Hatlen et al. provide an integrative analysis of the mutational landscape of mouse and human AML and identify functionally relevant cooperation between AML1-ETO and PTPN11 D61Y. Based on these findings, they generate a novel mouse model of t(8;21)+ AML.
Abstract
t(8;21) is one of the most frequent chromosomal abnormalities observed in acute myeloid leukemia (AML). However, expression of AML1-ETO is not sufficient to induce transformation in vivo. Consistent with this observation, patients with this translocation harbor additional genetic abnormalities, suggesting a requirement for cooperating mutations. To better define the genetic landscape in AML and distinguish driver from passenger mutations, we compared the mutational profiles of AML1-ETO–driven mouse models of leukemia with the mutational profiles of human AML patients. We identified TET2 and PTPN11 mutations in both mouse and human AML and then demonstrated the ability of Tet2 loss and PTPN11 D61Y to initiate leukemogenesis in concert with expression of AML1-ETO in vivo. This integrative genetic profiling approach allowed us to accurately predict cooperating events in t(8;21)+ AML in a robust and unbiased manner, while also revealing functional convergence in mouse and human AML.
4–12% of acute myeloid leukemia (AML) patients present with a translocation between chromosomes 8 and 21 (Müller et al., 2008). However, transgenic mice expressing AML1-ETO only develop AML after treatment with mutagenic agents, suggesting a requirement for cooperating events (Higuchi et al., 2002). Two mouse models of AML1-ETO–driven AML have been generated that may depend on the acquisition of such events for leukemogenesis. Expression of AML1-ETO in Cdkn1a-null cells or expression of AML1-ETO9a, a splice variant of AML1-ETO, in WT cells can induce fully penetrant AML after a prolonged latency (Yan et al., 2006; Peterson et al., 2007b). CDKN1A has never been found to be disrupted in t(8;21)+ AML (Shiohara et al., 1997), suggesting that its loss does not functionally cooperate with AML1-ETO. Instead, it is thought that loss of CDKN1A in vivo prevents the repair of damaged DNA and allows for the accumulation of mutations and cooperating alleles in AML1-ETO+ hematopoietic progenitors (McDonald et al., 1996). Studies of AML1-ETO9a–driven leukemia have found dysregulated expression of DNA repair genes (Alcalay et al., 2003), which might facilitate a similar acquisition of cooperating secondary events. We hypothesized that we could identify and credential disease alleles in t(8;21)+ AML through integrative genomic studies of human AML and the mouse AMLs that arise from these models.
RESULTS AND DISCUSSION
Identification of phenotypically similar mouse models of t(8;21)+ AML
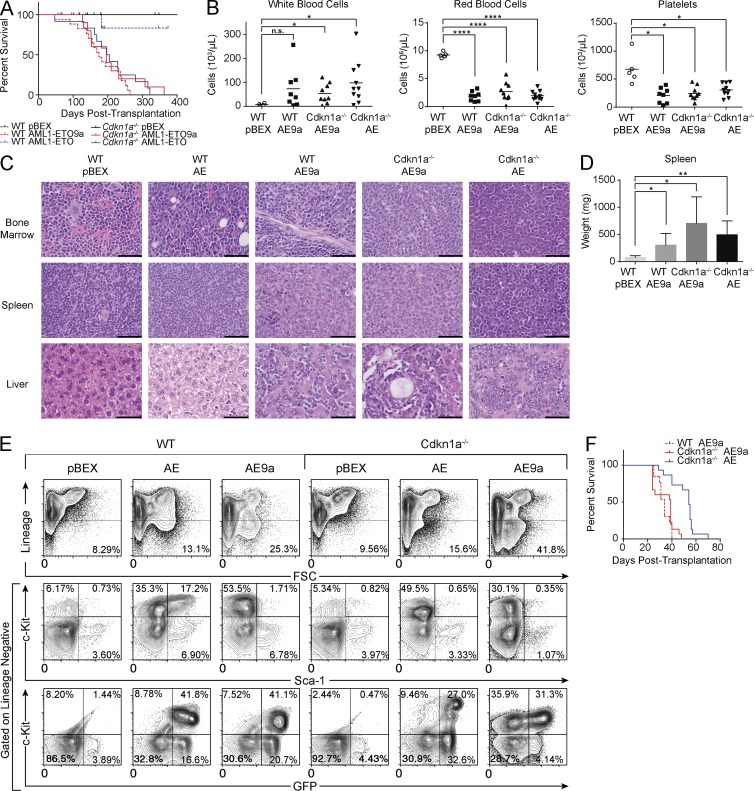
We first expressed AML1-ETO or AML1-ETO9a in fetal liver cells isolated from WT or Cdkn1a−/− mice. All transplant recipients of AML1-ETO–expressing Cdkn1a−/− cells developed lethal AML with a median latency of 201 d. Expression of AML1-ETO9a in WT or Cdkn1a−/− cells resulted in a fully penetrant, lethal leukemia after a median latency of 175 and 196 d, respectively (Fig. 1 A). At euthanasia, leukemic mice had anemia, thrombocytopenia, and elevated white blood cell counts (Fig. 1 B). We observed blast cell accumulation in the bone marrow, liver, and spleen, resulting in splenomegaly (Fig. 1, C and D). The GFP+ blast cells display an immature immunophenotype; they are negative for lineage markers and SCA-1 but are c-KIT and MPO positive (Fig. 1 E and not depicted). Leukemia with an identical immunophenotype develops within secondary recipients (Fig. 1 F and not depicted). The observations that these mouse models are phenotypically similar and develop AML with similar penetrance and latency suggest that they have acquired similar secondary events.
