Abstract
Arabidopsis thaliana is widely accepted as a model species in plant biology. Its genome, due to its small size and diploidy, was the first to be sequenced among plants, making this species also a reference for plant comparative genomics. Nevertheless, the evolutionary mechanisms that shaped the Arabidopsis genome are still controversial. Indeed, duplications, translocations, inversions, and gene loss events that contributed to the current organization are difficult to be traced. A reliable identification of paralogs and single-copy genes is essential to understand these mechanisms. Therefore, we implemented a dedicated pipeline to identify paralog genes and classify single-copy genes into opportune categories. PATsi, a web-accessible database, was organized to allow the straightforward access to the paralogs organized into networks and to the classification of single-copy genes. This permits to efficiently explore the gene collection of Arabidopsis for evolutionary investigations and comparative genomics.
Keywords: Arabidopsis thaliana, paralogs, singletons
Introduction
Arabidopsis thaliana, belonging to the Brassicaceae family, is widely exploited as a reference in plant biology thanks to its short generation time, small size, that limited the requirement for growth facilities, prolific seed production through self-pollination, and its small diploid genome.1,2 This is also the reason why it was the first plant with a genome completely sequenced in 2000. Since its first release, the Arabidopsis genome has been thoroughly investigated, providing a better assessment of the structure and the functionality of plant genomes2–5 and creating the potential for a deeper understanding of the plant development and environmental responses. However, a deeper analysis of the genome sequence of A. thaliana also revealed a high complexity due to several events of whole-genome duplications, the occurrence of large-scale duplications, and reshufflings.6 In particular, these studies showed evidence of at least three rounds of whole-genome duplications followed by diploidization events.7–15 Moreover, the high frequency of gene reduction, ie, gene loss after each duplication event, translocations, inversions,16,17 and probable chromosome losses,18–20 further contributed to reshuffle the retained portions of the genome shaping the current organization.
Gene duplication plays a key role in the origin of novel gene functions.21–25 A clear assessment of the duplicated gene content in a genome, accompanied by a suitable description of those genes that are in single copy, is fundamental for a reliable validation of gene expression,26 of the complexity of gene regulatory networks27 and also to support functional and evolutionary analyses in a species.28
Although several collections of ortholog genes are available today,29–36 only one reference web-accessible database, Eukaryotic Paralog Group Database (EPGD),37 is exclusively dedicated to describe paralogs in 26 available eukaryotic genomes. Indeed, EPGD is a web resource for integrating and displaying eukaryotic paralog information, in terms of paralog families and intragenome segmental duplications. However, the collections of Arabidopsis paralog genes can also be accessed from some of the collections available worldwide which are related to orthologs, such as Ensembl Compara31 and NCBI Homologene,30 which include both animals and plants, and Plaza,38 which is exclusively dedicated to plants genomes. These databases, being mainly dedicated to comparative interspecies analyses, usually refer to the clusters of duplicated genes, where orthologs between species and paralogs are both included. These collections do not provide an overview of those genes that are associated with similarity relationships in a cluster, and therefore, it is rather difficult to identify subgroups in each cluster. Moreover, none of the existing collections enable a graphical visualization of Arabidopsis duplicated genes through a network view, which facilitates a direct and an easy exploitation of the information about paralog relationships.
PATsi, a database to organize paralog and singleton gene in A. thaliana, organizes the protein-coding gene collection of A. thaliana in sets of paralogs and singleton genes identified according to a dedicated pipeline described in Sangiovanni et al.28 Gene association, mainly defined by protein similarity, is assigned using two different cutoffs. Similar genes at each cutoff are organized in the networks of paralogs, accessible as lists but also by graphs through a suitable web interface, with the aim of allow immediate access to genes that share direct similarity relationships in a network shaped at a given threshold. This supports further investigations on the structural and evolutionary relationships among genes.
A detailed classification and immediate download of genes that are not grouped into network of paralogs represent another novelty in this database. Moreover, collections of single-copy genes are not available for A. thaliana, though the wide interest in tracing gene reduction during diploidization events and/or in understanding speciation.
