Abstract
The demand for phenomics, a high-dimensional and high-throughput phenotyping method, has been increasing in many fields of biology. The budding yeast Saccharomyces cerevisiae, a unicellular model organism, provides an invaluable system for dissecting complex cellular processes using high-resolution phenotyping. Moreover, the addition of spatial and temporal attributes to subcellular structures based on microscopic images has rendered this cell phenotyping system more reliable and amenable to analysis. A well-designed experiment followed by appropriate multivariate analysis can yield a wealth of biological knowledge. Here we review recent advances in cell imaging and illustrate their broad applicability to eukaryotic cells by showing how these techniques have advanced our understanding of budding yeast.
INTRODUCTION
The morphological features (i.e., shapes and sizes) of organisms and cells have long been of interest to biologists. Driven by advances in microscopic technologies and the increasing availability of fluorescence microscopy, cell biology and genetics have benefited substantially from the observation of cell and organelle morphology. Indeed, automated image acquisition systems in microscopy and image-analysis technologies have recently been developed for the high-dimensional phenotyping of many model organisms (Collinet et al., 2010; Neumann et al., 2010; Sozzani and Benfey, 2011; Pardo-Martin et al., 2013). Since the development of high-throughput microscopy (Rimon and Schuldiner, 2011; Tkach et al., 2012), methods to reinforce the automated image acquisition of subcellular structures have been sought in cell biology to better understand the complex cellular processes in eukaryotic cells. The budding yeast Saccharomyces cerevisiae is especially interesting because it is the leading model organism for studies of global regulation in cells (Costanzo et al., 2010). Several methods have been developed to analyze single-cell phenomics in S. cerevisiae (Ohya et al., 2005; Vizeacoumar et al., 2010; Handfield et al., 2013; Okada et al., 2015). Of importance, these methods are superior to previous ready-made image-analysis packages in terms of accuracy and reproducibility because they are designed for budding yeast. Here we focus not on high-throughput microscopy or quantitative analysis, but on current advances in the field of yeast single-cell high-dimensional phenotyping by considering the spatial and temporal attributes of subcellular structures. We also discuss how this technique can be applied to further our understanding of budding yeast, as well as of animal and plant species.
EXTRACTION OF SUBCELLULAR STRUCTURES FROM DIGITAL IMAGES
As a model system for eukaryotic cells, budding yeast contain organelles that are both shared among higher organisms and are ideally suited to the study of subcellular morphology, using either fluorescence staining or the expression of green fluorescent protein (GFP)–labeled proteins. Huh et al. (2003) were able to localize >4000 proteins by systematically constructing strains expressing GFP-tagged proteins, and they visually categorized 21 subcellular localizations or organelles. Recently, high-content screening using a machine learning approach was performed using GFP-tagged proteins to measure protein abundance and localization changes in a systematic and quantitative manner (Chong et al., 2015). The results of that study revealed that subcellular localization can be classified into 16 subcellular compartments, achieving a high level of precision and recall based on a single experiment using automated analysis. The next step after identifying subcellular compartments is to extract the morphological features of subcellular structures. The number of such defined structures, including protein supercomplexes and subcellular components, is increasing rapidly. To date, 12 subcellular structures have been successfully extracted for morphological analysis from digital images, including cell shape (cell wall and plasma membrane), nuclear DNA, mitochondria, mitochondrial DNA, actin structures, spindle microtubules, spindle pole bodies, septin rings, the vacuole, the cis- and trans-Golgi, and lipid droplets (Ohya et al., 2005; Negishi et al., 2009; Vizeacoumar et al., 2010; Wolinski et al., 2011; Rafelski et al., 2012; Osman et al., 2015).
