Abstract
Since the cytogenetic identification of azoospermia factor regions 40 years ago, the Y chromosome has dominated research on the genetics of male infertility. We hypothesized that hotspots of structural rearrangement, which are dispersed across the genome, may mediate rare, recurrent copy number variations (CNVs), leading to severe infertility. We tested this hypothesis by contrasting patterns of rare CNVs in 970 Han Chinese men with idiopathic nonobstructive azoospermia and 1661 ethnicity-matched controls. Our results strongly support our previous claim that sperm production is modulated by genetic variation across the entire genome. The X chromosome in particular was enriched for loci modulating spermatogenesis—rare X-linked deletions larger than 100 kb were twice as common in patients compared with controls (odds ratio [OR] = 2.05, P = 0.01). At rearrangement hotspots across the genome, we observed a 2.4-fold enrichment of singleton CNVs in patients (P < 0.02), and we identified 117 testis genes, such as SYCE1, contained within 47 hotspots that may plausibly mediate genomic disorders of fertility. In our discovery sample we observed 3 case-specific duplications of the autosomal gene MAST2, and in a replication phase we found another 11 duplications in 1457 patients and 1 duplication in 1590 controls (P < 5 × 10−5, combined data). With a large, polygenic genetic basis, new ways of establishing the pathogenicity of rare, large-effect mutations will be needed to fully reap the benefit of genome data in the management of azoospermia.
Keywords: aneuploidy, genetics, genomics, male infertility
INTRODUCTION
Genomic disorders are a class of rare diseases caused by segmental copy number variation and mediated by nonallelic homologous recombination between large segmental duplications [1]. In seminal work published nearly 15 yr ago, a first-generation map of potential genomic disorder loci was generated by analyzing the duplication structure of the human genome [2]. The functional consequence of copy number variations (CNVs) at these 169 putative rearrangement hotspots has been extensively investigated since then, and at least 45 hotspots have now been associated with a genomic disorder [3]. Although some hotspots may be recurrently mutated without functional consequences, many hotspots remain to be characterized and have been observed to produce little or no CNV in control populations. As we describe below, there are dozens of rearrangement hotspots that harbor genes important for spermatogenesis. We hypothesized that some of the “missing” CNVs at these hotspots may cause severe infertility, and thus are purged rapidly from the population.
To test this hypothesis we have assembled a cohort of men that received a diagnosis of nonobstructive azoospermia (NOA), more than 2500 men in total, and have performed genome-wide screens for novel, recurrent CNVs causing NOA. This cohort of Han Chinese was initially used in a single-nucleotide polymorphism (SNP)-based genome-wide association study (GWAS) for common NOA risk variants [4] and, more recently, as a replication cohort for a single-locus CNV analysis [5]. The present study provides the first description of genome-wide CNV discovery data from these samples.
The first genetic cause of NOA ever identified was large-scale deletion of sequence on the Y chromosome, in the so-called azoospermia factor (AZF) regions [6]. Since this initial discovery, research into the genetics of sperm production has been dominated by investigation of the Y chromosome. In this era of readily accessible DNA sequencing, it is crucial to clarify whether genes outside the Y chromosome are important for human sperm production prior to conducting large-scale investment in whole-genome sequencing studies. The results of our present study strengthen our earlier report that rare CNVs throughout the genome, and especially on the X and Y chromosomes, can impair spermatogenesis [5]. We identify a novel, rare CNV association in our discovery samples that replicates in an independent case-control cohort. More urgently, we observe a large number of extremely rare, patient-specific CNVs that are highly plausible causes of NOA. Novel approaches to assessing the significance of these, and making precise statistical statements about their disease relevance, are essential for integrating genomes into the management of infertility and other diseases with an etiology tied to extremely rare, large-effect mutations.
MATERIALS AND METHODS
Study Populations
We performed a two-stage (discovery and replication) case-control study. Subject recruitment for the discovery cohort has been previously described in our report of an SNP-based GWAS for NOA risk factors [4]. The discovery cohort included 1000 NOA patients (mean age: 32.12 ± 4.56 yr) recruited from the Center of Reproductive Medicine 2005–2011, and 1703 male controls (873 healthy men and 830 male lung cancer patients; mean age, 60.26 ± 9.67 yr). The replication cohort included 1500 patients (mean age, 31.92 ± 3.07 yr) and 1600 apparently healthy controls (mean age, 56.81 ± 10.80 yr) recruited from southeastern China: Nanjing and Shanghai. The patients were genetically unrelated Han Chinese men determined to have idiopathic NOA and selected on the basis of comprehensive andrological testing, including examination of medical history, physical examination, semen analysis, scrotal ultrasound, hormone analysis, karyotyping, and Y chromosome microdeletion screening. Those with a history of cryptorchidism, vascular trauma, orchitis, obstruction of the vas deferens, vasectomy, abnormalities in chromosome number, or microdeletions of the azoospermia factor region on the Y chromosome were excluded from the study. Semen analysis for sperm concentration, motility, and morphology was performed following World Health Organization criteria [7]. Patients with NOA had no detectable sperm in the ejaculate after evaluation of the centrifuged pellet. To ensure the reliability of the diagnosis, each individual was examined twice, and the absence of spermatozoa from both replicate samples was taken to indicate azoospermia. All controls had fathered one or more healthy children and were frequency matched to the patients on the basis of age and area of residence. At recruitment, informed consent was obtained from each participant, and the study was approved by the Institutional Review Boards from Nanjing Medical University, Shanghai Renji Hospital, and Nanjing Jinling Hospital.
