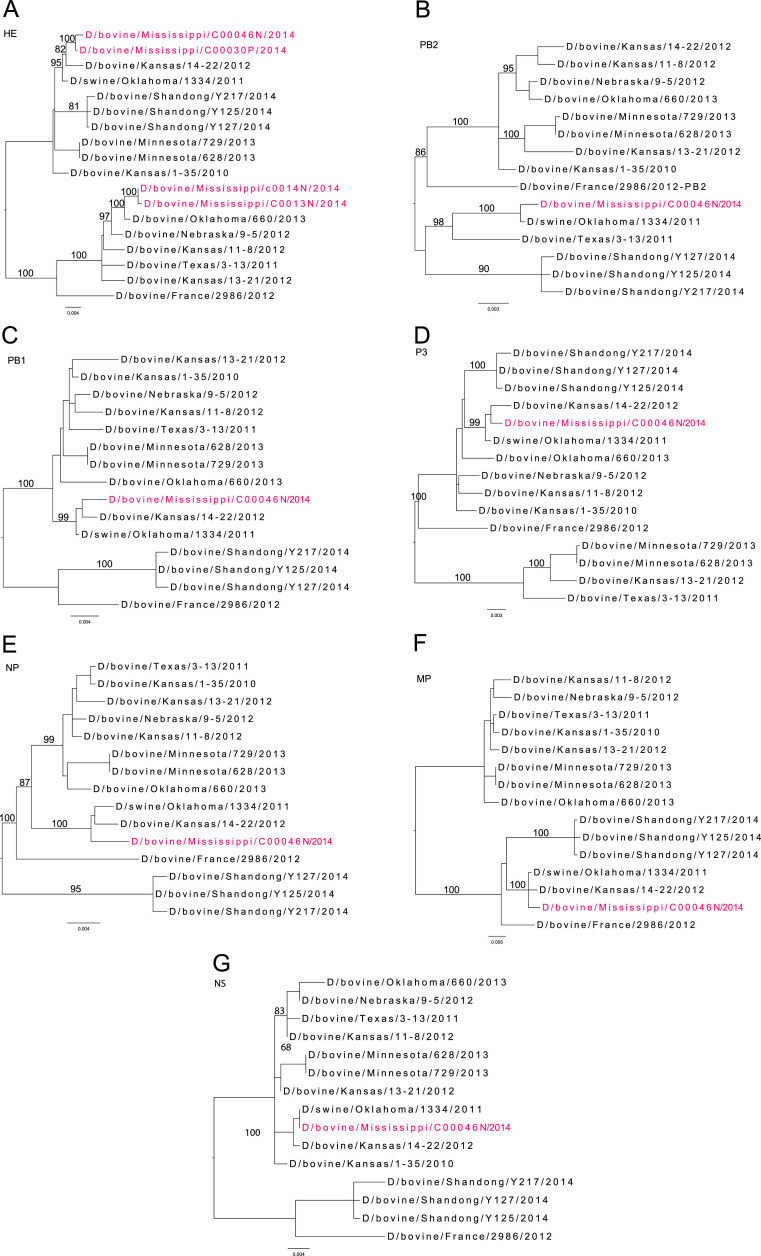
The HE gene of IDV isolates D/bovine/Mississippi/C00046N/2014, D/bovine/Mississippi/C00013N/2014, D/bovine/Mississippi/C00030P/2014, and D/bovine/Mississippi/C00014N/2014 were sequenced. Phylogenetic analyses of the HE sequence aligned the IDV isolates from Mississippi into two genetic clusters: D/bovine/Mississippi/C00046N/2014 and D/bovine/Mississippi/C00030P/2014's HE sequence were genetically close to D/swine/Oklahoma/1334/2011 (D/OK), and D/bovine/Mississippi/C00013N/2014 and D/bovine/Mississippi/C00014N/2014 were genetically close to D/bovine/Oklahoma/660/2013 (D/660) ( Fig. 1A).
Fig. 1.