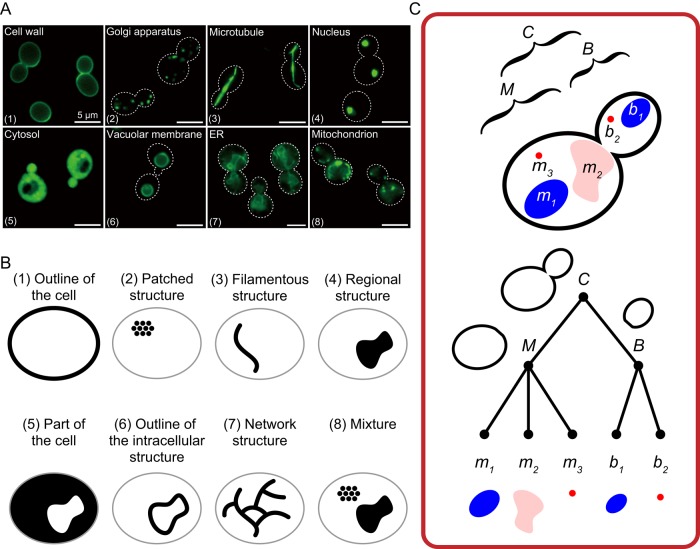
To extract subcellular structures from digital images, it is essential to first define the typical geometry of the organelles in a given image. Membrane-bound organelles naturally occupy certain regions within the cell, and considering that the resolution of a conventional optical microscope is 0.2 μm, small organelles and granular structures almost always appear as patches in microscopic images. Similarly, when viewed under a conventional optical microscope, tubular cytoskeletal structures appear as filaments. Therefore, the subcellular structures and organelles of budding yeast (Figure 1A) are recognized as “regions,” “patches,” or “filaments,” and more complex structures are recognized as combinations of these elements and network structures (Figure 1B).
FIGURE 1:
Extraction of the morphological features of subcellular structures. (A) Fluorescence images of subcellular structures in budding yeast. Images of the cell wall (fluorescein isothiocyanate–labeled concanavalin A), Golgi apparatus (Vrg4-GFP), microtubules (anti-tubulin), nucleus (Htb2-GFP), cytosol (Ssb1-GFP), vacuolar membrane (Vma4-GFP), ER (Sec61-GFP), and mitochondria (Atp5-GFP) are shown with the category number indicated. (B) Categorization of subcellular structures. The geometry of yeast subcellular structures can be classified into eight categories: 1) The cell periphery is classified as the outline of the cell. 2) Actin, spindle pole body, bud neck, cis-Golgi, trans-Golgi, endosome, and peroxisome are classified as patched structures. 3) The spindle is classified as a filamentous structure. 4) The nucleus, vacuole, and nucleolus are classified as regional structures. 5) The nuclear cytoplasm, cytoplasm, nucleus, and bud are classified as parts of the cell. 6) The nuclear periphery and vacuolar membrane are classified as the outline of intracellular structures. 7) The ER is classified as a network. 8) Mitochondria and mitochondrial DNA are classified as a mixture. (C) Schematic view of a budded cell with two nuclei (top) and a tree structure diagram of the subcellular structures (bottom). The whole structure (or the whole cell) is labeled as C, the mother cell as M, and the bud as B. The organelles in M and B are labeled as m and b with numbering, respectively. The addition of spatial attributes to subcellular structures is achieved manually (Rafelski et al., 2012) or automatically (Ohya et al., 2005; Handfield et al., 2013) in the indicated pipelines. Multiple subcellular structures of interest are shown for their inclusion relation using a tree structure diagram. Different kinds of features and inclusions can also be expressed in a similar manner. Each node has morphometric information, including name of node, size of area, and length of perimeter.
In those situations in which morphological features cannot be extracted (i.e., “regions,” “patches,” and “filaments”), use of free open-source image analysis software tools for morphological filtering based on top-hat transformation (Kimori, 2011) is effective. A structuring element that is the equivalent of a filter kernel is optimized for the shapes and sizes of the features using an automated or semiautomated approach. The extraction of subcellular structures provides rich material for single-cell phenotyping but does not lead to a direct understanding of their dynamic aspects. For more refined purposes, imaging using other microscopic methods such as optical sectioning by confocal microscopy and time-lapse imaging can be used. Optical sectioning by confocal microscopy makes it possible to construct three-dimensional (3D) images of mitochondria (Rafelski et al., 2012). Time-lapse imaging has been used to produce sets of images to clarify the dynamics and movement of lipid droplets (Wolinski et al., 2011) and to monitor the kinetics of Far1, a cyclin-dependent kinase inhibitor (Doncic et al., 2015).