CNV Discovery
All participants were genotyped on the Affymetrix 6.0 genotyping platform. As described previously, the population structure of these samples was assessed using the SNP genotype data and found to be negligible for our purposes (lambda = 1.073; Supplemental Fig. S1; Supplemental Data are available online at www.biolreprod.org). We created a high-quality set of CNV calls for all individuals using our own internal pipeline (Affy6CNV, a wrapper for the Birdsuite package [8]) for data processing and quality control (QC; Supplemental Methods). Affy6CNV is available online from http://sourceforge.net/projects/affy6cnv/files/. The models used by Birdsuite assume a mixture of both male and female samples within each batch of arrays. Because our discovery samples were exclusively male, we used a second pipeline for CNV calling from the X and Y chromosomes and based on the GADA algorithm (Supplemental Methods). Samples failing CNV QC were removed from the study, leaving 970 NOA cases and 1661 controls.
Burden Analysis
We used a permutation-based testing framework for assessing CNV frequency differences between cases and controls [9]. The power to discover CNVs from a given array experiment is dictated by the noise in that experiment (e.g., the variance in probe intensities). To properly account for differential data quality between cases and controls, we adopted a conditional permutation strategy where case and control are first matched into bins of equivalent array noise, and then case-control labels are permuted within bins. We used two different statistics for summarizing the noise in each array experiment: 1) the median absolute deviation of probe intensities, and 2) the number of QC-passed CNV calls per sample. Permutation results with each statistic were highly similar, and thus we report results using 1) in the main text. Statistical significance was established by 10 000 rounds of permutation.
Association Analysis
Copy number variation discovery is performed on one individual at a time, which can lead to groups of CNVs with overlapping genomic locations but with estimated break points that are slightly different in different individuals. Prior to association analysis, we defined CNV events (CNVEs) as groups of CNV calls that all share the property of having 50% reciprocal overlap. Fisher exact test was performed at each rare CNVE, and CNVtools [10] analysis was performed at each common CNVE for detection of association signal. Five categories of CNVEs, namely: 1) rare, case-enriched CNVEs (nominal P value <0.05); 2) rare, case-specific large recurrent CNVEs; 3) rare, case-specific small recurrent CNVEs; 4) rare, case-specific large singleton deletions; and 5) case-enriched common CNVEs (nominal P value <0.05) were kept as candidate loci for manual examination of calling and local annotations.
Validation
Using quantitative PCR (qPCR), we performed experimental validation of several high-interest CNV calls made in the discovery cohort. These experiments were strictly to confirm the existence of specific CNVs predicted from the array data. The qPCR experiments were conducted using SYBR Green Realtime PCR Master (Toyobo) in triplicate reactions on an ABI 7900 Real-Time PCR System machine (Applied Biosystems Inc.) according to the manufacturer's instructions. The following reagents were used for amplification in 10 μl: 2 μl of DNA, 5 μl of SYBR Realtime Green PCR Master Mix (2×), 0.4 μl of forward primer (10 μM), and 0.4 μl of reverse primer (10 μM). Thermal cycling was initiated with conditions consisting of one cycle of 30 sec at 95°C, followed by 40 cycles of 5 sec at 95°C, 10 sec at 55°C, and 15 sec at 72°C. Primers were designed to target the likely phenotype-causing gene encompassed by the CNV if the gene is not embedded in segmental duplications; otherwise they were designed to target a gene in the same CNV interval but not overlapping segmental duplications. The primers are listed in Supplemental Table S1. A threshold for gene dosage of <0.75 (loss) and >1.30 (gain) was applied for CNV validation. Full data are available as Supplemental Table S2.
Replication
An advanced qPCR assay called Accucopy [11] was used to replicate promising association signals with an independently recruited cohort of 1500 infertile men and 1600 apparently healthy men used as common controls in a variety of GWA studies. Both case and control groups were composed of completely different sets of individuals from the ones studied in the discovery cohort. The cases of this replication cohort were determined to be idiopathic after a thorough andrological workup, whereas the controls were evidently fertile from fathering at least one child. Briefly, the method applies multiplex competitive amplification to both known diploid regions and regions of unknown copy number to determine the DNA dosage of regions of interest. In addition to known diploid regions, a segment in gene SRY was chosen as a haploid reference for the candidate CNVs on the Y chromosome. We designed assays for five genomic loci identified as of interest in the discovery phase: MAST2, TUBA3E, SYCE1, USP9Y, and DDX9Y. The primers are listed in Supplemental Table S3. The intensities of the target region were converted to copy number by dividing the intensities of the reference region with matching size, normalizing by the plate median, and multiplying by the copy number of the reference region (Supplemental Figs. S2–S5). Thresholds for assigning copy number were determined manually based on the positions of copy number clusters for each target region.
Hotspot Regions with Possible Reproductive Association
To define potential rearrangement hotspots, we downloaded the “genomicSuperDups” track from the UCSC genome browser for assembly hg18. We created a table of all intrachromosomal duplication pairs that met the following criteria: greater than 95% sequence identity, greater than 50 kb apart, less than 10 Mb apart, and in tandem orientation (as opposed to inverted). We further consolidated this list by merging pairs with similar start and end points, creating a final list of 381 pairs.
Data Availability
We are in the process of submitting all CNV calls from the study to dbVAR (http://www.ncbi.nlm.nih.gov/dbvar/).
RESULTS
Following call-level and sample-level QC, our CNV discovery set consisted of 49 806 CNVs in 970 cases and 85 173 CNVs in 1661 controls. Previously, we have assessed CNV burden in a smaller cohort of men with spermatogenic impairment (azoospermia and oligozoospermia) and reported several novel features regarding the genetic architecture of sperm production [5]. Here, we first attempted replication of our finding that men with spermatogenic failure have a genome-wide burden of CNVs that involves the Y chromosome, the X chromosome, and the autosomes (Supplemental Tables S4–S7). We address each genomic region in turn.