Figure 1.
Mouse models of AML1-ETO–driven leukemogenesis are phenotypically similar. (A) Kaplan-Meier plot of mouse survival after transplantation with cells transduced with the given vector. WT pBEX (n = 12); WT AE9a (AML1-ETO9a; n = 17); WT AE (AML1-ETO; n = 17); Cdkn1a−/− pBEX (n = 11); Cdkn1a−/− AE9a (n = 10); Cdkn1a−/− AE (n = 12). (B) Peripheral blood counts of leukemic mice (from left to right, n = 8, n = 9, and n = 10) and controls (n = 5). Horizontal bars represent mean. (C) Representative hematoxylin and eosin staining (n = 3). Bars, 50 µm. (D) At euthanasia, leukemic mice exhibit significant splenomegaly compared with controls. WT pBEX (n = 2); WT AE9a (n = 8); Cdkn1a−/− AE9a (n = 5); Cdkn1a−/− AE (n = 7). Error bars represent the standard deviation from the mean. (B and D) P-values were generated using the Student's t test: *, P < 0.05; **, P < 0.01; ****, P < 0.0001; n.s., not significant. (E) Representative flow cytometry analysis of the bone marrow of transplant recipients (n = 10). (F) Kaplan-Meier plot of mouse survival after transplantation with 2 × 106 leukemic spleen cells. WT AE9a (n = 15); Cdkn1a−/− AE9a (n = 13); Cdkn1a−/− AE (n = 15).
Mouse models of AML reproduce the genomic landscape of human AML
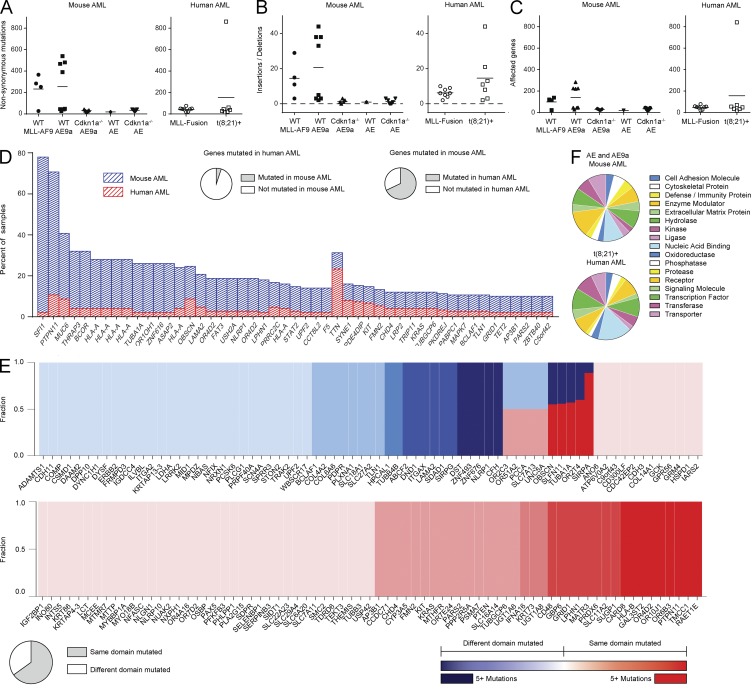
We next investigated the mutational landscape of the mouse AMLs and compared it with somatic mutational analyses of other mouse AML models and AML patients (Stubbs et al., 2008; Cancer Genome Atlas Research Network, 2013). We did not observe a significant difference in the number of variants detected or genes targeted in the mouse models compared with AML patient samples (Fig. 2, A–C). When we restricted our analysis to genes for which human–mouse orthology is known, we observed a significant enrichment in mouse AML for specific genes (hypergeometric P ≤ 4.26 × 10−20; Fig. 2 D) and protein domains (hypergeometric P ≤ 4.23 × 10−3) mutated in human AML. The domains mutated in both human and mouse AML were similarly affected by recurrent mutations (Spearman correlation of domain p-values r = 0.53, P ≤ 2.73 × 10−8; Fig. 2 E). We next investigated the frequency of mutations in different protein classes and found no differences in the protein classes targeted in AML1-ETO/AML1-ETO9a mouse AML versus human t(8;21)+ AML (P = 0.327; Fig. 2 F).