SPATIAL ATTRIBUTES OF SUBCELLULAR STRUCTURES
Budding yeast are typically divided into mother cells and buds, which adds a new attribute to subcellular components. Of note, many proteins are synthesized, accumulated, and degraded asymmetrically (Nyström and Liu, 2014; Zhou et al., 2014), which further differentiates daughter cells from mother cells. To identify precisely intracellular compartments and monitor the differentiation of bud from mother, it is important not only to classify each structure but also to know its intracellular location (i.e., the part of the cell to which each structure belongs). For instance, mitochondria in the mother cell should be considered distinct from mitochondria in the bud. Rafelski et al. (2012) analyzed images of live budding cells using the MitoGraph pipeline and found dramatic asymmetry in mitochondrial accumulation between the mother and the bud. By considering the spatial attributes of a subcellular structure and quantifying them as different morphological features, it is possible to precisely describe and analyze subcellular components. The inclusion relationship of the morphological features of subcellular structures can be represented in the form of a tree (Figure 1C). This is useful for understanding and communicating the morphological features of subcellular structures.
TEMPORAL ATTRIBUTES OF CELLULAR FEATURES
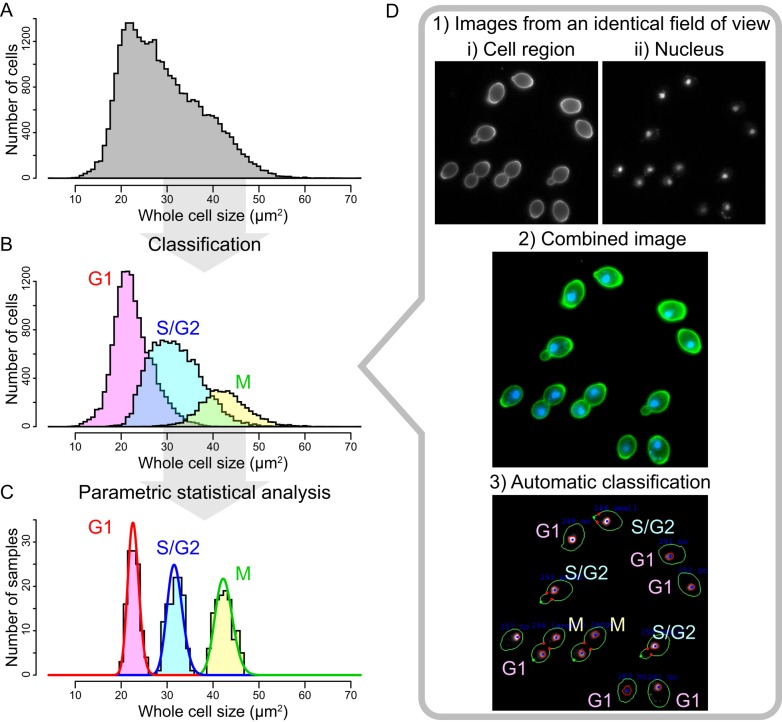
Variables in nature do not always have unimodal distributions; in addition, due to cell-to-cell variation (Liberali et al., 2015), many features of yeast cell morphology have a multimodal distribution (e.g., whole cell size; Figure 2A). Thoughtful consideration should be given to the possible multimodal distribution of any given variable of interest, especially in phenotyping asynchronous yeast cell populations. In contrast to from unimodal distributions, it is difficult to analyze multimodal distributions (for which parametric statistical analyses are quite limited), suggesting a need for alternative methods. One efficient strategy for handling complex data sets with multimodal distribution involves classification. After applying temporal classification according to cell cycle stage, size distribution becomes nearly unimodal (Figure 2B). It is now possible to identify a probability distribution model (Figure 2C) and perform a parametric statistical analysis with high power.
FIGURE 2:
Unimodal distribution of morphological data extracted by classification. (A) Distribution of whole cell size. Cell sizes of 30,583 wild-type yeast cells (BY4743) were quantified under fluorescence microscopy after staining with fluorescein isothiocyanate–labeled concanavalin A (FITC-Con A). The distribution is multimodal because of a mixture of cells at different stages of the cell cycle. (B) Distribution of cell size at each stage of the cell cycle. Magenta, cyan, and yellow boxes indicate the distribution of cell sizes at G1, S/G2, and M, respectively. The mean cell sizes at each stage were distributed differently but overlapped. (C) Distribution of the mean cell size at each stage of the cell cycle. The distributions of 114 wild-type replicates in terms of cell size were distinguishable at different stages of the cell cycle. Red, blue, and green curves indicate the gamma distribution approximated by a maximum likelihood estimation for the mean values at G1, G2/S, and M, respectively. Because the distribution of mean values in each stage is unimodal, approximation by a unimodal probability distribution (e.g., gamma distribution) is applicable (Yang et al., 2014). (D) Automatic classification of cells by simultaneously processing multiple images of the same cell. 1) Microscopic images of (i) cell shape and (ii) nuclear DNA were acquired in the same field of view after staining with FITC-Con A and 4′,6-diamidino-2-phenylindole, respectively. 2) The two images were combined to identify the nuclear cycle stages. 3) Cells were automatically classified by cell cycle stage using the CalMorph image-processing system (Ohya et al., 2005).