Construction and Content
Data source
The entire A. thaliana genome, intergenic regions, and gene family information were downloaded from The Arabidopsis Information Resource (TAIR) web server.39 A. thaliana expressed sequence tag (EST) sequences to confirm gene expression were downloaded from GenBank.
Collection of paralogs and singletons
A dedicated pipeline to identify paralogs and singleton genes was implemented and applied to the Arabidopsis protein-coding gene collection. Moreover, all the other nonprotein-coding genes available in the collection were filtered out from the subsequent investigations.28 The analysis was based on an all-against-all protein sequence similarity search performed with the BLASTp software,40 using two different cutoff settings: a more stringent expected value (E ≤ 10−10) was used to select the alignments with higher similarity, and a less stringent one was set at E ≤ 10−5.26,41 Then, the two collections were filtered applying the Rost’s formula,42,43 to discriminate significant similarity relationships from less reliable sequence alignments. This formula asserts that all the alignments with an identity percentage lower than 30% and a length shorter than 150 amino acids should be discarded, due to the ambiguity of their relationships. The analysis resulted in 22,522 and 21,843 structurally related genes defined as paralogs and organized in two different datasets, a more stringent one (dataset A), with higher similarity levels between the genes, and a less stringent one (dataset B), including more genes sharing lower similarities.
Several filtering steps were also used to filter out genes with some similarity, lower than the threshold, with other protein-coding genes, or with nucleotide sequence similarity with any genic or intergenic region of the entire genome, permitting to collect genes that could be reliably classified as true singletons with respect to the whole-genome content. Therefore, all the genes are organized into two major groups: those that are in network at the less stringent threshold and those that are not in networks. Accordingly, this last group is classified considering several distinct features based on the pipeline described by Sangiovanni et al.28 Therefore, all the Arabidopsis genes are organized in classes, which are summarized in Table 1 together with the number of associated genes.
Table 1.
Details of the Arabidopsis gene collection and classification.
| CLASSIFICATION | GENE NUMBER | ANALYSIS |
|---|---|---|
| Nonprotein-coding genes | 6070 | miscRNAs, tRNAs, rRNAs, ncRNAs, pseudogenes, transposons and unknown genes |
| Paralogs classified into networks | 22522 | All-against-all BLASTp E ≤ 10−5 |
| Unassigned genes due to the Rost’s formula | 405 | Filtering with Rost’s formula |
| Unassigned genes due to the masking filter | 213 | All-against-all BLASTp E ≤ 10−5 without masking filter |
| Unassigned genes due to loose protein similarity | 440 | All-against-all BLASTp E ≤ 10−3 of protein-coding genes |
| Unassigned genes due to the ORF annotation error | 2 | Transcripts BLASTx E ≤ 10–5 versus proteins for ORF validation |
| Unassigned genes due to similarities with nonprotein-coding genes | 178 | Full genes BLASTn E ≤ 10−5 versus nonprotein-coding genes |
| Unassigned genes due to similarities with intergenic regions | 0 | Full genes BLASTn E ≤ 10−5 versus intergenic regions |
| Singletons not confirmed by ESTs (no EST trace) | 24 | Transcripts BLASTn (free E-value cutoff) versus ESTs |
| Singletons not confirmed by ESTs (discarded by E-value cutoff) | 688 | Filtering of BLASTn results by E ≤ 10−5 |
| Singletons not confirmed by ESTs (discarded by coverage and identity requirements) | 201 | Filtering of BLASTn versus EST results by coverage and identity |
| Singletons not confirmed by ESTs | 100 | Filtering by Delta ≥ 20 (EST length ≥ 20 nt than the transcript) |
| Singletons confirmed by ESTs | 9 | 0 < Delta < 20 (EST length greater than transcript but less than 20 nt) |
| Singleton confirmed by ESTs | 2387 | Delta ≤ 0 (Transcript longer than the EST) |
Notes: A summary of the classes of genes defined in pATsi. The analyses performed to obtain genes in each class are also reported.