Y chromosome rearrangements have been recognized as important causes of azoospermia for nearly 40 yr [6], and Y deletion screening is a standard part of the clinical workup for male infertility. However, perhaps because of the technical challenges of screening for duplications by fluorescent in situ hybridization or PCR, the role of Y duplications as risk factors for azoospermia has been poorly studied. Isodicentric Y chromosomes, known causes of azoospermia, can create signals in array data that mimic large duplications. We identified array profiles consistent with an isodicentric Y chromosome in 16 cases and 1 control, and removed these individuals from our subsequent Y chromosome analyses (Fig. 1).
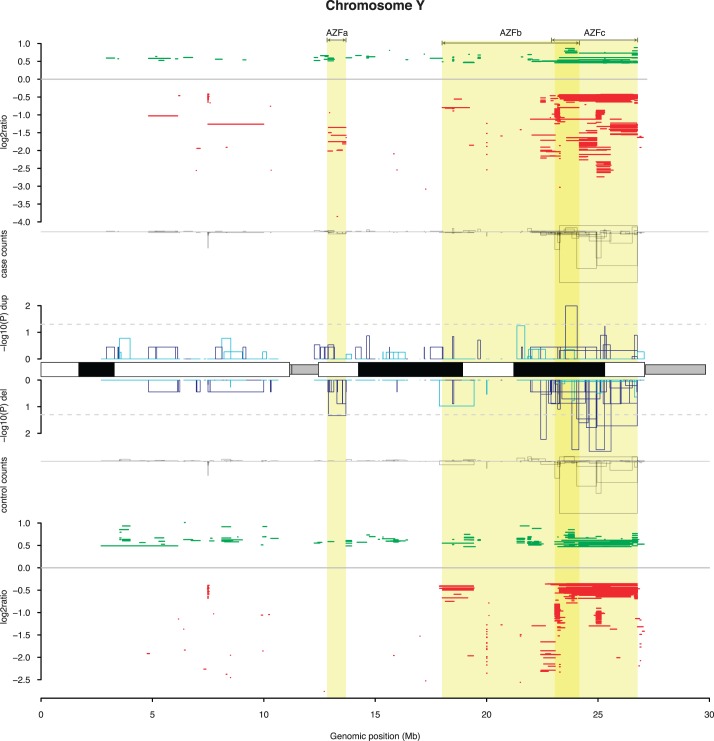
FIG. 1.
Overview of CNVs on the Y chromosome in cases and controls. Prior to plotting, all CNV calls were merged into a smaller number of CNV regions (Materials and Methods), and samples with isodicentric Y chromosome were removed. The plot is divided into five tracks. From top to bottom: (i) location and average log2 ratio for all duplications (green) and deletions (red) detected in cases. (ii) A bar plot of counts of case CNV alleles grouped by CNV region, with the height of each histogram above the x-axis corresponding to duplication counts, and heights below the x-axis corresponding to deletion counts. (iii) A bar plot of log10 P values for a single-locus test of association between cases and controls, testing each CNV region separately, with the histograms above the ideogram indicating tests for duplications, whereas histograms below the ideogram indicate tests for deletions. The color of the bars indicates the direction of the association: dark blue bars indicate an enrichment of the CNV in cases, whereas light blue bars indicate an enrichment in controls. (iv) A bar plot of control CNV counts, as in (ii). (v) The map of control CNV calls, as in (i). The locations of the AZFa, AZFb, and AZFc regions are highlighted in yellow and labeled at the top of the figure.
We carefully characterized the patterns of deletion and duplication in the AZF regions—historically referred to as AZFa, AZFb, and AZFc. The complex structure of segmental duplications in these regions leads to a variety of recurrent deletions that confer risk for impaired spermatogenesis. Recent estimates based on large sample sizes [12] suggest that about 7% of all cases of spermatogenic failure can be attributed to these deletions (6% in AZFc and 1% in AZFa and AZFb). We identified 5 AZFa deletions, 5 AZFb deletions, 1 large deletion spanning AZFb and AZFc, and 19 full deletions of AZFc (b2/b4 deletions). All of these events were in cases, accounting for 3% of the total cohort. Other deletions of the AZFc region could be observed in cases and controls but showed no association with case/control status in our cohort. Because our patient samples had been prescreened by PCR for full AZFc deletions prior to this study, the numbers reported here reflect ascertainment and are an underestimate of the true frequency of AZFc microdeletions in azoospermic Han Chinese men (Supplemental Methods).
Despite this, after removing AZFa, AZFb, and b2/b4 deletions, we still observed a strongly significant case enrichment of Y-linked deletions between 200 and 500 kb (P < 1 × 10−4), indicating that large Y deletions outside the canonical AZF regions confer risk for azoospermia (Fig. 2 and Supplemental Table S5).
FIG. 2.
Burden of large and rare CNVs in cases of NOA relative to controls. In order to understand the genetic architecture of azoospermia risk, we performed a variety of case-control comparisons, contrasting the frequency of CNVs in cases versus controls using different groupings of CNVs. All contrasts use CNVs greater than 100 kb in size and rarer than 1% in frequency unless otherwise specified. For each comparison the observed rate ratio is plotted along with a 95% CI obtained from permutation (Materials and Methods). From top to bottom of the plot, we have examined the impact of CNV size on burden across the entire genome (“Whole genome”), the relative impact of CNVs on the autosomes, X chromosome, and Y chromosome (“Whole chromosome(s)”), and the impact of CNVs specifically affecting genes thought to be involved in human sperm production.
We and others have previously reported an association between Y duplications and spermatogenic impairment, although conflicting reports do exist [5, 13–15]. In our cohort, AZFc duplications were more frequent in cases, but the difference was not significant (odds ratio [OR], 1.36; P = 0.08; Supplemental Fig. S6). When considering the entire Y chromosome, we found a higher rate of large, singleton Y-linked duplications in cases compared with controls, a difference that increased with increasing duplication size threshold; none of these enrichments were significant (Supplemental Tables S4 and S5).