Automatic temporal classification, used in the CalMorph pipeline (Ohya et al., 2005; Okada et al., 2015), is performed by superimposing cell images on nucleus images from an identical field of view (Figure 2D). This approach provides attribute information pertaining to time points before and after cell division. The addition of temporal attributes to morphological features does not increase the number of independent features, but it does contribute to a fully analytical imaging system. An alternative and more reliable way is to use synchronized yeast cells; however, this requires extra time and is unsuitable for large-scale phenotyping. In addition to the nuclear cycle, other cell cycle landmarks, such as bud size, can be used for temporal segregation (Handfield et al., 2013). This method is superior in terms of monitoring cell cycle–dependent changes in the localization of cellular proteins.
Cell morphology is a complex phenotype that is controlled via a number of pathways and complex interplay between growth and division. The temporal classification of cell morphology in accordance with cell cycle progression is therefore valuable for understanding the molecular mechanisms of cell morphogenesis. Two major points of cell cycle control occur during G1 (cell size control) and G2 (entry into mitosis). Owing to this temporal segregation, these two processes have been investigated separately (McNulty and Lew, 2005; Di Talia et al., 2007; Charvin et al., 2009). A recent study identified cell size–regulatory mutants by considering birth size and growth rate (Soifer and Barkai, 2014).
STATISTICAL ANALYSIS FOR CELL PHENOTYPING
A well-designed experiment followed by appropriate statistical analysis can provide a wealth of biologically meaningful information. In this context, the analysis of phenotypes of numerous sets of mutants is a powerful tool to extract genetically and biologically important information. To illustrate this point, we address here some of the findings to which high-dimensional morphometric analysis has contributed significantly.
High-dimensional phenotyping of yeast strains
Because each cellular image contains numerous morphological features, a single experiment usually provides a high-dimensional morphological data set. Quantification of morphological features, including cell shape, actin, nuclear DNA, and microtubules, was completed for a catalogued mutant collection, including nonessential deletion mutants (Ohya et al., 2005; Vizeacoumar et al., 2010), temperature-sensitive mutants of essential genes (Li et al., 2011), and decreased abundance by mRNA perturbation mutants of essential genes (Bauer et al., 2015). The results revealed a close relationship between morphological phenotype and the functional annotation of a gene; thus, a functional analysis of genetics became possible by evaluating similarities in mutant morphology.
Cell-to-cell morphological variation has been analyzed using CalMorph to understand system robustness. The genes responsible for morphological robustness were identified from both nonessential (Levy and Siegal, 2008) and essential (Bauer et al., 2015) genes and shown to function as phenotypic stabilizers for nongenetic sources of variation, such as environmental perturbation. Because a large amount of cell-to-cell morphological variation is observed in natural isolates (Yvert et al., 2013), the evolutionary costs and benefits of robustness can be argued in nature. Strain-to-strain morphological variation has been investigated, revealing essential genes in a natural environment (Yang et al., 2014). Quantitative trait loci (Nogami et al., 2007) and the structure of phenotypic diversity (Skelly et al., 2013) in natural isolates have also been assessed.
A high-dimensional data set alone does not intuitively provide compelling biological information; it is ambiguous as to which traits should be of focus. When the data set is sparsely structured (i.e., the morphological phenotype is not statistically significant in most of the tested elements), dimensionality reduction procedures such as principal component analysis, and partitioning around medoids should be applied to define the biologically informative phenotype for effective analysis (Levy and Siegal, 2008; Yvert et al., 2013; Yang et al., 2014; Bauer et al., 2015).
