Takeuchi et al. demonstrate that CRTAM identifies CD4 T cells with cytotoxic function, and present new insights into CD4+CTL development.
Abstract
Naive T cells differentiate into various effector T cells, including CD4+ helper T cell subsets and CD8+ cytotoxic T cells (CTL). Although cytotoxic CD4+ T cells (CD4+CTL) also develop from naive T cells, the mechanism of development is elusive. We found that a small fraction of CD4+ T cells that express class I–restricted T cell–associated molecule (CRTAM) upon activation possesses the characteristics of both CD4+ and CD8+ T cells. CRTAM+ CD4+ T cells secrete IFN-γ, express CTL-related genes, such as eomesodermin (Eomes), Granzyme B, and perforin, after cultivation, and exhibit cytotoxic function, suggesting that CRTAM+ T cells are the precursor of CD4+CTL. Indeed, ectopic expression of CRTAM in T cells induced the production of IFN-γ, expression of CTL-related genes, and cytotoxic activity. The induction of CD4+CTL and IFN-γ production requires CRTAM-mediated intracellular signaling. CRTAM+ T cells traffic to mucosal tissues and inflammatory sites and developed into CD4+CTL, which are involved in mediating protection against infection as well as inducing inflammatory response, depending on the circumstances, through IFN-γ secretion and cytotoxic activity. These results reveal that CRTAM is critical to instruct the differentiation of CD4+CTL through the induction of Eomes and CTL-related gene.
The T cell precursors differentiate into CD4+ and CD8+ T cells during thymic development, a process tightly regulated by several key transcription factors such as RUNX3, ThPOK/cKrox, GATA-3, and Tox (Hernández-Hoyos et al., 2003; Pai et al., 2003; He et al., 2005; Sun et al., 2005; Wang et al., 2008; Aliahmad et al., 2011). Runx3 is a transcription factor of the RUNX family and binds to the CD4 silencer element, which down-regulates CD4 expression and promotes differentiation to the cytotoxic T cells (CTL) linage (Taniuchi et al., 2002; Woolf et al., 2003). CTLs play critical roles in protection from viral infection and tumor growth. CD8+ T cells recognize and respond to antigen (Ag) peptides displayed by MHC class I on APCs and target cells, and function to exert cytotoxicity or recruit and activate other immune cells. These CTL effector functions are critically controlled by two T-box transcription factors, T-bet and Eomesodermin (Eomes; Pearce et al., 2003; Eshima et al., 2012). On the other hand, ThPOK, GATA3, and Tox inhibit the differentiation to CD8+ T cells and induce CD4+ helper T cell development.
Naive CD4+ T cells differentiate into various effector T helper (Th) cells such as Th1, Th2, and Th17 cells, which produce IFN-γ, IL-4/IL-5/IL-9/IL-13, and IL-17/IL-22, respectively (O’Shea and Paul, 2010). Functional differentiation into different Th subsets is regulated by environmental factors, mainly by cytokines; Th1 by IL-12/IFN-γ, Th2 by IL-4, and Th17 by IL-6 and TGFβ. IFN-γ and IL-12 are important for Th1 differentiation, and IFN-γ production is regulated by various transcription factors, such as T-bet, Eomes, Runx3, and STAT4. T-bet in particular is the leading player in Th1 differentiation and regulates not only induction of IFN-γ production but also suppression of the expression of GATA-3, the master regulator of Th2 differentiation. Although the differentiation of these CD4+ Th subsets has been well defined, little is known about regulation of the development of the CD4+ subset with cytotoxic function, the CD4+CTL.
Cytotoxic CD4+ T cells (CD4+CTL) were identified as T cells that have the ability to acquire cytotoxic activity and directly kill infected, transformed, or allogeneic MHC class II–expressing cells. Many studies have described CD4+CTL cell lines and clones from both humans (Wagner et al., 1977; Feighery and Stastny, 1979) and mice (Lukacher et al., 1985; Maimone et al., 1986), and CD4+CTL have also been identified among the peripheral blood mononuclear cells (PBMCs) of humans seropositive after chronic viral infections such as human cytomegalovirus (HCMV; van Leeuwen et al., 2004; Zaunders et al., 2004), HIV-1 (Appay et al., 2002; Zaunders et al., 2004), and hepatitis virus (Aslan et al., 2006), as well as in mice infected by lymphocytic choriomeningitis virus (LCMV; Jellison et al., 2005) or γ-herpes virus (Stuller and Flaño, 2009). It has been suggested that CD4+CTL could have a potential therapeutic role for antitumor immunity (Quezada et al., 2010; Xie et al., 2010).
We have previously identified MHC class I–restricted T cell–associated molecule (CRTAM) as an Ig domain–containing and activation-induced surface receptor predominantly expressed on activated CD8+ T cells and NK/NKT cells, and cell adhesion molecule 1 (CADM1)/Necl2/TSLC1 as its ligand (Kennedy et al., 2000; Kuramochi et al., 2001; Arase et al., 2005; Boles et al., 2005; Galibert et al., 2005). The CRTAM–CADM1 binding results from a heterotypic interaction between different cell types. CRTAM is transiently expressed in the early phase of T cell activation, and CRTAM+ T cells mediate cell adhesion with CADM1+ cells. The association between CRTAM+ CD8+ T cells and CADM1+ CD8+ DCs in LNs is critical for the accumulation of antigen-specific CTLs and their subsequent proliferation within the draining LNs (Takeuchi et al., 2009).
Here, we show that a small fraction of activated CD4+ T cells also express CRTAM and have characterized these unique CD4+ T cells. We found that the CRTAM+ CD4+ T cells have the characteristics of both CD4+ and CD8+ T cells and that these cells particularly express CTL-related genes such as Granzyme B (gzmB), IFN-γ, and Eomes, and exhibit cytotoxicity after cultivation. Furthermore, ectopic expression of CRTAM in vivo can induce CD4+CTL differentiation. This unique population is notably observed in the mucosal tissue and inflammatory sites and likely plays a role in protection from infection and in immune responses.
RESULTS
CRTAM expression on a small fraction of CD4+ T cells
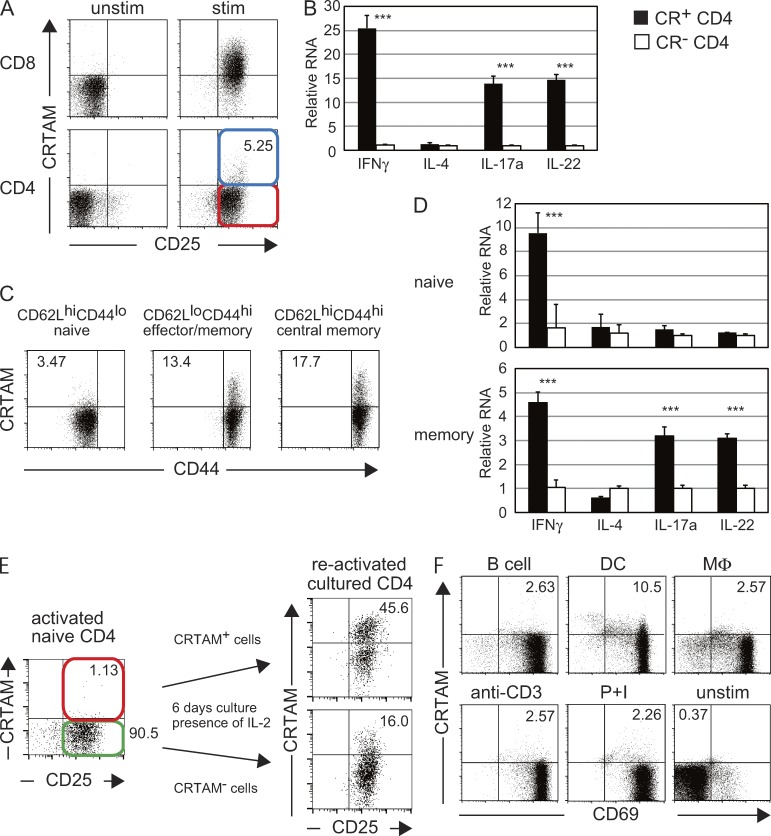
We previously reported that almost all CD8+ T cells transiently express CRTAM at the early stage of T cell activation. Yeh et al. (2008) first reported that a small fraction of activated CD4+ T cells also express CRTAM and suggested that the CRTAM-expressing cells might be a distinct T cell subpopulation. We now confirm that CRTAM is expressed on the surface of ∼2–5% of splenic CD4+ T cells after TCR stimulation (Fig. 1 A). To characterize this unique population of CRTAM-expressing CD4+ T cells, CRTAM+ and CRTAM− cells were sorted after stimulation, and the production of various cytokines was analyzed. The CRTAM+ T cells produced high levels of effector cytokines, such as IFN-γ, IL-17, and IL-22, but not IL-4 (Fig. 1 B). We assumed that the CRTAM+ population contains effector–memory T cells that produce high levels of effector cytokines. To confirm this possibility, naive (CD4+CD62LhiCD44lo), effector memory (CD4+CD62LloCD44hi), and central memory (CD4+CD62LhiCD44hi) CD4+ T cells were purified and stimulated, and the percentage of CRTAM+ cells was analyzed (Fig. 1 C). Interestingly, CRTAM+ cells were detected in each subset, although naive cells generated fewer than memory cells. As expected, CRTAM+ effector memory CD4+ T cells produce much higher amounts of IFN-γ and IL-17a than CRTAM− cells (Fig. 1 D), and restimulation of CRTAM+ cells induced higher levels of CRTAM expression (Fig. 1 E). In contrast, only a small percentage of CRTAM− T cells become CRTAM+ cells after stimulation. These data suggest that a majority of CRTAM-expressing CD4+ T cells are memory-type T cells that produce high levels of cytokines. However, we noticed that a small fraction of naive CD4+ T cells also express CRTAM upon stimulation (Fig. 1 C). We found that the CRTAM+ activated naive T cells produce high amount of IFN-γ but not other effector cytokines (Fig. 1 D). Because this population is different from the effector memory population, this observation indicates that activated naive CD4+ T cells already contain some T cells producing IFN-γ immediately after stimulation. Next, we tested whether the expression of CRTAM on activated naive CD4+ T cells is constant or flexibly changed by the interaction with different APC populations (Fig. 1 F). Naive OT-II Tg CD4+ T cells were stimulated by peptide-pulsed various APCs, including B cells, DCs, and macrophages. B cells and macrophages induced CRTAM in a similar level to those stimulated by anti-CD3 Ab or P+I. In contrast, more than fourfold of CRTAM-expressing cells were induced by stimulation with DCs. These data indicate that CRTAM expression is flexibly induced by environmental situation, most efficiently upon DC stimulation.