There is an increasing appreciation that genes specialized for enhancing sperm production [16] and genetic variation modulating male fertility [5, 17, 18] are enriched on the X chromosome. To our knowledge, the data we present here represent the largest whole-chromosome CNV call set for the X chromosome generated from a cohort of men with NOA. Simple burden testing comparing counts of rare X-linked CNVs between cases and controls shows an excess of both duplications and deletions at multiple size thresholds (Supplemental Table S6). For example, when considering all X-linked deletions with <1% frequency we observed a 2-fold enrichment in cases (2.7% frequency vs. 1.3% frequency; P < 0.03). This enrichment increased to a 3.7-fold difference when considering just the subset of loss-of-function deletions (P = 1.1 × 10−3).
As a class, autosomal CNVs were the most modestly enriched in patients. Singleton deletions were 19% more frequent in cases compared with controls (P = 0.05), but no significant enrichment was observed for duplications (Supplemental Table S7). This discrepancy between the signals on the sex chromosomes and autosomes likely reflects a higher density of genes essential for reproduction on the X and Y compared with elsewhere in the genome.
CNVs Affecting Genes Known to Be Involved in Spermatogenesis
Our findings clearly show that there was an increased load of rare deleterious CNVs in cases compared with controls, affecting genes other than those on the Y chromosome. We next searched for additional information on the nature of this burden by characterizing the type of genes and genomic regions affected by this CNV load—an exercise that we hoped would shed light on the biological processes being perturbed, but at the same time add further credibility to our claim of a pathogenic load in cases (Fig. 2).
We created a list of 190 genes identified in a recent literature review as having a causal role in mammalian spermatogenic output; many of these are derived from mouse models that manifest spermatogenic impairment (Supplemental Table S8) [19]. We intentionally excluded Y chromosome genes from this list. Even so, there was a strong association between large deletions and duplications of these genes and case status (e.g., a 1.7-fold enrichment when considering CNVs >100 kb; P = 0.02). At least 1 CNV overlapped 43 of the genes in this list: 7 covered by deletions alone, 24 with duplications alone, and 12 with deletions and duplications. Genes affected include BRDT, SMC1B, DAZAP, TAF7L, ATM, FANCA, and PARP2 (Supplemental Table S9).
We noted that there were a number of DNA damage repair genes on this list with case-specific deletions, including FANCA. We then tested for additional CNVs in the Fanconi Anemia Complementation group and found three coding deletions (all in cases) and six coding duplications (three in cases and three in controls). These involved the genes FANCL, FANCD2, FANCF, FANCI, and FANCA. The association with the set of coding deletions in these genes was marginally significant (P < 0.05), but there was no significant association with the duplications.
Previously, we reported that rare CNVs of X-linked cancer-testis antigen genes (CT-X genes) were associated with spermatogenic impairment. The CT-X genes contain many ampliconic gene families: our total CT-X gene list consisted of 229 annotated ORFs, only 4 of which are not part of a multicopy gene family. However, these gene families showed striking heterogeneity in their rates of CNV: Some members of the SSX, CT-45, and SPANX gene clusters showed rates of CNV approaching 5% of our population, whereas the CT-47, MAGE, GAGE, and XAGE gene families showed virtually no CNVs. We confirm previous reports of an association between CT-X CNVs and azoospermia, observing 13 cases and 8 controls carrying deletions of a CT-X gene greater than 100 kb in size and less than 5% frequency (P < 0.02). No association was observed for duplications.
Single-Gene Rare CNV Analyses
For all genes overlapping three or more CNVs, we tested for an association between deletion or duplication and case-control status. The strongest evidence for association was seen at the genes DMRT1, SYCE1, TUBA3E, and MAST2. At each locus we observed a pattern of three deletions or duplications in cases that were absent from controls (Figs. 3 and 4 and Supplemental Figs. S7 and S8).
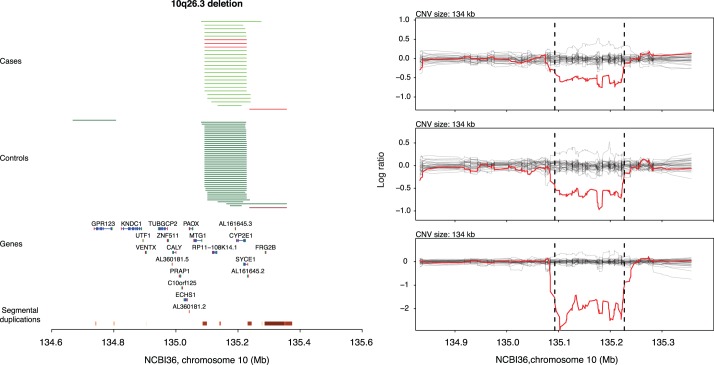
FIG. 3.
Recurrent case-specific deletions affecting SYCE1. In the left subpanel, red and green horizontal lines indicate deletions and duplications, respectively. Case CNVs are grouped in the top half and colored in light green and light red, whereas control CNVs are grouped in the bottom half and colored in dark green and dark red. Segmental duplications annotated on NCBI36 are plotted at the very bottom. In the right subpanel, the normalized and smoothed log2 ratio data for each deletion carrier is plotted in red, along with a random sample of log2 ratio profiles from other samples genotyped on the same plate, in gray.