Phylogenetic analyses of influenza D virus from Mississippi cattle: (A) HE, (B) PB2, (C) PB1, (D) P3, (E) NP, (F) MP, and (G) NS. The isolates from Mississippi are shown in red.
The genome of D/bovine/Mississippi/C00046N/2014 was fully sequenced. Phylogenetic analyses aligned the PB2, NP, and NS genes of D/bovine/Mississippi/C00046N/2014 (Fig. 1) to D/swine/Oklahoma/1334/2011 with 99.27–100% nucleotide sequence identities ( Table 3) and the PB1, P3, HE, and MP genes to D/bovine/Kansas/14-22/2012 with 99.07–99.50% identity (Table 3). Predicted key residues in the receptor binding and antibody binding domains were conserved between D/bovine/Mississippi/C00046N/2014 and D/swine/Oklahoma/1334/2011 (data not shown) (Hause et al., 2013).
Table 3.
Sequence identity of D/bovine/Mississippi/C00046N/2014 compared to closely related IDVs in public database.
| Gene segment | Virus | Identity percentile (amino acid/nucleotide) |
|---|---|---|
| HE | D/bovine/Kansas/14–22/2012 | 98.80%/99.07% |
| PB2 | D/swine/Oklahoma/1334/2011 | 99.87%/99.79% |
| PB1 | D/bovine/Kansas/14–22/2012 | 99.60%/99.44% |
| P3 | D/bovine/Kansas/14–22/2012 | 99.58%/99.50% |
| NP | D/swine/Oklahoma/1334/2011 | 99.46%/99.27% |
| MP | D/bovine/Kansas/14–22/2012 | 99.74%/99.34% |
| NS | D/swine/Oklahoma/1334/2011 | 100%/100% |
Sampling at two time-points showed that IDV infection could occur after admission to the conditioning yards
Sampling was performed from two lots of calves to determine the dynamics of IDV infection at an order-buying facility. Convenience samples were collected on the first day of arrival at the facility on March 26th at Lot 1 (n=19) and May 16th at Lot 2 (n=27). Seroprevalence was 21.1% with a GMT of 1:80 (±42.9) and 22.2% with GMT of 1:127.0 (±76.7) upon arrival at Lots 1 and 2, respectively. One week after arrival, seroprevalence was similar at 22.2% seropositive with GMT of 56.6 (±27.3) and 33.3% seropositive with GMT of 1:113.1 (±66.9) in Lots 1 (n=18) and Lot 2 (n=6), respectively. Five calves from Lot 1 (calf ID 1–5; Table 4), and two from Lot 2 (calf ID 6 and 7) were sampled twice, upon entry and one-week post-arrival. Among these seven calves, calf ID 2, 3, 5, and 6 were seronegative to IDV; calf ID 4 maintained an HAI titer of 1:80 throughout the study; calf ID 7 had an increase in HI titer from 0 to 1:80 one week post arrival; and calf ID 1 dropped in titer from 1:160 to 1:80. These samples make up Table 1, dataset I and came from calves without apparent clinical signs.
Table 4.
Serological results for the longitudinal serum samples collected in Mississippi cattle against D/swine/Oklahoma/1334/2011 using HAI assay.
| Calf ID | Sample ID | Sampling datea | HAI titer | Sample ID | Sampling date | HAI titer |
|---|---|---|---|---|---|---|
| 1 | C00047 | March 26, 2014 | 1:160 | C00066 | April 2, 2014 | 1:80 |
| 2 | C00050 | March 26, 2014 | 0 | C00073 | April 2, 2014 | 0 |
| 3 | C00054 | March 26, 2014 | 0 | C00068 | April 2, 2014 | 0 |
| 4 | C00057 | March 26, 2014 | 1:80 | C00066 | April 2, 2014 | 1:80 |
| 5 | C00065 | March 26, 2014 | 0 | C00075b | April 2, 2014 | 0 |
| 6 | C00107 | May 16, 2014 | 0 | C00132 | May 23, 2014 | 0 |
| 7 | C00118 | May 16, 2014 | 0 | C00140 | May 23, 2014 | 1:80 |
This date is when the cattle enter the conditioning yard.