Figure 1.
A small fraction of CD4+ T cells expresses CRTAM. (A) Comparison of CRTAM expression between CD8+ and CD4+ T cells. Splenic T cells were unstimulated (left) or stimulated (right) with anti-CD3/CD28 Abs, and then stained with anti-CRTAM and anti-CD25 Abs. Cells were analyzed 14 h after stimulation. The numbers indicate the percentages of CRTAM+ CD25+ cells among CD4+ T cells. (B) Quantitative analysis of effector cytokine production between CRTAM+ CD4+ T cell (CR+ CD4, closed column) and CRTAM− CD4+ T cell (CR− CD4, open column). Both populations were purified from activated CD4 splenic T cells, which were activated with anti-CD3 and CD28 Abs for 14 h. (C) CRTAM expression on naive and effector memory T cells. Each population was sorted from splenic T cells and the expression of CRTAM was analyzed after stimulation. The numbers indicate the percentage of CRTAM+ cells. (D) Naive (top) and effector memory (bottom) cells were isolated, stimulated, and sorted for CRTAM+ or CRTAM− cells. Cytokine expression in each population was quantified. Closed and open columns are CRTAM+ and CRTAM− CD4+ T cells, respectively. (E) CRTAM expression upon restimulation. Activated naive CD4+ T cells were sorted into CRTAM+ and CRTAM− cells, and the isolated populations were incubated for 6 d in the presence of IL-2, and then restimulated by anti-CD3/CD28 Abs for 14 h, after which the cell surface expression of CRTAM was analyzed. The numbers indicate the percentages of CRTAM+ CD25+ cells among CD4+ T cells. (F) CRTAM expression in various types of cells. OT-II Tg CD4+ T cells were stimulated by various peptide-loaded APCs, antibody, or PMA + ionomycin for 14 h, and expression level of CRTAM was quantified. The numbers indicate the percentages of CRTAM+ cells. All data are representative of at least two independent experiments. Error bars are SD. ***, P < 0.001, Student’s t test.
CRTAM+ CD4+ T cells have characteristics of both CD4+ and CD8+ T cells
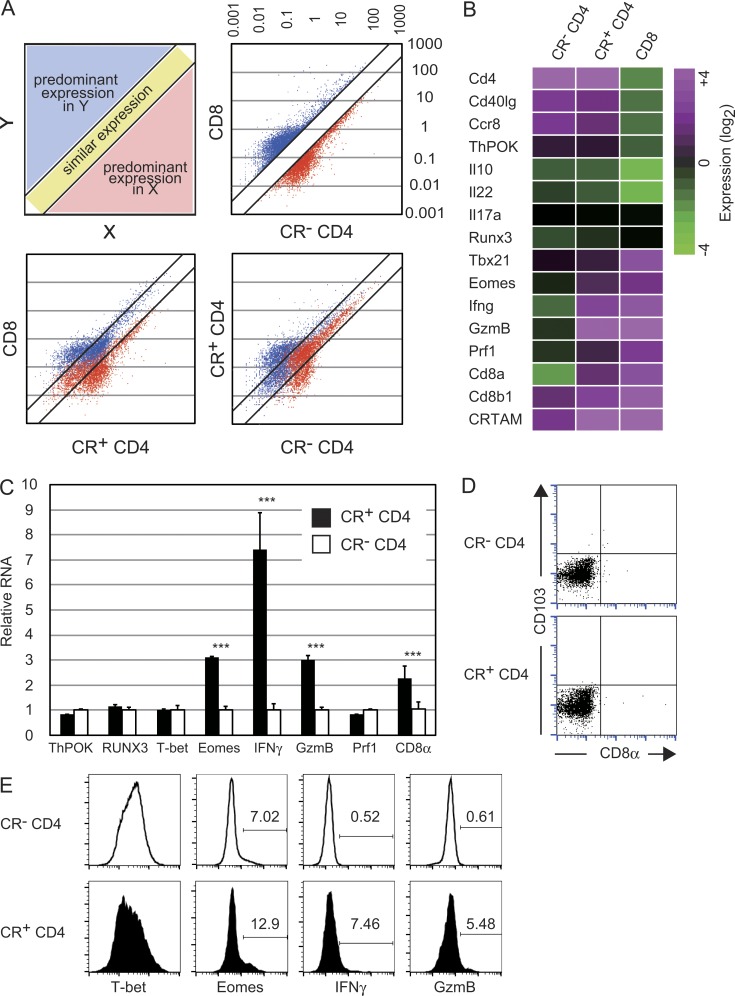
After the observation that CRTAM+ naive CD4+ T cells are high producers of IFN-γ, we analyzed the expression of transcription factors related to IFN-γ production such as RUNX3, T-bet, and Eomes (Fig. 2 C). The expression of T-bet and Runx3 was comparable to that in CRTAM− CD4+ T cells but, interestingly, Eomes expression was clearly up-regulated in the CRTAM+ CD4+ T cells. Considering that Eomes is predominantly expressed in CD8+ T cells and induces IFN-γ production, and that T-bet directly activates IFN-γ transcription and is considered to be the master regulator of Th1 differentiation (Szabo et al., 2000, 2003; Pearce et al., 2003; Glimcher et al., 2004), these results suggest that CRTAM+ CD4+ T cells are not typical Th1 cells but rather CD8+ T-like cells. Gene expression profiles of CRTAM+ versus CRTAM− naive CD4+ T cells were analyzed by microarray (Fig. 2 A). Genes predominantly expressed in either CD4+ or CD8+ T cells (more than threefold higher expression) were depicted and compared with those in CRTAM+ CD4+ T cells. Whereas >70% of genes were similarly expressed between CRTAM+ and CRTAM− CD4+ T cells, 68% of genes were expressed at a comparable level between CRTAM+ CD4+ T cells and CD8+ T cells. The expression level of the majority of genes in CRTAM+ CD4+ T cells was found to be intermediate between CD4+ and CD8+ T cells. Whereas both CRTAM+ and CRTAM− CD4+ T cells similarly express CD4+ T cell–related genes, including CD4 and ThPOK, they also express CTL-related genes, such as IFN-γ, CD8α, gzmB, and Eomes (Fig. 2, B and C). These data strongly suggest that CRTAM+ CD4+ T cells have the characteristics of both CD4+ and CD8+ T cells. Interestingly, CD8α expression was only observed at the mRNA level, but was not detectable on the cell surface. CD8α mRNA expression was not a result of contaminating CD8+ T cells because CD8+ T cells were extensively eliminated during the purification of naive CD4+ T cells. Indeed, we could not detect the cell surface expression of CD8α even 6 d after stimulation (Fig. 2 D). In contrast, the expression of Eomes, IFN-γ, and gzmB were slightly but significantly increased at the protein levels in CRTAM+ CD4+ T cells, whereas CRTAM+ T cells tend to express less T-bet (Fig. 2 E). From these data, we confirmed that CTL-related genes are up-regulated in CRTAM+ CD4+ T cells at both mRNA and protein levels, except for CD8α expression.
Figure 2.
CRTAM+ CD4+ T cells possess the potential of both CD4+ and CD8+ T cells. (A) Comparison of the gene expression pattern among three populations; CRTAM+ (CR+ CD4+) T cells, CRTAM− (CR− CD4+) T cells, and CD8+ T cells. Blue and red dots indicate genes predominantly expressing in CD8+ or CD4+ T cells, respectively. (bottom) Scatter plots of CD8+ and CD4+ T cell predominant genes. (B) Heat map of the microarray analysis data of the three populations in A. (C) Quantitative real-time PCR analysis for CTL-related genes in CR+ and CR− CD4+ T cells. GzmB, Granzyme B; Prf1, perforin 1. (D) Surface expression of CD8α on CRTAM+ CD4+ T cells. Sorted CRTAM+ or CRTAM− cells were incubated for 5 d in the presence of IL-2. CR+ CD4+ and CR− CD4+ T cells were stained for CD8α. (E) Protein expression of CTL-related genes in CRTAM+CD4+ T cells. CRTAM+ and CRTAM− CD4+ T cells were prepared similarly as in C were subjected to intracellular staining, and were analyzed by flow cytometry using specific Abs. Microarray analysis was performed once with three mice from each sample. The numbers indicate the percentages of positive cells expressing each gene. Data (C–E) are representative of at least two independent experiments. Error bars are SD. ***, P < 0.001, Student’s t test.