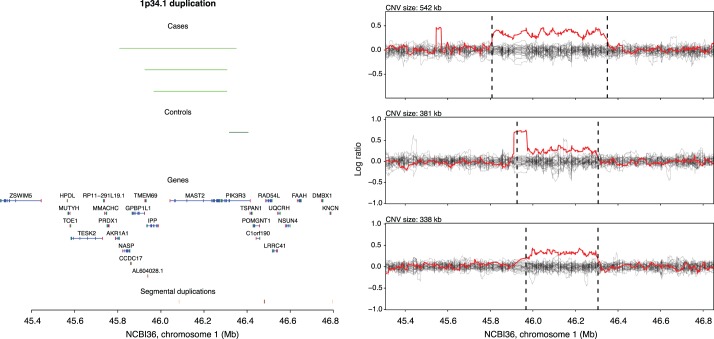
FIG. 4.
Recurrent case-specific duplications affecting MAST2. In the left subpanel the genomic locations of three case-specific duplications are annotated in light green. Segmental duplications annotated on NCBI36 are plotted at the very bottom. In the right subpanel, the normalized and smoothed log2 ratio data for each duplication carrier is plotted in red, along with a random sample of log2 ratio profiles from other samples genotyped on the same plate, in gray.
We have previously reported that deletions of DMRT1 are associated with NOA, and it is well known that DMRT1 loss-of-function can lead to a variety of derangements in gonad development [5, 20, 21]. SYCE1 encodes synaptonemal complex element 1, a gene with high and specific testis expression. Mice that are homozygous for a SYCE1 null allele exhibit failure to repair double-strand breaks during meiosis, leading to apoptosis of germ cells and infertility [22]. Interestingly, one of our three cases was clearly homozygous for the deletion, despite an overall frequency of 4 deletion alleles out of 5552 in the entire case-control cohort—an exceptionally unlikely observation under the Hardy-Weinberg equilibrium. The TUBA3E gene encodes for Tubulin alpha 3e and shows high and specific expression in testis. A mouse model has not been characterized for this gene. Finally, MAST2 encodes microtubule-associated serine/threonine kinase. The mouse ortholog of MAST2 is only transcribed in testis and increases during postnatal testicular development coincident with the onset of meiosis and spermiogenesis [23, 24]. Immunofluorescence microscopy localized the protein product to microtubules associated with the sperm manchette.
Candidate Loci for New “Genomic Disorders” and Risk Factors Mediated by NAHR
We assessed the potential for fertility-related genomic disorders by searching for rearrangement hotspots that contain genes with key roles in spermatogenesis. We created a list of genes that are specifically expressed in testis, differentially expressed in spermatogenesis, and/or are known to cause azoospermia when deleted in mice, and mapped their location with respect to 381 pairs of highly homologous duplications with spacing amenable to NAHR. We found 117 spermatogenesis genes that appear to be highly vulnerable to recurrent deletion or duplication because of their location in regions of genome instability (Supplemental Table S10). In total, 47 of the segmental pairs (12%) interrogated flanked at least one spermatogenesis gene. Interestingly, 44% of spermatogenesis genes in rearrangement hotspots were located on the X chromosome—a chromosome previously noted to harbor a large number of ampliconic gene families involved in spermatogenesis [16]. This analysis is not meant to provide a comprehensive catalog of all hotspots that may mediate infertility; rather, it is a first pass to provide some sense of the extent to which genes important for testis function may be vulnerable to NAHR-mediated mutation.
Crucially, the SYCE1 and TUBA3E deletions described above occur at rearrangement hotspots (Fig. 3 and Supplemental Fig. S7). Although a large number of duplications are segregating at these two loci in cases and controls, the reciprocal deletions were observed only in cases. Another hotspot of interest contains the cancer-testis antigen SSX6. We previously reported recurrent deletions of SSX6 to be modestly associated with spermatogenic impairment in white individuals [5]. These deletions, caused by NAHR between two 16-kb segmental duplications on Xp11.23, were found at a frequency of 1.6% in 724 cases and 0.55% in 3245 controls in our study of white individuals (OR, 3.0; 95% confidence interval [95% CI], 1.31–6.62; P = 0.007). Here, we observed seven deletions in cases and seven in controls, consistent with a modest association but not statistically significant (OR, 1.8; 95% CI, 0.54–6.03; P = 0.27).
Validation and Replication of Important Findings
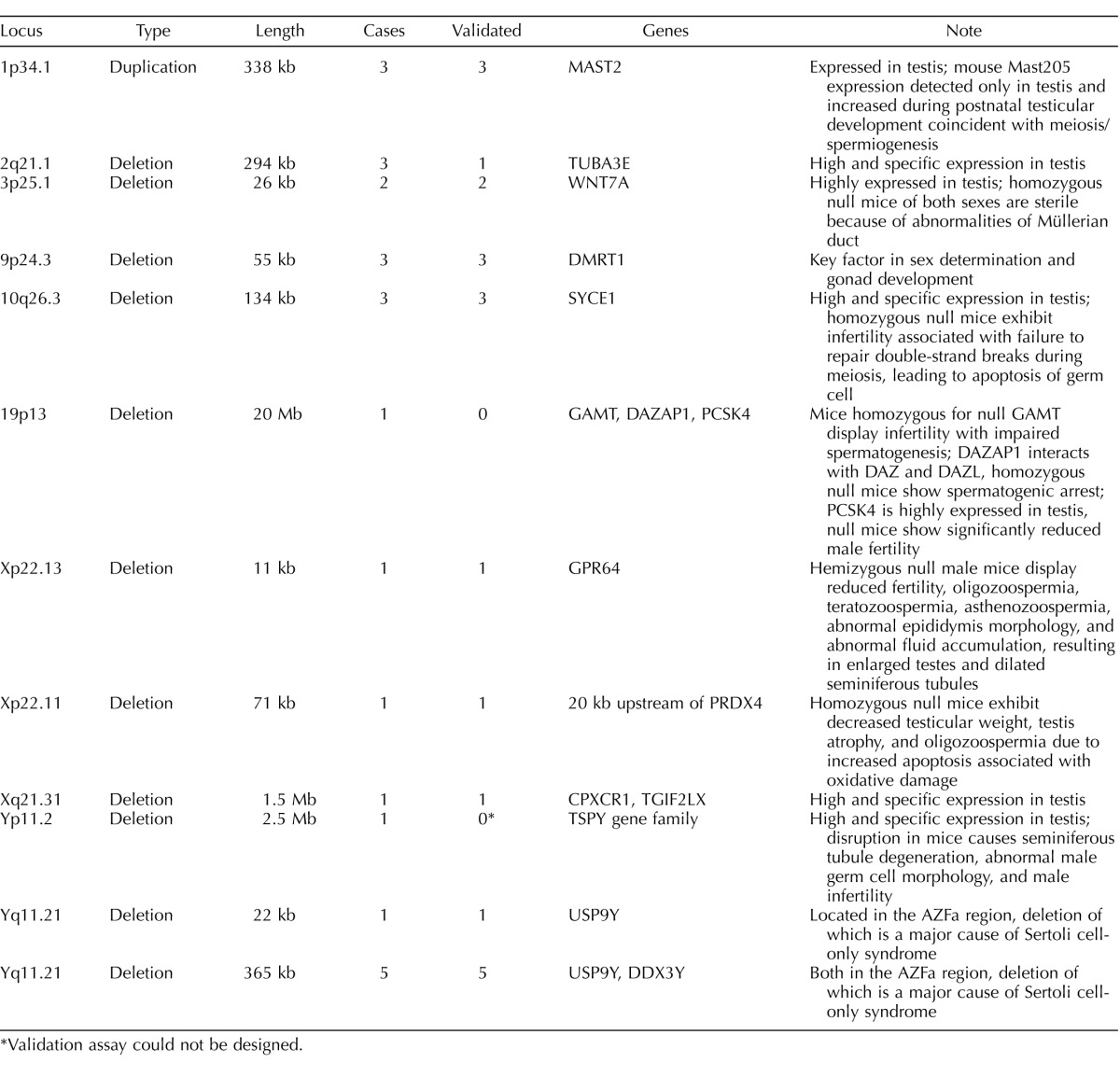