Phenotypic similarity used for the prediction of drug targets
High-dimensional morphological information makes it possible to search for a set of mutants with a similar morphology (Ohnuki et al., 2010). This approach was applied successfully to predict drug targets in a cell, using the morphology of a drug-treated cell as the query and matching it to the morphology of the mutant collection. Of note, this method was used to predict and identify the target of a new antifungal agent, poacic acid, which binds to β-1,3-glucan (Piotrowski et al., 2015), an antifungal agent with a new mode of action, echinocandin B (Okada et al., 2014), and a fermentation inhibitor, vanillin, which affects the large subunit of cytoplasmic ribosomes (Iwaki et al., 2013). Although this system uses mutant information, another chemical-genetic phenotype profiling approach is possible without relying on any mutant information in advance (Gebre et al., 2015).
Automated classification of mutants by morphological phenotype
When dealing with genetic mutants, it is common to classify the mutants according to their morphology. Traditionally, such classification has relied on subjective judgment, but cluster analysis has turned out to be a powerful tool for automating classification procedures when working with high-dimensional morphological datasets. Genes responsible for Ca2+ tolerance have been classified into seven functional groups based on Ca2+-dependent morphological changes (Ohnuki et al., 2007). In addition, the catalytic subunit of β-1,3-glucan synthase was functionally dissected on the basis of high-dimensional phenotyping and the clustering analysis of temperature-sensitive mutants (Okada et al., 2010).
Quantitative analysis of genetic interactions
Quantitative readout is useful for the analysis not only of single mutants but also of double mutants. The use of an endoplasmic reticulum (ER) stress–sensing reporter in yeast double mutants can reveal functional interdependencies of ER folding processes (Jonikas et al., 2009). With morphometric data, systematic analyses of genetic interactions were achieved during Drosophila development (Fischer et al., 2015). Similar phenotypic analyses of double mutants and the gene–gene, or genetic, interactions of the mutations will also be examined in yeast.
Simulation and modeling
The quantification of morphology has the potential to facilitate the simulation or modeling of how the biophysical and chemical characteristics of gene products affect morphogenetic events and organelle dynamics, although this has not yet been attempted. Such models enable us to predict unknown interactions or links as new testable hypotheses, which can later be confirmed using experimental validation. In one relevant study, mathematical modeling was used to predict that the yeast polarity establishment circuit involves negative feedback (Howell et al., 2012).
CONCLUSIONS
Automatic image extraction, classification based on temporal and spatial information, and the quantification of subcellular structures ensure the accuracy, objectivity, and reproducibility of high-dimensional cell phenotyping. With rapid advances in microscopic technology, unconventional cell phenotyping procedures will inevitably continue to improve. Optical sectioning using confocal microscopy and microscopy with high-content time-lapse imaging, superresolution confocal live imaging (Nakano, 2013), light sheet microscopy (Mohan et al., 2014), and use of microfluidics systems (Taylor et al., 2009) will expand our view of 3D structures and the dynamic movements and fusion of organelles. To accommodate an ever-increasing number of images from such methodologies, systematic and universal procedures will need to be developed. The engagement of a broad group of scientists in stimulating discussions regarding sharing image data sets, standardizing protocols for image analysis, and enabling standard statistical analysis will greatly contribute to single-cell phenomics in the future.
Acknowledgments
We thank researchers in the Laboratory of Signal Transduction, Department of Integrated Bioscience, University of Tokyo, for fruitful discussions, Andreas Doncic for valuable comments on the manuscript, and the anonymous reviewers for their suggestions on improving the review article. This work was supported by a grant for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (15H04402 to Y.O.).
Abbreviations used:
- ER
endoplasmic reticulum
- GFP
green fluorescent protein.