CRTAM+ CD4+ T cells differentiate into CTL
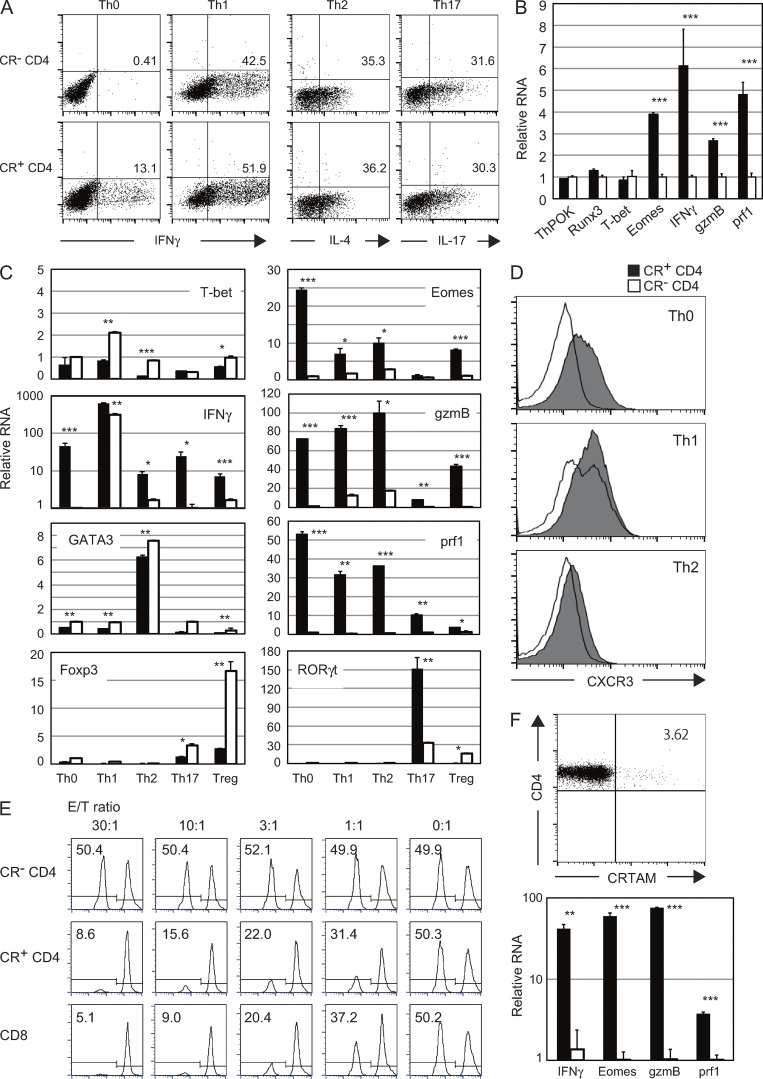
Next, we examined whether CRTAM+ CD4+ T cells exhibit functions of both CD4+ and CD8+ T cells. Naive CD4+ T cells were stimulated and CRTAM+ or CRTAM− T cells were isolated and incubated under the optimal conditions for each type of Th cell polarization: Th1 with IL-12, Th2 with IL-4, Th17 with IL-6/TGFβ, and iTreg with TGFβ. CRTAM+ CD4+ T cells differentiated normally into Th1, Th2, Th17, and iTreg cells, similar to the CRTAM− population (Fig. 3 A and not depicted), suggesting that CRTAM+ CD4+ T cells have a normal capacity to differentiate to each of the Th lineages. However, we noted that under nonskewed conditions without any additional cytokines, a significant proportion of CRTAM+ CD4+ T cells differentiated to IFN-γ–producing cells. These cells are not typical Th1 cells because they express high levels of CTL-related genes but not T-bet (Fig. 3 B). In contrast, no IFN-γ–producing cells developed from CRTAM− CD4+ T cells under this condition. It is interesting because anti-CD3/CD28 stimulation in general induces certain levels of IFN-γ under Th0 condition. It is possible that the IFN-γ–producing T cells under Th0 condition are predominantly CRTAM+ CD4+ T cells. Alternatively, this could be attributed to experimental conditions. Because CRTAM is expressed only upon stimulation, CRTAM+CD4+ T cells were sorted after stimulation. Thereafter, the sorted T cells were returned to the culture for restimulation. Such slightly modified stimulation/culture condition may have reduced population producing IFN-γ. We also confirmed the expression of the transcription factors that are critical for each Th subset differentiation under the each skewing conditions (Fig. 3 C). Under Th1-skewing condition, both CRTAM+ and CRTAM− populations showed high levels of IFN-γ, but T-bet expression was not increased in CRTAM+ CD4+ T cells. However, other lineage specification transcription factors clearly up-regulated under the relevant skewing conditions. Interestingly, CTL-related genes such as Eomes, gzmB, and perforin were still increased in CRTAM+ CD4+ T cells under the skewing conditions for Th0, Th1, and Th2. Next, we analyzed the CXCR3 expression level as a marker of the Th1 cells (Fig. 3 D). In all situations, the CXCR3 expression was clearly up-regulated in CRTAM+ CD4+ T cells. These data suggest that CRTAM+ CD4+ T cells seem to be able to differentiate into each Th subsets; however, they are atypical Th subsets with remaining unique features of CTL. Whereas these activated naive CRTAM+ T cells expressed Eomes, IFN-γ, and gzmB, but not perforin (Fig. 2 C), the CRTAM+ effector T cells clearly showed elevated expression of perforin after 6 d of culture in the presence of IL-2. These results suggest that CRTAM+ CD4+ T cells may also have cytotoxic function. This possibility was indeed demonstrated in a retargeting cytotoxicity assay (Fig. 3 E) where anti-CD3 Ab–coated A20 target cells were incubated with CRTAM+ CD4+ effector T cells that had been cultured for 6 d under the nonskewing condition. The CTL activity was clearly observed with CRTAM+ CD4+ T cells, but not CRTAM− CD4+ T cells, and was similar to that of effector CD8+ T cells. These data clearly indicate that CRTAM+ CD4+ T cells have both the functional potential of CD4+ and CD8+ T cells and can produce effector Th cytokines and differentiate into CTLs, depending on the environmental conditions.
Figure 3.
CRTAM+ CD4+ T cells differentiate into CD4+ helper T cells and CD4+CTL. (A) CRTAM+ or CRTAM− cells were sorted and incubated under the optimal conditions for Th1, Th2, Th17, or Th0 differentiation. The numbers indicate the percentages of each cytokine-producing cell. (B) Quantitative analysis of the expression of CTL-related genes under nonpolarizing conditions. Closed column: CRTAM+ CD4+ T cells, open column: CRTAM− CD4+ T cells. (C and D) Expression of Th- and CTL-related genes and transcription factors (C) and surface expression of CXCR3 (D) in T cells under each Th differentiation condition. (E) Retargeting cytotoxicity assay using CRTAM+ CD4+ T cells. Effector cells were prepared from nonskewed conditions. CFSE-labeled cells that were a 1:1 mixture of target cells A20 (low CSFE) and Jurkat internal control cells (high CSFE) were co-cultured with anti-CD3 Ab and effector cells. 4 h later, living target cells were quantified by flow cytometry. Percentage of living target cells and the E/T ratio were indicated. (F) Differentiation of CD4+CTL in human T cells. A small fraction of human CD4+ T cells also express CRTAM (top) and the CRTAM+ but not CRTAM− T cells express CTL-related genes after 5 d of culture (bottom). The numbers indicate the percentage of CRTAM positive cells. All data are representative of at least two independent experiments. Error bars are SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
We next analyzed whether human CRTAM+ CD4+ T cells also preferentially differentiate into CTL or not (Fig. 3 F). A small fraction of human CD4+ T cells (1–5%) also express CRTAM after stimulation, similar to mouse CD4+ T cells. Similar to mouse T cells, these human CRTAM+ CD4+ T cells express high levels of CTL-related genes after culture with IL-2, strongly suggesting that human CRTAM+ CD4+ T cells can also differentiate into CTL and exhibit cytotoxic function.
Eomes does not regulate CRTAM expression
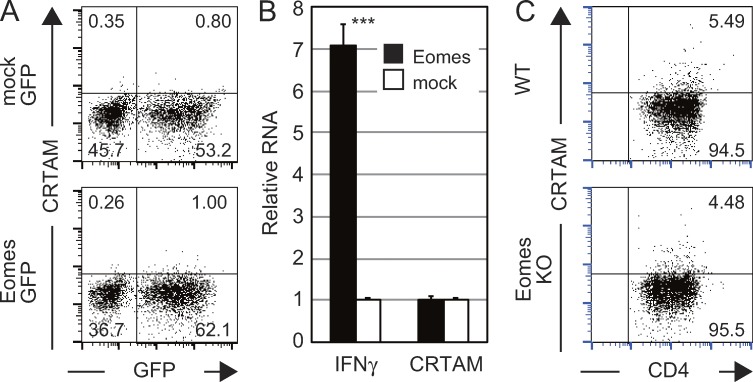
Unlike differentiated Th1 cells, activated naive CRTAM+ CD4+ T cells already express IFN-γ and Eomes before differentiation. Because it is well known that Eomes activates IFN-γ transcription, we examined the possibility that CRTAM expression is also induced in naive T cells by Eomes. For this purpose, the Eomes–IRES–eGFP genes were introduced into activated CD4+ T cells and the surface expression of CRTAM was analyzed after restimulation (Fig. 4 A). However, no CRTAM expression was observed on the surface of Eomes-introduced T cells. We could not detect CRTAM mRNA, even though IFN-γ expression was clearly enhanced by the transfection of Eomes (Fig. 4 B). We further analyzed CRTAM expression by using Eomes-deficient CD4+ T cells (Fig. 4 C). The same level of CRTAM expression was observed on the Eomes-deficient T cells after stimulation. These results indicate that CRTAM expression is not regulated by Eomes.
Figure 4.
CRTAM induces Eomes expression, but Eomes does not regulate CRTAM expression. (A) The Eomes-IRES-GFP (Eomes-GFP) or control (mock-GFP) expression vectors were transfected to activated naive CD4+ T cells. T cells were restimulated 4 d later, and then analyzed for the surface expression of CRTAM. The numbers indicate the percentages of each population among CD4+ T cells. (B) GFP+ cells from Eomes- (filled column) or mock- (open column) transfected cells in A were analyzed for of IFN-γ and CRTAM mRNA expression by qPCR. (C) CRTAM expression in Eomes-deficient T cells. CRTAM expression was analyzed in naive CD4+ T cells from WT and Eomes-deficient T cells 14 h after stimulation. The numbers indicate the percentages of CRTAM+ or CRTAM− population among CD4+ T cells. All data are representative of at least two independent experiments. Error bars are SD. ***, P < 0.001, Student’s t test.