Nasal swab for a cattle was quantitative RT-PCR positive for bovine influenza D virus at the time of sampling.
Nasal swabs were also collected throughout the study and evaluated for presence of IDV PCR products. No calves were quantitative RT-PCR positive at arrival, but 2 calves were positive at one-week post arrival. Calf ID 5 was negative for IDV upon arrival but was positive upon sampling one week after arrival. Another calf (sample ID C00067) sampled only on April 2nd one week post-entry to the site, was also quantitative RT-PCR positive; both quantitative RT-PCR positive calves were housed at Lot 2 (Table 4).
Maternal antibodies highly prevalent in calves
To determine the level of passively acquired maternal antibody, we tested serum samples collected from neonatal calves 24–36 h after birth for D/bovine/Mississippi/C00046N/2014 specific antibodies. After 24–36 h of age, calves can no longer absorb antibody through the intestinal epithelia (Quigley et al., 2002). Of the 448 samples from 2013 (n=284) and 2014 (n=164), 95.1% and 92.1% of calves were seropositive against IDV with a GMT of 1:345.6 (±491.5), and GMT of 1:557.7 (±498.6), respectively. Altogether, of the 448 serum samples, there was a 94.0% IDV seropositive rate among neonatal calves with a GMT of 1:410.3 (±499.5) (Table 1; Fig. 2A).
Fig. 2.
Influenza D virus specific antibody responses in different ages of healthy cattle: (A) calves which are younger than 1 year from 2013 to 2014; (B) cattle which is at least 1 year old from 2004–2006. The serological analysis was performed by hemagglutination inhibition (HI) assay. The serum with a HAI titer of ≥1:40 was defined as seropositive.
Results from archived serum samples suggests that IDV is prevalent in some Mississippi cow–calf herds
To explore the historical seroprevalence of IDV in Mississippi cattle, we tested 605 samples collected from 484 six to eight month-old calves and 121 cows aged >1 year from a single farm from 2004 to 2006. The seroprevalence of cattle sampled in 2004, 2005, and 2006 was 18.3% (n=241), 14.8% (n=223), and 13.5% (n=141), respectively with an overall rate of 15.9%. The IDV GMT was 71.8 (±43.1) over all years and 1:63.5 (±30.2), 1:146.9 (45.9), and 1:83.0 (±53.9) for 2004, 2005, and 2006, respectively (Table 1, dataset III). These results demonstrated that IDV exposure has been present in some Mississippi cattle herds since at least 2004.
To analyze the effect of age on IDV seroprevalence, the sera were categorized into six groups according to age at time of sampling, including 6 month (n=52), 7 month (n=244), 8 month (n=188), 1–3 year old heifers (n=64), 3–9 year old cows (n=33), and 9–14 year old cows (n=24). The seroprevalence in these groups was 11.5%, 3.7%, 6.9%, 54.7%, 60.6%, 54.2%, respectively (Fig. 2B and Table 1). Furthermore, in the 1–3 year old age group (n=64), the cattle at one year of age (n=42) had a seroprevalence of 66.7% against IDV with a GMT of 1:67.3 (±46.3).
Discussion
Bovine IDV has been reported previously in either healthy cattle or sick cattle from China, France, and seven states in the U.S. (Collin et al., 2015, Ducatez et al., 2015, Hause et al., 2013, Jiang et al., 2014, Ng et al., 2015). Detection rates of IDV in healthy calves and calves with respiratory disease were higher in this study than previously reported. In China, the quantitative RT-PCR detection rates of IDV in healthy cattle from Shandong, China was 0.7% of 453 cattle (Jiang et al., 2014). Our sampling of healthy weaned calves showed 2.4% of 82 calves IDV were RT-PCR positive.