Networks construction
Paralog genes were organized into two different sets of networks, depending on the expected value used. The less stringent cutoff (E ≤ 10−5) led to a set of 2,754 networks, including 22,522 paralog genes, while the more stringent cutoff (E ≤ 10−10) led to a set of 3,017 networks containing 21,843 paralogs. Each gene (represented by a node) is connected by at least one paralogy relationship (represented by an edge) to at least another gene in the same network. The networks have various sizes depending on the gene content, ranging from 2 to 6,834 genes, this last number reflecting the maximum number of genes in a network and corresponding to the biggest network (Fig. 1) obtained with the less stringent threshold (E ≤ 10−5).
Figure 1.
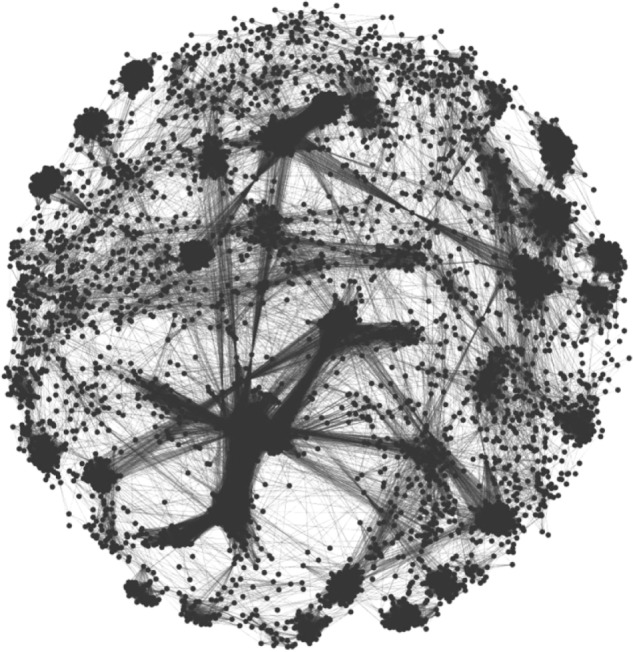
Example view of the largest network of paralogs. Network of paralogs consisting of 6,834 genes.
Notes: Each dot in dark gray represents a single gene, and each line in light gray represents a paralogy relationship between two genes.
To keep track of the relationships between networks defined at different cutoffs, the network naming is assigned as follows: the networks at the less stringent threshold were named as NETxGy_z, where x indicates a number assigned when sorting the total amount of networks by decreasing network size; G stands for genes; y indicates the network size, ie, the number of included genes; and z is the number of networks and singletons in which the network is split when the more stringent cutoff is applied.
Results from the two cutoffs, both considered significant in similar approaches,26,41 are provided here as they can be useful for an approximated estimate of stable or varying network organizations at the two settings.
Database development
The relational database used in this platform was designed under MySql v.5.5.31 and the InnoDb storage engine. For higher efficiency and lower execution time, all tables are indexed using a normal BTree indexing, based on individual and multiple index keys, according to the queries defined in the system. User interface and usage pATsi can be accessed at http://cab.unina.it/athparalog/main.html (Fig. 2A).
Figure 2.