CRTAM induces Eomes, IFN-γ production, and CTL function
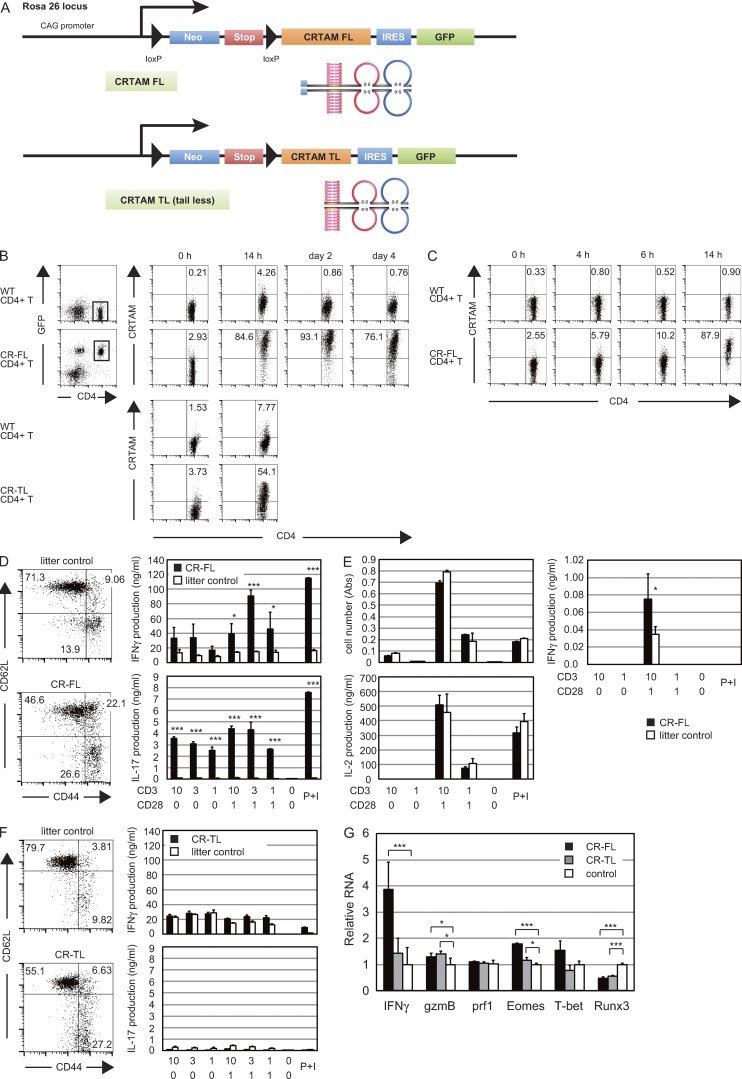
To analyze the function of CRTAM, we intended to prepare mice whose T cells all expressed CRTAM. For this purpose, we generated CRTAM knock-in (KI) transgenic (Tg) mice. A full-length CRTAM (CR-FL) cDNA attached to IRES-GFP was located downstream of a LoxP-Stop-LoxP cassette under the control of the CAG promoter and integrated into the Rosa26 locus, and the Tg mice were crossed with Lck-cre Tg mice (Fig. 5 A). In the Tg mouse, even though all T cells constitutively expressed GFP, the constitutive expression of CRTAM on the cell surface was not detected, but all CD4+ T cells immediately expressed cell surface CRTAM upon stimulation (Fig. 5 B). These results suggest that CRTAM expression is also regulated at the translational or posttranslational level. To distinguish these two possibilities, naive T cells from the Tg mice were treated with MG132, a potent proteasome inhibitor, and there was clear induction of surface expression of CRTAM without stimulation (Fig. 5 C). These results indicate that CRTAM expression is tightly regulated both transcriptionally and posttranslationally.
Figure 5.
Expression of CRTAM induces CD4+CTL in CRTAM-Tg mice. (A) Vector construction for the CRTAM-Tg mice. Two loxP sites were inserted upstream and downstream of Neomycin-resistant gene (Neo) and the stop codon cassette (Stop), and they were conjugated upstream of CRTAM (FL or TL)-IRES-eGFP cording sequences. These constructs were under the control of CAG promoter, and inserted in Rosa26 locus target sequence (as knock-in transgenic). (B) CRTAM expression in CRTAM Tg mice. CD4+ splenic T cells from CRTAM-FL Tg mice were stimulated with anti-CD3/CD28 Abs for the indicated periods. The numbers indicate the percentages of CD4+CRTAM+ cells. (C) Regulation of CRTAM expression. T cells from CR-FL Tg mice were cultured without stimulation in the presence of MG132. The numbers indicate the percentages of CD4+CRTAM+ cells. (D) Naive and effector memory cells in the spleen from full-length CRTAM knock-in Tg mice (CR-FL) and littermate controls were analyzed by staining for CD62L and CD44 (left). Whole splenic CD4+ T cells from CR-FL Tg mice (filled column) and WT mice (open column) were stimulated with anti-CD3/CD28 Abs for 48 h, and the cytokines produced were measured by ELISA. The numbers indicate the percentages of each quadrant among CD4+ T cells. (E) Cell proliferation and cytokine production by naive CD4+ T cells prepared from CR-FL. IL-17 production was not detected. (F) T cells in the spleen from tail-less mutant CRTAM knock-in Tg mice (CR-TL) were analyzed as in D. The numbers indicate the percentages of each quadrant among CD4+ T cells. (G) Quantitative real-time PCR analysis of CTL-related gene expression. Naive CD4+ T cells were stimulated with anti-CD3/CD28 Abs and mRNA samples were collected 14 h after stimulation and subjected to qPCR. The results shown are representative of at least two independent experiments. Error bars are SD. *, P < 0.05; ***, P < 0.001, Student’s t test.
In the CRTAM-FL Tg mouse, CD44hi effector memory cells were dramatically increased both in CD4+ and CD8+ T cell compartments, and the production of effector cytokines was clearly enhanced (Fig. 5 D and not depicted), confirming that CRTAM expression induces further maturation of effector memory cells and the production of effector cytokines. However, naive T cells in the Tg mice showed normal proliferation and IL-2 production upon stimulation (Fig. 5 E). The production of IFN-γ was clearly elevated, though at a low level, upon activation (Fig. 5, E and G). Interestingly, although IFN-γ production was enhanced, the expression of CTL-related genes was not induced (Fig. 5 G). These results suggest that naive CRTAM Tg CD4+ T cells do not yet have CTL competence at the early stage of T cell activation.
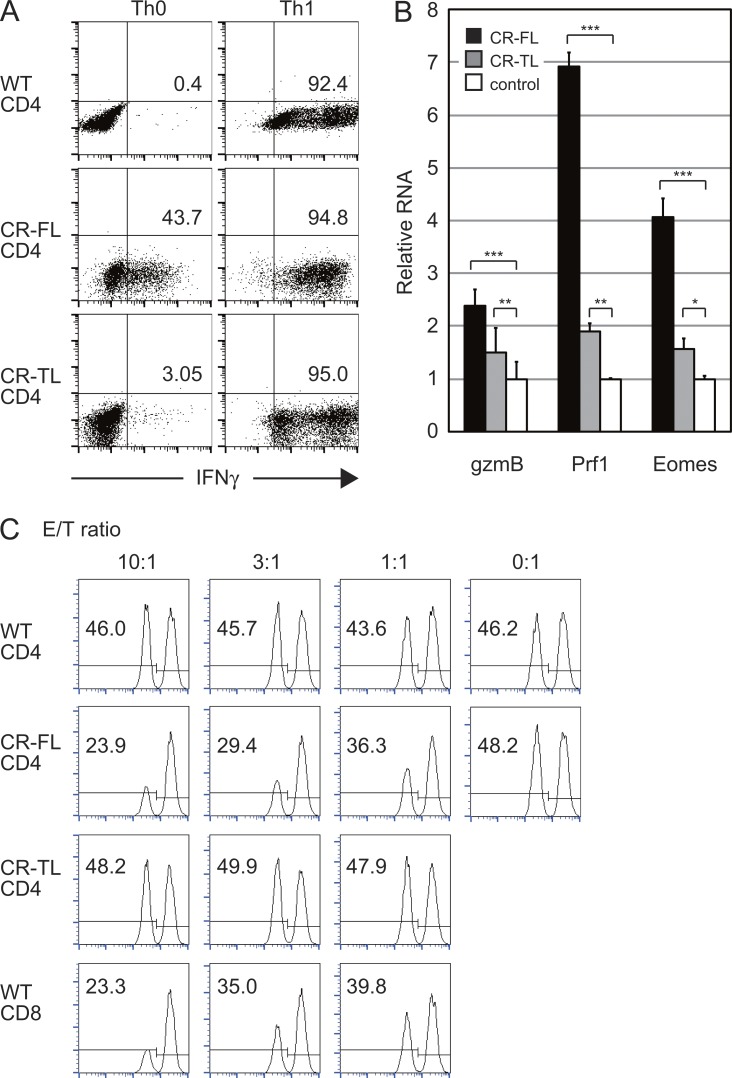
Next, we analyzed the ability of the Tg CD4+ T cells to differentiate into Th1, Th2, and Th17 cells (Fig. 6 A and not depicted). They could differentiate into all Th subsets under optimal conditions after 5–6 d of culture. We also noted that a high proportion of the Tg T cells differentiate into IFN-γ–producing cells under nonskewing conditions, similar to the situation in CRTAM+ WT T cells. These IFN-γ–producing T cells also express high levels of Eomes, gzmB, and perforin (Fig. 6 B), and they acquired cytotoxic function against target cells (Fig. 6 C). These CTL-related proteins were clearly induced after their stimulation-induced differentiation, whereas their expression was not enhanced in activated naive T cells (Fig. 5 G). These data indicate that CRTAM expression resulted in the induction of the expression of IFN-γ, CTL-related genes, and the acquisition of cytotoxic function.
Figure 6.
T cells from the CRTAM-Tg mice efficiently induce CD4+CTL. (A) Naive CD4+ T cells from CR-FL Tg, CR-TL Tg, and WT mice were stimulated and cultured for 6 d under Th1-skewing (Th1) or nonskewed (Th0) conditions, and cells were subjected to intracellular staining for IFN-γ. The numbers indicate the percentages of IFN-γ–producing cells among CD4+ T cells. (B) Quantitative real-time PCR analysis of T cells from CR-FL Tg (filled column), CR-TL (gray column) and WT mice (open column) under nonskewed condition as in A. (C) Retargeting cytotoxicity assay of CRTAM-Tg mice. Each effector cells were prepared from nonskewed condition cultures as in Fig. 3 C. Percentage of living target cells is indicated in each profile. The results shown are representative of at least two independent experiments. Error bars are SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, Student’s t test.
We then analyzed whether CRTAM-mediated signaling was involved in the CD4+CTL differentiation by analyzing KI-Tg mice expressing a truncated form of CRTAM lacking its cytoplasmic domain (tail-less mutant; CR-TL; Fig. 5 A). In this mouse, CD44hi effector–memory T cells increased, similar to the case in CR-FL Tg (Fig. 5 F, left). However, unlike CR-FL Tg, the production of effector cytokines such as IFN-γ and IL-17 was not enhanced at all (Fig. 5 F, right). Even though these T cells could differentiate into each Th subset under the appropriate differentiation conditions (Fig. 6 A and not depicted), unlike CR-FL T cells, these CR-TL T cells did not become IFN-γ–producing cells under nonskewed conditions and also did not develop cytotoxic functions (Fig. 6, B and C). Collectively, these results indicate that the cytoplasmic region of CRTAM is critical for inducing intracellular signaling for IFN-γ production and differentiation of CD4+CTLs, whereas the extracellular domain is involved in maturation of effector memory T cells.