Figure 2.
Mouse AML reflects the mutational landscape of human AML. (A–C) The number of nonsynonymous mutations (A), insertion and deletion events (B), and the number of genes harboring nonsynonymous, insertion, or deletion events in leukemic mice and human AML patients (C). Horizontal bars represent mean. (D) A stacked plot of the genes that are mutated in ≥2% of human AML samples and ≥8% of mouse AML samples. 67.90% of genes mutated in leukemic mice are also mutated in human AML patients, whereas only 5.07% of genes with a protein-coding mutation in AML patients are also mutated in leukemic mice. This enrichment is significant (hypergeometric P ≤ 4.26 × 10−20). (E) Of the genes that harbor protein-coding mutations in both leukemic mice and human AML patients, the fraction of protein domains that are mutated in both species (red) or in only one species (blue) is depicted. The color intensity increases with the number of mutations in a given protein domain. 65.07% of these protein domains are mutated in both species, demonstrating enrichment (hypergeometric P ≤ 4.23 × 10−3). Domains similarly affected are recurrently mutated (Spearman correlation of domain p-values r = 0.53, binomial P ≤ 2.73 × 10−8). (F) The frequencies with which various protein classes are mutated in t(8;21)+ mouse and human AML are not significantly different (P = 0.327, a transformed value of the Pillai-Bartlett statistic from a MANOVA model).
Integrated genomic approach identifies potential cooperating events in t(8;21)+ AML
We next sought to mine the mutational data to identify mutations capable of cooperating with AML1-ETO, focusing our efforts on the 424 genes mutated in both human and mouse AML and then further narrowing our focus to those genes that were also significantly mutated (defined as Genome MuSiC significantly mutated gene [SMG] false discovery rate [FDR] ≤ 30%). This reduced the number of potential candidates from 424 to 38 human genes, representing 45 mouse orthologues (Fig. 3 A). We focused on the mouse orthologues that were significantly mutated (FDR ≤ 10%) in AML1-ETO–driven leukemias, and not in MLL-AF9–driven leukemia, and identified 38 mouse genes that met these criteria (Fig. 3 B). These 38 genes correspond to 32 human orthologues, which include three alleles annotated in COSMIC as cancer genes (Futreal et al., 2004): TET2, PTPN11, and THRAP3.
Figure 3.
SMGs in mouse AML identify SMGs in human AML that are potential AML1-ETO cooperating partners. (A) The patients that harbor nonsynonymous mutations in genes that are significantly mutated in human AML (FDR ≤ 30%). (B) For the genes significantly mutated in human AML, the types of mutations present in mouse AML samples are depicted. Those genes that are significantly mutated in a given mouse AML group (FDR ≤ 10%) are highlighted in blue under the given group. (C) TET2 mutations identified in t(8;21)+ AML patients. (D) PTPN11 mutations identified in AML patients.
Expression of AML1-ETO in Tet2-null cells generates a novel model of t(8;21)+ AML
The variants detected in TET2 in AML patients include nonsense, frameshift, and missense mutations within conserved domains, consistent with loss of TET2 enzymatic function (Table S1). Notably, 3 of the 29 t(8;21)+ AML patients enrolled in the ECOG E1900 trial (Patel et al., 2012) harbored TET2 mutations (Fig. 3 C and Table S2). In vivo, monoallelic or biallelic loss of Tet2 in the hematopoietic compartment of mice results in myeloproliferation and extramedullary hematopoiesis but not AML (Li et al., 2011; Moran-Crusio et al., 2011).