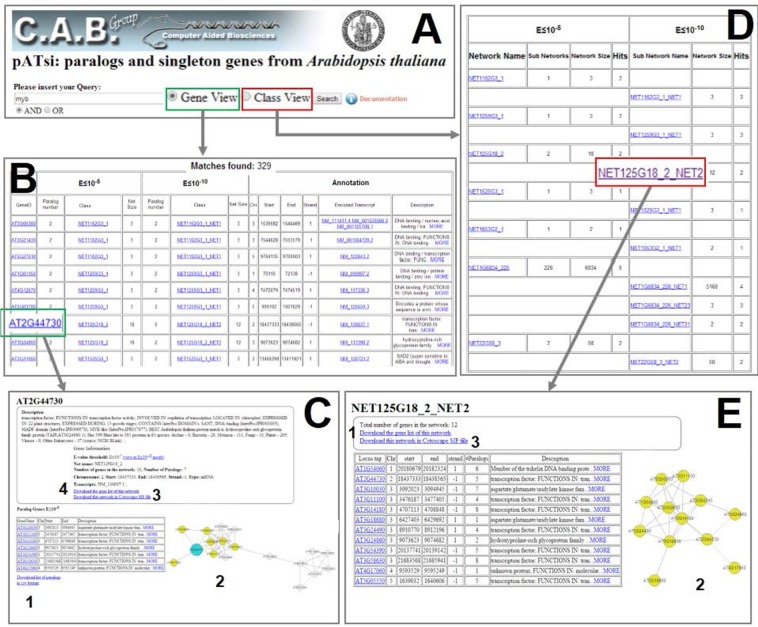
Possible queries workflow in pATsi web interface. (A) Main page of the pATsi database browser; for each query, the user can switch from the gene view (bordered in green) to the class view (bordered in red). (B) List of genes associated with a query. (C) Gene information page. In (C2), a network graph is shown; each circle is a GeneID, with the light blue-circled one representing the selected GeneID and the yellow-circled one(s) representing the paralog(s) of the selected GeneID; gray lines represent paralogies between genes. (D) List of networks associated with a query. (E) Network information page.
All A. thaliana genes and networks can be browsed or searched using keywords. The keyword searching in pATsi can accept a gene locus ID, a network name, or every string to search the annotation content. Two different views are offered. The gene view (Fig. 2A, in green) results in the list of loci associated with the query (Fig. 2B). Each row of the list contains the following information:
GeneID represents the official TAIR classification for A. thaliana. By clicking on the GeneID, it is possible to browse the GeneID-related page, containing information about the gene investigated and the two networks organization in which the gene may be included. In case the gene is a singleton, no network organization is shown.
Paralog number shows the number of paralogs of the gene at the two different cutoffs or zero in the case of singletons or non-messenger RNA (mRNA) genes.
Class: if the gene has paralogs at one of the e-value cutoffs, the network name is shown; for genes without paralogs, the name of the class is reported. In the case of unassigned genes, the classification field contains a brief explanation of the reason that led to that specific category.
Net size: this field shows the number of all paralog genes contained into the network, zero if the corresponding gene is a singleton or a non-mRNA gene.
Chr: the chromosome on which the locus maps.
Start/end: starting/ending position of the locus on the chromosome.
Strand: direction of the locus transcription.
Encoded transcript: the RefSeq or the encoded transcript. Each RefSeq has a link to GenBank.
Description: the TAIR functional annotation39 for each of the RefSeq.
By clicking on the GeneID in the results table, the user will be redirected to a new page (Fig. 2C). In the topmost part of the page, the GeneID, several details about the gene annotation and possible network information are reported. In the bottom left part of the page, the list of paralogs of the selected gene is shown. Each gene is also cross-linked to its specific description in the database. The list of paralogs can be downloaded (Fig. 2[C1]). In the bottom center part of the page, an interactive network graph is displayed (Fig. 2[C2]) using CytoscapeWeb, a web-based network visualization tool.44 Users are enabled to interact with the displayed network by selecting nodes and edges and modeling the network view accordingly. For each network, it is possible to download a file (Fig. 2[C3]), which can be easily imported into Cytoscape, for onsite visualization or for managing more complex networks.45 The list of genes that are included in the displayed network can also be downloaded (Fig. 2[C4]).
Selecting the class view (Fig. 2A in red), the query process organizes the genes associated with the query into classes, separating the genes that are not included in the network from those included in networks. The resulting page also provides the list of networks associated with the query (Fig. 2D), indicating the following information in each row of the list of networks:
Network: the name assigned to the network at the lowest cutoff (E ≤ 10−5). By clicking on the network name, it is possible to browse the network ID-related page, containing information about the network investigated and the genes included in it. For each network, a list of one or more subnetworks is also shown.
Subnetworks: it shows the number of subnetworks or singletons in which the network is split when the cutoff of E ≤ 10−10 is applied.
Network size: ie, the number of genes included in the network.