CRTAM+ CD4+ T cells traffic to the inflammatory and mucosal sites
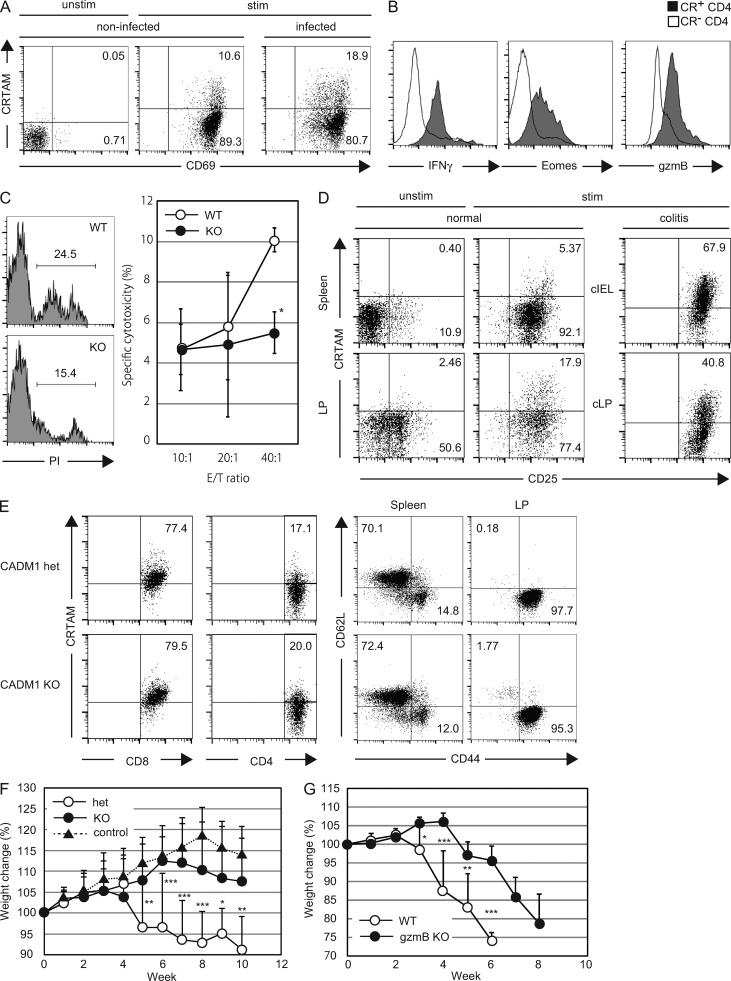
To analyze the function of CRTAM+ CD4+ T cells in vivo, we first examined the tissue distribution of CRTAM+ CD4+ T cells in various secondary lymphoid tissues and mucosal tissues. Whereas the CD4+ T cells isolated from spleen, peripheral LNs, and Peyer’s patch showed a similar frequency of CRTAM+ T cells upon stimulation, T cells from the lung and intestinal lamina propria (iLP) contain higher percentage of CRTAM+ CD4+ T cells compared with other tissues (Fig. 7, A and D), indicating that CRTAM+ CD4+ T cells have a tendency of traffic to mucosal tissues. We also examined the possibility that CRTAM+ T cells could be observed in the inflammatory sites upon infection. It was recently reported that CD4+ T cells that are activated by influenza virus infection could acquire CTL activity and contribute to protection against influenza virus infection (Brown et al., 2012). Thus, we analyzed the CRTAM expression level on CD4+ T cells that reside in the lung after influenza virus infection (Fig. 7 A). As expected, a higher percentage of CRTAM+ CD4+ T cells were detected in the virus-infected lung compared with noninfected control. CRTAM+ CD4+ T cells exhibited high expression of Eomes and gzmB, as well as IFN-γ production (Fig. 7 B). More importantly, these CD4+ T cells from the lung exhibited influenza-specific cytotoxicity, whereas CD4+ T cells from virus-infected CRTAM-KO mice showed very diminished killing activity (Fig. 7 C). These data indicate that after the influenza virus infection, high proportion of CRTAM+ T cells were detected in the infected inflammatory sites, and they develop into Ag-specific CD4+CTL.
Figure 7.
Accumulation of CRTAM+ CD4+ T cells in the inflammatory and mucosal tissues. (A) CRTAM+ CD4+ T cells in the lung of influenza virus–infected mice. CD4+ T cells were prepared from the lung of influenza virus–infected mice, and simulated with anti-CD3/CD28 Abs for 14 h. CRTAM expression was analyzed by flow cytometry. The numbers indicate the percentages of CRTAM+ and CRTAM− population among CD4+ CD69+ T cells. (B) Protein expression of CTL-related genes in CRTAM+ CD4+ T cells residing in the lung. (C) Influenza-specific cytotoxicity by lung CD4+ T cells from virus-infected mice. Lung CD4+ T cells from WT and CRTAM-KO mice were analyzed for influenza-specific cytotoxicity against NP-peptide pulsed LPS-activated B cells as the target. Representative FACS profiles of cytotoxic analysis are shown by PI-staining of dead cells at E:T ratio 40:1 (left), and specific cytotoxicity at various E:T ratios (right). The numbers indicate the percentages of PI+ dead cells. (D) CRTAM+ CD4+ T cells in the intestine. CD4+ T cells from the spleen and intestinal lamina propria (LP) were unstimulated (left) or simulated with anti-CD3/CD28 Abs (middle) for 14 h. Experimental colitis was induced by transferring naive CD4+ T cells into RAG-deficient mice. CD4+ T cells from colonic LP (cLP) and intraepithelial lymphocyte (cIEL) in colitis-induced mice (right). CRTAM expression was quantified by flow cytometry. The numbers indicate the percentages of CRTAM+ and CRTAM− population among CD4+CD25+ T cells. (E) CRTAM expression in CADM1-deficient mice. CRTAM expression was analyzed on T cells from iLP of CADM1+/− and CADM1−/− mice after stimulation (left). CRTAM–CADM1 interaction influences on effector memory differentiation (right). The numbers indicate the percentages of CRTAM+ cells (left) or CD62L+ cells (right). (F) Time course of body weight loss under colitis induction. Naive CD4+ T cells from CRTAM-deficient (KO), CRTAM-heterozygous (Het) mice (or no transfer control) were transferred into RAG-deficient mice. Body weight loss was measured every week. (G) Colitis induction in Granzyme B–deficient mice. Naive CD4+ T cells prepared from gzmB-KO or WT mice were transferred into RAG-deficient mice. The results shown are representative of at least two independent experiments. Statistical significance was determined by a two-tailed unpaired Student’s t test. Error bars are SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
In addition to the lung infection, we found that iLP contains a relatively high proportion of CRTAM+ CD4+ T cells. This is consistent with the idea that CRTAM+ T cells traffic into inflammatory sites. Because the CRTAM ligand CADM1 is widely expressed in the gut (not depicted), we next used CADM1-KO mice to address the question of whether increasing the percentage of CRTAM+ T cells is dependent on the CRTAM–CADM1 interaction. The number of CRTAM+ cells in iLP was comparable between CADM1-KO and WT mice (Fig. 7 E, left), indicating that the CRTAM–CADM1 itself is not involved in the induction of CRTAM expression in iLP. However, we found that the CRTAM–CADM1 interaction is involved in effector–memory differentiation (Fig. 7 E, right). In CADM1-KO mice, effector–memory T cells are slightly decreased in spleen. Furthermore, even though almost all iLP CD4+ T cells showed effector–memory phenotype in CADM-1 heterozygous mouse, naive cells were 10 times higher in CADM1-KO mouse (Fig. 7 E). These results are consistent with those of CRTAM Tg in Fig. 5, and support the idea that CRTAM–CADM1 interaction is involved in maturation of effector memory T cells, but not in the development of CD4+CTL.
CRTAM+ T cells contribute to induction of intestinal colitis
To clarify the in vivo function of CRTAM+ CD4+ T cells, we analyzed their role in the induction of colitis using a T cell–mediated colitis model. Purified naive CD4+CD45RBhiCD25− T cells were transferred into RAG-deficient mice to induce colitis. After the induction of colitis, infiltrating cells were isolated from inflamed colon lamina propria (cLP) and epithelia (cIEL), and the percentage of CRTAM+ CD4+ T cells was quantified (Fig. 7 D). Considering that CRTAM+ CD4+ T cells are present only at 1–4% in the spleen and LN, interestingly, >40% of CD4+ T cells in cLP and 67% of cIEL in the inflamed area expressed CRTAM, indicating that CRTAM+ CD4+ T cells are enriched in colonic inflammatory sites. To clarify the contribution of CRTAM expression, we compared colitis symptoms induced by CRTAM−/− T cells (Fig. 7 F). Analysis of colitis-induced body weight loss clearly showed that CRTAM−/− CD4+ T cells almost failed to induce colitis. These data indicate that CRTAM+ CD4+ T cells may be involved in the efficient induction of inflammation and also in the defense against pathogens in the gut.
Our data clearly demonstrated that CRTAM+ CD4+ T cells are able to produce high level of IFN-γ (Fig. 2 C), and CRTAM expression efficiently induce differentiation to IFN-γ and IL-17–secreting cells in vivo (Fig. 5 D). These cytokines are known to play key roles in the induction of colitis in this model (Powrie et al., 1994; Ito and Fathman, 1997; O’Connor et al., 2009; Sujino et al., 2011). In addition, our data suggested the possibility that CD4+CTL activity is also involved in inflammation in the gut. This was supported by the analysis of gzmB−/− T cells (Fig. 7 G). Whereas the naive T cells from gzmB-KO mice could induce colitis, the induction of body weight loss was much slower by gzmB−/− T cells although they eventually induce colitis. When compared with CRTAM−/− T cells, the induction of colitis by the gzmB−/− T cells appeared more severe than induction by CRTAM−/− T cells (Fig. 7, F and G), suggesting that CD4+CTL activity may also contribute to the induction of colitis in this model, together with inflammatory cytokines. These results strongly suggest that CRTAM+ CD4+ T cells play critical roles in inflammationthrough cytokine production and CTL function.