Footnotes
REFERENCES
- Bauer CR, Li S, Siegal ML. Essential gene disruptions reveal complex relationships between phenotypic robustness, pleiotropy, and fitness. Mol Syst Biol. 2015;11:773. doi: 10.15252/msb.20145264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvin G, Cross FR, Siggia ED. Forced periodic expression of G1 cyclins phase-locks the budding yeast cell cycle. Proc Natl Acad Sci USA. 2009;106:6632–6637. doi: 10.1073/pnas.0809227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong YT, Koh JL, Friesen H, Duffy SK, Cox MJ, Moses A, Moffat J, Boone C, Andrews BJ. Yeast proteome dynamics from single cell imaging and automated analysis. Cell. 2015;161:1413–1424. doi: 10.1016/j.cell.2015.04.051. [DOI] [PubMed] [Google Scholar]
- Collinet C, Stöter M, Bradshaw CR, Samusik N, Rink JC, Kenski D, Habermann B, Buchholz F, Henschel R, Mueller MS, et al. Systems survey of endocytosis by multiparametric image analysis. Nature. 2010;464:243–249. doi: 10.1038/nature08779. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- Doncic A, Atay O, Valk E, Grande A, Bush A, Vasen G, Colman-Lerner A, Loog M, Skotheim JM. Compartmentalization of a bistable switch enables memory to cross a feedback-driven transition. Cell. 2015;160:1182–1195. doi: 10.1016/j.cell.2015.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Sandmann T, Horn T, Billmann M, Chaudhary V, Huber W, Boutros M. A map of directional genetic interactions in a metazoan cell. Elife. 2015;4:e05464. doi: 10.7554/eLife.05464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebre AA, Okada H, Kim C, Kubo K, Ohnuki S, Ohya Y. Profiling of the effects of antifungal agents on yeast cells based on morphometric analysis. FEMS Yeast Res. 2015;15:fov040. doi: 10.1093/femsyr/fov040. [DOI] [PubMed] [Google Scholar]
- Handfield L-F, Chong YT, Simmons J, Andrews BJ, Moses AM. Unsupervised clustering of subcellular protein expression patterns in high-throughput microscopy images reveals protein complexes and functional relationships between proteins. PLoS Comput Biol. 2013;9:e1003085. doi: 10.1371/journal.pcbi.1003085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu C-F, Zyla TR, Elston TC, Lew DJ. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh W-K, Falvo J V, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Iwaki A, Ohnuki S, Suga Y, Izawa S, Ohya Y. Vanillin inhibits translation and induces messenger ribonucleoprotein (mRNP) granule formation in Saccharomyces cerevisiae: application and validation of high-content, image-based profiling. PLoS One. 2013;8:e61748. doi: 10.1371/journal.pone.0061748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimori Y. Mathematical morphology-based approach to the enhancement of morphological features in medical images. J Clin Bioinformatics. 2011;1:33. doi: 10.1186/2043-9113-1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberali P, Snijder B, Pelkmans L. Single-cell and multivariate approaches in genetic perturbation screens. Nat Rev Genet. 2015;16:18–32. doi: 10.1038/nrg3768. [DOI] [PubMed] [Google Scholar]
- McNulty JJ, Lew DJ. Swe1p responds to cytoskeletal perturbation, not bud size, in S. cerevisiae. Curr Biol. 2005;15:2190–2198. doi: 10.1016/j.cub.2005.11.039. [DOI] [PubMed] [Google Scholar]
- Mohan K, Purnapatra SB, Mondal PP. Three dimensional fluorescence imaging using multiple light-sheet microscopy. PLoS One. 2014;9:e96551. doi: 10.1371/journal.pone.0096551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano A. Super-resolution confocal live imaging microscopy (SCLIM)—cutting-edge technology in cell biology. Conf Proc IEEE Eng Med Biol Soc. 2013;2013:133–135. doi: 10.1109/EMBC.2013.6609455. [DOI] [PubMed] [Google Scholar]
- Negishi T, Nogami S, Ohya Y. Multidimensional quantification of subcellular morphology of Saccharomyces cerevisiae using CalMorph, the high-throughput image-processing program. J Biotechnol. 2009;141:109–117. doi: 10.1016/j.jbiotec.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Hériché JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S, Ohya Y, Yvert G. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 2007;3:e31. doi: 10.1371/journal.pgen.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyström T, Liu B. The mystery of aging and rejuvenation—a budding topic. Curr Opin Microbiol. 2014;18:61–67. doi: 10.1016/j.mib.2014.02.003. [DOI] [PubMed] [Google Scholar]