Hits: the number of matching genes with the user query.
By clicking on the network name in the resulting table, the user will be redirected to a new page (Fig. 2E). In the topmost part of the page, the network name and the number of genes are shown. In the left part of the page, the list of genes of the selected network is reported. It is also possible to download the list of genes (Fig. 2[E1]). In the right part of the page, the network graph is displayed (Fig. 2[E2]). The file of the network, in.sif format, can also be downloaded (Fig. 2[E3]).
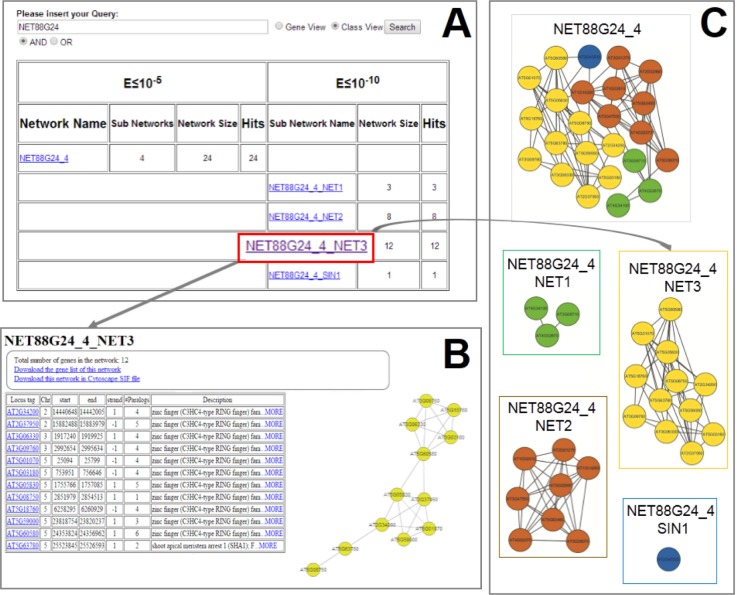
As mentioned earlier, networks presented here are classified according to two different thresholds. The use of a more stringent threshold defines a lower number of paralogy relationships between genes, hence obtaining a larger number of networks in comparison with the ones obtained with the less stringent threshold. This is explained considering the effects of the less stringent cutoff that permits to also include genes in a network which are otherwise excluded when the more stringent threshold is used (Fig. 3).
Figure 3.
Network organization. (A) List of subnetworks associated with a network query. (B) Network information page. (C) Graphic representation of a network of 24 genes (NET88G24_4) splitted into three subnetworks (NET88G24_4 NET1-NET2-NET3) and one singleton (NET88G24_4 SIN1).
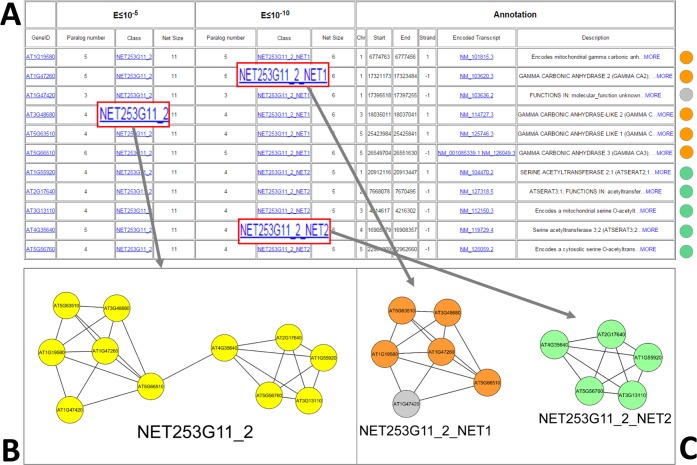
An example query in pATsi using serine acetyltransferase as keyword and selecting the gene view option resulted in six hits: one annotated as a pseudogene and five as protein-coding genes. These five genes are grouped into one network, NET253G11_2, together with other six genes (Fig. 4) not reflecting the same annotation.
Figure 4.