DISCUSSION
We show here that a small fraction of activated CD4+ T cells expressing CRTAM contain the immediate precursor of CD4+ cytotoxic T cells and that CRTAM expression induces the process of differentiation into CD4+CTL.
CD4+CTLs mediate their killing function by the directed exocytosis of cytotoxic granules toward target cells, such as CD8+CTL, to induce apoptosis. Degranulation of the perforin and Granzyme B–containing granules is required for the killing of target cells (Marshall and Swain, 2011), and the cytotoxic activity is further enhanced under nonskewed conditions in the presence of IL-2 (Brown et al., 2009). Together with these studies, our data suggest that the CRTAM+ CD4+ T cells with cytotoxic function are identical to the previously described CD4+CTL. Our findings clearly demonstrated that the CD4+CTL precursor already exists at the early stage of T cell activation and that precursor cells express CRTAM. CRTAM expression is also regulated by cellular interaction, and it is particularly efficiently induced by the interaction with DC. After T cell activation, CRTAM-mediated signaling induces the expression of CTL-related genes, and the CRTAM+ T cells differentiate into CD4+CTL in the presence of IL-2. We confirmed that this system is also functioning in human T cells; human T cells contain a small fraction of CRTAM+ T cells, which generate CD4+CTL similar to mouse T cells. This observation was also confirmed by CRTAM knock-in (KI) Tg mice. T cells from the full-length CRTAM-FL, but not from the tail-less mutant CRTAM-TL Tg mice, differentiated into CD4+CTL in vivo. Therefore, CD4+CTL development is dependent on CRTAM-induced signals, which are mediated through the intracellular domain. The intracellular domain of CRTAM contains a PDZ-binding motif at the C terminus; one family of PDZ-containing protein, the Discs Large (DLG), selects this sequence (Kornau et al., 1995; Songyang et al., 1997). It has been shown that Scrib, one member of this protein family, binds to CRTAM and regulates T cell polarity and cytokine production, and that knockdown of Scrib results in the reduction of IFN-γ production (Yeh et al., 2008). Together with the aforementioned findings, our results suggest that the differentiation of CD4+CTL is also regulated by CRTAM-Scrib–mediated signaling.
Although CRTAM is critical for the development of CD4+CTL, the requirement for the CRTAM ligand CADM1 is complex. Although CADM1 is highly expressed on epithelial cells and CD8+ dendritic cells (Shingai et al., 2003; Galibert et al., 2005), because there were no CADM1-expressing cells in our in vitro experiments, CD4+CTL can be differentiated in the absence of the interaction between CRTAM and CADM1. However, because of several reports suggesting weak expression of CADM1 by T cells (Yeh et al., 2008; Takeuchi et al., 2009; Kim et al., 2011), we examined the involvement of CADM1 on T cells for the induction of CD4+CTL. To this end, interestingly, CADM1−/− T cells express CRTAM normally and are able to differentiate into CD4+CTL similar to WT cells (not depicted). These data suggest that CADM1 on T cells, if any, does not have a significant effect, and that CRTAM may mediate signals to induce CD4+CTL development without ligand interaction, probably through the dimerization of CRTAM on the cell surface. In contrast to CD4+CTL differentiation, CRTAM signaling appears to be dispensable for the cytotoxic function of CD8+ T cells, because CRTAM−/− CD8+ T cells are normal in their ability to kill target cells (Takeuchi et al., 2009). The major signaling cascade for CD8+CTL development, including RUNX3 induction, may be sufficient to induce CD8+CTL without additional signals through CRTAM.
However, our results suggested that the CRTAM–CADM1 interaction is involved in the expansion of memory phenotype cells in vivo, because the tailless CRTAM-TL Tg mice increases memory-type cells to the level similar to WT mice even in the absence of the CRTAM-mediated signals. In this case, similar to CRTAM-mediated CD8+ T cell development, as previously shown (Takeuchi et al., 2009), it is speculated that the CRTAM–CADM1 interaction is important to enhance the retention and maturation of CD4+ T cells to effector memory cells within LNs.
We also demonstrated that CRTAM+ CD4+ T cells have in vivo function in the colitis induction model. CRTAM+ CD4+ T cells were clearly increased at the inflammation sites. This is consistent with a recent study indicating that CRTAM+ CD4+ T cells accumulate in the intestine (Cortez et al., 2014), and our data strongly suggests that CRTAM+ CD4+ T cells function at the inflamed site to induce colitis through both CTL activity and cytokine production. When CRTAM−/− CD4+ T cells were transferred, inflammation became milder than observed with WT cells, suggesting that CRTAM-mediated signals are important for differentiation into CD4+CTL and secretion of inflammatory cytokines. In our influenza virus infection model, CRTAM+ T cells accumulated in the infection sites, and developed into CD4+CTL mediating virus-specific cytotoxicity. These results support previous studies that CD4+CTL can function as a compensatory mechanism when CD8+CTL activity is impaired in the case as chronic viral infections (Stuller and Flaño, 2009; Zhou and McElhaney, 2011). Because CD8+ T cells are absent in the colitis model, CD4+CTL may predominantly function similar to the chronic infection case. Because CD4+CTL are restricted by MHC class II, class II expression is critical for CD4+CTL function. Whereas MHC class II is normally expressed only on APCs, such as DCs, macrophages, and B cells, the treatment with IFN-γ or radiation induces class II expression on epithelial or tumor cells (Quezada et al., 2010; Xie et al., 2010; Thibodeau et al., 2012; Thelemann et al., 2014). Because IFN-γ is an essential factor for the induction of inflammation in the colitis model, it is likely that secreted IFN-γ induces class II expression on intestinal epithelia, which could accelerate CD4+CTL activity.
The observation that CRTAM−/− CD4+ T cells failed to efficiently induce inflammation may reflect the likely multiple functions of CRTAM at several points of colitis induction, which may synergistically induce exacerbation of symptoms. First, CRTAM-mediated induction of CD4+CTL and their production of inflammatory cytokines would directly induce inflammation. Second, based on the finding that the number of T cells in the gut was clearly decreased during the colitis when CRTAM−/− T cells were transferred, CRTAM likely enhances the recruitment of T cells in the gut (Cortez et al., 2014). Third, based on our previous observation that CRTAM−/− CD8+ T cells cannot proliferate well within the draining LN, CRTAM appears to play a role in retention and functional maturation of CD4+ T cells in LNs, similar to CD8+ T cells (Takeuchi et al., 2009).
Recently, two papers reported a unique population of T cells that express CD4+CD8α+ and reside in the gut (Mucida et al., 2013; Reis et al., 2013). This population has CTL function and can be generated from CD4+CD8− peripheral T cells by treatment with TGFβ and retinoic acid (RA), which induce up-regulation of RUNX3 and down-regulation of ThPOK expression. We also confirmed the presence of CD4+CD8α+ T cells in the colitis induction model. Interestingly, all CD4+CD8α+ T cells express CRTAM after stimulation. However, >80% of CRTAM-expressing cells in the gut lamina propria were CD4+CD8α− T cells (not depicted), indicating that some of the CRTAM-expressing cells are CD4+CD8α+ T cells. Furthermore, in the case of splenic CRTAM+ CD4+ T cells, CD8α expression was not observed on the cell surface and the expression of ThPOK and RUNX3 were almost the same as in CRTAM− CD4+ T cells (Fig. 2, C and D). Nevertheless, because CTL function was clearly observed after cultivation (Fig. 3 E), these data indicate that CD4+CTL are not equivalent to the CD8α-expressing T cells.
Because the perforin expression was induced only after incubation, CRTAM+ CD4+ T cells do not have CTL function initially but differentiate into CTL after incubation. These data strongly suggest that peripheral CRTAM+ CD4+CD8α− T cells are the precursor of CD4+CD8α+ T cells in the gut. After TCR-mediated activation, these cells would gain killing function, migrate to the gut, and further differentiate into CD4+CD8α+ T cells in the gut. A recent study demonstrated that intestinal CD4+CD8+ T cells are severely reduced in both CRTAM−/− and CADM1−/− mice (Cortez et al., 2014), suggesting that the maturation from CD4+CD8− CTL in LNs into CD4+CD8+ CTL, as well as their maintenance in the gut, is induced through the CRTAM–CADM1 interaction. This speculation suggests that the CTL have already determined the fate to differentiate into CD4+CD8α+ cells before down-regulation of ThPOK. Therefore, CRTAM expression defines the lineage of CD4+CTL after stimulation. Consistently, the expression of CTL-related genes is induced in the CRTAM+ CD4+ T cells, and the cells acquire characteristics similar to CD8+CTL and the CTL activity. Thus, CRTAM expression is critical for differentiation of the CD4+ T cells into the CTL linage, and CRTAM is thus a useful and functional marker to define CTL-inducible cells. These characteristics might be able to control CD4+CTL functions and should be applicable for therapeutic aims. CD4+CTLs enriched in infectious/inflammatory sites may function for protective immunity, especially in chronic virus infection or antitumor responses, and the CD4+CTLs can now be generated and expanded using CRTAM as a defined marker. Alternatively, blockade of CRTAM may become a target for the treatment of inflammatory diseases.
MATERIALS AND METHODS
Mouse
C57BL/6 mice were purchased from CLEA Japan. CRTAM- and CADM1-deficient mice have been previously described (Takeuchi et al., 2009). Eomes-deficient mice were provided by S. Reiner (University of Pennsylvania, Philadelphia, PA) courtesy of T. Nakayama (Chiba University, Chiba, Japan). CRTAM-Tg targeting vectors were constructed using the following method. Two loxP sites were inserted up- and downstream of a Neomycin-resistant gene and STOP codon cassette, and they were ligated to upstream of a CRTAM-IRES-eGFP cording sequence. This construct, which is under the transcriptional control of the CAG promoter, was inserted into the Rosa26 locus target sequence. Targeting vectors were introduced in Bruce4 ES cells, and homologous recombinant ES cells were injected into blastocysts of BALB/c mice. Chimeric mice were crossed with C57BL/6 J mice to obtain mice with germ line transmission. T cell–specific CRTAM-Tg mice were obtained by crossing with Lck-Cre Tg mice. All animal experiments were performed in compliance with the institutional guidelines of the animal facility of Institute of Physical and Chemical Research Yokohama Institute (Yokohama, Japan).