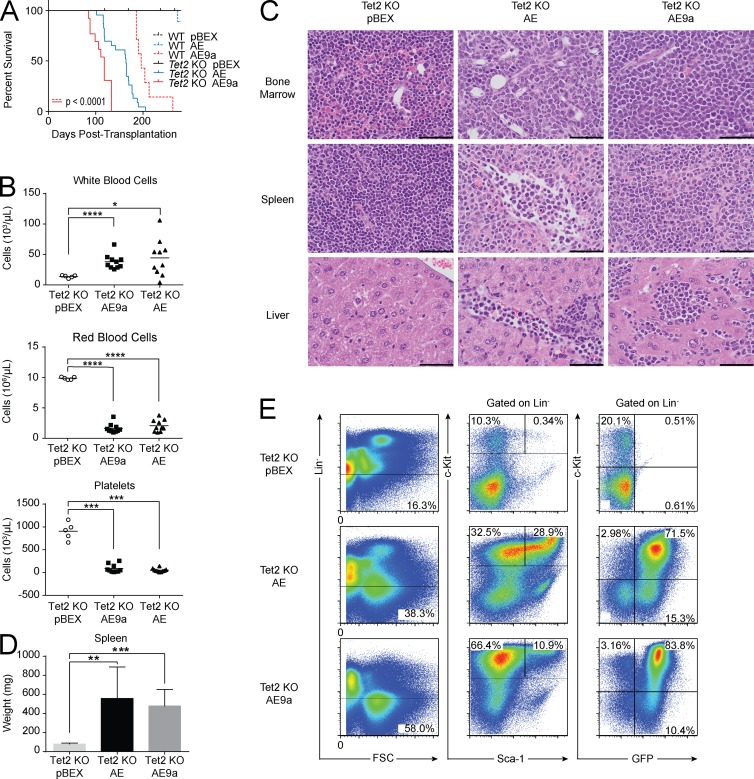
To determine whether loss of Tet2 can cooperate with AML1-ETO in vivo, we transduced bone marrow cells from Vav-Cre+Tet2f/f (Tet2 KO) mice with AML1-ETO or AML1-ETO9 and transplanted the cells into lethally irradiated recipients. Mice transplanted with AML1-ETO–expressing Tet2 KO cells developed fully penetrant leukemia, whereas mice transplanted with AML1-ETO9a–expressing Tet2 KO cells developed a significantly accelerated leukemia with a median latency of 118 d (P < 0.0001; Fig. 4 A). At euthanasia, leukemic recipients had anemia, thrombocytopenia, elevated white blood cell counts (Fig. 4 B), and blast cell infiltration into the liver and spleen, with consequent splenomegaly (Fig. 4, C and D). The GFP+ blast cells are lineage negative, SCA-1 positive or negative, and c-Kit positive (Fig. 4 E).
Figure 4.
Tet2 loss cooperates with AML1-ETO to induce leukemia in vivo. (A) Kaplan-Meier plot of mouse survival after transplantation with cells transduced with the given vector. WT pBEX (n = 5); WT AE (n = 10); WT AE9a (n = 7); Tet2 KO pBEX (n = 12); Tet2 KO AE (n = 23); Tet2 KO AE9a (n = 13). Tet2 loss significantly accelerates AE9a leukemias (P < 0.0001 using the Log-rank test). (B) Peripheral blood counts of mice transplanted with Tet2 KO cells expressing AE9a (n = 10), AE (n = 10), or pBEX (n = 5). Horizontal bars represent mean. (C) Representative hematoxylin and eosin staining (n = 3). Bars, 50 µm. (D) At euthanasia, leukemic mice exhibit significant splenomegaly compared with controls. Tet2 KO pBEX (n = 2); Tet2 KO AE (n = 9); Tet2 KO AE9a (n = 9). Error bars represent the standard deviation from the mean. (B and D) P-values were generated using the Student’s t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (E) Representative flow cytometry analysis of the bone marrow of transplant recipients (n = 10).
The observed cooperativity between Tet2 loss and AML1-ETO in vivo implicates TET2 loss-of-function mutations in t(8;21)+ AML patients as functional cooperating events and challenges the traditional “two-hit hypothesis” of AML, which posits that AML1-ETO commonly cooperates with events that activate oncogenic signaling (Kelly and Gilliland, 2002). Our data support an expanded version of this hypothesis, in which a third class of events capable of enhancing self-renewal are capable of cooperating with AML1-ETO. Given that neomorphic mutations in IDH1/2 and loss-of-function mutations in WT1 also attenuate TET2 function and reduce DNA hydroxymethylation in vivo (Figueroa et al., 2010; Rampal et al., 2014), it will be important to determine whether altered DNA hydroxymethylation is a common pathogenetic event in AML1-ETO+ AML and whether AML1-ETO+ leukemias with concurrent mutations in epigenetic regulators have a distinct epigenetic profile and/or a distinct response to epigenetic therapies.