Example of pATsi usage. (A) List of genes associated with the NET253G11_2 network. (B) Graph view of NET253G11_2 network. (C) Graph view of NET253G11_2_NET1 and NET253G11_2_NET12 subnetworks.
Notes: Orange circles represent genes annotated as gamma carbonic anhydrases; yellow circles represent genes annotated as serine acetyltransferases; gray circle represents a gene with an unknown function.
The family of serine acetyltransferases catalyzes the limiting reaction in cysteine biosynthesis.46–48 These enzymes are of great interest to the scientific community because of their active role in creating nutritionally essential sulfur amino acids.49 Analyzing the network (NET253G11_2) organization at E5, we noted that the more stringent cutoff (E ≤ 10−10) identified two subnetworks. The first one (NET253G11_2_NET1) is formed by the five serine acetyltransferase enzymes previously discussed, while the other (NET253G11_2_NET2) is formed by five genes annotated as gamma carbonic anhydrases. Moreover, a gene with an unknown function falls in the second network (Fig. 4C). The function of the unknown gene can be more straightforward inferred thanks to the more stringent cutoff, since all the associated paralogs in the subnetwork have a similar function. Moreover, serine acetyltransferases and carbonic anhydrases belong to the same trimeric LpxA-like superfamily, a set of enzymes with trimeric repeats of hexapeptide motifs. This shows the reliability of their grouping at a less stringent threshold. Beyond the usage of the platform, this example also highlights that different thresholds can support the identification of not only well known but also unexpected relationships among genes in a network.28
Conclusion
The collection described here is useful for promoting an efficient exploitation of the Arabidopsis gene collection in terms of intragenome similarities, contributing to the identification of structurally related genes, to their functional assignment, and to the classification of singleton genes within this reference genome.28 In addition, pATsi provides a novel approach to the classification of protein-coding genes from A. thaliana, based on a similarity defined at both protein and genome level, focusing on paralogs to build gene networks and on an appropriate classification of singleton genes.
Today, there are several collections available for paralog genes in plants35,50 including data from A. thaliana. However, none of these manifold available resources allow to fully access descriptions of both the genes having at least one paralog and the ones being singletons in A. thaliana. Moreover, many of the currently available platforms are based on methods that are not sufficiently described and/or are not easily reproducible, defining often incomparable collections hard to be further exploited in associated efforts. Some independent studies also aimed to identify singleton genes from A. thaliana,51 but no dataset is provided to the scientific community to support validations or further investigations, despite the importance of addressing the origin of single-copy genes in a highly duplicated genome, as they can be a trace of mechanisms of evolutions, of speciation events, or even annotation limits.28,52
One of the novelties of this database is to provide immediate access to gene classes and possible paralogy relationships, supported by graphical approaches, offering at the same time a detailed description of the methods and parameters used for each class.28 Being fully reproducible, the database may provide a common reference and a shared framework for related efforts.
The database represents a permanent resource for studies that need a reference collection for gene family analysis and comparative genomics based on A. thaliana and may be used as an effective and powerful tool to address the process of deciphering the complexity of its genome.
Acknowledgments
The authors thank the anonymous reviewers for their comments and suggestions, which were helpful in improving the manuscript, and also they acknowledge Dr. Gennaro Oliva for technical suggestions and support.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
PEER REVIEW: Two peer reviewers contributed to the peer review report. Reviewers’ reports totaled 641 words, excluding any confidential comments to the academic editor.