- Ohnuki S, Nogami S, Kanai H, Hirata D, Nakatani Y, Morishita S, Ohya Y. Diversity of Ca2+-induced morphology revealed by morphological phenotyping of Ca2+-sensitive mutants of Saccharomyces cerevisiae. Eukaryot Cell. 2007;6:817–830. doi: 10.1128/EC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnuki S, Oka S, Nogami S, Ohya Y. High-content, image-based screening for drug targets in yeast. PLoS One. 2010;5:e10177. doi: 10.1371/journal.pone.0010177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y, Sese J, Yukawa M, Sano F, Nakatani Y, Saito TL, Saka A, Fukuda T, Ishihara S, Oka S, et al. High-dimensional and large-scale phenotyping of yeast mutants. Proc Natl Acad Sci USA. 2005;102:19015–19020. doi: 10.1073/pnas.0509436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Abe M, Asakawa-Minemura M, Hirata A, Qadota H, Morishita K, Ohnuki S, Nogami S, Ohya Y. Multiple functional domains of the yeast l,3-β-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics. 2010;184:1013–1024. doi: 10.1534/genetics.109.109892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Ohnuki S, Ohya Y. Quantification of cell, actin, and nuclear DNA morphology with high-throughput microscopy and CalMorph. Cold Spring Harb Protoc. 2015;2015:408–412. doi: 10.1101/pdb.prot078667. [DOI] [PubMed] [Google Scholar]
- Okada H, Ohnuki S, Roncero C, Konopka JB, Ohya Y. Distinct roles of cell wall biogenesis in yeast morphogenesis as revealed by multivariate analysis of high-dimensional morphometric data. Mol Biol Cell. 2014;25:222–233. doi: 10.1091/mbc.E13-07-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Noriega TR, Okreglak V, Fung JC, Walter P. Integrity of the yeast mitochondrial genome, but not its distribution and inheritance, relies on mitochondrial fission and fusion. Proc Natl Acad Sci USA. 2015;112:E947–E956. doi: 10.1073/pnas.1501737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Martin C, Allalou A, Medina J, Eimon PM, Wählby C, Fatih Yanik M. High-throughput hyperdimensional vertebrate phenotyping. Nat Commun. 2013;4:1467. doi: 10.1038/ncomms2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski JS, Okada H, Lu F, Li SC, Hinchman L, Ranjan A, Smith DL, Higbee AJ, Ulbrich A, Coon JJ, et al. Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc Natl Acad Sci USA. 2015;112:E1490–E1497. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski SM, Viana MP, Zhang Y, Chan Y-HM, Thorn KS, Yam P, Fung JC, Li H, Costa L da F, Marshall WF. Mitochondrial network size scaling in budding yeast. Science. 2012;338:822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimon N, Schuldiner M. Getting the whole picture: combining throughput with content in microscopy. J Cell Sci. 2011;124:3743–3751. doi: 10.1242/jcs.087486. [DOI] [PubMed] [Google Scholar]
- Skelly DA, Merrihew GE, Riffle M, Connelly CF, Kerr EO, Johansson M, Jaschob D, Graczyk B, Shulman NJ, Wakefield J, et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res. 2013;23:1496–1504. doi: 10.1101/gr.155762.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soifer I, Barkai N. Systematic identification of cell size regulators in budding yeast. Mol Syst Biol. 2014;10:761. doi: 10.15252/msb.20145345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzani R, Benfey PN. High-throughput phenotyping of multicellular organisms: finding the link between genotype and phenotype. Genome Biol. 2011;12:219. doi: 10.1186/gb-2011-12-3-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RJ, Falconnet D, Niemistö A, Ramsey SA, Prinz S, Shmulevich I, Galitski T, Hansen CL. Dynamic analysis of MAPK signaling using a high-throughput microfluidic single-cell imaging platform. Proc Natl Acad Sci USA. 2009;106:3758–3763. doi: 10.1073/pnas.0813416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizeacoumar FJ, van Dyk N, FS Vizeacoumar, Cheung V, Li J, Sydorskyy Y, Case N, Li Z, Datti A, Nislow C, et al. Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J Cell Biol. 2010;188:69–81. doi: 10.1083/jcb.200909013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124:3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- Yang M, Ohnuki S, Ohya Y. Unveiling nonessential gene deletions that confer significant morphological phenotypes beyond natural yeast strains. BMC Genomics. 2014;15:932. doi: 10.1186/1471-2164-15-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvert G, Ohnuki S, Nogami S, Imanaga Y, Fehrmann S, Schacherer J, Ohya Y. Single-cell phenomics reveals intra-species variation of phenotypic noise in yeast. BMC Syst Biol. 2013;7:54. doi: 10.1186/1752-0509-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Slaughter BD, Unruh JR, Guo F, Yu Z, Mickey K, Narkar A, Ross RT, McClain M, Li R. Organelle-based aggregation and retention of damaged proteins in asymmetrically dividing cells. Cell. 2014;159:530–542. doi: 10.1016/j.cell.2014.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]