Expression of AML1-ETO with PTPN11 D61Y generates a novel model of t(8;21)+ AML
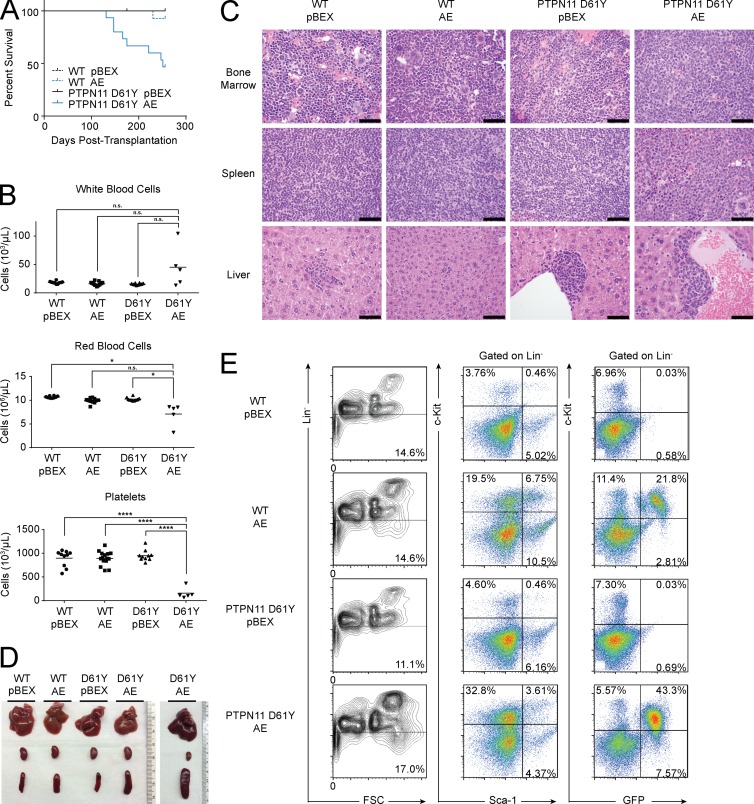
The PTPN11 mutations that we observe in human and mouse AML occur within highly conserved regions of PTPN11 critical to phosphatase activity and the stimulation of RAS signaling (Fig. 3 D and Table S3; Yu et al., 2014). Previous studies have shown that mice expressing the constitutively active D61Y mutation (Ptpn11D61Y) develop a fatal myeloproliferative neoplasm. However, bone marrow cells from these mice fail to engraft beyond 20 wk in transplant recipients and do not induce AML (Chan et al., 2009). We therefore transduced bone marrow cells from Ptpn11D61Y mice with AML1-ETO and transplanted the cells into lethally irradiated recipients. Mice expressing both alleles developed fatal AML (Fig. 5 A). At euthanasia, leukemic mice had elevated white blood cell counts, anemia, thrombocytopenia (Fig. 5 B), and blast cell accumulation in the bone marrow, liver, and spleen (Fig. 5, C and D). These GFP+ blasts were Sca-1 negative and c-Kit positive (Fig. 5 E) and capable of transmitting the disease into secondary recipients (not depicted).
Figure 5.
PTPN11 D61Y cooperates with AML1-ETO to induce leukemia in vivo. (A) Kaplan-Meier plot of mouse survival after transplantation with cells transduced with the given vector. WT pBEX (n = 10); WT AE (n = 15); PTPN11 D61Y pBEX (n = 10); PTPN11 D61Y AE (n = 15). (B) Peripheral blood counts at euthanasia of leukemic PTPN11 D61Y AE mice (n = 5), as well as controls (from left to right, n = 10, n = 15, and n = 10). Horizontal bars represent mean. P-values were generated using the Student’s t test: *, P < 0.05; ****, P < 0.0001; n.s., not significant. (C) Representative hematoxylin and eosin staining (n = 3). Bars, 50 µm. (D) At euthanasia, leukemic mice exhibit splenomegaly. From top to bottom: liver, kidney, and spleen. (E) Representative flow cytometry analysis of the bone marrow of transplant recipients (n = 3).
The observation that AML1-ETO can cooperate with PTPN11 D61Y is more consistent with the two-hit hypothesis and implicates a novel member of the RAS signaling cascade in leukemogenesis. Although RAS mutations occur in 8–10% of AML1-ETO+ AML patients (Peterson et al., 2007a), PTPN11 mutations may be a rare event (Loh et al., 2004). Previous studies pairing AML1-ETO with activating RAS mutations in vivo have had mixed results (Flotho, C. et al. 2004. 46th Annual Meeting of the American Society of Hematology. Poster #770-II; Chou et al., 2011; Zhao et al., 2014). Our data showing that PTPN11 D61Y is capable of cooperating with AML1-ETO suggests that MAPK-directed therapies should be investigated in these genetically defined t(8;21)+ AML subsets.