Cells and reagents
The mouse B cell line A20.2J (A20) and human T cell line Jurkat E6.1 (Jurkat) were cultured in RPMI-1640 and 10% FCS. The eomes expression vector was provided by K. Eshima (Kitasato University, Tokyo, Japan; Eshima et al., 2012). The following fluorochrome-labeled Abs (purchased from BD, BioLegend, or eBioscience) were used: Abs against CD4 (GK1.5), CD8 (Ly2), CD62L (MEL-14), CD44 (IM7), CD45RB (C363.16A), CD25 (PC61), CD69 (H1.2F3) and TCRβ (H57-597), B220 (RA3-6B2), IL4 (11B11), IFN-γ (XMG1.2), IL-17a (TC11-18H10), and Foxp3 (FJK-16S).
Quantitative PCR
Total RNA was prepared from sorted cells by RNeasy Mini kit (QIAGEN) and treated with DNase (Nippongene). cDNA was synthesized using SuperScript II reverse transcription (Invitrogen). qPCR was performed with the Fast Syber Green Master Mix (Applied Biosystems). Data were collected and calculated by using the StepOnePlus real-time PCR system (Applied Biosystems).
Helper T cell differentiation
CD4+CD62LhiCD44loCD25− (naive) T cells were isolated from spleens using a FACSAria cell sorter (BD). For Th0 cells, cells were stimulated with plate-bound anti-CD3ε (2C11; 10 µg/ml) and anti-CD28 (PV-1; 1 µg/ml) Abs in the presence of the indicated ligands. For Th1 cells, cells were cultured in the presence of IL-12 (10 ng/ml) and anti–IL-4 Abs (10 ng/ml). For Th2 cells, cells were similarly cultured in the presence of IL-4 (10 ng/ml) and anti–IFN-γ (10 ng/ml). For Th17, IL-6 (20 ng/ml), TGFβ (10 ng/ml), anti–IL-4 Abs (10 ng/ml), and anti–IFN-γ Abs (10 ng/ml).
Intracellular cytokine staining
CD4+ T cells were restimulated with immobilized anti-CD3ε and anti-CD28 for 6 h in the presence of 2 µM monensin (Sigma-Aldrich). Cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. After blocking with 3% BSA-PBS, cells were stained with antibodies to each cytokine. Flow cytometric analysis was performed on a FACSCalibur (BD) and data were analyzed with Cell Quest (BD).
Isolation and analysis of human T cells
PBMCs were isolated from healthy donors by centrifugation over Ficoll density gradient. CD4+ T cells were isolated by anti–human CD4 MACS beads (Miltenyi Biotec) and were stimulated by 10 µg/ml of anti–human CD3 antibody (OKT3) for 14 h. After stimulation, T cells were stained with anti–human CRTAM mAb (Cr24.1; BioLegend), and CRTAM+ and CRTAM− cells were sorted by FACSAria and incubated with 2,000 U/ml of human IL-2 (Ajinomoto) for 5 d. These experiments were performed in compliance with the institutional guidelines of the Tokyo University of Science (Tokyo, Japan), and all subjects provided informed consent as approved by the ethical committee. Healthy volunteers were recruited after obtaining informed consent.
Influenza virus infection
Influenza A virus (H1N1) A/PR8 was obtained from ATCC. Infection was performed by intranasal injection of virus suspension in PBS with the sublethal dose, which was defined as causing 20% weight loss (200–400 pfu). CD4+ T cells were purified from the lung at 6 d after infection by using gentleMACS (Miltenyi Biotec). For influenza virus–specific killing assay, NP peptide (NP 264–279; LILRGSVAHKSCLPAC; Gao et al., 1989) was used to pulse to LPS-activated B cells from C57BL/6 mice as the target cells.
Induction of colitis
CD4+ T cells were enriched from spleen and LNs of WT or CRTAM-deficient mice by using magnetic beads (Bio-Mag; QIAGEN), and CD4+CD25−CD45RBhi naive CD4+ T cells were sorted by flow cytometry. 5 × 105 cells were injected i.v. into Rag1-deficient mice, and body weight loss was monitored weekly as a clinical sign of colitis. Mice were euthanized when they had lost 20% of their initial weight.
In vitro cytotoxicity assay
For retargeting cytotoxic assay, naive CD4+ T cells were stimulated by plate-coated anti-CD3ε and anti-CD28 for 14 h, and then sorted into CRTAM+ and CRTAM− cells. Sorted T cells were further incubated for 5 d in the presence of IL-2 and were differentiated into effector cells. CFSE-labeled A20 cells (target cells: low intensity) and Jurkat cells (internal control: high intensity) were mixed at a one-to-one ratio and coincubated with 105 target cells for 4 h in the presence of anti-CD3ε antibody (10 µg/ml). After the incubation, living target cells were quantified by flow cytometry. For influenza-specific cytotoxic assay, CD4+ T cells were isolated from the lungs of mice that were infected with influenza virus using autoMACS. Wild-type and CRTAM−/− T cells were labeled with different concentrations of CMTPX, and graded numbers of T cells were mixed with the target B cells which had been activated by LPS for 12 h and pulsed with Influenza virus NP peptide 264–279 (LILRGSVAHKSCLPAC) for 6 h (Gao et al., 1989). The mixture was centrifuged and incubated for 6 h, and the cytotoxicity was analyzed by flow cytometry using FACSCanto (BD) after staining with PI.
Gene expression profiling
Naive CD4+ and CD8+ T cells (CD25−CD62LhiCD44lo) were purified from spleen and LNs by flow cytometry. Cells were stimulated by plate-coated anti-CD3ε (10 µg/ml) and anti-CD28 (1 µg/ml) antibody for 14 h. The activated cells were stained by anti-CRTAM antibody and resorted into CRTAM+ and CRTAM− cells. RNA was isolated, labeled, and hybridized to a Mouse Genome 430 2.0 array (Affymetrix). Expression values for each probe set were calculated using the GC-RMA method in the GeneSpring GX 7.3 software package (Agilent Technologies).
The microarray data are available in the Institute of Physical and Chemical Research database (http://refdic.rcai.riken.jp/welcome.cgi). Sample numbers are RSM14569, RSM14571, and RSM14572.
Acknowledgments
We thank Ms. M. Yoshioka and H. Yamaguchi for secretarial assistance.
This work was supported by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan (JSPS KAKENHI, grant numbers 23790551 for A. Takeuchi and 24229004 for T. Saito).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- Ag
- antigen
- CADM1
- cell adhesion molecule 1
- CRTAM
- MHC class I–restricted T cell–associated molecule
- CTL
- cytotoxic T cells
- eomes
- eomesodermin
- gzmB
- granzyme B
- HCMV
- human cytomegalovirus
- IEL
- intraepithelial lymphocyte
- LCMV
- lymphocytic choriomeningitis virus
- LP
- lamina propria
- PBMC
- peripheral blood mononuclear cell
- prf
- perforin
References
- Aliahmad P., Kadavallore A., de la Torre B., Kappes D., and Kaye J.. 2011. TOX is required for development of the CD4 T cell lineage gene program. J. Immunol. 187:5931–5940. 10.4049/jimmunol.1101474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appay V., Zaunders J.J., Papagno L., Sutton J., Jaramillo A., Waters A., Easterbrook P., Grey P., Smith D., McMichael A.J., et al. 2002. Characterization of CD4+ CTLs ex vivo. J. Immunol. 168:5954–5958. 10.4049/jimmunol.168.11.5954 [DOI] [PubMed] [Google Scholar]
- Arase N., Takeuchi A., Unno M., Hirano S., Yokosuka T., Arase H., and Saito T.. 2005. Heterotypic interaction of CRTAM with Necl2 induces cell adhesion on activated NK cells and CD8+ T cells. Int. Immunol. 17:1227–1237. 10.1093/intimm/dxh299 [DOI] [PubMed] [Google Scholar]
- Aslan N., Yurdaydin C., Wiegand J., Greten T., Ciner A., Meyer M.F., Heiken H., Kuhlmann B., Kaiser T., Bozkaya H., et al. 2006. Cytotoxic CD4 T cells in viral hepatitis. J. Viral Hepat. 13:505–514. 10.1111/j.1365-2893.2006.00723.x [DOI] [PubMed] [Google Scholar]
- Boles K.S., Barchet W., Diacovo T., Cella M., and Colonna M.. 2005. The tumor suppressor TSLC1/NECL-2 triggers NK-cell and CD8+ T-cell responses through the cell-surface receptor CRTAM. Blood. 106:779–786. 10.1182/blood-2005-02-0817 [DOI] [PubMed] [Google Scholar]
- Brown D.M., Kamperschroer C., Dilzer A.M., Roberts D.M., and Swain S.L.. 2009. IL-2 and antigen dose differentially regulate perforin- and FasL-mediated cytolytic activity in antigen specific CD4+ T cells. Cell. Immunol. 257:69–79. 10.1016/j.cellimm.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D.M., Lee S., Garcia-Hernandez M.L., and Swain S.L.. 2012. Multifunctional CD4 cells expressing γ interferon and perforin mediate protection against lethal influenza virus infection. J. Virol. 86:6792–6803. 10.1128/JVI.07172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez V.S., Cervantes-Barragan L., Song C., Gilfillan S., McDonald K.G., Tussiwand R., Edelson B.T., Murakami Y., Murphy K.M., Newberry R.D., et al. 2014. CRTAM controls residency of gut CD4+CD8+ T cells in the steady state and maintenance of gut CD4+ Th17 during parasitic infection. J. Exp. Med. 211:623–633. 10.1084/jem.20130904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshima K., Chiba S., Suzuki H., Kokubo K., Kobayashi H., Iizuka M., Iwabuchi K., and Shinohara N.. 2012. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4+ Th cells cytotoxic by activating both perforin- and FasL-pathways. Immunol. Lett. 144:7–15. 10.1016/j.imlet.2012.02.013 [DOI] [PubMed] [Google Scholar]
- Feighery C., and Stastny P.. 1979. HLA-D region-associated determinants serve as targets for human cell-mediated lysis. J. Exp. Med. 149:485–494. 10.1084/jem.149.2.485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galibert L., Diemer G.S., Liu Z., Johnson R.S., Smith J.L., Walzer T., Comeau M.R., Rauch C.T., Wolfson M.F., Sorensen R.A., et al. 2005. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 280:21955–21964. 10.1074/jbc.M502095200 [DOI] [PubMed] [Google Scholar]