Prevalence of IDV in cattle with signs of respiratory disease in France and the United States were 4.5% of 134 bovine and 4.8% of 208 bovine, respectively (Collin et al., 2015, Ducatez et al., 2015), whereas we found 23.6% of 55 weaned calves with respiratory disease were IDV quantitative RT-PCR positive. The higher rates of detection we observed might be due to our targeted sampling of six to eight-month old calves that were immunologically naïve, comingled, and recently weaned. Although we were unable to document active infection among the 65 calves at arrival to the order-buying facility, active infections were observed 1 week after arrival in two of the 24 calves sampled. Seroprevalence was 21.5% on arrival and did not appear to change within the first week following arrival among healthy calves. This data suggests the transmission of IDV occurred after arrival in this population of highly stressed, comingled, and immunologically naïve calves based on the proportion of IDV positive respiratory swabs among calves with respiratory disease being greater than healthy calves. However, an epidemiological study is needed to test the hypothesis that IDV infection would be associated with BRD. Sick calves that were IDV positive had been at the facility for 19.4 days (±7.5), suggesting that active infection could occur as early as one week post-entry to the order-buying facility. The two apparently healthy calves that were IDV positive had both been maintained in the same lot, but had only been at the facility for seven days.
BRD is a multi-agent, multi-factorial disease with numerous causative agents. The contribution of IDV in respiratory disease in the sick calves of this study is unknown; however, IDV is present in cattle with respiratory disease signs and potentially could be one of the numerous agents that alter host defenses and contribute to the pathogenesis of respiratory disease in bovines. A metagenomics analysis of the respiratory specimens from 50 dairy calves with BRD demonstrated that contig reads of bovine adenovirus 3, bovine rhinitis A virus, and IDV were identified among in 62% of these specimens, either alone or altogether (Ng et al., 2015). On the other hand, it is possible that other respiratory diseases could contribute to higher IDV positive rate in sick herds than in healthy herds.
Within the first 24 h of life, calves acquire passive immunity through maternal immunoglobulins in colostrum (Quigley et al., 2002). The half-life of passively acquired IgG is typically 21.2–35.9 days; therefore, calves maintain passive immunity for roughly three to four months (Fulton et al., 2002). However, the dynamics of maternal antibodies are pathogen dependent. Our results demonstrated that 94% of neonatal cattle sampled in 2013 and 2014 obtained maternal immunity against IDV via maternal colostrum, which appeared to decrease with age. At the age of six to eight months, only 3.7–11.5% of calves were seropositive. Thus, after six months, many cattle were likely susceptible to IDV infection. Furthermore, when we look at the seroprevalence from youth to one year, we found an increase from 11.7% at 8 months (n=188) to 66.7% at 1 year (n=42). It is understood that management style can affect cattle overall health. In our study, we found that six to eight-month old healthy calves held in an order-buyer facility had a seropositive rate of 21.5% at entry (n=65) and 25.0% at one week (n=24); whereas six to eight months calves in a cow-calf operation had a seropositive rate of 5.8% (n=484). We attribute these differences to the fact that cattle held at the order-buyer facilities were all weaned, transported, auctioned, and comingled among cattle of similar ages and similar susceptibilities. However, our observations are limited based on differences in sample size, sampling style, and sampling time.
The HE gene segments that were sequenced four isolates collected from sick calves sampled on two occasions: February 7th, 2014 (D/bovine/Mississippi/C00013N/2014 and D/bovine/Mississippi/C00014N/2014) and February 19th, 2014 (D/bovine/Mississippi/C00030P/2014 and D/bovine/Mississippi/C00046N/2014) (Fig. 1A). Two genetic clusters (D/OK-like and D/660-like viruses) for IDV have been identified to be antigenically distinct. In this study, D/OK and D/660 were chosen as the two prototype viruses for these two representative clusters (Fig. 1). Our study found both D/OK-like and D/660-like viruses co-circulating in the same Mississippi order-buying facility. The four isolates shown in the Fig. 1A were randomly chosen, and the fully sequenced isolate was preferentially chosen because it cultured well in HRT-18G cells. Future work will involve further surveillance, deep-sequencing of all recovered isolates recovered from this order-buyer location, antigenic characterization, and viral growth kinetics.