In each of these newly developed mouse models of AML1-ETO–driven AML, the possibility remains that a third hit has been acquired that is necessary for disease initiation. Fortunately, our integrated approach can be applied to these leukemias such that those events are identified and credentialed in an iterative manner. Like previous studies in mouse solid tumors (Maser et al., 2007) and acute promyelocytic leukemia (Wartman et al., 2011), our data suggest that systematic comparisons of mutational patterns in mouse and human malignancies represent a powerful means of identifying cooperating disease alleles with mechanistic and therapeutic relevance.
MATERIALS AND METHODS
Construction of expression vectors and virus production
The cDNAs encoding AML1-ETO or AML1-ETO9a were digested with NotI and cloned into the NotI multiple cloning site of the MSCV-IRES-BEX plasmid (pBEX), upstream of the IRES and blue-excited GFP motifs. pBEX differs from the previously described MigR1 plasmid by featuring cDNA for enhanced GFP that has been altered such that it can be distinguished from violet-excited GFP (Anderson et al., 1996). To produce retrovirus capable of expressing AML1-ETO or AML1-ETO9a, 293T cells were cotransfected with a pBEX plasmid and MCV-Ecopac via calcium phosphate precipitation.
Fetal liver and bone marrow transduction and transplantation
Fetal liver cells were harvested from embryonic day (E) 14.5 WT (The Jackson Laboratory) or Cdkn1a−/− (provided by T. Jacks, The David H. Koch Institute for Integrative Cancer Research at MIT, Cambridge, MA) C57BL/6 embryos, whereas bone marrow cells from adult Vav-Cre+ or Vav-Cre+Tet2f/f C57BL/6 mice (Moran-Crusio et al., 2011) were harvested 6 d after intraperitoneal administration of 200 mg/kg 5-fluorouracil (Sigma-Aldrich). Bone marrow cells were harvested from 9-wk-old Mx1-Cre+ or Ptpn11D61Y mice that had been treated at 3 wk with poly(I:C) as previously published (Chan et al., 2009). Harvested cells were cultured in RPMI media supplemented with 10% fetal bovine serum, 10 ng/ml IL-3, 10 ng/ml IL-6, and 100 ng/ml stem cell factor (PeproTech) before transplantation, with 8 µg/ml polybrene (Sigma-Aldrich) added during spinoculation. Transduced cells were transplanted into 8–10-wk-old female C57BL/6 recipient mice by tail vein injection after the recipient mice had been lethally irradiated with 950 cGy, given in a split dose separated by 3 h. Procedures performed on these mice were approved by the Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center.
Exome sequencing
Genomic DNA from GFP+ mouse spleen cells or mouse tail tissue was isolated using phenol-chloroform extraction. Exome capture was performed using the SureSelect Mouse All Exon kit (Agilent Technologies), and sequencing was performed using an Illumina platform. Approximately 80 million reads per sample were acquired after a paired-end run with 75 or 100 bp. Paired-end reads were aligned to the NCBIM37.67/mm9 mouse reference genome using the Burrows-Wheeler Aligner (BWA) aln v0.6.2 and processed using the best practices pipeline that included marking of duplicate reads by the use of Picard tools and realignment around indels and base recalibration via Genome Analysis Toolkit (GATK) v3.1.1. Somatic single nucleotide variants (SNVs) were called using muTect v1.1.4, Virmid v1.1.0, and Strelka v1.0.12 and somatic indels via SomaticIndelDetector v2.3.9 and Strelka. SNVs and indels were filtered using the default filtering criteria as implemented in each of the callers and annotated via snpEff and snpSift. In addition, all germline SNVs and indels identified in the 17 key mouse strains as part of the Mouse Genome Project (Keane et al., 2011) were filtered out of the call set. To compare our results with what has been previously discovered in human patients, we downloaded the BAM files for the 150 human AML exomes from The Cancer Genome Atlas project (Cancer Genome Atlas Research Network, 2013). For consistency, we extracted the reads from human exomes, reprocessed them, and called somatic variants using the same pipeline as for mouse exomes.
SMGs and domain analysis
SMGs were identified in mouse and human samples using the SMG module from Genome MuSiC v0.4. Homology between mouse and human genes was based on the mouse–human homologue list downloaded from The Jackson Laboratory website. FDRs were determined using the likelihood ratio test from the Genome MuSiC SMG module. We annotated proteins with protein domains from Pfam-A, a database of high-quality, manually curated protein families. We used the hmmscan algorithm from HMMER3.0 package to match protein sequences against the Pfam-A database.