We selected 25 individual CNV calls of high interest and used qPCR to validate the original array findings in each sample (Table 1). Twenty-two of these calls showed signals in the qPCR confirming our initial discoveries; curiously, the three that failed validation were calls that all looked very convincing in the primary array data: a 20-Mb deletion on chromosome 19, and two deletions of TUBA3E apparently mediated by NAHR between pairs of large segmental duplications. Notably, a third TUBA3E deletion was validated cleanly by qPCR, showing a consistent 50% reduction in copy number.
TABLE 1.
Summary of validation experiments.
*Validation assay could not be designed.
Epidemiological replication of rare variant associations is exceptionally hard because of the large sample sizes needed to achieve adequate power. Nonetheless, we attempted to replicate five rare variant associations in an independent cohort of 1457 cases and 1590 controls. We designed qPCR-based assays for MAST2, TUBA3E, SYCE1, USP9Y, and DDX3Y (Materials and Methods). We included SRY as a haploid control locus. After QC of the resulting raw data, we were left with data on at least 1258 cases and 944 controls for each assay (Supplemental Figs. S2, S3, S9, and S10, and Table 2). Two of our initial associations showed robust replication signals. First, we tested for an association between NOA and duplication of each of the two genes in the AZFa region, USP9Y and DDX3Y. Each gene showed strong association (USP9Y: OR, 21.6; P < 2 × 10−6; DDX3Y: OR, 4.9; P < 0.006). Because the replication cohort has not been screened for karyotype anomalies, it is impossible to define the extent of aneuploidy for each apparent AZFa duplication carrier. However, when we normalized our qPCR data for these loci against SRY copy number, the association at each locus disappeared, suggesting that the majority of the observed signal is being driven by large aneuploidies containing AZFa as well as SRY, for instance, isodicentric Y chromosomes (Table 2).
TABLE 2.
Summary of replication experiments.
P value for fisher exact test of association of CNV carrier status with case-control status.
Normalized against SRY.
The second association that we report as replicable involves duplications of the MAST2 locus. We observed clear evidence for replication (OR, 12; 95% CI, 1.75–519; P < 3 × 10−3). Deletions and duplications were called at both TUBA3E and SYCE1, but no significant associations were observed at these loci after accounting for multiple testing.
DISCUSSION
The human genome is rife with large, highly identical segmental duplications. Judging from the genes involved in these duplications and the timing of their evolutionary origin, it has been widely proposed that they are responsible for some hominid-specific traits. They are also irrefutably a source of recurrent CNV, leading to a diverse set of developmental and neurological phenotypes. To investigate the role of CNVs generated by NAHR, we created a map of rare CNVs in cases of NOA from Nanjing, China, and ethnicity-matched controls. In terms of the number of individuals involved, it is, to date, the largest map of CNVs generated from people with infertility.
In the discovery phase of our study, we identified a number of promising new candidate loci as being affected by recurrent CNVs in cases of NOA, specifically TUBA3E, SYCE1, and MAST2. Of these three, only the MAST2 locus showed clear signs of replication. SYCE1 encodes a germ cell-specific protein that localizes to the synaptonemal complex of spermatocytes and oocytes. In mouse, monoallelic deletion of SYCE1 produces no overt effects on fertility, but SYCE1 null mice of both sexes are infertile [22]. In our replication cohort, we found heterozygous SYCE1 deletion carriers in equal frequencies in cases and controls. We did observe one SYCE1 deletion homozygote among the discovery cases and another in the replication cases, but none in the discovery and replication controls, consistent with SYCE1 loss-of-function being a large-effect, recessive mutation causing infertility.