FUNDING: This work is supported by the Genopom Pro and HORT Projects (Ministero dell’Istruzione, dell’Università e della Ricerca, Italy) and is in the frame of the Cost Action FA1106. HB is supported by the Solanaceae Pollen Thermotolerance – Marie Curie Initial Training Network project (Grant agreement no. 289220). The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Wrote the manuscript and implemented the network visualization in the web pages: LA. Organized the database and implemented the query interfaces and the web-based resource: HB. Contributed to the web page implementation: PDS. Developed the pipeline and extracted the results collected in the database: MS, AV. Conceived the analysis and the system, supported, drove, and directed all the work, and wrote the manuscript: MLC. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Koornneef M, Meinke D. The development of Arabidopsis as a model plant. Plant J. 2010;61(6):909–21. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- 2.The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 3.Bevan M, Walsh S. The Arabidopsis genome: a foundation for plant research. Genome Res. 2005;15(12):1632–42. doi: 10.1101/gr.3723405. [DOI] [PubMed] [Google Scholar]
- 4.Meinke DW, Cherry JM, Dean C, et al. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282(5389):662, 679–82. doi: 10.1126/science.282.5389.662. [DOI] [PubMed] [Google Scholar]
- 5.Somerville C, Koornneef M. A fortunate choice: the history of Arabidopsis as a model plant. Nat Rev Genet. 2002;3(11):883–9. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- 6.Simillion C, Vandepoele K, Van Montagu MC, et al. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2002;99(21):13627–32. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blanc G, Barakat A, Guyot R, et al. Extensive duplication and reshuffling in the Arabidopsis genome. Plant Cell. 2000;12(7):1093–101. doi: 10.1105/tpc.12.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanc G, Hokamp K, Wolfe KH. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 2003;13(2):137–44. doi: 10.1101/gr.751803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16(7):1679–91. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui L, Wall PK, Leebens-Mack JH, et al. Widespread genome duplications throughout the history of flowering plants. Genome Res. 2006;16(6):738–49. doi: 10.1101/gr.4825606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiao Y, Leebens-Mack J, Ayyampalayam S, et al. A genome triplication associated with early diversification of the core eudicots. Genome Biol. 2012;13(1):R3. doi: 10.1186/gb-2012-13-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao Y, Wickett NJ, Ayyampalayam S, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473(7345):97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 13.Van de Peer Y. A mystery unveiled. Genome Biol. 2011;12(5):113. doi: 10.1186/gb-2011-12-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vision TJ, Brown DG, Tanksley SD. The origins of genomic duplications in Arabidopsis. Science. 2000;290(5499):2114–7. doi: 10.1126/science.290.5499.2114. [DOI] [PubMed] [Google Scholar]
- 15.Wolfe KH. Yesterday’s polyploids and the mystery of diploidization. Nat Rev Genet. 2001;2(5):333–41. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]
- 16.Gaut BS. Patterns of chromosomal duplication in maize and their implications for comparative maps of the grasses. Genome Res. 2001;11(1):55–66. doi: 10.1101/gr.160601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ku HM, Vision T, Liu J, et al. Comparing sequenced segments of the tomato and Arabidopsis genomes: large-scale duplication followed by selective gene loss creates a network of synteny. Proc Natl Acad Sci U S A. 2000;97(16):9121–6. doi: 10.1073/pnas.160271297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conner JA, Conner P, Nasrallah ME, et al. Comparative mapping of the Brassica S locus region and its homeolog in Arabidopsis. Implications for the evolution of mating systems in the Brassicaceae. Plant Cell. 1998;10(5):801–12. doi: 10.1105/tpc.10.5.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston JS, Pepper AE, Hall AE, et al. Evolution of genome size in Brassicaceae. Ann Bot. 2005;95(1):229–35. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lysak MA, Koch MA, Pecinka A, et al. Chromosome triplication found across the tribe Brassiceae. Genome Res. 2005;15(4):516–25. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183(3):557–64. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 22.Hughes AL. Gene duplication and the origin of novel proteins. Proc Natl Acad Sci U S A. 2005;102(25):8791–2. doi: 10.1073/pnas.0503922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaessmann H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010;20(10):1313–26. doi: 10.1101/gr.101386.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magadum S, Banerjee U, Murugan P, et al. Gene duplication as a major force in evolution. J Genet. 2013;92(1):155–61. doi: 10.1007/s12041-013-0212-8. [DOI] [PubMed] [Google Scholar]