The molecular characterization suggested that the genes of D/bovine/Mississippi/C00046N/2014 were 99.27–100% identical at nucleotide level to D/bovine/Oklahoma/1334/2011. There were no mutations identified among the receptor binding and antibody binding sites between D/bovine/Mississippi/C00046N/2014 and D/swine/Oklahoma/1334/2011. Previous work has yet to define the nucleotide substitution rate of IDV due to the lack of sequenced isolates, however, it has been suspected that IDV would evolve at rates slower than IAV and IBV (Sheng et al., 2014).
This study suggests that IDV has been present in at least some Mississippi cattle herds at least since 2004 and that weaned, comingled calves can support transmission of IDV infections. At least two viruses, phylogenetically similar to two antigenically distinct clusters, are co-circulating in the Mississippi cattle population. Lastly, we are limited in our representation of antigenic diversity of serum samples since only D/bovine/C00046N/Oklahoma/2014 was used in serological surveillance. D/bovine/C00046N/Oklahoma/2014 only represents one of two co-circulating antigenic clusters in Mississippi, and thus the actual seropositive rates against IDV could be potentially higher. Continuous surveillance of IDV in the bovine population would require increased sampling, locations, and time points to adequately monitor the evolution and epidemiology of IDV in Mississippi.
Materials and methods
Sample collection
Blood was collected from the jugular vein using individual disposable 18 gauge needles and blood tubes under vacuum, and serum was separated via centrifugation. Nasal and nasopharyngeal swabs were collected using Starswabs II (Starplex Scientific, Cleveland, TN) and 33″ Double Guarded Culture Swabs (Santa Cruz Animal Health, Dallas, TX), respectively. Swabs were transported in 1 mL of Media 199 (Gibco® Life Technologies, Grand Island, NY) with 1% PenStrep (Gibco® Life Technologies) in 4 mL Corning® Cryogenic Vials (Corning®, Tewksbury, MA). Swabs and blood specimens were kept on ice in a cooler after sampling. Swabs were stored at −80 °C until further analysis, and serum was stored at −20 °C.Three populations of beef cattle were examined in this study.Population 1 (Table 1, dataset I): from February to May 2014, sera, nasal swabs, and nasopharyngeal swabs were collected from 137 six to 9 month old calves at a cattle order-buying facility in Mississippi. The order-buying facility's role in cattle production is to buy recently weaned calves from auction yards to fill orders from backgrounding and finishing operations. To complete the orders, calves are comingled, sorted by weight and other characteristics. Calves purchased by order-buyers are usually immunologically naïve calves at increased risk to develop BRD. Pre-conditioning programs offered by this order-buyer are designed to reduce BRD incidence by strategic use of vaccines, antibiotics, and nutritious rations for 60–90 days. These calves are subsequently moved either to extensive grazing operations known as a stocker operation, or to intensive feeding facilities called feedlots, for finishing prior to harvest for beef.
Calves were housed and maintained in purchase lots and individually moved to a hospital barn for treatment if any signs of illness occurred. In our surveillance of Population 1, a total of 144 samples were collected from 137 individual calves. Of the 144 samples, 55 were from calves showing signs of clinical BRD (referred to as “sick”) whose samples were collected in February (n=45) and May (n=10) of 2014 in the order-buyer facility's hospital barn. Sick calves that were sampled at the hospital barn were not all held in the same purchase lot prior to sampling. A total of 55 sick came from a total of 25 different purchase lots with an average of 2.2 (±2.2) calves per purchase lot. Sera and nasal swabs were collected from all sick calves and nasopharyngeal swabs from 46 of the 55 calves. Eighty-two calves were sampled while without apparent illness and were held in four separate purchase lots (referred to as “healthy”). Serum and nasal swab specimens were obtained from 65 animals within 24 h of arrival at the facility with a further 24 sera and nasal swab specimens collected from calves one week after entry (seven animals were sampled twice). Healthy calves were sampled in March (n=19), April (n=37), and May (n=33).