Statistical analysis
Statistical significance between two groups was assessed using the Student’s t test with Welch’s correction: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant. The hypergeometric test was used to determine whether there was significant overlap in the genes that were mutated in mouse and human AML, as well as whether there was significant overlap in the protein domains that were affected in genes mutated in both mouse and human AML. The significance of recurrently mutated protein domains was determined using the binomial test, where p = length of domain (in AA)/length of protein (in AA). To determine whether mouse leukemias were significantly different from human leukemias with respect to the frequency with which all protein classes were affected by gene mutations, protein classes annotated using PANTHER (Mi et al., 2013) were compared across the two groups using the Pillai-Bartlett statistic from a multivariate ANOVA (MANOVA) model. The corresponding p-values were computed from a transformed value of the test statistic which has an approximate F distribution. All calculations were performed using R version 3.0.
Flow cytometry
To determine the immunophenotype of normal and leukemic populations, cells were stained with a panel of antibodies against lineage markers, each of which was conjugated to APC-Cy7: CD3, CD4, CD11b, CD19, CD45R, Gr-1, NK1.1, and TER119. Cells were also stained with anti-CD117 (c-KIT) and anti–SCA-1, conjugated to PE and PE-Cy7, respectively (BioLegend). Stained cells were analyzed using an LSR Fortessa or LSR II flow cytometer (BD), and data were analyzed using FlowJo software.
Online supplemental material
Table S1 lists the TET2 mutations identified in AML patients from studies by the Cancer Genome Atlas Research Network (2013) and Patel et al. (2012). Table S2 provides statistics on the TET2 mutations identified in AML patients from the Patel et al. (2012) study. Table S3 lists the PTPN11 mutations identified in AML patients from the Cancer Genome Atlas Research Network (2013) study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20150524/DC1.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank members of the Nimer, Levine, and Kharas laboratories for their suggestions and helpful comments regarding this work.
This work was funded by National Institutes of Health R01 grant CA 166835 to S.D. Nimer, by grant CA172636-01 to R.L. Levine, and by Memorial Sloan Kettering Cancer Center Support Grant/Core Grant P30 CA008748. R.L. Levine is a Scholar and A.H. Shih is a Special Fellow of the Leukemia and Lymphoma Society.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AML
- acute myeloid leukemia
- FDR
- false discovery rate
- SMG
- significantly mutated gene
- SNV
- single nucleotide variant
References
- Alcalay M., Meani N., Gelmetti V., Fantozzi A., Fagioli M., Orleth A., Riganelli D., Sebastiani C., Cappelli E., Casciari C., et al. . 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Invest. 112:1751–1761. 10.1172/JCI17595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M.T., Tjioe I.M., Lorincz M.C., Parks D.R., Herzenberg L.A., Nolan G.P., and Herzenberg L.A.. 1996. Simultaneous fluorescence-activated cell sorter analysis of two distinct transcriptional elements within a single cell using engineered green fluorescent proteins. Proc. Natl. Acad. Sci. USA. 93:8508–8511. 10.1073/pnas.93.16.8508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G., Kalaitzidis D., Usenko T., Kutok J.L., Yang W., Mohi M.G., and Neel B.G.. 2009. Leukemogenic Ptpn11 causes fatal myeloproliferative disorder via cell-autonomous effects on multiple stages of hematopoiesis. Blood. 113:4414–4424. 10.1182/blood-2008-10-182626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou F.-S., Wunderlich M., Griesinger A., and Mulloy J.C.. 2011. N-RasG12D induces features of stepwise transformation in preleukemic human umbilical cord blood cultures expressing the AML1-ETO fusion gene. Blood. 117:2237–2240. 10.1182/blood-2010-01-264119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa M.E., Abdel-Wahab O., Lu C., Ward P.S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H.F., et al. . 2010. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 18:553–567. 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., and Stratton M.R.. 2004. A census of human cancer genes. Nat. Rev. Cancer. 4:177–183. 10.1038/nrc1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M., O’Brien D., Kumaravelu P., Lenny N., Yeoh E.-J., and Downing J.R.. 2002. Expression of a conditional AML1-ETO oncogene bypasses embryonic lethality and establishes a murine model of human t(8;21) acute myeloid leukemia. Cancer Cell. 1:63–74. 10.1016/S1535-6108(02)00016-8 [DOI] [PubMed] [Google Scholar]
- Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M., et al. . 2011. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 477:289–294. 10.1038/nature10413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly L.M., and Gilliland D.G.. 2002. Genetics of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 3:179–198. 10.1146/annurev.genom.3.032802.115046 [DOI] [PubMed] [Google Scholar]
- Li Z., Cai X., Cai C.L., Wang J., Zhang W., Petersen B.E., Yang F.C., and Xu M.. 2011. Deletion of Tet2 in mice leads to dysregulated hematopoietic stem cells and subsequent development of myeloid malignancies. Blood. 118:4509–4518. 10.1182/blood-2010-12-325241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh M.L., Reynolds M.G., Vattikuti S., Gerbing R.B., Alonzo T.A., Carlson E., Cheng J.W., Lee C.M., Lange B.J., and Meshinchi S.. Children’s Cancer Group . 2004. PTPN11 mutations in pediatric patients with acute myeloid leukemia: results from the Children’s Cancer Group. Leukemia. 18:1831–1834. 10.1038/sj.leu.2403492 [DOI] [PubMed] [Google Scholar]
- Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.-K., Protopopov A., O’Neil J., Gutierrez A., Ivanova E., Perna I., et al. . 2007. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 447:966–971. 10.1038/nature05886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald E.R. III, Wu G.S., Waldman T., and El-Deiry W.S.. 1996. Repair defect in p21 WAF1/CIP1 −/− human cancer cells. Cancer Res. 56:2250–2255. [PubMed] [Google Scholar]
- Mi H., Muruganujan A., and Thomas P.D.. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41:D377–D386. 10.1093/nar/gks1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K., Reavie L., Shih A., Abdel-Wahab O., Ndiaye-Lobry D., Lobry C., Figueroa M.E., Vasanthakumar A., Patel J., Zhao X., et al. . 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 20:11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A.M., Duque J., Shizuru J.A., and Lübbert M.. 2008. Complementing mutations in core binding factor leukemias: from mouse models to clinical applications. Oncogene. 27:5759–5773. 10.1038/onc.2008.196 [DOI] [PubMed] [Google Scholar]
- Patel J.P., Gönen M., Figueroa M.E., Fernandez H., Sun Z., Racevskis J., Van Vlierberghe P., Dolgalev I., Thomas S., Aminova O., et al. . 2012. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N. Engl. J. Med. 366:1079–1089. 10.1056/NEJMoa1112304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L.F., Boyapati A., Ahn E.-Y., Biggs J.R., Okumura A.J., Lo M.-C., Yan M., and Zhang D.-E.. 2007a Acute myeloid leukemia with the 8q22;21q22 translocation: secondary mutational events and alternative t(8;21) transcripts. Blood. 110:799–805. 10.1182/blood-2006-11-019265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson L.F., Yan M., and Zhang D.-E.. 2007b The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood. 109:4392–4398. 10.1182/blood-2006-03-012575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R., Alkalin A., Madzo J., Vasanthakumar A., Pronier E., Patel J., Li Y., Ahn J., Abdel-Wahab O., Shih A., et al. . 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Reports. 9:1841–1855. 10.1016/j.celrep.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiohara M., Koike K., Komiyama A., and Koeffler H.P.. 1997. p21WAF1 mutations and human malignancies. Leuk. Lymphoma. 26:35–41. [DOI] [PubMed] [Google Scholar]
- Stubbs M.C., Kim Y.M., Krivtsov A.V., Wright R.D., Feng Z., Agarwal J., Kung A.L., and Armstrong S.A.. 2008. MLL-AF9 and FLT3 cooperation in acute myelogenous leukemia: development of a model for rapid therapeutic assessment. Leukemia. 22:66–77. 10.1038/sj.leu.2404951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartman L.D., Larson D.E., Xiang Z., Ding L., Chen K., Lin L., Cahan P., Klco J.M., Welch J.S., Li C., et al. . 2011. Sequencing a mouse acute promyelocytic leukemia genome reveals genetic events relevant for disease progression. J. Clin. Invest. 121:1445–1455. 10.1172/JCI45284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Kanbe E., Peterson L.F., Boyapati A., Miao Y., Wang Y., Chen I.-M., Chen Z., Rowley J.D., Willman C.L., and Zhang D.-E.. 2006. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 12:945–949. 10.1038/nm1443 [DOI] [PubMed] [Google Scholar]
- Yu Z.-H., Zhang R.-Y., Walls C.D., Chen L., Zhang S., Wu L., Liu S., and Zhang Z.-Y.. 2014. Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry. 53:4136–4151. 10.1021/bi5002695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Zhang Y., Sha K., Tang Q., Yang X., Yu C., Liu Z., Sun W., Cai L., Xu C., and Cui S.. 2014. KRAS (G12D) cooperates with AML1/ETO to initiate a mouse model mimicking human acute myeloid leukemia. Cell. Physiol. Biochem. 33:78–87. 10.1159/000356651 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.