We have identified MAST2 duplications as a novel risk factor for NOA. The protein encoded by MAST2, microtubule-associated serine/threonine kinase, associates with microtubules in the spermatid manchette [23]. The manchette is a transient subcellular structure that forms around the posterior spermatid nucleus during spermiogenesis, and it appears to act as a storage and sorting area for proteins needed in the formation of the sperm tail. MAST2 has not been well studied in the 20 yr since its discovery, with fewer than 20 papers published specifically on the gene, but we were able to find a few noteworthy results supporting its further characterization. In a study of adaptive protein evolution in all testis-expressed proteins, Turner et al. [25] found MAST2 to show the third strongest signal of selection when comparing Mus musculus and Rattus norvegicus, with a remarkable dN:dS ratio of 3.95. The dN:dS ratio, the ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site, is an indicator of selective pressure acting on a protein-coding gene, where a value great than one implies positive selection and a value less than one implies purifying selection. In humans, MAST2 has been reported to be under recent positive selection in a recent survey of southeast Asian populations [26]. This selective sweep was apparently restricted to a Philippine Negrito population, and it was unlikely to be confounding our association study, which is strictly composed of Han Chinese. Inspection of the location of the MAST2 duplication carriers in the principal components plot of our discovery samples further indicates that stratification is not driving this association (Supplemental Fig. S4). Finally, a human-specific subtype of the SVA transposon family, called SVA F1, is the result of a 5′ transduction of MAST2 exon 1 and cis-regulatory sequence onto an ancestral SVA element [27]. This transduced MAST2 sequence has been shown to drive reporter gene expression in human germ cells and is proposed to be the source of the exceptional success of this novel SVA subfamily [28].
Performing well-powered association studies, like the one presented here, requires exceptionally large sample sizes. The ideal controls for our study would be a large number of men with a complete andrological workup and found to be in the normal range regarding all semen parameters; however, because of the costs involved in phenotyping and data generation, we have opted to use for our controls a set of individuals shared with other genome-wide association studies. Previous high-profile genome-wide association studies have successfully used shared controls as an economic and useful alternative to ideal controls, in the process generating a large number of reproducible disease associations (see the Wellcome Trust Case Control Consortium [29] for an example and further discussion of shared controls). One risk of using a shared control group is the chance for misclassification bias, whereby a proportion of the controls may have NOA, or the frequencies of deleterious genetic variation in controls may be slightly modulated because of the high frequency of lung cancer in this group. We believe our primary discoveries are not artifacts of the study design: 1) our findings relate to an excess of rare deleterious mutations in the cases, not the controls, 2) our burden results have been reported previously using a completely independent cohort [5], and 3) our MAST2 association was replicated using a completely independent cohort. We believe the most important consequence of our use of shared controls is a bias in the effect sizes (ORs) that we report for these associations—the true effect sizes may be larger than we estimate here.
As we enter the era of high-throughput genome sequencing, it is important to clarify the genetic architecture of sperm production, a trait that undeniably has a substantial genetic component but has had little systematic investigation beyond Y chromosome assays. Specifically, we are interested in demonstrating that genetic variation outside the Y chromosome can substantially influence sperm production before we invest in genome-wide sequencing of large cohorts of men with well-defined semen parameters. As we have reported earlier, the results presented here clearly confirm that mutations throughout the genome, not just the Y chromosome, can impair sperm production: based on tests of overall burden, Y chromosome variants appear responsible for the most disease liability, followed by the X, and then the autosomes.
We identified several sets of genes or genomic regions that showed an enrichment of rare CNVs in cases compared with controls. Beyond highlighting important biology underlying sperm production, the most important use of these burden results is to provide evidence that individually rare mutations in these cases are associated with disease. Many of the variants in these burden categories were singletons, seen in only one member of the cohort. Traditional epidemiological methods, which look for differences in variant frequency between cases and controls, or coinheritance of a variant with a phenotype, will struggle to assign convincing statistical significance to such rare variants. We are actively working on novel methods, leveraging data sets of tens of thousands of individuals, to provide more sensitive detection of highly unusual singleton genotypes or diplotypes [30]. Such methods hold promise for not only accelerating our understanding of sperm production, but identifying variants with diagnostic value or variants that are therapeutic targets in the clinic.
ACKNOWLEDGMENTS
We thank Dr. Michiel Noordam for helpful discussions regarding Y chromosome rearrangements, and Dr. Katinka Vigh-Conrad for assistance preparing figures.
Footnotes
This work was supported by National Key Basic Research Program grants 2011CB944304 and 2013CB911400; National Nature Science Foundation for Distinguished Young Scholars of China grant 81225020; the National Program for Support of Top-notch Young Professionals from the Organization Department of the CPC Central Committee; Distinguished Professor of Jiangsu Province; and the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the U.S. National Institutes of Health (grant R01HD078641 to D.F.C.). The content of this paper is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. All CNV calls from the study are being submitted to dbVAR ( http://www.ncbi.nlm.nih.gov/dbvar/).
These authors are joint first authors.
These authors are joint senior authors.