Population 2 (Table 1, dataset II): sera was collected for serologic analysis from healthy 24–36 h-old calves from two Mississippi farms, one in 2013 (n=284) and another in 2014 (n=164).Population 3 (Table 1, dataset III): 605 archived sera collected from 484 six to eight-month old calves and 121 cows age >1 year were serologically analyzed. Serum samples were collected from a single farm from 2004 to 2006, and were stored at −80 °C following collection.Populations 2 and 3 are known as cow–calf farms, and are relatively stable populations of adult cows kept for breeding to produce and grow calves for three to twelve months. Calves reared in cow–calf operations are typically sold directly or through auction markets to feedlots or stocker operations.RNA extraction, quantitative RT-PCR, PCR, and genomic sequencingGeneJET Viral DNA/RNA Purification Kit was used for RNA extraction using the manufacture's protocol (Thermo Scientific, Pittsburgh PA). Viral RNA was tested for the presence of IDV using quantitative RT-PCR with TaqMan Fast Virus 1-step Master Mix (Life Technology, Carlsbad, CA) and the PB1 specific primer set: Forward 5′-GCTGTTTGCAAGTTGATGGG-3′; reverse 5′-TGAAAGCAGGTAACTCCAAGG-3′; and FAM-Probe 5′-TTCAGGCAAGCACCCGTAGGATT-3′ (Hause et al., 2013). Gene specific amplification primers were designed according to the D/swine/1334/Oklahoma/2011 sequence and a reverse transcription primer based upon the conserved non-coding regions (Table 2) (Ducatez et al., 2015). IDV sequencing was performed using Super Script® III Reverse Transcriptase (Life Technologies, Carlsbad, CA), followed by PCR using Phusion High-Fidelity PCR Master Mix with HF Buffer (New England BioLab Inc., Ipswich, MA, USA). PCR products were purified by agarose gel electrophoresis and bands extracted using the QIAquick Gel Extraction Kit (Qiagen Inc., Valencia, CA, USA). Sanger sequencing was performed by Eurofin (Huntsville, AL) using publically available primer sequences (Ducatez et al., 2015). The GenBank accession numbers are KT581409 to KT581418.
Table 2.
Primers used in the reverse transcription and amplification of IDV isolates.
| Gene segment | Primer name | Primer sequence |
|---|---|---|
| PB2 | 1-Forward | 5′-GGC ATA AGC AGA GGA TGT C-3′ |
| 2364-Reverse | 5′-AGC AGT AGC AAG AGG ATT TTT TCA ATG T-3′ | |
| PB1 | 1-Forward | 5′-GGC ATA AGC AGA GGA TTT TAT-3′ |
| 2330-Reverse | 5′-AGC AGT AGC AAG AGG ATT TTT CTG TTA T-3′ | |
| P3 | 1-Forward | 5′-GGC ATA AGC AGG AGA TTT A-3′ |
| 2195-Reverse | 5′-AGC AGT AGC AAG GAG ATT TTT AAC A-3′ | |
| HE | 1-Forward | 5′-AGC ATA AGC AGG AGA TTT TCA AAG-3′ |
| 2049-Reverse | 5′-AGC AGT AGC AAG GAG ATT TTT TCT AA-3′ | |
| NP | 1-Forward | 5′-GGC ATA AGC AGG AGA TTA TTA AGC-3′ |
| 1764-Reverse | 5′-AGC AGT AGC AAG GAG ATT TTT TGT TAA-3′ | |
| M | 1-Forward | 5′-GCA TAA GCA GAG GAT ATT TTT GA-3′ |
| 1219-Reverse | 5′-AGC AGT AGC AAG AGG ATT TTT TCG CG-3′ | |
| NS | 1-Forward | 5′-AGC ATA AGC AGG GTG TAC AAT TTC A-3′ |
| 868-Reverse | 5′-AGC AGT AGC AAG GGG TTT TTT CAT ACT A-3′ | |
| Reverse transcription | 5′-CTC CTT GCT ACT GCT-3′ | |