- 25.Rensing SA. Gene duplication as a driver of plant morphogenetic evolution. Curr Opin Plant Biol. 2014;17c:43–8. doi: 10.1016/j.pbi.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 26.He X, Zhang J. Gene complexity and gene duplicability. Curr Biol. 2005;15(11):1016–21. doi: 10.1016/j.cub.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 27.Teichmann SA, Babu MM. Gene regulatory network growth by duplication. Nat Genet. 2004;36(5):492–6. doi: 10.1038/ng1340. [DOI] [PubMed] [Google Scholar]
- 28.Sangiovanni M, Vigilante A, Chiusano ML. Exploiting a reference genome in terms of duplications: the network of paralogs and single copy genes in Arabidopsis thaliana. Biology (Basel) 2013;2(4):1465–87. doi: 10.3390/biology2041465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen F, Mackey AJ, Stoeckert CJ, Jr, et al. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34(Database issue):D363–8. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.NCBI Resource Coordinators Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013;41(Database issue):D2–8. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flicek P, Ahmed I, Amode MR, et al. Ensembl 2013. Nucleic Acids Res. 2013;41(Database issue):D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33(Database issue):D476–80. doi: 10.1093/nar/gki107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Powell S, Forslund K, Szklarczyk D, et al. eggNOG v4.0: nested orthology inference across 3686 organisms. Nucleic Acids Res. 2014;42(Database issue):D231–9. doi: 10.1093/nar/gkt1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouard M, Guignon V, Aluome C, et al. GreenPhylDB v2.0: comparative and functional genomics in plants. Nucleic Acids Res. 2011;39(Database issue):D1095–102. doi: 10.1093/nar/gkq811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Bel M, Proost S, Wischnitzki E, et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Physiol. 2012;158(2):590–600. doi: 10.1104/pp.111.189514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waterhouse RM, Tegenfeldt F, Li J, et al. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 2013;41(Database issue):D358–65. doi: 10.1093/nar/gks1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding G, Sun Y, Li H, et al. EPGD: a comprehensive web resource for integrating and displaying eukaryotic paralog/paralogon information. Nucleic Acids Res. 2008;36(Database issue):D255–62. doi: 10.1093/nar/gkm924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Proost S, Van Bel M, Sterck L, et al. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21(12):3718–31. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamesch P, Berardini TZ, Li D, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40(Database issue):D1202–10. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 41.Rubin GM, Yandell MD, Wortman JR, et al. Comparative genomics of the eukaryotes. Science. 2000;287(5461):2204–15. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24(3):319–24. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 43.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12(2):85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 44.Lopes CT, Franz M, Kazi F, et al. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26(18):2347–8. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tavares S, Wirtz M, Beier MP, et al. Characterization of the serine acetyltransferase gene family of Vitis vinifera uncovers differences in regulation of OAS synthesis in woody plants. Front Plant Sci. 2015;6:74. doi: 10.3389/fpls.2015.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yi H, Dey S, Kumaran S, et al. Structure of soybean serine acetyltransferase and formation of the cysteine regulatory complex as a molecular chaperone. J Biol Chem. 2013;288(51):36463–72. doi: 10.1074/jbc.M113.527143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nguyen HC, Hoefgen R, Hesse H. Improving the nutritive value of rice seeds: elevation of cysteine and methionine contents in rice plants by ectopic expression of a bacterial serine acetyltransferase. J Exp Bot. 2012;63(16):5991–6001. doi: 10.1093/jxb/ers253. [DOI] [PubMed] [Google Scholar]
- 49.Tabe L, Wirtz M, Molvig L, et al. Overexpression of serine acetlytransferase produced large increases in O-acetylserine and free cysteine in developing seeds of a grain legume. J Exp Bot. 2010;61(3):721–33. doi: 10.1093/jxb/erp338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kinsella RJ, Kahari A, Haider S, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database (Oxford) 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duarte JM, Wall PK, Edger PP, et al. Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels. BMC Evol Biol. 2010;10:61. doi: 10.1186/1471-2148-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahalak KK, Chamberlin HM. Orphan genes find a home: interspecific competition and gene network evolution. PLoS Genet. 2015;11(6):e1005254. doi: 10.1371/journal.pgen.1005254. [DOI] [PMC free article] [PubMed] [Google Scholar]