REFERENCES
- Stankiewicz P, Lupski JR. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H, Bader P, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Xia Y, Guo X, Dai J, Li H, Hu H, Jiang Y, Lu F, Wu Y, Yang X, Yao B, Lu C, et al. A genome-wide association study in Chinese men identifies three risk loci for non-obstructive azoospermia. Nat Genet. 2012;44:183–186. doi: 10.1038/ng.1040. [DOI] [PubMed] [Google Scholar]
- Lopes AM, Aston KI, Thompson E, Carvalho F, Goncalves J, Huang N, Matthiesen R, Noordam MJ, Quintela I, Ramu A, Seabra C, Wilfert AB, et al. Human spermatogenic failure purges deleterious mutation load from the autosomes and both sex chromosomes, including the gene DMRT1. PLoS Genet. 2013;9:e1003349. doi: 10.1371/journal.pgen.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolo L, Zuffardi O. Localization of factors controlling spermatogenesis in the nonfluorescent portion of the human Y chromosome long arm. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- World Health Organization. WHO Laboratory Manual for the Examination of Human Semen and Sperm-Cervical Mucus Interaction. Cambridge, UK: Cambridge University Press; 1999. [Google Scholar]
- Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, Hubbell E, Veitch J, Collins PJ, Darvishi K, Lee C, Nizzari MM, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, Saeed S, Hamilton-Shield J, Clayton-Smith J, O'Rahilly S, Hurles ME, Farooqi IS. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C, Plagnol V, Fitzgerald T, Redon R, Marchini J, Clayton D, Hurles ME. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008;40:1245–1252. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du R, Lu C, Jiang Z, Li S, Ma R, An H, Xu M, An Y, Xia Y, Jin L, Wang X, Zhang F. Efficient typing of copy number variations in a segmental duplication-mediated rearrangement hotspot using multiplex competitive amplification. J Hum Genet. 2012;57:545–551. doi: 10.1038/jhg.2012.66. [DOI] [PubMed] [Google Scholar]
- Rozen SG, Marszalek JD, Irenze K, Skaletsky H, Brown LG, Oates RD, Silber SJ, Ardlie K, Page DC. AZFc deletions and spermatogenic failure: a population-based survey of 20,000 Y chromosomes. Am J Hum Genet. 2012;91:890–896. doi: 10.1016/j.ajhg.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giachini C, Laface I, Guarducci E, Balercia G, Forti G, Krausz C. Partial AZFc deletions and duplications: clinical correlates in the Italian population. Hum Genet. 2008;124:399–410. doi: 10.1007/s00439-008-0561-1. [DOI] [PubMed] [Google Scholar]
- Lin YW, Hsu LC, Kuo PL, Huang WJ, Chiang HS, Yeh SD, Hsu TY, Yu YH, Hsiao KN, Cantor RM, Yen PH. Partial duplication at AZFc on the Y chromosome is a risk factor for impaired spermatogenesis in Han Chinese in Taiwan. Hum Mutat. 2007;28:486–494. doi: 10.1002/humu.20473. [DOI] [PubMed] [Google Scholar]
- Lu C, Zhang F, Yang H, Xu M, Du G, Wu W, An Y, Qin Y, Ji G, Han X, Gu A, Xia Y, et al. Additional genomic duplications in AZFc underlie the b2/b3 deletion-associated risk of spermatogenic impairment in Han Chinese population. Hum Mol Genet. 2011;20:4411–4421. doi: 10.1093/hmg/ddr369. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Skaletsky H, Brown LG, Zaghlul S, Rock S, Graves T, Auger K, Warren WC, Wilson RK, Page DC. Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet. 2013;45:1083–1087. doi: 10.1038/ng.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Giacco D, Chianese C, Ars E, Ruiz-Castane E, Forti G, Krausz C. Recurrent X chromosome-linked deletions: discovery of new genetic factors in male infertility. J Med Genet. 2014;51:340–344. doi: 10.1136/jmedgenet-2013-101988. [DOI] [PubMed] [Google Scholar]
- Chianese C, Gunning AC, Giachini C, Daguin F, Balercia G, Ars E, Lo Giacco D, Ruiz-Castane E, Forti G, Krausz C. X chromosome-linked CNVs in male infertility: discovery of overall duplication load and recurrent, patient-specific gains with potential clinical relevance. PLoS One. 2014;9:e97746. doi: 10.1371/journal.pone.0097746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzuk MM, Lamb DJ. The biology of infertility: research advances and clinical challenges. Nat Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Ledig S, Hiort O, Wunsch L, Wieacker P. Partial deletion of DMRT1 causes 46,XY ovotesticular disorder of sexual development. Eur J Endcrinol. 2012;167:119–124. doi: 10.1530/EJE-12-0136. [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, de Massy B, Benavente R, Cooke HJ. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden PD, Cowan NJ. A novel 205-kilodalton testis-specific serine/threonine protein kinase associated with microtubules of the spermatid manchette. Mol Cell Biol. 1993;13:7625–7635. doi: 10.1128/mcb.13.12.7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden PD, Millette CF. Increased activity associated with the MAST205 protein kinase complex during mammalian spermiogenesis. Biol Reprod. 1996;55:1039–1044. doi: 10.1095/biolreprod55.5.1039. [DOI] [PubMed] [Google Scholar]
- Turner LM, Chuong EB, Hoekstra HE. Comparative analysis of testis protein evolution in rodents. Genetics. 2008;179:2075–2089. doi: 10.1534/genetics.107.085902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Deng L, Lu D, Xu S. Genome-wide landscapes of human local adaptation in Asia. PLoS One. 2013;8:e54224. doi: 10.1371/journal.pone.0054224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damert A, Raiz J, Horn AV, Lower J, Wang H, Xing J, Batzer MA, Lower R, Schumann GG. 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabolotneva AA, Bantysh O, Suntsova MV, Efimova N, Malakhova GV, Schumann GG, Gayfullin NM, Buzdin AA. Transcriptional regulation of human-specific SVAF(1) retrotransposons by cis-regulatory MAST2 sequences. Gene. 2012;505:128–136. doi: 10.1016/j.gene.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Manolio TA, Dimmock DP, Rehm HL, Shendure J, Abecasis GR, Adams DR, Altman RB, Antonarakis SE, Ashley EA, Barrett JC, Biesecker LG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We are in the process of submitting all CNV calls from the study to dbVAR (http://www.ncbi.nlm.nih.gov/dbvar/).