Hemagglutination assay (HA) and hemagglutination inhibition (HAI) assayHA and HAI assays were performed according to the World Health Organization manual on animal influenza diagnosis and surveillances (http://www.who.int/vaccine_research/diseases/influenza/WHO_manual_on_animal-diagnosis_and_surveillance_2002_5.pdf) using 0.5% turkey red blood cells in U-bottom 96 well plates (USA Scientific, Ocala, FL). Sera were treated 1:3 with receptor-destroying enzyme (RDE) (Denka Seiken Co., Tokyo, Japan) at 37 °C for 18–20 h, followed by heat inactivation at 55 °C for 30 min. Inactivated serum was diluted to a final concentration of 1:10 with 1×PBS. The assay was conducted at room temperature for detection of D/bovine/Mississippi/C00046N/2014 specific antibodies. HAI results were confirmed by HAI for D/swine/Oklahoma/1334/2011 specific antibodies for a subset of serum samples. A serum with a HAI titer ≥1:40 was defined as seropositive.Cell line and viral isolationHuman Rectal Tumor cells (HRT-18G) (ATCC, Manassas, VA) were propagated in 1x DMEM (Gibco® Life Technologies) with 1% Pen/Strep (Gibco® Life Technologies) and 5% Fetal Bovine Serum (Gibco® Life Technologies). IDV isolates were recovered from nasal or nasopharyngeal swabs in 90–95% confluent HRT-18G cells in T-25 cell culture flasks. Cells were gently washed twice with sterile PBS and media was replaced with 5 mL of 1x Opti MEM cell culture media (Gibco® Life Technologies) with 8% of Bovine Serum Albumin (Gibco® Life Technologies), 1% of Pen/Strep (Gibco® Life Technologies), and 1:2000 TPCK-trypsin. Swab media (120 μL) was incubated at 4 °C with 80 μL Pen/Strep and 800 μL media for one hour to reduce bacterial contamination prior to addition to the culture flask, at 1:50 the total volume. Culture was maintained at 37 °C and 5% CO2 and virus was harvested 5 days post infection. Subsequent viral passage was performed at 1:50 inoculation and were screened by HA prior to confirmation of positive isolates by PB1 gene based quantitative RT-PCR. Four isolates, which includes the isolate D/bovine/Mississippi/C00046N/2014 to be used in HAI assays, were further tested at the Mississippi Veterinary Diagnostic Laboratory in Pearl, MS, for common BRD including bovine viral diarrhea virus (BVDV), bovine parainfluenza virus-3 (PI3), adenovirus, coronavirus, bovine herpesvirus (BHV), and bovine respiratory syncytial virus (BRSV). The goals of these tests were be sure the HA positive isolates were clear of coinfection after cell culture prior to application in downstream experiments. No swabs or other IDV isolates was subjected to these tests.Molecular characterization, phylogenetic analyses, and statistical analysesSequence assembly was conducted using Lasergene version 8.0.4. Multiple sequence alignments were conducted by the MUSCLE software package (Edgar, 2004). Maximum-likelihood phylogenetic analyses were performed by using GARLI version 0.96 (Zwickl, 2006) and maximum-likelihood with bootstrap resampling analyses with 1000 runs by using PAUP* 4.0 Beta (Swofford, 1998) with a neighbor-joining method as previously described (Wan et al., 2008). Geometric mean titer (GMT), standard deviation, and percent positive were calculated using Microsoft Excel. For GMT, only positive samples were used in the calculation.
Acknowledgment
We are grateful for the technical assistance from Dr. Hailiang Sun, Dr. Nan Zhao, and Dr. Jianli Xue. We thank Karen Nyguen for assisting in serological testing, and we thank Dr. Maria Piccone and Dr. Chun Kai Yang for helpful discussion. The laboratory trainings for undergraduate sophomore Lucas Ferguson was supported by 1R15AI107702 from National Institutes of Health.