- Gao X.M., Liew F.Y., and Tite J.P.. 1989. Identification and characterization of T helper epitopes in the nucleoprotein of influenza A virus. J. Immunol. 143:3007–3014. [PubMed] [Google Scholar]
- Glimcher L.H., Townsend M.J., Sullivan B.M., and Lord G.M.. 2004. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 4:900–911. 10.1038/nri1490 [DOI] [PubMed] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., and Kappes D.J.. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833. 10.1038/nature03338 [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G., Anderson M.K., Wang C., Rothenberg E.V., and Alberola-Ila J.. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94. 10.1016/S1074-7613(03)00176-6 [DOI] [PubMed] [Google Scholar]
- Ito H., and Fathman C.G.. 1997. CD45RBhigh CD4+ T cells from IFN-γ knockout mice do not induce wasting disease. J. Autoimmun. 10:455–459. 10.1016/S0896-8411(97)90152-9 [DOI] [PubMed] [Google Scholar]
- Jellison E.R., Kim S.K., and Welsh R.M.. 2005. Cutting edge: MHC class II-restricted killing in vivo during viral infection. J. Immunol. 174:614–618. 10.4049/jimmunol.174.2.614 [DOI] [PubMed] [Google Scholar]
- Kennedy J., Vicari A.P., Saylor V., Zurawski S.M., Copeland N.G., Gilbert D.J., Jenkins N.A., and Zlotnik A.. 2000. A molecular analysis of NKT cells: identification of a class-I restricted T cell-associated molecule (CRTAM). J. Leukoc. Biol. 67:725–734. [DOI] [PubMed] [Google Scholar]
- Kim H.R., Jeon B.H., Lee H.S., Im S.H., Araki M., Araki K., Yamamura K., Choi S.C., Park D.S., and Jun C.D.. 2011. IGSF4 is a novel TCR ζ-chain-interacting protein that enhances TCR-mediated signaling. J. Exp. Med. 208:2545–2560. 10.1084/jem.20110853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornau H.C., Schenker L.T., Kennedy M.B., and Seeburg P.H.. 1995. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 269:1737–1740. 10.1126/science.7569905 [DOI] [PubMed] [Google Scholar]
- Kuramochi M., Fukuhara H., Nobukuni T., Kanbe T., Maruyama T., Ghosh H.P., Pletcher M., Isomura M., Onizuka M., Kitamura T., et al. 2001. TSLC1 is a tumor-suppressor gene in human non-small-cell lung cancer. Nat. Genet. 27:427–430. 10.1038/86934 [DOI] [PubMed] [Google Scholar]
- Lukacher A.E., Morrison L.A., Braciale V.L., Malissen B., and Braciale T.J.. 1985. Expression of specific cytolytic activity by H-2I region-restricted, influenza virus-specific T lymphocyte clones. J. Exp. Med. 162:171–187. 10.1084/jem.162.1.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimone M.M., Morrison L.A., Braciale V.L., and Braciale T.J.. 1986. Features of target cell lysis by class I and class II MHC-restricted cytolytic T lymphocytes. J. Immunol. 137:3639–3643. [PubMed] [Google Scholar]
- Marshall N.B., and Swain S.L.. 2011. Cytotoxic CD4 T cells in antiviral immunity. J. Biomed. Biotechnol. 2011:954602 10.1155/2011/954602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucida D., Husain M.M., Muroi S., van Wijk F., Shinnakasu R., Naoe Y., Reis B.S., Huang Y., Lambolez F., Docherty M., et al. 2013. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat. Immunol. 14:281–289. 10.1038/ni.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor W. Jr., Kamanaka M., Booth C.J., Town T., Nakae S., Iwakura Y., Kolls J.K., and Flavell R.A.. 2009. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10:603–609. 10.1038/ni.1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J.J., and Paul W.E.. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 327:1098–1102. 10.1126/science.1178334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai S.Y., Truitt M.L., Ting C.N., Leiden J.M., Glimcher L.H., and Ho I.C.. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875. 10.1016/S1074-7613(03)00328-5 [DOI] [PubMed] [Google Scholar]
- Pearce E.L., Mullen A.C., Martins G.A., Krawczyk C.M., Hutchins A.S., Zediak V.P., Banica M., DiCioccio C.B., Gross D.A., Mao C.A., et al. 2003. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 302:1041–1043. 10.1126/science.1090148 [DOI] [PubMed] [Google Scholar]
- Powrie F., Leach M.W., Mauze S., Menon S., Caddle L.B., and Coffman R.L.. 1994. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1:553–562. 10.1016/1074-7613(94)90045-0 [DOI] [PubMed] [Google Scholar]
- Quezada S.A., Simpson T.R., Peggs K.S., Merghoub T., Vider J., Fan X., Blasberg R., Yagita H., Muranski P., Antony P.A., et al. 2010. Tumor-reactive CD4+ T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 207:637–650. 10.1084/jem.20091918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis B.S., Rogoz A., Costa-Pinto F.A., Taniuchi I., and Mucida D.. 2013. Mutual expression of the transcription factors Runx3 and ThPOK regulates intestinal CD4+ T cell immunity. Nat. Immunol. 14:271–280. 10.1038/ni.2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai T., Ikeda W., Kakunaga S., Morimoto K., Takekuni K., Itoh S., Satoh K., Takeuchi M., Imai T., Monden M., and Takai Y.. 2003. Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J. Biol. Chem. 278:35421–35427. 10.1074/jbc.M305387200 [DOI] [PubMed] [Google Scholar]
- Songyang Z., Fanning A.S., Fu C., Xu J., Marfatia S.M., Chishti A.H., Crompton A., Chan A.C., Anderson J.M., and Cantley L.C.. 1997. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 275:73–77. 10.1126/science.275.5296.73 [DOI] [PubMed] [Google Scholar]
- Stuller K.A., and Flaño E.. 2009. CD4 T cells mediate killing during persistent gammaherpesvirus 68 infection. J. Virol. 83:4700–4703. 10.1128/JVI.02240-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino T., Kanai T., Ono Y., Mikami Y., Hayashi A., Doi T., Matsuoka K., Hisamatsu T., Takaishi H., Ogata H., et al. 2011. Regulatory T cells suppress development of colitis, blocking differentiation of T-helper 17 into alternative T-helper 1 cells. Gastroenterology. 141:1014–1023. 10.1053/j.gastro.2011.05.052 [DOI] [PubMed] [Google Scholar]
- Sun G., Liu X., Mercado P., Jenkinson S.R., Kypriotou M., Feigenbaum L., Galéra P., and Bosselut R.. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373–381. 10.1038/ni1183 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Kim S.T., Costa G.L., Zhang X., Fathman C.G., and Glimcher L.H.. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 100:655–669. 10.1016/S0092-8674(00)80702-3 [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Sullivan B.M., Peng S.L., and Glimcher L.H.. 2003. Molecular mechanisms regulating Th1 immune responses. Annu. Rev. Immunol. 21:713–758. 10.1146/annurev.immunol.21.120601.140942 [DOI] [PubMed] [Google Scholar]
- Takeuchi A., Itoh Y., Takumi A., Ishihara C., Arase N., Yokosuka T., Koseki H., Yamasaki S., Takai Y., Miyoshi J., et al. 2009. CRTAM confers late-stage activation of CD8+ T cells to regulate retention within lymph node. J. Immunol. 183:4220–4228. 10.4049/jimmunol.0901248 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., and Littman D.R.. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633. 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Thelemann C., Eren R.O., Coutaz M., Brasseit J., Bouzourene H., Rosa M., Duval A., Lavanchy C., Mack V., Mueller C., et al. 2014. Interferon-γ induces expression of MHC class II on intestinal epithelial cells and protects mice from colitis. PLoS One. 9:e86844 10.1371/journal.pone.0086844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau J., Bourgeois-Daigneault M.C., and Lapointe R.. 2012. Targeting the MHC Class II antigen presentation pathway in cancer immunotherapy. OncoImmunology. 1:908–916. 10.4161/onci.21205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen E.M., Remmerswaal E.B., Vossen M.T., Rowshani A.T., Wertheim-van Dillen P.M., van Lier R.A., and ten Berge I.J.. 2004. Emergence of a CD4+CD28− granzyme B+, cytomegalovirus-specific T cell subset after recovery of primary cytomegalovirus infection. J. Immunol. 173:1834–1841. 10.4049/jimmunol.173.3.1834 [DOI] [PubMed] [Google Scholar]
- Wagner H., Starzinski-Powitz A., Jung H., and Röllinghoff M.. 1977. Induction of I region-restricted hapten-specific cytotoxic T lymphocytes. J. Immunol. 119:1365–1368. [PubMed] [Google Scholar]
- Wang L., Wildt K.F., Castro E., Xiong Y., Feigenbaum L., Tessarollo L., and Bosselut R.. 2008. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 29:876–887. 10.1016/j.immuni.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736. 10.1073/pnas.1232420100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Akpinarli A., Maris C., Hipkiss E.L., Lane M., Kwon E.K., Muranski P., Restifo N.P., and Antony P.A.. 2010. Naive tumor-specific CD4+ T cells differentiated in vivo eradicate established melanoma. J. Exp. Med. 207:651–667. 10.1084/jem.20091921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J.H., Sidhu S.S., and Chan A.C.. 2008. Regulation of a late phase of T cell polarity and effector functions by Crtam. Cell. 132:846–859. 10.1016/j.cell.2008.01.013 [DOI] [PubMed] [Google Scholar]
- Zaunders J.J., Dyer W.B., Wang B., Munier M.L., Miranda-Saksena M., Newton R., Moore J., Mackay C.R., Cooper D.A., Saksena N.K., and Kelleher A.D.. 2004. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 103:2238–2247. 10.1182/blood-2003-08-2765 [DOI] [PubMed] [Google Scholar]
- Zhou X., and McElhaney J.E.. 2011. Age-related changes in memory and effector T cells responding to influenza A/H3N2 and pandemic A/H1N1 strains in humans. Vaccine. 29:2169–2177. 10.1016/j.vaccine.2010.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]