References
- Collin E.A., Sheng Z., Lang Y., Ma W., Hause B.M., Li F. Cocirculation of two distinct genetic and antigenic lineages of proposed influenza D virus in cattle. J. Virol. 2015;89:1036–1042. doi: 10.1128/JVI.02718-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez M.F., Pelletier C., Meyer G. Influenza D virus in cattle, France, 2011–2014. Emerg. Infect. Dis. 2015;21:368–371. doi: 10.3201/eid2102.141449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl. Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton R.W., Cook B.J., Step D.L., Confer A.W., Saliki J.T., Payton M.E., Burge L.J., Welsh R.D., Blood K.S. Evaluation of health status of calves and the impact on feedlot performance: assessment of a retained ownership program for postweaning calves. Can. J. Vet. Res. 2002;66:173–180. [PMC free article] [PubMed] [Google Scholar]
- Guo Y.J., Jin F.G., Wang P., Wang M., Zhu J.M. Isolation of influenza C virus from pigs and experimental infection of pigs with influenza C virus. J. Gen. Virol. 1983;64(Pt 1):177–182. doi: 10.1099/0022-1317-64-1-177. [DOI] [PubMed] [Google Scholar]
- Hause B.M., Collin E.A., Liu R., Huang B., Sheng Z., Lu W., Wang D., Nelson E.A., Li F. Characterization of a novel influenza virus in cattle and Swine: proposal for a new genus in the Orthomyxoviridae family. MBio. 2014;5 doi: 10.1128/mBio.00031-14. e00031–00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hause B.M., Ducatez M., Collin E.A., Ran Z., Liu R., Sheng Z., Armien A., Kaplan B., Chakravarty S., Hoppe A.D., Webby R.J., Simonson R.R., Li F. Isolation of a novel swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.M., Wang S.C., Peng C., Yu J.M., Zhuang Q.Y., Hou G.Y., Liu S., Li J.P., Chen J.M. Identification of a potential novel type of influenza virus in Bovine in China. Virus Genes. 2014;49:493–496. doi: 10.1007/s11262-014-1107-3. [DOI] [PubMed] [Google Scholar]
- Matsuzaki Y., Katsushima N., Nagai Y., Shoji M., Itagaki T., Sakamoto M., Kitaoka S., Mizuta K., Nishimura H. Clinical features of influenza C virus infection in children. J. Infect. Dis. 2006;193:1229–1235. doi: 10.1086/502973. [DOI] [PubMed] [Google Scholar]
- Ng T.F., Kondov N.O., Deng X., Van Eenennaam A., Neibergs H.L., Delwart E. A metagenomics and case-control study to identify viruses associated with bovine respiratory disease. J. Virol. 2015;89:5340–5349. doi: 10.1128/JVI.00064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley J.D., 3rd, Kost C.J., Wolfe T.M. Absorption of protein and IgG in calves fed a colostrum supplement or replacer. J. Dairy Sci. 2002;85:1243–1248. doi: 10.3168/jds.S0022-0302(02)74188-X. [DOI] [PubMed] [Google Scholar]
- Sheng Z., Ran Z., Wang D., Hoppe A.D., Simonson R., Chakravarty S., Hause B.M., Li F. Genomic and evolutionary characterization of a novel influenza-C-like virus from swine. Arch. Virol. 2014;159:249–255. doi: 10.1007/s00705-013-1815-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford D.L. Sinauer; Sunderland, Massachusetts: 1998. PAUP*: Phylogenic Analysis Using Parsimony. [Google Scholar]
- Wan X.F., Nguyen T., Davis C.T., Smith C.B., Zhao Z.M., Carrel M., Inui K., Do H.T., Mai D.T., Jadhao S., Balish A., Shu B., Luo F., Emch M., Matsuoka Y., Lindstrom S.E., Cox N.J., Nguyen C.V., Klimov A., Donis R.O. Evolution of highly pathogenic H5N1 avian influenza viruses in Vietnam between 2001 and 2007. PLoS One. 2008;3:e3462. doi: 10.1371/journal.pone.0003462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwickl D.J. The University of Texas at Austin; Austin: 2006. Genetic Algorithm Approaches for the Phylogenetic Analysis of Large Biological Sequence Datasets Under the Maximum Likelihood Criterion. [Google Scholar]